Background: NAD(P)H cytochrome b5 oxidoreductase (Ncb5or) participates in a variety of metabolic conversions.
Results: A new Ncb5or acts as a redox partner of Δ12 fatty acid desaturase.
Conclusion: Ncb5or null mutant suffers from impaired linoleate synthesis leading to oxidative stress and cell death.
Significance: Our results have exposed a novel fatty acid synthesis pathway in L. major that differs from the mammalian system.
Keywords: Apoptosis, Cell Death, Cytochrome b, Enzyme Catalysis, Flavoproteins, Leishmania, Lipid Metabolism, Oxidative Stress, Polyunsaturated Fatty Acids
Abstract
NAD(P)H cytochrome b5 oxidoreductase (Ncb5or), comprising cytochrome b5 and cytochrome b5 reductase domains, is widely distributed in eukaryotic organisms. Although Ncb5or plays a crucial role in lipid metabolism of mice, so far no Ncb5or gene has been reported in the unicellular parasitic protozoa Leishmania species. We have cloned, expressed, and characterized Ncb5or gene from Leishmania major. Steady state catalysis and spectral studies show that NADH can quickly reduce the ferric state of the enzyme to the ferrous state and is able to donate an electron(s) to external acceptors. To elucidate its exact physiological role in Leishmania, we attempted to create NAD(P)H cytochrome b5 oxidoreductase from L. major (LmNcb5or) knock-out mutants by targeted gene replacement technique. A free fatty acid profile in knock-out (KO) cells reveals marked deficiency in linoleate and linolenate when compared with wild type (WT) or overexpressing cells. KO culture has a higher percentage of dead cells compared with both WT and overexpressing cells. Increased O2 uptake, uncoupling and ATP synthesis, and loss of mitochondrial membrane potential are evident in KO cells. Flow cytometric analysis reveals the presence of a higher concentration of intracellular H2O2, indicative of increased oxidative stress in parasites lacking LmNcb5or. Cell death is significantly reduced when the KO cells are pretreated with BSA bound linoleate. Real time PCR studies demonstrate a higher Δ12 desaturase, superoxide dismutase, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA with a concomitant fall in Δ9 desaturase mRNA expression in LmNcb5or null cell line. Together these findings suggest that decreased linoleate synthesis, and increased oxidative stress and apoptosis are the major consequences of LmNcb5or deficiency in Leishmania.
Introduction
Fatty acid desaturases (FADs)2 produce a variety of essential fatty acids (FA) for the biphasic parasitic life cycle of trypanosomatids. Earlier it was established that Leishmania promastigotes can grow in selective medium that lacks lipid moieties (1). Later works reveal that trypanosomatids possess the capacity to synthesize FAs de novo (2, 3). These organisms have unique endoplasmic reticulum (ER) based elongases (4). A few years ago the polyunsaturated FA synthesis pathway was completely revealed in trypanosomatids (5, 6). Among the trypanosomatids, only Leishmania has the ability to convert C18:0 to arachidonate or to even larger and more unsaturated FAs (up to 22:6); therefore, this organism can synthesize all of the polyunsaturated FAs (PUFAs) it requires from stearate (6, 7). Trypanosomes have to take up precursors from the host to produce FAs beyond linoleate (18:2) as they apparently lack the desaturases that are required for processing linoleate further (7, 8). All FADs require a redox partner for transferring electron from NAD(P)H (9). But until now nothing is known regarding the redox partner of FADs in Leishmania.
NAD(P)H cytochrome b5 oxidoreductase (Ncb5or) belongs to the cytochrome b5 family of flavohemoprotein that is widely expressed in eukaryotic organisms (10). Ncb5or is an endoplasmic reticulum-bound protein that has been implicated in lipid metabolism and diabetes (11, 12). Ncb5or contains two redox domains; the amino-terminal portion is highly homologous to classical microsomal cytochrome b5, and the carboxyl-terminal portion of the protein is highly similar to cytochrome b5 reductase domain (10). The carboxyl-terminal cytochrome b5 reductase (cb5r) domain contains a single flavin adenine dinucleotide prosthetic group and catalyzes the transfer of reducing equivalents from NAD(P)H to cytochrome b5 heme (cb5) domain. The cb5 domain participates in a variety of metabolic conversions including the desaturation and elongation of FAs (13, 14), cholesterol biosynthesis (15), and cytochrome P450-mediated mono-oxygenation (16). Specific examples of these various electron acceptors are Δ9-, Δ6-, and Δ5-acyl CoA desaturases (17), FA elongation enzyme (13), alkyl acylglycero-3-phosphoryl ethanolamine desaturase (18), phospholipid desaturase (19), Δ7-sterol Δ9-desaturase (15), N-hydroxylamine reductase (19), 4-methylsterol oxidase (20), prostaglandin synthase reductase (21), and cytochrome P450 4A7 (22). In addition, cb5 domain is found in other unrelated proteins such as nitrate reductase (23), sulfite oxidases (24, 25), and lactate dehydrogenase (26).
The cb5 domain, a six-coordinate heme protein, conducts electron transfer in desaturation reaction by two possible modes. The classical cb5 along with cb5r protein forms a complex and resides in ER membrane with desaturase enzyme. The membrane-bound cb5 transfers electron from cb5r to the desaturase (27). In the other mode, cb5 is itself included as an extra domain in the amino-terminal (9, 28) or in the carboxyl-terminal end of desaturase (23, 29) and mediates the electron transfer from cb5r to the desaturase.
A novel ER-based soluble Ncb5or has been identified from mouse liver (30). Ncb5or deficiency in hepatocytes results in an impaired Δ9 desaturation pathway, and saturated FA induced lipotoxicity is observed (12). Ncb5or knock-out mice have usual embryonic and fetal growth, and normal viability at birth with no gross anatomic abnormalities (31). However, the null mice develop diabetes at ∼7 weeks of age due to progressive loss of insulin-producing β-cells but maintain usual insulin sensitivity (31). The transcription factor X-box-binding protein-1 (XBP-1) regulating the proapoptotic gene C/EBP-homologous protein (CHOP) in the ER stress pathway appears to be involved in β-cell loss in knock-out mice as CHOP knock-out delays onset of diabetes by 2 weeks (32). Ncb5or knock-out cells reveal increased susceptibility to the oxidant streptozotocin-induced diabetogenic effects in vivo (31) and in vitro (33). Furthermore, β-cells of 12-week-old (diabetic) Ncb5or knock-out mice have mitochondrial hypertrophy and hyperplasticity (31). Recently Xu et al. (11) suggest that increased free FA accumulation and catabolism and oxidative stress are the major consequences of Ncb5or deficiency in liver.
The wide diversity of cytochrome b5 genes and large number of this b5-type cytochrome in Leishmania genome sequence are indicative of their important role in parasite survivability and infectivity within the host (34). However, the potential role of Ncb5or in lipid metabolism and mitochondrial hyperactivity in Leishmania as well as in other trypanosomatids has not been explored to date. The presence of two plant-like Δ12-fatty acid desaturase (Δ12-FAD) genes (35) (see genedb online) and the polyunsaturated FA components in Leishmania coupled with the absence of the genes in host macrophage indicates that the unsaturated FA synthesis pathway differs from that found in the typical mammalian system, making it an interesting subject of investigation. In this study we have established for the first time that Ncb5or deficiency results in impaired Δ12 desaturation in Leishmania, which provides new insights into the regulation of a crucial step in poly unsaturated FA biosynthesis. We have shown that Ncb5or deletion results in dramatic increments in both the Δ12-FADs mRNA level, O2 consumption, ROS generation, ATP production, and apoptosis in Leishmania. Wild type characteristics are restored in KO cells on pretreatment with BSA-bound linoleate. We report LmNcb5or to be the redox partner of Δ12-FADs in linoleate biosynthesis of Leishmania, and the most important consequences of knock-out of the gene are manifested by decreased growth rate and increased oxidative stress.
EXPERIMENTAL PROCEDURES
Reagents
The Mitoprobe JC-1 assay kit for flow cytometry, Vibrant apoptosis assay kit no. 3 for annexin V and propidium iodide (PI), and the ATP determination kit were purchased from Molecular Probes. All other chemicals were obtained from Sigma or from sources mentioned previously (36–38).
Cloning of LmNcb5or
Genomic DNA was isolated from Leishmania major promastigotes using a Qiagen genomic DNA isolation kit. The entire ORF of LmNcb5or was amplified by PCR using primers 1 and 2 (supplemental Table S1) to get a 1611-bp fragment that was digested and cloned within pET15B vector in NdeI/BamHI restriction sites. DNA sequencing was performed to confirm the ORF.
Expression and Purification of Proteins
Recombinant pET15B/LmNcb5or were used to transform Escherichia coli BL21(DE3) cells. Transformed cells were grown overnight in 100 ml Luria-Bertani broth containing 200 μg/ml ampicillin at 37 °C in a shaker. Cultures grown overnight were then inoculated in 1.0 liter of terrific broth. When the culture reached an absorbance of 0.8 at 600 nm, 0.5 mm isopropyl β-d-thiogalactoside and 0.4 mm δ-aminolevulinic acid were added, and the bacteria were further grown for 18 h at 20 °C. Cells were then harvested by centrifugation at 6000 × g for 10 min, and pellets were washed twice with PBS. Pellets were resuspended in 10 ml of 50 mm potassium phosphate buffer, pH 7.5, containing 150 mm NaCl, 1.0 mm PMSF, and 1.0 mg/ml lysozyme, a protein inhibitor mixture tablet without EDTA from Roche Applied Science, 1.0 mm EDTA, and 10% glycerol. The resuspended solution was kept for 45–50 min in ice, and the cells were freeze-thawed in liquid nitrogen followed by a 25-s pulse sonication (Sonicator Cell Disrupter, model no. W-220F, Heat Systems, Ultrasonics, Inc.) with 1 min of rest on ice in between. The lysate was centrifuged at 14,000 × g for 60 min at 4 °C. The cell-free supernatant, designated as the crude extract, was precipitated by adding 50% (w/v) solid ammonium sulfate at 4 °C. The ammonium sulfate precipitate was centrifuged at 4 °C for 1.0 h at 15,000 rpm and kept at −80 °C and then resuspended in a minimum volume of resuspension buffer of 50 mm potassium phosphate buffer, pH 7.5, having 150 mm NaCl, 1.0 mm PMSF, 10% glycerol, 20 mm imidazole, and loaded onto a Ni2+-NTA column. The column was washed with equilibration buffer (50 mm phosphate buffer, pH 7.5, containing 150 mm NaCl, 1.0 mm PMSF, 10% glycerol, 20 mm imidazole) followed by 50 mm phosphate buffer, pH 7.5, 150 mm NaCl, 10% glycerol, 40 mm imidazole. The enzyme was eluted with 50 mm phosphate, pH 7.5, 150 mm NaCl, 10% glycerol, 100 mm imidazole, and then dialyzed 3 times against 50 mm phosphate buffer, pH 7.5, having 10% glycerol. The eluted protein was concentrated by YM-50 (Amicon, molecular weight cutoff 50 kDa) and at 4 °C loaded onto a G-200 column (120 × 10 cm) pre-equilibrated with 50 mm potassium phosphate buffer, pH 7.5, and protein fractions were collected after passing 11.0 ml of buffer. Desired protein fractions were concentrated and kept at −80 °C for storage.
UV-Visible Spectroscopy
All kinetic and spectral studies were performed at 25 °C on a Shimadzu UV-2550 spectrophotometer using quartz cuvette of 1.0-cm path length. Wavelength and extinction coefficient used to measure NADH oxidation was at 340 nm (6.2 mm−1cm−1), and NADH-dependent reduction of cytochrome c, ferricyanide, and 2,6-dichloroindophenol (DCIP) were at 550 nm (21 mm−1cm−1), 420 nm (1.2 mm−1cm−1), and 600 nm (20.6 mm−1cm−1), respectively. The assay mixture was composed of 50 mm potassium phosphate buffer, pH 7.5, 4 μm flavin adenine dinucleotide, and 10 units/ml superoxide dismutase. NADH, horse heart cytochrome c, DCIP, and ferricyanide concentration were used at 1.0, 0.1, 0.1, and 1.0 mm, respectively.
Protein Concentration Determination
The heme was identified and quantified by the pyridine-hemochrome method (39). The molar absorption coefficient of the protein at 412 nm was 78 mm−1cm−1.
Determination of Bound Flavin Adenine Dinucleotide
Bound flavin adenine dinucleotide was released from the protein by heat-denaturing the enzyme (95 °C for 5 min in dark) in a sealed vial (40). Subsequently the sample was cooled at 4 °C and filtered to remove denatured protein. Fluorescence intensity was measured by fluorescence spectroscopy, PTI fluorometer (Photon Technology International), with excitation at 450 nm and emission at 530 nm and quantified by comparing with the standard curve prepared from a freshly prepared flavin adenine dinucleotide stock solution. The flavin and protein ratio suggests that the LmNcb5or contained ∼1.0 flavin adenine dinucleotide per monomer protein (data not shown).
Size Exclusion Chromatography
LmNcb5or was studied by gel exclusion chromatography at room temperature using a column namely protein Pak SW 125 (Waters) in an HPLC system pre-equilibrated with 50 mm potassium phosphate buffer, pH 7.5, containing 150 mm NaCl with a flow rate 0.8 ml/min at A280. Column calibration was done using standard protein mixture of ovotransferrin (78 kDa), albumin (66 kDa), ovalbumin (43 kDa), carboanhydrase (30 kDa), myoglobin (16 kDa), and cytochrome c (12 kDa).
Parasite Culture
L. major (strain 5ASKH) promastigotes were routinely cultured at 22 °C in M199 medium (Invitrogen) supplemented with 40 mm HEPES, pH 7.4, 200 μm adenine, 1% penicillin-streptomycin (v/v), 50 μg/ml gentamycin, and 10% heat-inactivated fetal bovine serum (Invitrogen).
Constructing Over-expression System for LmNcb5or in Leishmania
Primers 11 and 12 (supplemental Table S1) were used to amplify the ORF of LmNcb5or gene by PCR. Then the amplified portion was cloned into the SmaI/BamHI site of pXG-B2863 vector, which was transfected in L. major by electroporation (41). Overexpressed cells were maintained at 200 μg/ml neomycin.
Generation of Knock-out Strain for LmNcb5or Alleles
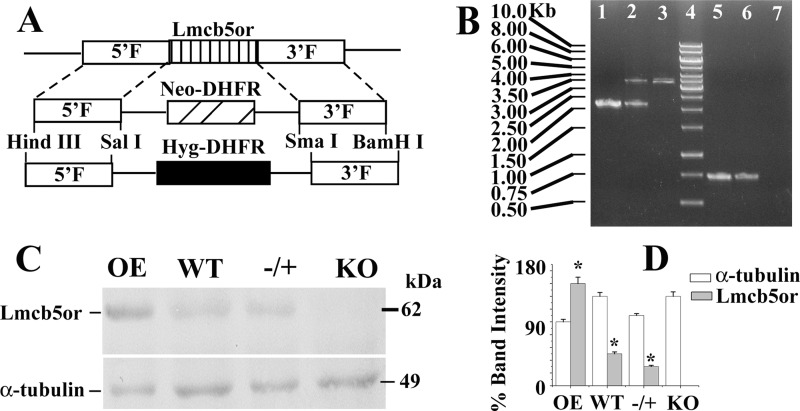
Modified pXG-neo and pXG-hyg (38) vectors were used to generate the knock-out constructs of LmNcb5or gene. 1.91- and 1.876-kb DNA fragments were amplified by PCR from 5′- and 3′-flanking regions of the LmNcb5or gene, respectively. Primers 3 and 4 were used for amplifying 5′-flank, and primers 5 and 6 were used for amplifying 3′-flank of the gene. The 5′-flank and 3′-flank DNA fragments were cloned on either side (at HindIII/SalI and SmaI/BamHI site) of the neomycin and hygromycin gene of pXG-neo and pXG-hyg vectors, respectively (Fig. 1A). Both constructs were then digested with HindIII and BamHI to get linear fragments of gene deletion constructs LmNcb5or::NEO and LmNcb5or::HYG, which were transfected into wild type Leishmania sequentially as described previously (38). The knock-out strain was maintained in 60 μg/ml neomycin and 100 μg/ml hygromycin drug. Gene knock-out was confirmed by PCR using primers 7/8 as external to the gene and primers 9/10 as internal (Fig. 1B). KO cells were further confirmed by Western blotting (Fig. 1, C and D).
FIGURE 1.
Targeted gene replacement of LmNcb5or alleles. A, shown is a schematic representation of the LmNcb5or locus and the plasmid constructs used for gene replacement. DHFR, dihydrofolate reductase. B, shown is agarose gel analysis of PCR-amplified products of LmNcb5or gene. Lane 4 shows the molecular mass markers, lanes 1 and 5, 2 and 6, and 3 and 7 correspond to PCR with genomic DNA from WT, −/+ heterozygous mutants, and KO mutants, respectively, with primers external (lanes 1–3) and internal (lanes 5–7) to the LmNcb5or gene. External forward and reverse primers were generated from 454 bp upstream of the LmNcb5or gene and position 1715 bp downstream of the start codon of gene, respectively. The expected size of the LmNcb5or, NEO, and HYG gene PCR product are 2.17, 3.25, and 3.372 kb, respectively. Internal forward and reverse primers were generated from positions 718 and 1450 bp of the gene, respectively. The expected size of the PCR product is 0.733 kb. C, Western blot results using rabbit anti-LmNcb5or and mouse anti-α-tubulin antibody are shown. 200 μg of L. major lysate were used for Western blotting. All the data are representative of at least three independent experiments. D, shown is a bar diagram depicting the % band intensities in the Western blot. Band intensity was quantified by ImageJ software (NIH), and error bars represent the S.D. from three independent experiments. Band intensity of OE α-tubulin was considered as 100%.
Western Blotting
Proteins were resolved by 13% SDS-PAGE and then transferred to nitrocellulose membrane by Bio-Rad semidry transfer apparatus. The membrane was incubated with rabbit anti-LmNcb5or polyclonal antibody (1:50) followed by anti-rabbit secondary (1:5000) antibody. ECL kit (Amersham Biosciences) was used for band detection. α-Tubulin was used as the endogenous control.
Fatty Acid Analysis
Mid-log phase cells (3 × 108) were harvested by centrifugation and washed twice with ice-cold PBS. FA extraction and analysis were carried out as described earlier (42). Briefly, cells were suspended in 20 ml of chloroform:methanol (2:1) solvent containing 0.1% butylhydroxytoluene as antioxidant and homogenized by a homogenizer. After homogenization, the whole mixture is agitated for 1.0 h in an orbital shaker at room temperature. Cell debris was separated by a glass-filter unit. 0.2 ml (4 ml for 20 ml) of 0.9% NaCl solution was added to 1.0 ml of solvent for washing. The mixture was agitated for a few minutes, and then it was centrifuged at 2000 rpm to separate the two phases. The upper aqueous phase was removed, and the interface was washed twice with methanol/water (1/1). The lower chloroform phase-containing lipids was reduced to dryness under a nitrogen stream. FA methyl esters were obtained by incubation with 1.0 ml of 0.5 m sodium methoxide in methanol for 30 min at room temperature. The samples were neutralized by 6 m HCl and extracted with 2 ml of hexane. Again the solvent was evaporated under the nitrogen stream, and the residue was dissolved in isohexane for GC-MS analysis. FA methyl esters of OE, WT, and KO cells were analyzed by GC-MS Shimadzu QP5050A using ZB-5 column (30 m × 0.25 mm × 0.25 μm) (ZebronTM). Gas chromatographic analysis was performed at an oven temperature of 50 °C for 2 min, gradually going up to 330 °C at 10 °C/min. Injector and interface temperatures were 240 and 250 °C, respectively. Helium was used as the carrier gas. The GC-MS was carried out using a mass detector at an ionization voltage of 70 eV. Supelco37 FA methyl ester mix standards (sigma) and also the internal database (National Bureau of Standards) were used to identify each peak of FA methyl esters. Percentages of FAs were calculated by measuring the peak area of the chromatogram peaks. The FA content was normalized by measuring the protein content of each cell type.
In Vitro Promastigote Growth Profile Analysis
106 mid-log phase L. major promastigote cells were inoculated in 10 ml of M199 media and 10% FCS. Growth rates were measured at a 24-h interval by counting cell number in an improved Neubauer chamber (hemocytometer) for 8 consecutive days. Experiments were done in triplicate for each cell type.
Fatty Acid Treatment
FA-free bovine serum albumin (BSA), oleic acid complexed with BSA, and linoleic acid complexed with BSA were purchased from Sigma. Concentration of FAs was measured using a non-esterified FA detection kit from Zenbio. Promastigote cells were washed twice in PBS and incubated for 2 h at 22 °C in chemically defined M199+ medium (43). Then cells were treated with 0.5 mm oleate complexed with BSA or linoleate complexed with BSA for 12 h. FA-free BSA (1%) was added to cells as the control experiment. This protocol was followed in a cell viability test, P/O assay, mitochondrial membrane potential measurement, intracellular H2O2 determination, intracellular ATP analysis, and the measurement of oxygen consumption. For the Leishmania promastigote growth profile analysis, FA treatment was carried out in 10 ml of medium and for 8 consecutive days.
Oxygen Consumption Measurement
Exponentially growing 0.5 × 108 mid-log phase cells were harvested and washed twice with cold PBS. Oxygen consumption was measured by Oxytherm system (Hansatech Instruments, Penteny, King's Lynn, Norfolk, England) using Oxygraph plus software (44). The electrode was calibrated with air-saturated water and dithionite. O2 consumption by 0.5 ml of resuspended cells in a 1.0-ml chamber at 25 °C was measured, and for each set of experiments base-line oxygen consumption was monitored with only PBS. Each reading was taken after stabilization of the O2 consumption rate and continued for 15 min. All experiments were done in triplicate to minimize the experimental error. 10% sodium azide solution was used to stop respiration of cells and used as control for experiments.
P/O Ratio Assays
P/O ratio (the relationship between ATP synthesis and oxygen consumption) in digitonin-permeabilized cells were carried out by measuring the oxygen consumption during the rapid burst of state 3 respiration after adding 0.1 mm ADP (45). The measurements were made on 1.0 × 108 cells permeabilized with 30 μg of digitonin/mg of protein for 5 min at 28 °C in an assay buffer consisting of 200 mm sucrose, 10 mm phosphate buffer, pH 7.4, 1.0 mm EDTA, 2 mm MgCl2, and succinate (2.5 mm) as respiratory substrate.
Measurement of Cellular ATP Content
Cellular ATP content was determined by a luciferin-luciferase bioluminescence assay using an ATP determination kit (46) (Molecular Probes). In brief, 106 cells were mixed with a standard reaction buffer containing DTT (1.0 mm), D-luciferin (0.5 mm), and firefly luciferase (1.25 μg/ml). The luminescence intensity was measured in a Modulus luminometer (Turner Biosystems). A standard curve of ATP was made by a series of ATP concentrations ranging from 1.0 to 1.0 μm, and ATP concentrations of different cell types were compared and calculated against this standard curve. ATP content was expressed in nmol/106 cells.
Measurement of Cellular Hydrogen Peroxide
Cellular hydrogen peroxide levels were determined by staining with 2′,7′-dichlorofluorescin diacetate (Molecular Probes). 106 cells were taken and washed twice with PBS. Fluorescence was measured subsequently using a FACS Canto flow cytometer (BD Biosciences) at an excitation wavelength of 502 nm and emission wavelength of 530 nm.
Mitochondrial Membrane Potential (ΔΨ) Measurement
5,5,6,6-Tetrachloro-1,1,3,3-tetraethylbenzimidazole carbocyanide iodide (JC-1) was used as a probe for the measurement of ΔΨ by flow cytometry (37, 46). JC-1 is a lipophilic, cationic dye that enters mitochondria and emits green fluorescence (510–520 nm) at a low membrane potential and bright red-orange fluorescence (590 nm) at a relatively high membrane potential. Experiments were done according to instructions from the MitoProbeTM JC-1 assay kit (Molecular Probes). 106 cells were taken and washed twice in PBS, then incubated with 6 μm JC-1 dye for 20 min at 26 °C. For a positive control, 50 μm carbonyl cyanide 3-chlorophenylhydrazone, a mitochondrial uncoupler, was added to the non-treated cells 15 min before JC-1 treatment. Measurements were done on FacsCanto flow cytometer (BD Biosciences) at an excitation wavelength of 488 nm and emissions at 530 nm for green and 590 nm for red fluorescence after appropriate fluorescence compensation. Data were analyzed with FACSDiva.
FITC Annexin V-PI Assay for Apoptosis
Phosphatidylserine exposure of apoptotic cells can be seen by staining with FITC-conjugated annexin V (Vybrant apoptosis assay kit 3, Molecular Probes) as they show green fluorescence. PI is impermeable to live, but stains dead cells that show red fluorescence thus emitting both red and green fluorescence. 106 cells of different types were harvested, washed twice in cold PBS, and resuspended in binding buffer for 25 min followed by incubation with 5.0 μl of annexin V and 1.0 μg/ml PI for 20 min at room temperature. Finally 300 μl of binding buffer was added, and samples were analyzed by flow cytometry at an excitation wavelength of 488 nm and emissions at 530 nm for green and 590 nm for red fluorescence after appropriate fluorescence compensation. Measurements were completed within an hour. To eliminate background fluorescence, compensation was done with unstained and single-stained (with either only FITC or PI) cells.
For microscopic examination, promastigote cells were adhered on polylysine-coated slide and visualized under Olympus IX81 microscope. For each sample 20 fields were observed.
Quantitative Real Time PCR
Total RNA was extracted from WT, OE, and KO Leishmania cells using an RNAqueous®-4PCR kit (Ambion) according to manufacturer's protocol. RNA quantification was done by comparing absorbance at 260- and 280-nm wavelengths. Then 2 μg of RNA from each cell type was taken for cDNA synthesis by using a High Capacity cDNA reverse transcription kit (Applied Biosystems). Quantitative real time PCR was then performed with the cDNA on StepOne™ instrument (Applied Biosystems) using SYBR® Green dye (Applied Biosystems) as the reporter dye following the manufacturer's protocol. The mRNA quantification was done by comparative CT method using 18 S rRNA as the reference gene. Primer design was carried out by primer express 2 software.
Statistical Analysis
All results were expressed as the means ± S.D. from at least three independent experiments. Statistical analysis for parametric data was calculated using Student's t test or analysis of variance wherever applicable using Origin 6.0 software (Microcal Software, Northampton, MA). A p value of less than 0.05 was considered statistically significant.
RESULTS
Primary Structure of LmNcb5or Fusion Protein
A sequence (systematic no. LmjF30.0610) in the L. major genome database was identified as containing an ORF related to putative nitrate reductase (genedb). The translated protein sequence comprised 536 amino acid residues and exhibited 32% identity with Sus scrofa domestica cytochrome b5 (Ssdcb5, accession number 1106188C). It has 60% identity with Pichia stipitis NADH-cytochrome b5 reductase (Pscb5r, accession no. NP_001153064). Sequence alignments with both the S. scrofa domestica cytochrome b5 (Ssdcb5) and P. stipitis NADH-cytochrome b5 reductase (Pscb5r) confirmed that the fusion protein primary structure retained a tripartite arrangement consisting of a 74-residue amino-terminal b5-type cytochrome fragment and a 234-residue carboxyl-terminal flavin-containing fragment that were tethered by an ∼170 amino acid fragment residue “linker” region (Fig. 2). We amplified a 1611-bp fragment with the potential to encode a ∼58-kDa protein (designated LmNcb5or).
FIGURE 2.
Sequence alignment of LmNcb5or. The sequence of LmNcb5or was aligned with S. scrofa domestica cytochrome b5 (Ssdcb5, accession number 1106188C) and P. stipitis NADH-cytochrome b5 reductase (Pscb5r, accession number NP_001153064). The identical residues were denoted by asterisks.
LmNcb5or Fusion Protein Expression
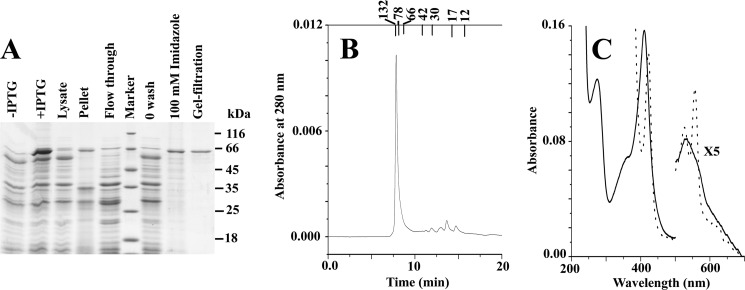
After IPTG induction, E. coli BL21(DE3) cells transformed with the full-length H6-tagged LmNcb5or expression construct resulted in the production of pink cells indicative of efficient expression of a heme containing protein. Isolation and purification of the recombinant LmNcb5or fusion protein to homogeneity was achieved using two chromatographic steps. The initial affinity chromatographic step utilized nickel-nitrilotriacetic acid-agarose and resulted in the rapid separation of the hemoflavoprotein from the majority of the other soluble E. coli proteins. The second gel filtration step resulted in the removal of both minor contaminating endogenous E. coli-derived proteins and degradation products resulting from LmNcb5or expression. The protein compositions of samples obtained from the various purification steps are shown in Fig. 3A using SDS-PAGE analysis. Both the soluble and membrane fractions obtained from the E. coli cells harboring the LmNcb5or expression vector showed the presence of a band at ∼60 kDa, corresponding to the mature LmNcb5or fusion protein and indicating that although significant quantities of a soluble form were produced, some of the fusion protein was sequestered in the insoluble fraction due to either incomplete cell lysis or protein aggregation. To investigate the native state molecular mass of LmNcb5or, purified proteins were subjected to gel filtration using HPLC. Results shown in Fig. 3B indicate that LmNcb5or eluted at a position expected of dimeric enzyme (∼120 kDa).
FIGURE 3.
Physical characterization of LmNcb5or. Panel A indicates SDS-PAGE. Panel B indicates the size exclusion chromatography of LmNcb5or after purification by using a Protein pak125 SW HPLC column. Panel C indicates electronic absorption spectra of 1.5 μm LmNcb5or in 50 mm phosphate buffer, pH 7.5. The solid line and dotted line represent the spectra before and after the addition of NADH on the native enzyme respectively.
Physical and Spectral Characteristics of LmNcb5or
The UV-visible spectra of LmNcb5or showed the presence of a Soret peak at 412 nm with a secondary peak α band at 560 nm (shoulder) and β band at 530 nm (Fig. 3C). These spectral peaks are characteristic of di-histidine-ligated cytochrome b5-type hemeprotein. When it is reduced by excess NADH, the Soret peak shifted to 423 nm, and the new β and α bands appeared at 525 and 555 nm, respectively, which is the characteristic of heme reduction. Subsequently 460-nm absorbance was decreased upon NADH addition, indicating the reduction of the protein-bound flavin adenine dinucleotide. To further characterize the steady state kinetics of enzyme, turnover of common electron acceptors was investigated in the presence of the excess NADH. Table 1 showed the steady state data of NADH oxidation, ferricytochrome c reduction, ferricyanide and DCIP reduction, and protein-exhibited Michaelis-Menten-type kinetics. All assays were done in the presence of iron superoxide dismutase (SOD) that actually inhibited electron transfer from the superoxide to external acceptor molecule and thus showed actual activity of the enzyme. The catalytic efficiency and substrate affinity with artificial electron acceptors for LmNcb5or have very similar kinetics behavior with previously reported human Ncb5or (30). Although NADPH is a better co-substrate than NADH in human Ncb5or, NADH is a better electron donor in case of LmNcb5or protein (data not shown).
TABLE 1.
Catalytic activities of LmNcb5or
The catalytic activity (kcat) is expressed as s−1. NADH oxidation, potassium ferricyanide, DCIP, and cytochrome c reduction rates were determined at 25 °C as described under “Experimental Procedures.” The values represent the mean and S.D. for three measurements each.
| Substrates | LmNcb5or enzyme |
||
|---|---|---|---|
| Km | kcat | kcat/Km | |
| μm | s−1 | μm−1 s−1 | |
| NADH | 3.3 ± 0.2 | 0.2 ± 0.05 | 0.06 |
| Cytochrome c | 12 ± 0.6 | 15 ± 0.8 | 1.25 |
| Potassium ferricyanide | 9 ± 0.7 | 26 ± 1.0 | 2.88 |
| DCIP | 14 ± 0.4 | 9 ± 0.9 | 0.64 |
Fatty Acid Analyses in OE, WT, and KO Leishmania Promastigotes
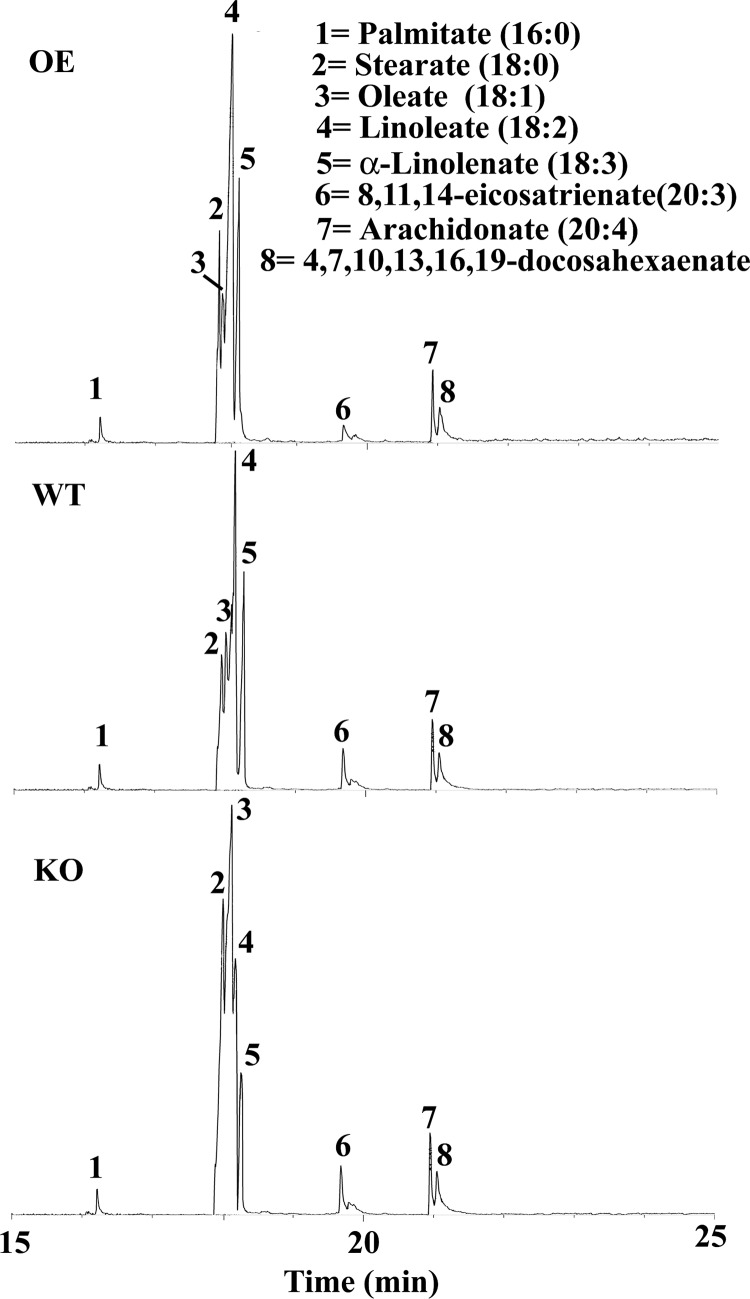
To investigate the role of LmNcb5or gene in L. major, a gene replacement technique was carried out. Western blotting confirmed that the resulting cells no longer expressed LmNcb5or (Fig. 1). Because Ncb5or contains cb5 and cb5r domains that could be the redox electron donor for FADs, we investigated whether the KO cells had an unbalanced FA desaturation pathway. Fig. 4 showed the FA profiles in OE, WT, and KO cells. Mass spectra of eight main peaks were shown in supplemental Fig. S1. Eight predominant FAs in wild type L. major cells were palmitate (16:0), stearate (18:0), oleate (18:1), linoleate (18:2), linolenate (18:3), eicosatrienate (20:3), arachidonate (20:4), docasahexaenate (22:6). As shown in Table 2, the KO cells were markedly deficient in linoleate and linolenate free FAs, whereas the oleate content was greater, indicating that the absence of Ncb5or expression impaired C18 Δ12 desaturation of oleate. This pathway is catalyzed by the Δ12 FADs. The results presented in Fig. 4 and Table 2 strongly suggest that KO cells have decreased ratios of 18:2/18:1, and Ncb5or is involved in Δ12 FA desaturation by the transferring electrons from NAD(P)H to the Δ12 FADs.
FIGURE 4.
Gas chromatographic analysis of main FA from L. major cells.
TABLE 2.
Comparative studies of main FAs content in OE, WT, and KO cells
The FA methyl esters analysis was described under “Experimental Procedures.” The values represent the mean and S.D. for three measurements each.
| FAs | OE | WT | KO | WT/KO |
|---|---|---|---|---|
| % | % | % | ||
| Total FAa | 1.1 ± 1.0 | 1.6 ± 1.8 | 4.5 ± 4.0 | 2.8 |
| 16:0 | 2.84 ± 0.15 | 2.44 ± 0.03 | 2.34 ± 0.08 | 1.04 |
| 18:0 | 14.87 ± 1.1 | 7.02 ± 0.4 | 23.11 ± 1.6 | 0.30 |
| 18:1 | 6.28 ± 0.5 | 9.68 ± 0.7 | 43.04 ± 3.1 | 0.22 |
| 18:2 | 50.45 ± 2.6 | 56.45 ± 4.2 | 14.62 ± 0.8 | 3.86 |
| 18:3 | 19.7 ± 1.4 | 17.17 ± 1.2 | 9.01 ± 0.3 | 1.91 |
| 20:3 | 1.42 ± 0.05 | 1.88 ± 0.08 | 2.07 ± 0.10 | 0.90 |
| 20:4 | 1.68 ± 0.03 | 2.88 ± 0.16 | 2.86 ± 0.17 | 1.00 |
| 22:6 | 2.76 ± 0.07 | 2.48 ± 0.21 | 2.95 ± 0.09 | 0.84 |
a Total FA was expressed as weight % FA based on lyophilized weight of L. major.
Characteristics of OE, WT, and KO Cells
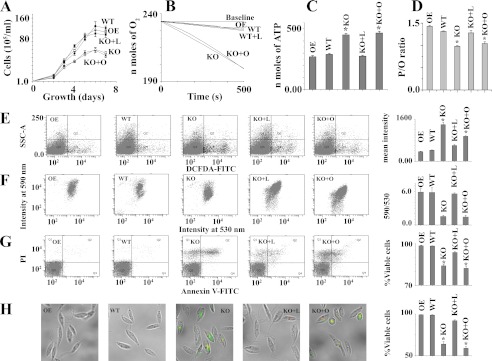
To investigate whether the growth rate of null mutants (KO) is similar to WT or OE, microscopically viable cell counting analysis was performed. The growth curve showed that KO population had a slower growth rate compared with WT or OE cells (Fig. 5A). To check the effect of linoleate or oleate in the recovery of the growth rate of KO, we added the BSA-bound FAs separately into growth media. Only on the addition of linoleate did the growth rate of KO cells become identical with WT cells. Fig. 5B showed the O2 consumption rates in OE, WT, and KO Leishmania promastigotes. It was found that KO cells consumed about 6 times more O2 than WT cells (Fig. 5B). This reflects a profound change in ATP generation (Fig. 5C), suggesting that the more oxidative phosphorylation occurred in KO cells compared with WT or OE cells. Interestingly, albeit there was an increase in both O2 consumption and total ATP generation in KO cells, the change in O2 consumption (6-fold) was much more pronounced than that observed for total ATP generation (1.5-fold) in KO cells. These results indicated that uncoupled oxidative phosphorylation is higher in KO cells compared with wild type cells. Furthermore, the O2 consumption rate and total cellular ATP content in linoleate-pretreated KO cells were similar to WT, but oleate-pretreated KO cells showed no reversal of condition. These results suggested that linoleate deficiency was the causative agent for growth retardation, higher O2 consumption, and ATP generation in KO cell. To investigate the uncoupled electron transfer among wild type, overexpression, and knock-out cells, we measure the P/O ratio (the relationship between ATP synthesis and oxygen consumption) in all types of cells with or without pretreatment of linoleate or oleate (supplemental Fig. S2). Ncb5or null Leishmania showed lower P/O ratio compared with wild type or overexpression cell (Fig. 5D). These results further indicate that a significant amount of the uncoupled electron transfer occurs with the Ncb5or null mutant. Pretreatment of KO cells with linoleate for 12 h maintained P/O ratio levels to 97% of WT cell, whereas the pretreatment with the oleate had no significant effect on P/O ratio compared with KO cells.
FIGURE 5.
Increased ROS and cell death in LmNcb5or null mutants. Panel A represents growth curve of OE, WT, and KO cells. L and O indicate BSA-bound linoleate and oleate-pretreated cells, respectively. Panel B indicates O2 consumption with time. Panel C denotes total ATP content in cells. Panel D represents the P/O ratio (ADP phosphorylation/oxygen atom consumption) of digitonin-permeabilized OE, WT, and KO cells during the rapid burst of state 3 respiration by the addition of 0.1 mm ADP. The cells were starved for 3 h and then supplemented with the substrate. 2.5 mm succinate was used as respiratory substrates. Panel E represents the flow cytometric determination of intracellular H2O2 by dichlorofluorescein diacetate (DCFDA). Dot plots of side-angle light scatter (SSC-A) versus dichlorofluorescein diacetate FITC is shown. Panel F denotes mitochondrial membrane potential (ΔΨm) in L. major cells. L. major cells (107/ml) were incubated with the potential sensitive probe JC-1 (6 μm) for 15 min at 25 °C to assess ΔΨm. A drop in ΔΨm was identified as a change in JC-1 properties from forming J-aggregates (emission at 590 nm) at high ΔΨm to producing J-monomers (emission at 530 nm) at low ΔΨm. Panel G represents cells double-stained with annexin V-FITC and PI to assess cell death. The lower left quadrant of each dot plot represents viable cells. H, shown are merged macroscopic images of the cells of panel G under bright-field. A bar diagram of % viable cells in 700 promastigotes is shown. Viable cells are the annexin V- and propidium iodide-negative cells. Error bars in all panels represent the mean ± S.D. from three independent experiments. *, Statistically significant value of less than 0.001. Linoleate or oleate treated WT and OE cells are identical with untreated WT and OE cells (data not shown). All the data are representative of three independent transfection experiments (three different KO clones).
Occasional electron leak in electron transport chain is highly associated with ROS generation. To evaluate the degree of ROS content in OE, WT, and KO cell lines, we measured intracellular levels of H2O2 by using the fluorescent probe dichlorofluorescein diacetate. The intracellular H2O2 concentration in the KO population was found to be higher than the WT or OE cells, indicating that KO cells produce more ROS (Fig. 5E). As expected, on pretreatment of cells with 0.5 mm linoleate, the generation of H2O2 was reduced in KO cells (Fig. 5E). Intracellular ROS buildup can induce mitochondrial dysfunction, which is highly coupled with a collapse in mitochondrial membrane potential (37). We examined mitochondrial membrane potential by flow cytometry assay using JC-1 dye. A shift in the fluorescence emission from green (535 nm) to red (595 nm) indicates accumulation of JC-1 in the mitochondria, which is dependent solely on the membrane potential of the mitochondria. Consequently, mitochondrial membrane depolarization is usually accompanied by a decrease in the fluorescence intensity ratio (red/green). KO cells revealed a shift in the fluorescence intensity from red (595 nm) to green (535 nm). The careful analysis of the data revealed that the red/green ratio for the KO cells was at least 3-fold lower than that for the wild type cells (Fig. 5F), indicating H2O2-induced mitochondrial membrane depolarization occurred in KO cells. Pretreatment of cells with 0.5 mm linoleate showed protection in the fluorescence intensity ratio (red/green) decline, indicating the potential role of linoleate in mitochondrial membrane depolarization of KO cells. Elevation of mitochondrial dysfunction is known to promote apoptosis (46). To investigate oxidative stress-mediated apoptosis in KO cells, we measured phosphatidylserine externalization as the marker of apoptosis. Representative FACS dot plots of FITC-conjugated annexin V- and PI-labeled samples are shown in Fig. 5G. KO cells have 18% more apoptotic cells compared with wild type cells. These results explain why the growth rate of KO cells is lower than wild type cells. However, critical analysis of the data revealed that cell death was prevented by linoleate pretreatment but not by oleate pretreatment, suggesting that the cell death of KO cells was due to deficiency of linoleate. Cell death was further visualized by microscopy, where higher PI red and FITC annexin V- green fluorescence was seen in KO cells (Fig. 5H).
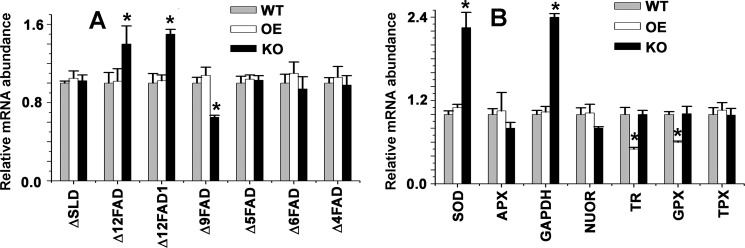
Deletion of LmNcb5or Alleles Leads to Changed Expression of Antioxidants, GAPDH, and FADs mRNA
To investigate the relative quantities of genes in KO, WT, and OE cells, we performed quantitative real time PCR to measure putative sphingolipid Δ4 desaturase (LmjF.26.1690), putative Δ12 desaturase (LmjF.10.0010), Δ12 desaturase (LmjF.33.3270), Δ9 desaturase (LmjF.24.2250), Δ6 desaturase (LmjF.36.6950), Δ5 desaturase (LmjF.07.1090), Δ4 desaturase (LmjF.14.1340), ascorbate peroxidase (LmjF.34.0070), type II (glutathione peroxidase-like) tryparedoxin peroxidase (LmjF.26.0810), putative NADH-ubiquinone oxidoreductase (LmjF.05.0980), putative iron superoxide dismutase (LmjF.30.2770), trypanothione reductase (LmjF.05.0350), tryparedoxin peroxidase (LmjF.15.1120), and glyceraldehydes-3-phosphate dehydrogenase (LmjF.30.2980). Quantitative real-time PCR results demonstrated a higher expression of Δ12FADs (1.5-fold), SOD (2.25-fold), and GAPDH (2.5-fold) mRNA in KO mutants compared with WT or OE cells (Fig. 6). These results indicated that Δ12FADs genes were induced due to decreased Δ12 polyunsaturated FAs (Δ12PUFAs) level. The expression of Δ9FADs was ∼2-fold reduced in KO mutants due to accumulation of oleate present in them compared with WT cells. Except SOD, the expression levels of the other anti-oxidant genes tested did not change significantly (Fig. 6B). A possible explanation for higher SOD expression in KO cells compared with WT cells might be due to increased superoxide generation in KO cells. However, the mRNA expression of type II (glutathione peroxidase-like) tryparedoxin peroxidase and trypanothione reductase in OE cells was 1.5- and 2.0-fold lower compared with WT cells, respectively, suggesting that lower H2O2 in OE might have suppressed the genes.
FIGURE 6.
mRNA status of different types of FA desaturase (A) and antioxidant (B) genes in WT, KO, and OE cells. Measurements of gene transcript abundance was analyzed by using quantitative RT-PCR as detailed under ”Experimental Procedures.” ΔSLD, sphingolipid Δ4 desaturase (LmjF.26.1690); Δ12FAD, Δ12 fatty acid desaturase (LmjF.10.0010); Δ12FAD1, Δ12 fatty acid desaturase (LmjF.33.3270); Δ9FAD, Δ9 fatty acid desaturase (LmjF.24.2250); Δ5FAD, Δ5 fatty acid desaturase (LmjF.07.1090); Δ6FAD, Δ6 fatty acid desaturase (LmjF.36.6950); Δ4FAD, Δ4 fatty acid desaturase (LmjF.14.1340). SOD (LmjF.30.2770); APX, ascorbate peroxidase (LmjF.34.0070); GAPDH (LmjF.30.2980); NUOR, NADH-ubiquinone oxidoreductase (LmjF.05.0980); TR, trypanothione reductase (LmjF.05.0350); GPX, non-selenium glutathione peroxidase (LmjF.26.0810); TPX, tryparedoxin peroxidase (LmjF.15.1120). All data were normalized using 18 S rRNA as the endogenous control. Data show the mean ± S.D. of three independent experiments. * p, 0.05, compared with WT sample. All the data are representative of three independent transfection experiments.
DISCUSSION
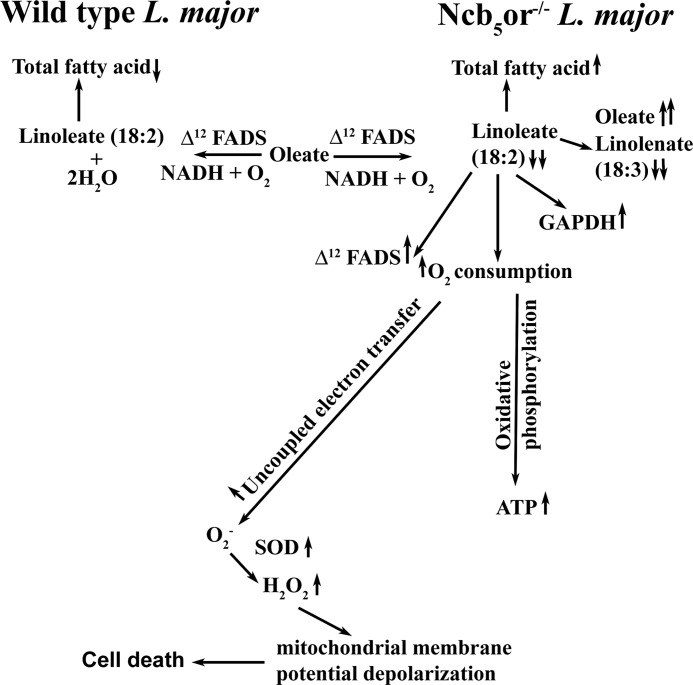
Although Δ12 FAD is well characterized in many other organisms, the redox partner of Δ12 FAD is still unknown in the literature. We for the first time demonstrate that Ncb5or acts as a redox partner (electron donor) for Δ12 FAD (electron acceptor). As shown in Fig. 4, free FA analysis in the Leishmania parasite of KO cells reveals marked reduction in linoleate concentration paralleled with a significant increment in oleate content resulting in an overall reduction in linoleate:oleate ratio. Abnormalities upon knock-out of the Ncb5or gene include increased total FA content, O2 consumption, and H2O2 and ATP generation and fall in mitochondrial membrane potential (Fig. 7). Finally knock-out cells undergo cell death and apoptosis. KO cells multiply slowly compared with wild type cells. A similar type of results has been reported in Δ12-desaturase mutant of Aspergillus parasiticus that shows a 2-fold reduction in growth compared with the wild type cells (47). Interestingly, 12- and 13-thiastearic acid, inhibitors of Δ12-desaturase, can inhibit the growth of the Trypanosoma brucei cells (48). Our findings from Ncb5or-deficient cells are similar to those reported for Δ12-FAD -deficient A. parasiticus, suggesting that both redox partner (Ncb5or) null mutant and redox acceptor (Δ12-FAD) null mutant show similar type of growth inhibition.
FIGURE 7.
Schematic diagram shows the possible role of Ncb5or in Leishmania. Ncb5or deficiency in Leishmania leads to a decrease in Δ12 FA desaturase efficiency and increase in oxidative stress and cell death. Upward or downward arrows indicate higher or lower levels than wild type cells, respectively.
The observed impairment in the Δ12 FA synthesis pathway in Ncb5or-deficient cells triggers marked increase in Δ12 FADs mRNA expression. Like our results, a number of researchers showed earlier that the desaturase gene transcription rate is stimulated in response to exogenous substrate FAs, whereas exposure to product FAs sharply reduces transcription (49, 50). They also proposed that the corresponding positive and negative response elements have been located in the gene upstream promoter region.
Now one question that immediately arises is which FA is apparently involved in KO cell death. One possibility is that the excessive oleate might be toxic to cell, whereas we can also suspect an essential requirement of linoleate for cell survival. The preincubation with BSA-conjugated oleate cannot exacerbate cell death in KO cells, but external addition of BSA-conjugated linoleate is able to protect from the cell death. Our experiments using external BSA-conjugated linoleate further indicates that perturbations of linoleate synthesis act as an upstream event of higher O2 consumption rate, ROS generation, and mitochondrial depolarization.
Linoleate appears to play a role in preventing mitochondrial membrane depolarization and uncoupling, but the reason behind these phenotypic observations is unknown. Table 2 shows that total FA content in Ncb5or null mutant is 2.8-fold greater than that seen for wild type cells. Our results are consistent with earlier reports, where they have shown five times higher total FA in Δ12 FAD-deficient cells compared with wild type cells (47, 51). They have demonstrated from their knock-out study that the linoleate down-regulates the FA biosynthesis genes. It has been well known for nearly 50 years that polyunsaturated FA exclusively suppresses lipid synthesis by coordinating a down-regulation of lipid synthesis and an up-regulation of lipid oxidation genes (52). In general FAs act as an uncoupler, which increases in mitochondrial state 4 respiration with a concomitant decrease in the P/O ratio (53, 54). Our results suggest that a higher amount of total FA content in Ncb5or null mutant may be responsible for a higher level of uncoupled electron transfer. The following possibilities have been proposed to explain the role of FA in uncoupled electron transfer. (a) FAs act as a protonophoric for dissipation of the mitochondrial inner membrane potential (53, 55). (b) FAs interfere with the proton-pumping machinery of the respiratory chain (56–60).
Kang et al. (61) show that free polyunsaturated FAs can reduce membrane electrical excitability of heart cells. Several reports suggest that the polyunsaturated FAs acts as the metabolic fuel “repartitioners” and may protect cells against the accelerated rates of apoptosis reportedly observed on excessive triglyceride accumulation (52, 62, 63). Like muscle cells, an increase in O2 consumption and ATP generation and induction of the energy-producing glyceraldehyde-3-phosphate dehydrogenase mRNA gene (64) are seen in Ncb5or-null Leishmania cells. Subsequently, an increase in cellular oxidative stress (H2O2) and mRNA expression of the oxidative stress-responsive gene (superoxide dismutase) are observed in KO cells. Thus, these results strongly suggest that linoleate in Leishmania protects cells against the accelerated rates of cell death by suppressing excessive generation of cellular oxidative stress.
We have suggested that the deficiency of linoleate in Ncb5or knock-out cell may cause increasing H2O2 and membrane depolarization, which is followed by apoptosis and cell death, but question obviously arise about the mechanism. Several workers have shown that H2O2 treatment of Leishmania cells results in mitochondrial dysfunction and is accompanied by cellular death (38, 46, 65–67). In mammalian cells an increase in cellular H2O2 generation has been claimed to be responsible for cell death (68). Thus, impaired linoleate synthesis-induced H2O2 generation in KO cells is one of the main causative agents for apoptosis and cell death. The other possible mechanism is that impaired linoleate synthesis may cause dysregulation in cardiolipin synthesis (69, 70), which is associated with release of cytochrome c from the mitochondrial membrane into the cytosol that may initiate apoptotic signals in heart tissue (41, 71, 72). The measurement of cardiolipin and reconstitution with tetralinoleoyl cardiolipin-rich vesicles in KO cells will be helpful in elucidating the exact molecular mechanisms of apoptosis in linoleate-deficient Leishmania in the near future.
Supplementary Material
Acknowledgments
We thank Dr. S. M. Beverley (Washington University, St. Louis, MO) for providing pXG-B2863, pXG-neo, and pXG-hyg vectors as gifts and Dr. Swati Pal for helpful discussions.
This work was supported by Council of Scientific and Industrial Research (CSIR) Project NWP 0038, UGC Fellowships (to S. M.), CSIR Fellowships (to S. D., M. B.), and CSIR-SPM Fellowships (to S. S. S.).

This article contains supplemental Table S1 and Figs. S1 and S2.
- FA
- fatty acid
- FAD
- FA desaturase
- cb5
- cytochrome b5
- cb5r
- cb5 reductase
- Ncb5or
- NAD(P)H cytochrome b5 oxidoreductase
- LmNcb5or
- Ncb5or from L. major
- OE
- LmNcb5or-overexpressing cell
- KO
- LmNcb5or knockout cell
- SOD
- superoxide dismutase
- PI
- propidium iodide
- DCIP
- 2,6-dichloroindophenol
- P/O
- phosphorylation/oxygen.
REFERENCES
- 1. Steiger R. F., Steiger E. (1977) Cultivation of Leishmania donovani and Leishmania braziliensis in defined media. Nutritional requirements. J. Protozool 24, 437–441 [DOI] [PubMed] [Google Scholar]
- 2. Paul K. S., Jiang D., Morita Y. S., Englund P. T. (2001) Fatty acid synthesis in African trypanosomes. A solution to the myristate mystery. Trends Parasitol. 17, 381–387 [DOI] [PubMed] [Google Scholar]
- 3. Morita Y. S., Paul K. S., Englund P. T. (2000) Specialized fatty acid synthesis in African trypanosomes. Myristate for GPI anchors. Science 288, 140–143 [DOI] [PubMed] [Google Scholar]
- 4. Lee S. H., Stephens J. L., Paul K. S., Englund P. T. (2006) Fatty acid synthesis by elongases in trypanosomes. Cell 126, 691–699 [DOI] [PubMed] [Google Scholar]
- 5. Tripodi K. E., Buttigliero L. V., Altabe S. G., Uttaro A. D. (2006) Functional characterization of front-end desaturases from trypanosomatids depicts the first polyunsaturated fatty acid biosynthetic pathway from a parasitic protozoan. FEBS J. 273, 271–280 [DOI] [PubMed] [Google Scholar]
- 6. Livore V. I., Tripodi K. E., Uttaro A. D. (2007) Elongation of polyunsaturated fatty acids in trypanosomatids. FEBS J. 274, 264–274 [DOI] [PubMed] [Google Scholar]
- 7. Lee S. H., Stephens J. L., Englund P. T. (2007) A fatty-acid synthesis mechanism specialized for parasitism. Nat. Rev. Microbiol. 5, 287–297 [DOI] [PubMed] [Google Scholar]
- 8. Tripodi K. E., Menendez Bravo S. M., Cricco J. A. (2011) Role of heme and heme-proteins in trypanosomatid essential metabolic pathways. Enzyme Res. 2011, 873230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Uttaro A. D. (2006) Biosynthesis of polyunsaturated fatty acids in lower eukaryotes. IUBMB Life 58, 563–571 [DOI] [PubMed] [Google Scholar]
- 10. Zhu H., Qiu H., Yoon H. W., Huang S., Bunn H. F. (1999) Identification of a cytochrome b-type NAD(P)H oxidoreductase ubiquitously expressed in human cells. Proc. Natl. Acad. Sci. U.S.A. 96, 14742–14747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu M., Wang W., Frontera J. R., Neely M. C., Lu J., Aires D., Hsu F. F., Turk J., Swerdlow R. H., Carlson S. E., Zhu H. (2011) Ncb5or deficiency increases fatty acid catabolism and oxidative stress. J. Biol. Chem. 286, 11141–11154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Larade K., Jiang Z., Zhang Y., Wang W., Bonner-Weir S., Zhu H., Bunn H. F. (2008) Loss of Ncb5or results in impaired fatty acid desaturation, lipoatrophy, and diabetes. J. Biol. Chem. 283, 29285–29291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oshino N., Imai Y., Sato R. (1971) A function of cytochrome b5 in fatty acid desaturation by rat liver microsomes. J. Biochem. 69, 155–167 [DOI] [PubMed] [Google Scholar]
- 14. Keyes S. R., Cinti D. L. (1980) Biochemical properties of cytochrome b5-dependent microsomal fatty acid elongation and identification of products. J. Biol. Chem. 255, 11357–11364 [PubMed] [Google Scholar]
- 15. Reddy V. V., Kupfer D., Caspi E. (1977) Mechanism of C-5 double bond introduction in the biosynthesis of cholesterol by rat liver microsomes. J. Biol. Chem. 252, 2797–2801 [PubMed] [Google Scholar]
- 16. Hildebrandt A., Estabrook R. W. (1971) Evidence for the participation of cytochrome b5 in hepatic microsomal mixed-function oxidation reactions. Arch. Biochem. Biophys. 143, 66–79 [DOI] [PubMed] [Google Scholar]
- 17. Oshino N. (1980) Hepatic Cytochrome P450 Monooxygenase Systems, pp. 407–447, Pergamon Press, New York [Google Scholar]
- 18. Paltuaf F., Prough R. A., Masters B. S., Johnston J. M. (1974) Evidence for the participation of cytochrome b5 in plasmalogen biosynthesis. J. Biol. Chem. 249, 2661–2662 [PubMed] [Google Scholar]
- 19. Pugh E. L., Kates M. (1977) Direct desaturation of eicosatrienoyl lecithin to arachidonoyl lecithin by rat liver microsomes. J. Biol. Chem. 252, 68–73 [PubMed] [Google Scholar]
- 20. Fukushima H., Grinstead G. F., Gaylor J. L. (1981) Total enzymic synthesis of cholesterol from lanosterol. Cytochrome b5 dependence of 4-methyl sterol oxidase. J. Biol. Chem. 256, 4822–4826 [PubMed] [Google Scholar]
- 21. Strittmatter P., Machuga E. T., Roth G. J. (1982) Reduced pyridine nucleotides and cytochrome b5 as electron donors for prostaglandin synthetase reconstituted in dimyristyl phosphatidylcholine vesicles. J. Biol. Chem. 257, 11883–11886 [PubMed] [Google Scholar]
- 22. Loughran P. A., Roman L. J., Miller R. T., Masters B. S. (2001) The kinetic and spectral characterization of the E. coli-expressed mammalian CYP4A7. Cytochrome b5 effects vary with substrate. Arch. Biochem. Biophys. 385, 311–321 [DOI] [PubMed] [Google Scholar]
- 23. Hyde G. E., Crawford N. M., Campbell W. H. (1991) The sequence of squash NADH:nitrate reductase and its relationship to the sequences of other flavoprotein oxidoreductases. A family of flavoprotein pyridine nucleotide cytochrome reductases. J. Biol. Chem. 266, 23542–23547 [PubMed] [Google Scholar]
- 24. Kisker C., Schindelin H., Pacheco A., Wehbi W. A., Garrett R. M., Rajagopalan K. V., Enemark J. H., Rees D. C. (1997) Molecular basis of sulfite oxidase deficiency from the structure of sulfite oxidase. Cell 91, 973–983 [DOI] [PubMed] [Google Scholar]
- 25. Rudolph M. J., Johnson J. L., Rajagopalan K. V., Kisker C. (2003) The 1.2 A structure of the human sulfite oxidase cytochrome b5 domain. Acta Crystallogr. D. Biol. Crystallogr. 59, 1183–1191 [DOI] [PubMed] [Google Scholar]
- 26. Xia Z. X., Shamala N., Bethge P. H., Lim L. W., Bellamy H. D., Xuong N. H., Lederer F., Mathews F. S. (1987) Three-dimensional structure of flavocytochrome b2 from bakers' yeast at 3.0-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 84, 2629–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thiede M. A., Ozols J., Strittmatter P. (1986) Construction and sequence of cDNA for rat liver stearyl coenzyme A desaturase. J. Biol. Chem. 261, 13230–13235 [PubMed] [Google Scholar]
- 28. Napier J. A., Sayanova O., Sperling P., Heinz E. (1999) A growing family of cytochrome b5 - domain fusion proteins. Trends Plant Sci. 4, 2–4 [Google Scholar]
- 29. Mitchell A. G., Martin C. E. (1995) A novel cytochrome b5-like domain is linked to the carboxyl terminus of the Saccharomyces cerevisiae Δ9 fatty acid desaturase. J. Biol. Chem. 270, 29766–29772 [DOI] [PubMed] [Google Scholar]
- 30. Zhu H., Larade K., Jackson T. A., Xie J., Ladoux A., Acker H., Berchner-Pfannschmidt U., Fandrey J., Cross A. R., Lukat-Rodgers G. S., Rodgers K. R., Bunn H. F. (2004) NCB5OR is a novel soluble NAD(P)H reductase localized in the endoplasmic reticulum. J. Biol. Chem. 279, 30316–30325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xie J., Zhu H., Larade K., Ladoux A., Seguritan A., Chu M., Ito S., Bronson R. T., Leiter E. H., Zhang C. Y., Rosen E. D., Bunn H. F. (2004) Absence of a reductase, NCB5OR, causes insulin-deficient diabetes. Proc. Natl. Acad. Sci. U.S.A. 101, 10750–10755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Y., Larade K., Jiang Z. G., Ito S., Wang W., Zhu H., Bunn H. F. (2010) The flavoheme reductase Ncb5or protects cells against endoplasmic reticulum stress-induced lipotoxicity. J. Lipid Res. 51, 53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Larade K., Jiang Z. G., Dejam A., Zhu H., Bunn H. F. (2007) The reductase NCB5OR is responsive to the redox status in beta cells and is not involved in the ER stress response. Biochem. J. 404, 467–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ivens A. C., Peacock C. S., Worthey E. A., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M. A., Adlem E., Aert R., Anupama A., Apostolou Z., Attipoe P., Bason N., Bauser C., Beck A., Beverley S. M., Bianchettin G., Borzym K., Bothe G., Bruschi C. V., Collins M., Cadag E., Ciarloni L., Clayton C., Coulson R. M., Cronin A., Cruz A. K., Davies R. M., De Gaudenzi J., Dobson D. E., Duesterhoeft A., Fazelina G., Fosker N., Frasch A. C., Fraser A., Fuchs M., Gabel C., Goble A., Goffeau A., Harris D., Hertz-Fowler C., Hilbert H., Horn D., Huang Y., Klages S., Knights A., Kube M., Larke N., Litvin L., Lord A. (2005) The genome of the kinetoplastid parasite Leishmania major. Science 309, 436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alloatti A., Uttaro A. D. (2011) Highly specific methyl-end fatty acid desaturases of trypanosomatids. Mol. Biochem. Parasitol. 175, 126–132 [DOI] [PubMed] [Google Scholar]
- 36. Adak S., Datta A. K. (2005) Leishmania major encodes an unusual peroxidase that is a close homologue of plant ascorbate peroxidase. A novel role of the transmembrane domain. Biochem. J. 390, 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dolai S., Pal S., Yadav R. K., Adak S. (2011) Endoplasmic reticulum stress-induced apoptosis in Leishmania through Ca2+-dependent and caspase-independent mechanism. J. Biol. Chem. 286, 13638–13646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pal S., Dolai S., Yadav R. K., Adak S. (2010) Ascorbate peroxidase from Leishmania major controls the virulence of infective stage of promastigotes by regulating oxidative stress. PLoS One 5, e11271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Paul K. G., Theorell H., Akeson A. (1953) The molar light absorption of pyridine ferroprotoporphyrin (pyridine haemochromogen). Acta Chem. Scand. 7, 1284–1287 [Google Scholar]
- 40. Adak S., Ghosh S., Abu-Soud H. M., Stuehr D. J. (1999) Role of reductase domain cluster 1 acidic residues in neuronal nitric-oxide synthase. Characterization of the FMN-FREE enzyme. J. Biol. Chem. 274, 22313–22320 [DOI] [PubMed] [Google Scholar]
- 41. Dolai S., Yadav R. K., Pal S., Adak S. (2008) Leishmania major ascorbate peroxidase overexpression protects cells against reactive oxygen species-mediated cardiolipin oxidation. Free Radic. Biol. Med. 45, 1520–1529 [DOI] [PubMed] [Google Scholar]
- 42. Folch J., Lees M., Sloane Stanley G. H. (1957) A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 43. McCarthy-Burke C., Bates P. A., Dwyer D. M. (1991) Leishmania donovani: use of two different, commercially available, chemically defined media for the continuous in vitro cultivation of promastigotes. Exp. Parasitol. 73, 385–387 [DOI] [PubMed] [Google Scholar]
- 44. Dolai S., Yadav R. K., Datta A. K., Adak S. (2007) Effect of thiocyanate on the peroxidase and pseudocatalase activities of Leishmania major ascorbate peroxidase. Biochim. Biophys. Acta 1770, 247–256 [DOI] [PubMed] [Google Scholar]
- 45. Santhamma K. R., Bhaduri A. (1995) Characterization of the respiratory chain of Leishmania donovani promastigotes. Mol. Biochem. Parasitol 75, 43–53 [DOI] [PubMed] [Google Scholar]
- 46. Dolai S., Yadav R. K., Pal S., Adak S. (2009) Overexpression of mitochondrial Leishmania major ascorbate peroxidase enhances tolerance to oxidative stress-induced programmed cell death and protein damage. Eukaryot. Cell 8, 1721–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wilson R. A., Calvo A. M., Chang P. K., Keller N. P. (2004) Characterization of the Aspergillus parasiticus Δ12-desaturase gene. A role for lipid metabolism in the Aspergillus-seed interaction. Microbiology 150, 2881–2888 [DOI] [PubMed] [Google Scholar]
- 48. Alloatti A., Gupta S., Gualdrón-López M., Igoillo-Esteve M., Nguewa P. A., Deumer G., Wallemacq P., Altabe S. G., Michels P. A., Uttaro A. D. (2010) Genetic and chemical evaluation of Trypanosoma brucei oleate desaturase as a candidate drug target. PLoS One 5, e14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Choi J. Y., Stukey J., Hwang S. Y., Martin C. E. (1996) Regulatory elements that control transcription activation and unsaturated fatty acid-mediated repression of the Saccharomyces cerevisiae OLE1 gene. J. Biol. Chem. 271, 3581–3589 [DOI] [PubMed] [Google Scholar]
- 50. Duplus E., Glorian M., Forest C. (2000) Fatty acid regulation of gene transcription. J. Biol. Chem. 275, 30749–30752 [DOI] [PubMed] [Google Scholar]
- 51. Calvo A. M., Gardner H. W., Keller N. P. (2001) Genetic connection between fatty acid metabolism and sporulation in Aspergillus nidulans. J. Biol. Chem. 276, 25766–25774 [DOI] [PubMed] [Google Scholar]
- 52. Clarke S. D. (2001) Nonalcoholic steatosis and steatohepatitis. I. Molecular mechanism for polyunsaturated fatty acid regulation of gene transcription. Am. J. Physiol. Gastrointest. Liver Physiol. 281, G865–G869 [DOI] [PubMed] [Google Scholar]
- 53. Wojtczak L., Schönfeld P. (1993) Effect of fatty acids on energy coupling processes in mitochondria. Biochim. Biophys. Acta 1183, 41–57 [DOI] [PubMed] [Google Scholar]
- 54. Hermesh O., Kalderon B., Bar-Tana J. (1998) Mitochondria uncoupling by a long chain fatty acyl analogue. J. Biol. Chem. 273, 3937–3942 [DOI] [PubMed] [Google Scholar]
- 55. Wojtczak L. (1976) Effect of long-chain fatty acids and acyl-CoA on mitochondrial permeability, transport, and energy-coupling processes. J. Bioenerg. Biomembr. 8, 293–311 [DOI] [PubMed] [Google Scholar]
- 56. Rottenberg H., Hashimoto K. (1986) Fatty acid uncoupling of oxidative phosphorylation in rat liver mitochondria. Biochemistry 25, 1747–1755 [DOI] [PubMed] [Google Scholar]
- 57. Rottenberg H. (1990) Decoupling of oxidative phosphorylation and photophosphorylation. Biochim. Biophys. Acta 1018, 1–17 [DOI] [PubMed] [Google Scholar]
- 58. Luvisetto S., Pietrobon D., Azzone G. F. (1987) Uncoupling of oxidative phosphorylation. 1. Protonophoric effects account only partially for uncoupling. Biochemistry 26, 7332–7338 [DOI] [PubMed] [Google Scholar]
- 59. Pietrobon D., Luvisetto S., Azzone G. F. (1987) Uncoupling of oxidative phosphorylation. 2. Alternative mechanisms. Intrinsic uncoupling or decoupling? Biochemistry 26, 7339–7347 [DOI] [PubMed] [Google Scholar]
- 60. Luvisetto S., Buso M., Pietrobon D., Azzone G. F. (1990) On the nature of the uncoupling effect of fatty acids. J. Bioenerg. Biomembr. 22, 635–643 [DOI] [PubMed] [Google Scholar]
- 61. Kang J. X., Xiao Y. F., Leaf A. (1995) Free, long-chain, polyunsaturated fatty acids reduce membrane electrical excitability in neonatal rat cardiac myocytes. Proc. Natl. Acad. Sci. U.S.A. 92, 3997–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kakuma T., Lee Y., Higa M., Wang Z., Pan W., Shimomura I., Unger R. H. (2000) Leptin, troglitazone, and the expression of sterol regulatory element binding proteins in liver and pancreatic islets. Proc. Natl. Acad. Sci. U.S.A. 97, 8536–8541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhou Y. T., Grayburn P., Karim A., Shimabukuro M., Higa M., Baetens D., Orci L., Unger R. H. (2000) Lipotoxic heart disease in obese rats. Implications for human obesity. Proc. Natl. Acad. Sci. U.S.A. 97, 1784–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Barber R. D., Harmer D. W., Coleman R. A., Clark B. J. (2005) GAPDH as a housekeeping gene. Analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol Genomics 21, 389–395 [DOI] [PubMed] [Google Scholar]
- 65. Gannavaram S., Vedvyas C., Debrabant A. (2008) Conservation of the pro-apoptotic nuclease activity of endonuclease G in unicellular trypanosomatid parasites. J. Cell Sci. 121, 99–109 [DOI] [PubMed] [Google Scholar]
- 66. Das M., Mukherjee S. B., Shaha C. (2001) Hydrogen peroxide induces apoptosis-like death in Leishmania donovani promastigotes. J. Cell Sci. 114, 2461–2469 [DOI] [PubMed] [Google Scholar]
- 67. Mukherjee S. B., Das M., Sudhandiran G., Shaha C. (2002) Increase in cytosolic Ca2+ levels through the activation of non-selective cation channels induced by oxidative stress causes mitochondrial depolarization leading to apoptosis-like death in Leishmania donovani promastigotes. J. Biol. Chem. 277, 24717–24727 [DOI] [PubMed] [Google Scholar]
- 68. Vander Heiden M. G., Chandel N. S., Williamson E. K., Schumacker P. T., Thompson C. B. (1997) Bcl-xL regulates the membrane potential and volume homeostasis of mitochondria. Cell 91, 627–637 [DOI] [PubMed] [Google Scholar]
- 69. Chicco A. J., Sparagna G. C. (2007) Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am. J. Physiol. Cell Physiol. 292, C33–C44 [DOI] [PubMed] [Google Scholar]
- 70. Schlame M., Rua D., Greenberg M. L. (2000) The biosynthesis and functional role of cardiolipin. Prog. Lipid Res. 39, 257–288 [DOI] [PubMed] [Google Scholar]
- 71. Ostrander D. B., Sparagna G. C., Amoscato A. A., McMillin J. B., Dowhan W. (2001) Decreased cardiolipin synthesis corresponds with cytochrome c release in palmitate-induced cardiomyocyte apoptosis. J. Biol. Chem. 276, 38061–38067 [DOI] [PubMed] [Google Scholar]
- 72. Mulligan C. M., Sparagna G. C., Le C. H., De Mooy A. B., Routh M. A., Holmes M. G., Hickson-Bick D. L., Zarini S., Murphy R. C., Xu F. Y., Hatch G. M., McCune S. A., Moore R. L., Chicco A. J. (2012) Dietary linoleate preserves cardiolipin and attenuates mitochondrial dysfunction in the failing rat heart. Cardiovasc Res. 94, 460–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.