Background: The interaction between methionine and aromatic residues in protein complexes is poorly understood.
Results: The Met-aromatic motif is prevalent in known protein structures and stabilizes TNF ligand-receptor binding interactions.
Conclusion: The Met sulfur-aromatic binding motif provides additional stabilization over purely hydrophobic interactions and at longer distances.
Significance: This motif is prevalent and may be associated with a number of mutation- and age-associated diseases.
Keywords: Bioinformatics, Computational Biology, Methionine, Molecular Modeling, Protein Structure, Protein-Protein Interactions, Quantum Chemistry, Tumor Necrosis Factor (TNF)
Abstract
Of the 20 amino acids, the precise function of methionine (Met) remains among the least well understood. To establish a determining characteristic of methionine that fundamentally differentiates it from purely hydrophobic residues, we have used in vitro cellular experiments, molecular simulations, quantum calculations, and a bioinformatics screen of the Protein Data Bank. We show that approximately one-third of all known protein structures contain an energetically stabilizing Met-aromatic motif and, remarkably, that greater than 10,000 structures contain this motif more than 10 times. Critically, we show that as compared with a purely hydrophobic interaction, the Met-aromatic motif yields an additional stabilization of 1–1.5 kcal/mol. To highlight its importance and to dissect the energetic underpinnings of this motif, we have studied two clinically relevant TNF ligand-receptor complexes, namely TRAIL-DR5 and LTα-TNFR1. In both cases, we show that the motif is necessary for high affinity ligand binding as well as function. Additionally, we highlight previously overlooked instances of the motif in several disease-related Met mutations. Our results strongly suggest that the Met-aromatic motif should be exploited in the rational design of therapeutics targeting a range of proteins.
Introduction
The biophysical and biochemical roles of methionine (Met) remain among the least well understood of the 20 amino acids. Mutations involving Met are associated with a number of pathological conditions, including Alzheimer, Creutzfield-Jacob, and von Willebrand disease. Despite its role in such pathologies, the structural and thermodynamic consequences of Met mutations are not well defined. Underlying this uncertainty, there remains an ambiguous biochemical classification of Met throughout the structural biology literature that reflects a poor understanding of its most basic biophysical interactions within structured proteins; at times Met can be classified as any one of a number of things, including nonpolar, polar, and weakly polar. Thus, in the context of protein folding and function, it remains fundamentally unclear how the sulfur atom differentiates Met from the other hydrophobic residues (e.g. valine, leucine, and isoleucine). In addition to the ambiguity of its role in stabilizing specific interactions within proteins and protein complexes, the quantum mechanical basis for its unique role has similarly remained elusive. Clarifying these ambiguities regarding Met is the focus of this study.
One potentially important finding regarding Met that has been largely overlooked is its propensity to interact with aromatic-containing residues, including tryptophan (Trp), tyrosine (Tyr), and phenylalanine (Phe). The higher-than-expected frequency of sulfur- and aromatic-containing residues in close proximity was first noted in a brief bioinformatics study of eight protein structures (1, 2). More extensive bioinformatics results extended these findings to include additional protein complexes as well as small molecules (3–5). Efforts to understand the chemistry of the sulfur-aromatic interaction have relied upon studies of small model compounds; in particular, quantum and in vacuo classical mechanical calculations of the interaction between dimethyl sulfide (DMS)2 and benzene were consistent with the bioinformatics findings, as were results from an experimental study of DMS and methylnaphthalene, which observed a 1:1 interaction between sulfur- and aromatic-containing compounds (6). Atomistic molecular dynamics (MD) simulations have previously shown the stability of a sulfur-aromatic contact in a model α-helix (7, 8). Collectively, these studies suggest that the energy associated with a sulfur-aromatic interaction is on the order of 1–3 kcal/mol; the intermolecular distance is ∼5.5 Å (between the sulfur and the ring center), and there is an orientational preference of 30–60° (between the sulfur and the normal vector defined by the plane of the aromatic ring), depending on the nature of the aromatic group. Despite their apparent promiscuity, the functionality of sulfur-aromatic interactions has not been explored deeply within the context of full-length protein structures. Herein, we make use of in vitro experiments, molecular simulations, and quantum computation to describe sulfur-aromatic interactions within protein complexes that are central targets of molecular therapy.
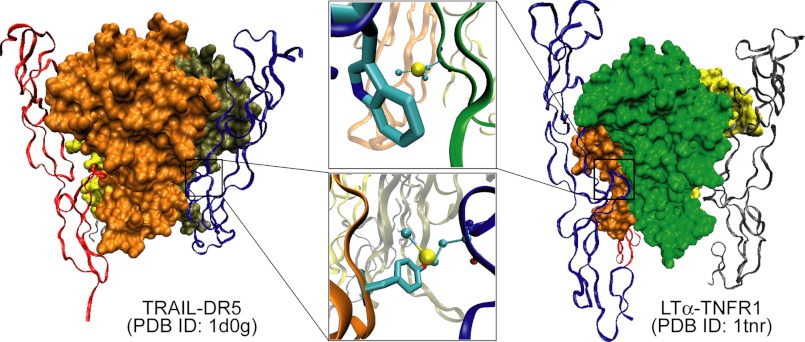
We have studied two distinct tumor necrosis factor (TNF) ligand-receptor complexes, the TNF-related apoptosis-inducing ligand (TRAIL) in complex with death receptor 5 (DR5 or TRAIL receptor 2) (9) and lymphotoxin-α (LTα or TNFβ) ligand in complex with TNF receptor 1 (TNFR1) (Fig. 1) (10), revealing potentially important Met sulfur-aromatic interactions. DR5 and TNFR1 are structurally homologous TNF receptors, having ∼40% sequence similarity (and ∼30% sequence identity) within their extracellular domains. Moreover, in their ligand-bound state, the structurally homologous complexes are formed via the high affinity engagement of a trimeric ligand by three noninteracting receptor monomers forming a symmetric trimer unit. TRAIL binds DR5 (and other TRAIL receptors, death receptor 4 and decoy receptors 1 and 2) with nanomolar affinity (11), and this high affinity, high specificity binding has been attributed to a number of noncovalent interactions largely clustered within two receptor loops that bury deep within the ligand (9, 12, 13). In DR5, these loops are known as the 50-s loop (residues 51–65) and the 90-s loop (residues 91–104), with interactions via the 90-s loop accounting for 85% of the buried surface area of DR5 in the ligand-receptor complex (Fig. 1, TRAIL-DR5, see boxed region) (9). Comparison of the TRAIL-DR5 and LTα-TNFR1 complexes, shown in Fig. 1, illustrates that a series of residues in TNFR1, the 100-s loop, residues 105–110 (previously called the d–e loop), has a well defined conformation resembling the 90-s loop of DR5 (Fig. 1, boxed region). Upon careful examination of the interactions mediated by this binding loop in each structure, we noted that both complexes contain a motif consistent with the previously described interaction between the sulfur-containing Met residue and aromatic-containing residues (Fig. 1, see enlargement). In the TRAIL-DR5 complex, the distance between the DR5/Met-99 sulfur atom and the TRAIL/Tyr-237 aromatic ring is 5.4 Å, and the separation between Met sulfur and Trp aromatic in the LTα-TNFR1 complex is 5.6 Å. From an evolutionary standpoint, it is conceivable that such an interaction may have been conserved across protein complexes of the same superfamily, such as the TNF ligand-receptor superfamily. However, perhaps most intriguingly, in the LTα-TNFR1 structure, the pairing is reversed; the aromatic residue, Trp-107, is found in the receptor, whereas the sulfur-containing residue, Met-120, is in the ligand, although the separation between sulfur and aromatic groups is comparable. Collectively, these observations suggest that the Met-aromatic interaction may not be an evolutionarily conserved motif in these TNF complexes, rather it is potentially a common motif that provides stabilization of protein structure as well as complex formation, such as high affinity and high specificity binding in the case of TNF ligand-receptor complexes.
FIGURE 1.
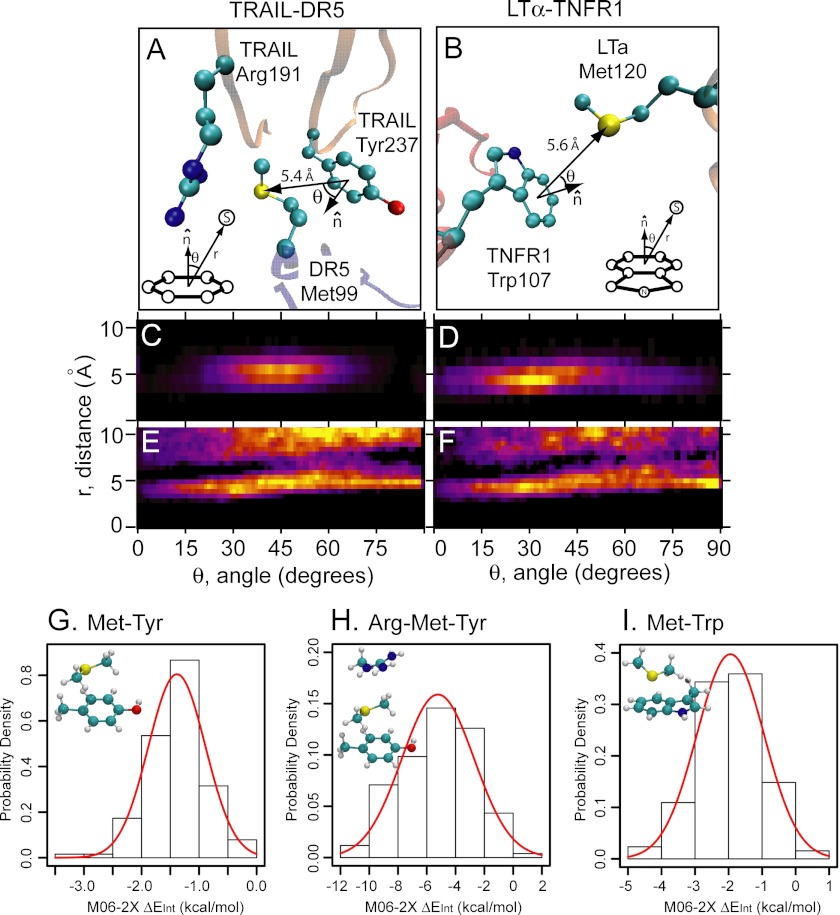
Crystal structures of the trimeric TRAIL-DR5 (left panel, PDB code 1d0g) and LTα-TNFR1 (right panel, PDB code 1tnr) complexes show a conserved binding loop in each receptor, the 90-s loop in DR5 and the 100-s loop in TNFR1, forming extensive contacts with the ligand (boxed region). Within this binding region, both structures show a single methionine aromatic interaction: DR5-Met-99/TRAIL-Tyr-237 (bottom box) and TNFR1-Trp107/LTα-Met-120 (top box). Both interactions are at a ∼5-Å separation.
Although there has been no specific discussion of the role of a sulfur-aromatic interaction in either complex, previous results suggest that these two residues are important for ligand binding in the TRAIL-DR5 complex (11, 14). Single amino acid mutation of DR5/Met-99 almost completely abolished the ligand-receptor interaction, as was observed by co-immunoprecipitation studies (14). Sequence alignment of DR5 with other TRAIL receptors show this Met residue to be conserved across all TRAIL receptors (data not shown), further highlighting the importance of this residue and potentially the motif in the formation of the complex, although DR5 is the only receptor for which a structure exists in complex with TRAIL. In a separate study, surface plasmon resonance measurements showed that mutation of Tyr-237 increases TRAIL-DR5 dissociation 5-fold (11). However, despite their physical proximity and, in the case of TRAIL receptors, sequence identity, no relationship between these mutants, or these residues in general, has been proposed. Furthermore, there is no empirical evidence to suggest that the sulfur- or aromatic-containing residues in LTα and TNFR1 (Met-120 and Trp-107, respectively) play any role in the ligand receptor interaction.
We demonstrate here that both Met and aromatic residues are necessary for high affinity ligand-receptor binding and function in both the TRAIL-DR5 and LTα-TNFR1 complexes. We highlight the importance of this interaction using bioinformatics tools, MD simulations, and quantum mechanical calculations. Our results demonstrate that, in comparison with a purely hydrophobic interaction with structural similarity, the sulfur-aromatic interaction at an ∼5 Å separation is more stable and has an orientational preference, allowing for a stronger interaction at greater distances than is provided by the hydrophobic interaction via alkylic equivalent. Our quantum mechanical (QM) calculations indicate that dispersion effects are responsible for this behavior. In our model protein complexes, MD simulations produced a range of protein configurations consistent with those found within the Protein Data Bank (PDB). Collectively, the sulfur-aromatic interaction in TRAIL-DR5 and LTα-TNFR1 yields an interaction energy of ∼1–1.5 kcal/mol greater than a hydrophobic interaction, and at a specific longer distance, enough to disrupt the ligand-receptor interaction.
EXPERIMENTAL PROCEDURES
Reagents
HEK293 cells were maintained in DMEM supplemented with 10% fetal bovine serum (HyClone), penicillin/streptomycin, and l-glutamine. Jurkat cells were cultured in RPMI supplemented with 10% FBS, penicillin/streptomycin, l-glutamine, and nonessential amino acids. Cells were maintained at subconfluence at 37 °C and 5% CO2 in a water-jacketed incubator. Antibodies against DR5, TNFR1, and IκBα were purchased from Cell Signaling Technologies.
Cloning and Ligand Purification
Mutants of DR5, TNFR1, TRAIL, and LTα were generated by the two-step PCR-based process of targeted mutagenesis with primers obtained from the University of Minnesota BioMedical Genomics Center. Wild type and mutant DR5 (Met mutants M99A, M99V, M99I, and M99C) as well as wild type and mutant TNFR1 (W107A) were inserted into the pcDNA3.1(+) vector using EcoRI and XhoI restriction enzyme sites. All plasmids were verified by sequencing by the University of Minnesota Biomedical Genomics Center. We also note that the numbering of this residue varies within the literature; due to the presence of an ∼52-amino acid N-terminal signaling sequence, Met-99 is sometimes referred to as Met-152 (within the described 90-s loop); here we use the number of the mature protein as was used with the original TRAIL-DR5 structure (9).
Soluble TRAIL (residues 95–114) and LTα (residues 35–205) and mutants were inserted in-frame with the FLAG sequence (DYKDDDDK) in the pT7-FLAG-1 expression vector. Ligands were produced and purified as described previously (15). Briefly, the N-terminal FLAG-tagged ligands were expressed in BL21(DE3) bacteria using isopropyl 1-thio-β-d-galactopyranoside and purified using the FLAG-affinity column (M2 anti-FLAG-agarose resin). Expression and purification were verified using Coomassie stain; concentrations were estimated using BCA assay, and activity of ligands was verified.
Co-immunoprecipitation and Activity Assays
HEK293 cells were transiently transfected with DR5 (WT or Met-99 mutants) or TNFR1 (WT or W107A) using the calcium phosphate method. Twenty four to 48 h after transfection, FLAG-TRAIL was added to cells at 500 ng/ml for 30 min. Cells were washed three times in PBS to dilute excess unbound ligand and subsequently lysed in Nonidet P-40 lysis buffer (50 mm Tris, 75 mm NaCl, 4 mm EDTA, 1% Nonidet P-40) containing protease inhibitors on ice for 60 min. Lysate was recovered after centrifugation at 13,000 × g for 10 min at 4 °C. Equal amounts of total protein (estimated by BCA assay) were subjected to immunoprecipitation using anti-FLAG M2-agarose beads (Sigma) overnight at 4 °C with gentle rotation. Beads were then washed with Nonidet P-40 buffer three times, resuspended in 2× Laemmli sample buffer with DTT and β-mercaptoethanol, boiled for 10 min, and run on an SDS-polyacrylamide gel (10%). Additionally, equal amounts of total protein from whole cell extract were run on SDS-polyacrylamide gel to verify equal expression of DR5 constructs. SDS-polyacrylamide gels were transferred to nitrocellulose membrane and blotted for DR5.
TRAIL activity was measured by MTT assay. Cells were seeded at 2 × 10^5 cells per ml in a 96-well plate and treated with the indicated concentration of TRAIL for 24 h. Cell viability was measured by incubation with MTT (0.5 mg/ml) during the last 3 h of incubation. Percent cell viability was calculated relative to untreated wells. LTα activity was assessed by measured IκBα degradation, a common marker for NFκB activation. Cells were plated at subconfluence and treated with the indicated concentration of LTα for 15 min and lysed. Equal amounts of total protein were loaded on SDS-PAGE and analyzed for IκBα expression, the degradation of which indicates NFκB activity.
Bioinformatics
The Protein Data Bank (16, 17) was analyzed using Biopython toolset. A custom Python script utilizing the PDBParser was used to parse each PDB structural entity into its component parts (i.e. model, chain, residue, and atom) and access atomic coordinate data. The sulfur-aromatic distance is defined as the separation between the Met sulfur (SD atom) and the center of mass of the six-membered aromatic ring. The angle is defined as the angle between the sulfur-aromatic vector and the normal vector of the aromatic ring. All interactions were calculated within a 20-Å cutoff distance, but for clarity only those interactions within 10 Å are shown. As a control, all interactions between aromatic groups and carbon (CH2 and CH3), nitrogen, oxygen, and sulfur were calculated on a subset of the PDB representing a random collection of structures.
Analysis of the Biopython-generated data, including probability density, interactions by structure, and two-dimensional histograms, was performed using Perl and Gnuplot. The sulfur-aromatic distance and angle were normalized by the shell volume, taking into account the radius and angle, respectively. Interactions were parsed according to aromatic type based on the residue (phenylalanine, tyrosine, and tryptophan). Interactions were also categorized as inter- or intra-chain interaction, with an inter-chain interaction corresponding to Met and aromatic residues within the different PDB chains, and intra-chain interaction corresponding to Met and aromatic residues within the same PDB chain.
Molecular Dynamics Simulations
Two systems were constructed using the CHARMM (version 32) package. Crystal structures for TRAIL-DR5 (PDB code 1d0g) and LTα-TNFR1 (PDB code 1tnr) complexes were used as starting configurations. Missing TRAIL residues 132–143 were simulated briefly as in solution and inserted to form a continuous soluble TRAIL (residues 119–281). Hydrogen atoms were added, and all disulfide bonds present in the crystal structure were defined. To accommodate the zinc atom of the TRAIL complex, which interacts with the sulfur atom of cysteine 230 in each TRAIL chain as well as a chloride ion in a tetragonal geometry, a patch was created and implemented to define a rigid bond between the zinc ion and each of the three cysteine sulfur atoms as well as the chloride ion. The geometry of these four atoms was maintained throughout the simulation. Structures were solvated in a waterbox of ∼50,000 TIP3 water molecules, including 284 resolved structural water molecules for TRAIL-DR5 and 138 for LTα-TNFR1. Sodium and chloride ions were added, and counter ions were added to neutralize the overall charge of the system. The complete solvated TRAIL-DR5 and LTα-TNFR1 complexes, consisting of 138,163 and 136,995 atoms, respectively, were minimized and slowly heated to 300 K with harmonic restraints on α-carbon atoms. Systems were run completely unrestrained for 35–40 ns using NAMD version 2.6 in an isothermal, isobaric (NPT) ensemble. A cutoff of 10 Å was used for van der Waals interactions, and particle mesh Ewald summation was used for electrostatic interactions. The time step was 2 fs, and all bonds involving hydrogens were fixed using the SHAKE/RATTLE algorithm with a tolerance (relative deviation) of 10−8 Å. Simulation trajectories were visualized using Visual Molecular Dynamics program, and analysis was performed using CHARMM and Perl.
Quantum Calculations
Three aromatic rings representative of the side chains involved in the interactions with sulfur were selected as follows: benzene, phenol, and indole. For each ring, the relaxed potential energy curves for the interaction with DMS and propane were computed for two conformations as follows: one with the sulfur facing the ring (“down”), and one with the sulfur pointing away from it (“up”). As an additional control, the up and down conformations were additionally run for dimethyl ether (DME) in complex with benzene. Structures were optimized with Gaussian 09 through a Z-matrix constraining the sulfur to be on the axis perpendicular to the center of the six-membered ring and the two methyl groups to be symmetric with respect this axis. Geometry optimizations were performed at the M06-2X/6-311+G(d,p) (18, 19) and MP2/6-311+G(d,p) levels, and for selected distances single point calculations were performed at the CCSD(T)/6-311+G(d,p)//MP2/6-31G(d) level. For similar systems, it has been shown (20) that noncounterpoise-corrected triple-ζ M06-2X results are within 0.2 kcal/mol of the reference CCSD(T) extrapolated to the complete basis set limit. On the up minimum structure, we verified that expanding the basis set for M06-2X from 6-311+G(d,p) to aug-cc-pVTZ and aug-cc-pVQZ also results in less than 0.3 kcal/mol change.
To address how the DMS-aromatic interaction energy is affected by the wide distribution of distances and conformations observed in protein complexes, 128 structures were randomly sampled from the MD simulations for TRAIL-DR5 and LTα-TNFR1. The Cα–Cβ bond of the Met involved in the interaction was cut and saturated with hydrogen to yield DMS. Similarly, Tyr-237 and Trp-107 were truncated to p-cresol and 3-methylindole, respectively. In addition, for TRAIL-DR5 the effects of Arg-191 were also investigated by truncating its side chain to methyl guanidinium. For each structure, the DMS-aromatic interaction energy was computed at the HF, M06-2X, and MP2 levels using the aug-cc-pVDZ basis set (including basis set superposition error (21)) for all. Interaction energies were computed by subtracting the energy of the components at infinite separation (each fixed at the geometry in the complex) from the energy of the complex.
RESULTS
Sulfur-aromatic Interaction Is a Commonly Occurring Motif
Previous statistical analyses of protein structures and small molecule compounds have suggested that an interaction exists between divalent sulfur and aromatic rings. These include informatics studies of the Brookhaven Protein Data Bank (1, 2), a separate study of 36 protein structures (4), as well as a more recent analysis of the Cambridge Crystallographic Database of small molecule compounds (5). Collectively, these results show a statistical enrichment of sulfur-containing residues in proximity to aromatic residues and with a specific geometrical conformation. However, there exists no recent analysis of the PDB, which now contains ∼80,000 structures (16, 17), nor is the prevalence of these interactions known. Here, using Biopython (22), we analyzed a complete set of protein structures deposited in the PDB for close contacts between sulfur- and aromatic-containing residues to update the current bioinformatics dataset relating to sulfur-aromatic interactions. We define the sulfur-aromatic separation as the distance between the Met sulfur atom and the center of mass of the aromatic ring of tyrosine, tryptophan, and phenylalanine.
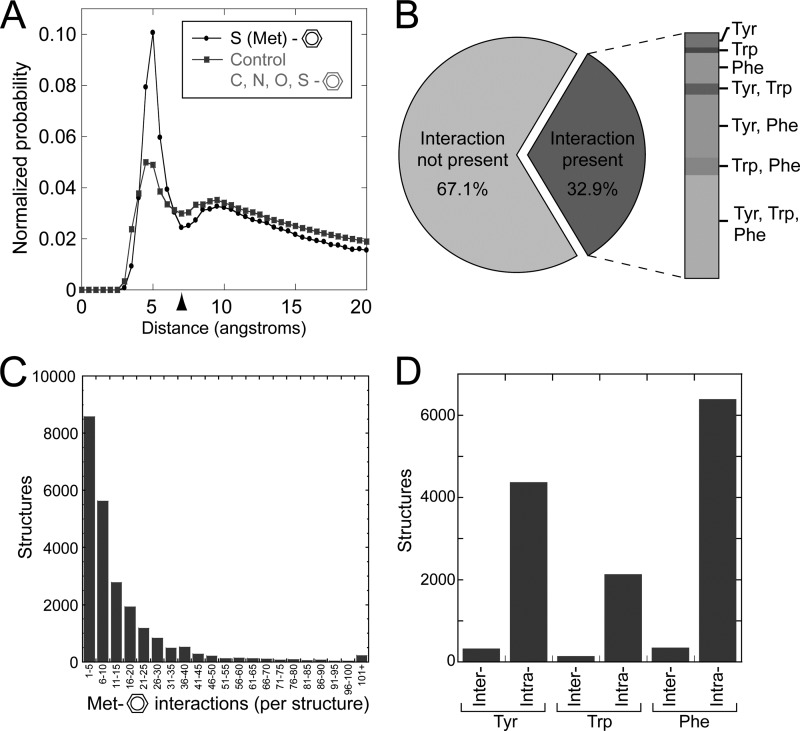
In agreement with previous smaller scale bioinformatics results (4), we observed an enrichment of Met sulfur atoms within 7 Å of any aromatic group, with a strong peak corresponding to a sulfur-aromatic separation of 5 Å (Fig. 2A). As a control, we compared the Met sulfur-aromatic distribution with the distribution of distances of carbon (both methyl and methylene carbon atoms), nitrogen, oxygen, and sulfur atoms to aromatic groups in a random subset of the PDB. Although the control group shows a modest peak at ∼5 Å separation, likely a result of hydrophobic packing, the probability of finding any non-hydrogen atom (CH2/CH3, nitrogen, oxygen, and sulfur) within 7 Å of an aromatic ring is notably lower than that of finding a Met sulfur within the same distance (Fig. 2A, compare black and gray probability distributions). This radial probability density of interaction distances normalized by shell volume was used to define a distance cutoff for the interaction; we consider the sulfur-aromatic interaction to be any Met sulfur atom within 7 Å of the aromatic ring center of mass, based on the first minimum in the distribution (Fig. 2A, arrowhead). Integration of the probability density distribution up to this cutoff reveals that Met sulfur atoms are almost twice as likely to be found within 7 Å of an aromatic compared with the control group, which also includes methyl and methylene carbon, nitrogen, and oxygen atoms within residue side chains. Analysis of high resolution crystal structures (the ∼20,000 PDB structures having an x-ray resolution less than or equal to 1.8 Å) yields an identical distribution (data not shown), and therefore the observed enrichment is not a result of poor structural resolution, under which the placement of atoms depends on empirical force fields that may not accurately capture the interaction. Further examination of the individual control atoms, CH2/CH3, nitrogen, and oxygen atoms, within residue side chains shows no difference between high resolution structures and the complete PDB (data not shown).
FIGURE 2.
Structural bioinformatics analysis of the Protein Data Bank demonstrates the prevalence of the Met sulfur-aromatic motif. A, probability density of Met sulfur-aromatic interaction distances relative to the control shows an enrichment of methionine sulfurs within 7 Å, indicated by the arrowhead. B, prevalence of Met-aromatic interaction in all protein structures using a 7-Å cutoff. C, number of Met-Aromatic interactions per structure. D, occurrence of inter- versus intra-chain Met-aromatic interactions.
Using the 7-Å cutoff based on the radial distribution of distances (Fig. 2A, arrowhead), we next tested the prevalence of these sulfur-aromatic interactions within all known protein structures in the PDB. Previous studies have shown an enrichment of sulfur atoms near aromatic residues (4, 5), with 8% of methionines interacting with aromatic groups (using a modest 3.6-Å cutoff) (23), and 22% of methionines interacting with π-electron donors (a group of amino acids broader than just aromatics that includes cation-π interactions in Arg, Lys, and His residues) (24). However it is unknown how many protein structures contain this motif, and of those structures, how many interactions are present. Using a 7-Å cutoff distance, we find that ∼33% of all known protein structures contain at least one Met sulfur-aromatic motif (Fig. 2B, left panel). Phenylalanine is the most common aromatic group involved in this interaction, followed by tyrosine and tryptophan (Fig. 2B, right panel), which follows the relative frequency of each aromatic amino acid. Of all protein structures with sulfur-aromatic motifs, it is most common to have interactions with multiple aromatic types than single aromatic types, and protein structures that contain the motif with all three aromatic groups are surprisingly likely. This result suggests that the majority of protein structures in fact have multiple motifs, either with different or identical aromatic groups.
We next analyzed the PDB for the number of interactions per structure. Within the PDB, ∼15,000 protein structures contain between 1 and 10 interactions, and the majority of protein structures involving Met-aromatic interactions have more than five interaction motifs (Fig. 2C). Surprisingly, several protein structures contain more than 100, and one structure, a 24-oligomeric hemocyanin (PDB code 3ixw), although large, has 388 interactions. Furthermore, we analyzed the PDB to determine whether this interaction is more commonly found within a single protein chain (i.e. intra-chain interaction) for stabilization of secondary or tertiary structure or between different protein chains thus stabilizing a complex of proteins (i.e. inter-chain interaction stabilizing quaternary structure). The results, shown in Fig. 2D, highlight that the majority of the Met sulfur-aromatic interactions in the PDB occur between residues of a single protein chain (i.e. intra-chain interaction). However, this could reflect that a majority of protein crystals, ∼83%, represent single chain protein structures and may skew the interpretation. Regardless, a number of these interactions occur between sulfur aromatic groups of different protein chains, such as the interactions found in TRAIL-DR5 and LTα-TNFR1. Moreover, the remarkably high occurrence of these motifs leads us to believe that interaction between Met and aromatic residues plays a critical role in protein structure, primarily through stabilization of secondary and tertiary structure, as well as protein function, through stabilization of protein complexes.
Sulfur and Aromatic Groups Are Critical for Binding and Function
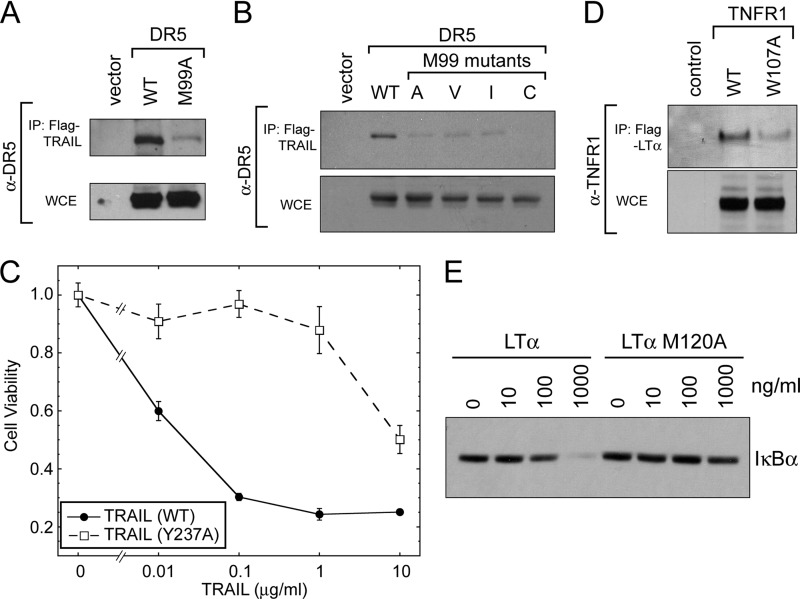
The structural comparison of the human TRAIL-DR5 and LTα-TNFR1 complexes reveals a similar interaction between Met and aromatic residues, tyrosine in TRAIL-DR5 and tryptophan in LTα-TNFR1 (Fig. 1). We wondered whether interactions via the Met-aromatic pair are driven through hydrophobic interactions between Met methyl and methylene groups and the hydrophobic ring. Single amino acid point mutations were generated to the ligand and receptor to disrupt the sulfur-aromatic interaction, and their activity was assessed by immunoprecipitation and downstream signaling (Fig. 3). DR5 Met-99 was mutated to alanine, which reduced ligand binding to ∼18% of wild type ligand-receptor interaction (Fig. 3A), as shown previously (14). To investigate if larger hydrophobic groups could restore the original Met activity, we further mutated DR5 Met-99 to valine and isoleucine, but the ability of these mutants to bind ligand did not increase with respect to the alanine mutant as shown by immunoprecipitation (Fig. 3, A and B). The observed relative binding capacity of valine and isoleucine is 29 and 18%, respectively, based on quantification of immunoprecipitation data. Mutation of Met-99 to cysteine, with a sulfur-containing thiol group, did not regain function and completely disrupted the interaction between ligand and receptor (<5% binding), which is potentially a result of misfolded receptor due to disulfide bridge formation between Cys-99 and the adjacent Cys-100, which in turn prevents the formation of a key structural disulfide bond between Cys-100 and Cys-86. Therefore, Met-99 is a critical residue within the 90-s loop of DR5 for its interaction with TRAIL, and stabilization of the ligand-receptor complex via this residue is not driven exclusively through hydrophobic interactions, as large hydrophobic residues do not regain the function of the sulfur-containing Met residue.
FIGURE 3.
A, mutation of DR5 Met-99 to alanine reduces ligand binding in transiently expressing HEK293 cells as measured by immunoprecipitation (IP). B, mutation of DR5 Met-99 to hydrophobic side chains of increasing size (Ala, Val, and Ile) does not recover the ligand-receptor interaction, measured by immunoprecipitation. The Met-99 to Cys mutation completely prevents ligand binding, likely due to disulfide bond formation and misfolding. C, Jurkat cells were treated with TRAIL, WT, and Y237A mutant, at various concentrations for 24 h and cell viability was measured by MTT assay. Mutation of the aromatic-containing residue within the ligand reduces the activity of TRAIL. D, mutation of the aromatic-containing residue within the structurally homologous loop in TNFR1 disrupts interaction with its cognate ligand, LTα, measured by immunoprecipitation. Whole cell lysates show equal expression of wild type and mutant receptor. E, cells were treated with various concentrations of LTα ligand, wild type and mutant. Mutation of the sulfur-containing methionine residue within LTα ligand disrupts the function, shown by degradation of IκBα at various concentrations of ligand.
The Met residue is playing a considerable role in this motif via interactions that cannot be regained through hydrophobic residues, and consequently we hypothesize that the aromatic residue in the TRAIL is also critical for ligand binding and function. Therefore, we next sought to confirm that mutation of the aromatic tyrosine residue within TRAIL disrupts the function of the ligand. Soluble TRAIL (residues 114–281) and a single amino acid point mutant coding for TRAIL Y237A were tagged with an N-terminal FLAG sequence and purified from Escherichia coli as described previously (15). We assessed the ability of TRAIL (both wild type and the tyrosine mutant) to induce cell death in a dose-dependent manner by MTT assay to measure cell viability. As expected, TRAIL induces cell death in Jurkat cells, which are expressing endogenous levels of DR5, with an ED50 of ∼20 ng/ml (Fig. 3C). Mutation of the tyrosine in the sulfur-aromatic binding motif decreases the activity of TRAIL by several orders of magnitude, having an ED50 of ∼10 μg/ml (Fig. 3C, compare solid circles with open squares). This confirms the importance of Tyr-237 in the TRAIL for its apoptosis-inducing activity.
Using similar methods, we tested the effect of mutations to the structurally homologous residues in the LTα-TNFR1 ligand-receptor complex, the function of which remains unknown. The aromatic residue (Trp-107) of TNFR1 was mutated to alanine and transfected into HEK293 cells. The ability of wild type and mutant receptors to interact with LTα ligand was again assessed by immunoprecipitation of the ligand-receptor complex. Mutation of Trp-107 in TNFR1 results in a noticeable decrease in the interaction between ligand and receptor relative to the wild type receptor, despite approximately equal expression of receptor in both the wild type and mutant transfections (Fig. 3D), demonstrating the importance of this aromatic residue.
Furthermore, we tested the function of LTα and mutants to the sulfur-containing Met-120 residue by measuring IκBα degradation after the addition of various doses of ligand. Soluble LTα ligands (both wild type and Met mutant) were purified, and Jurkat cells were treated with the indicated concentration of LTα for 20 min and subsequently lysed. Equal amounts of total protein lysate were analyzed by SDS-PAGE for IκBα expression, degradation of which is a common marker of NFκB activation (Fig. 3E). The addition of LTα results in noticeable degradation of IκBα as follows: ∼50% of IκBα degraded at 100 ng/ml, and nearly all of the IκBα is degraded at 1000 ng/ml. Addition of the mutated LTα, M120A, decreases the efficacy of the ligand both at low concentrations (causing no noticeable change in IκBα levels) and at high concentrations (with less than 40% degradation). These results demonstrate that Met-120 in the LTα ligand is critical for its functional activity. Thus, both sulfur- and aromatic-containing residues play a critical role in stabilizing the high affinity ligand-receptor interaction and greatly influence the ability of a TNF ligand to transduce its signal into the cell.
Quantum Mechanical Calculations of Sulfur- and Aromatic-containing Model Compounds
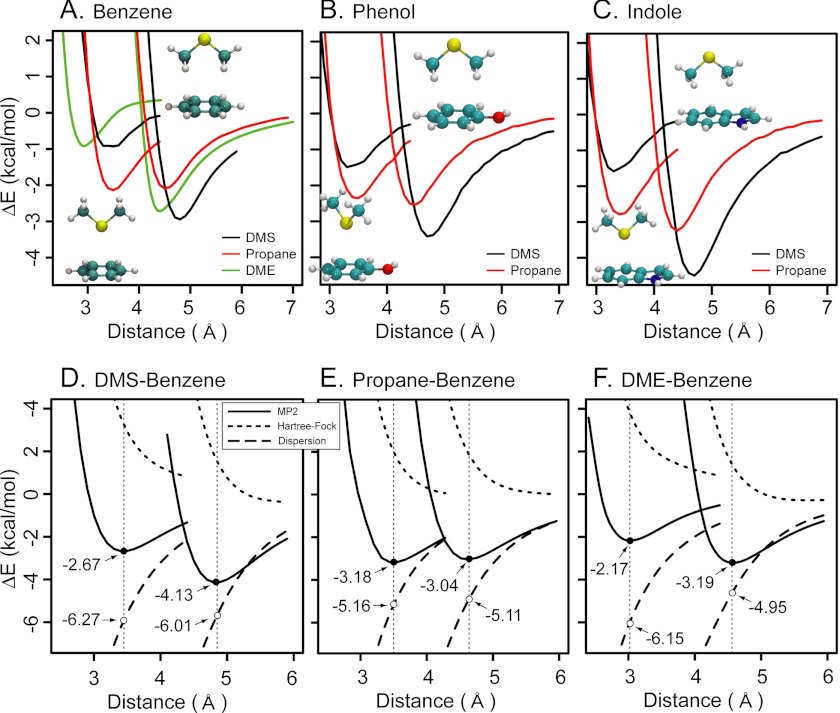
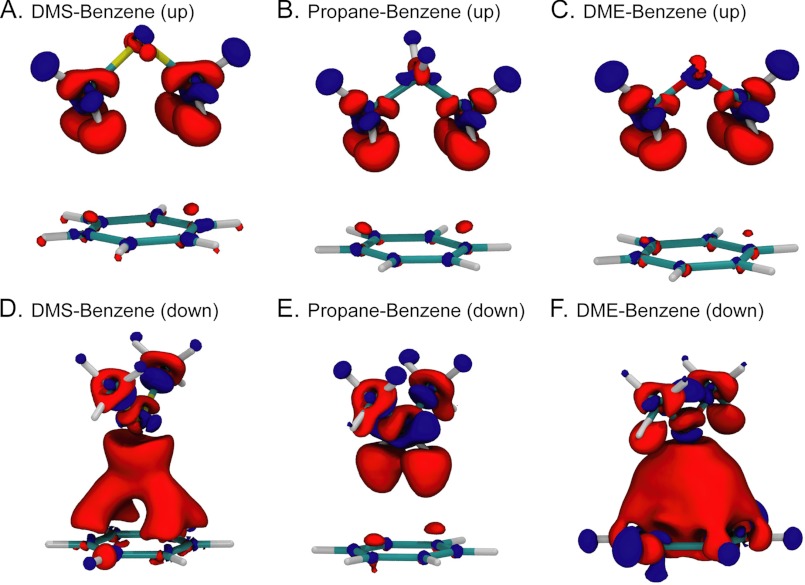
Bioinformatics and experimental results demonstrate a specific interaction involving the Met sulfur atom and aromatic rings, and this interaction is not found between other non-sulfur atoms and cannot be regained with larger hydrophobic residues (see Figs. 2 and 3). Therefore, we have used quantum mechanical electronic structure methods to investigate the origin of nonbonded interactions between sulfur-containing compounds and aromatic side chains and to differentiate these interactions from other non-sulfur-containing hydrophobic residues. In particular, HF, MP2, and density functional theory (M06-2X) QM calculations were performed on model complexes between the sulfur-containing DMS and the side chains of the aromatic amino acids Phe, Tyr, and Trp as follows: benzene (Fig. 4A), phenol (Fig. 4B), and indole (Fig. 4C), respectively. For comparison, QM calculations were also performed on the three aforementioned aromatics and propane, the sulfur-deficient methylene carbon-analog of DMS. Only for the benzene aromatic group was an additional set of calculations performed for the oxygen analog DME. For each complex, the relaxed potential energy curve for the interaction was computed by varying the distance between the aromatic ring and DMS/propane/DME. Similar to previous studies (25, 26), we investigated two conformations: DMS/propane/DME with the central atom (sulfur, CH2, or oxygen) pointing upwards (up conformation) or downwards (down conformation) shown in Fig. 4, A–C, top right and bottom left illustration insets, respectively.
FIGURE 4.
Sulfur-aromatic interaction provides additional stabilization over hydrophobic contacts due primarily to dispersion interactions. A–C, relaxed scans computed at the M06-2X/6–311+G(d,p) level for benzene (A), phenol (B), and indole (C) with DMS (black), propane (red), and DME (green). The first and second minima correspond to the down and up conformations, respectively, shown by the illustration inset. The distance is taken to be the separation between the center of the aromatic ring and the sulfur atom (or equivalent for propane, DME). D–F, contribution of dispersion energy to overall binding for benzene complexes with DMS (D), propane (E), and DME (F). The dispersion contribution (long dashed line) is computed as the difference between the MP2 (solid line) and HF (short dashed line) energies, both computed at the 6–311+G(d,p) level. Values at the MP2 minima (indicated by vertical dashed lines for the up and down configurations) are reported.
In each conformation, the three complexes involving DMS (Fig. 4, A–C, black line), propane (red line), and DME (green line) display a well defined minimum along the distance reaction coordinate, with a binding energy ranging between 1 and 5 kcal/mol. The down conformation minimum is characterized by a distance of about 3.4 Å for both DMS and propane, and for DME the equilibrium distance is around 3.0 Å. The up conformation minimum displays a larger separation for DMS (about 4.8 Å) than for propane or DME (about 4.4 Å). Particularly relevant to our study are the down and up conformation minima for propane, which are essentially isoenergetic (Fig. 4, A–C, red lines), whereas the presence of sulfur in DMS strongly skews the energy landscape, favoring the up conformation by 2–3 kcal/mol over the down conformation (Fig. 4, A–C, black line, and Table 1). It is also important to recognize that the up conformation minimum for DMS is about 1 kcal/mol more stable than either up or down minima for propane. Therefore, the presence of sulfur makes the interaction about 1 kcal/mol more stable and adds a conformational preference toward a longer distance. These results are further confirmed by ab initio calculations performed at the MP2 and CCSD(T) levels (see Table 1); although it seems that with respect to the reference CCSD(T), the complexes are slightly weakly bound using M06-2X and more strongly bound at the MP2 level; the relative energies of the minima are remarkably consistent across the different methods.
TABLE 1.
Interaction energy for DMS, propane, and DME in complex with benzene, phenol, and indole at the energy-scan minima computed at the M06-2X, MP2, and CCSD(T) levels with the 6-311+G(d,p) basis set
| Structure, conformation, and method | Aromatic |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Benzene |
Phenol |
Indole |
|||||||
| M06–2X | MP2 | CCSD(T) | M06–2X | MP2 | CCSD(T) | M06–2X | MP2 | CCSD(T) | |
| DMS | |||||||||
| Up | −2.94 | −4.13 | −3.62 | −3.42 | −4.89 | −4.25 | −4.50 | −6.57 | |
| Down | −0.93 | −2.67 | −2.16 | −1.49 | −3.36 | −2.80 | −1.58 | −4.14 | |
| Propane | |||||||||
| Up | −2.09 | −3.04 | −2.62 | −2.53 | −3.65 | −3.15 | −3.25 | −4.82 | |
| Down | −2.14 | −3.18 | −2.69 | −2.35 | −3.49 | −3.03 | −2.78 | −4.50 | |
| DME | |||||||||
| Up | −2.72 | −3.19 | |||||||
| Down | −0.92 | −2.17 | |||||||
To characterize the origin of these minima, we plotted the contribution of the dispersion energy as a function of the reaction coordinate for DMS, propane, and DME in complex with benzene (Fig. 4, D–F). We estimated the dispersion energy (Fig. 4, D–F, long dashed line) as the difference between the MP2 energies (Fig. 4, D–F, black line) and HF energies (Fig. 4, D–F, short dashed line). It is interesting to note that HF has a fully repulsive potential, and thus the attraction basin that characterizes the MP2 profile originates entirely from dispersion interactions. The dispersion contributions are on the order of 5–6 kcal/mol and are stronger in both down and up minima for DMS (−6.27 and −6.01 kcal/mol, respectively) than for propane (−5.16 and −5.11 kcal/mol). The down minimum for DME, which takes place at a shorter distance (3.0 Å), displays a dispersion contribution similar to DMS (−6.15 kcal/mol), but the contribution for the up minimum is −4.95 kcal/mol, similar to propane. This analysis suggests that most of the increased binding originating for DMS in the up minimum with respect to propane can be attributed to stronger dispersion interactions. This is qualitatively confirmed by electron density difference plots for the up conformation shown in Fig. 5. In these plots, we monitor the shift in electron density upon binding, which is an indicator of the combined effects of polarization and charge transfer. For the up conformation, the isocontours for all species appear to be similar, thus suggesting a similar contribution due to the polarization/charge transfer (Fig. 5, A–C). A different picture is found in the down conformation isocontours (Fig. 5, D–F). Here, the presence of the lone pairs on the sulfur and oxygen atoms is clearly reflected by a loss of electron density in the region where the lone pair orbitals overlap with the π-system (Fig. 5, D and F). Notice that the complex of DME, characterized by shorter bonding distance, shows the greatest electron density shift, followed by DMS. The isocontours for propane, on the contrary, are comparable in magnitude to those for the up conformation, thus suggesting a similar extent of electron rearrangement (Fig. 5, compare B and E).
FIGURE 5.
Electron density difference contours for each complex, DMS/propane/DME-benzene, in two different configurations, up and down. The electron density contours represent the difference between the electron density obtained at the complex geometry minimum and the species at infinite separation, each calculated at the M06-2X/6–311+G(d,p) level. The same isodensity value (0.0003) was used for all plots. Blue represents positive differences (i.e. gain of electron density in the complex state) and red represents negative differences (i.e. loss of electron density in the complex state).
To summarize, the origin of the difference between DMS and propane is the result of the sum of all pairwise interactions, as pointed out by Némethy and Scheraga (27). Although for propane this sum is almost the same for either conformation, for DMS the presence of the lone pairs on sulfur pointing toward the π-ring in the down conformation (28) makes this orientation less stable even if it is accompanied by a larger dispersion contribution than for propane. At the same time, the larger dispersion energy of DMS in comparison with propane can be attributed to the increased stability of the up conformation, i.e. the dispersion energy of DMS in the up conformation, unlike the down conformation, is not outweighed by the lone pair/π-ring electron repulsion. This explanation is further corroborated by the results for the DME complex as follows: in the up conformation DME displays a behavior more similar to propane, both in the depth of the minimum and in the dispersion contribution (see Fig. 4). This indicates that the origin of that minimum is best ascribed to dispersion contribution and not to effects due to the oxygen atom polarizing its neighboring atoms. Conversely, in the down conformation, the structure of DME complex behaves more similarly to that of DMS, although there is an even shallower minimum that can be possibly attributed to the “hardness” of its lone pairs when compared with the “softness” of the sulfur lone pairs.
Molecular Dynamics Simulations of the TRAIL-DR5 and LTα-TNFR1 Complexes
Our results from QM calculations characterize the origin of the nonbonded sulfur-aromatic interaction as well as the specific energetic advantage and conformational specificity compared with purely hydrophobic contacts (Figs. 4 and 5). Moreover, our experimental evidence demonstrates clearly that sulfur- and aromatic-containing residues in TRAIL-DR5 and LTα-TNFR1 are critical for ligand binding and function (Fig. 3), corroborating the increased probability of contacts between the sulfur-containing Met residue and aromatic residues throughout the Protein Data Bank (Fig. 2). That a single interaction within a large multiprotein complex has a profound ability to stabilize the structure led us to question the energy associated with the interaction between these side chains in biologically relevant structural configurations. Given the frequency with which such interactions occur in known protein complexes, including the number of complexes with more than 100 interactions, we aimed to characterize the biologically relevant interaction between Met sulfur and the aromatic-containing tyrosine and tryptophan side chains. To investigate a range of biologically relevant conformations, we performed all-atom molecular dynamics simulations of the TRAIL-DR5 and LTα-TNFR1 complexes. The geometrical configuration for the sulfur-aromatic interaction, including radial separation and angular orientation, is illustrated for TRAIL-DR5 and LTα-TNFR1 in Fig. 6, A and B, respectively, and was calculated at each step over the equilibrated portion of a simulation trajectory. Additionally, shown in the TRAIL-DR5 complex is a nearby residue in the ligand, Arg-191, shown previously to be important for binding (11), which forms an interesting Arg-Met-Tyr ligand-receptor motif that further stabilizes the complex via interactions with Met-99 (discussed further below).
FIGURE 6.
MD simulation of the TRAIL-DR5 and LTα-TNFR1 complexes. A, Met-99/Tyr-237 interaction in the TRAIL-DR5 complex. The nearby Arg-191 is also shown. B, Met-120/Trp-107 interaction in the LTα-TNFR1 complex. C and D, radial and angular population distribution of the conformations sampled by MD of the TRAIL/DR5 (C) and of the LTα-TNFR1 (D) complexes, with warm colors representing a more frequent geometry. E and F, radial and angular population distribution analysis of the PDB as follows: Met/Tyr (E) and Met/Trp (F) using the same color scheme. G–I, histograms of CP-corrected interaction energies for 128 structures taken from MD simulations corresponding to the Met-Tyr interaction in TRAIL-DR5 (G), the Arg-Met-Tyr interaction in TRAIL-DR5 (H), and the Met-Trp interaction in LTα-TNFR1 (I).
The radial and angular density distribution of the conformations sampled through MD for the Met-tyrosine interaction within the TRAIL-DR5 complex demonstrates the strong preference for an interaction at ∼5 Å and 45° from the ring normal (Fig. 6C). The LTα-TNFR1 complex, where the Met residue of LTα is interacting with a tryptophan residue in TNFR1, shows a peak in density at ∼5 Å and 30° from the ring normal (Fig. 6D). For comparison, the same density distributions calculated across the entire PDB for the Met-tyrosine and Met-tryptophan interactions are shown in Fig. 6, E and F, respectively. The ensemble of Met-Tyr and Met-Trp structures sampled by each MD simulation (TRAIL-DR5 and LTα-TNFR1, respectively) appear to be a subspace of the wider distribution obtained from the analysis of the PDB for each interaction. It is of interest to notice that although the radial peak around 5 Å is unambiguously defined both in the MD and bioinformatics analyses, the angular dependence is less marked, especially in the PDB results. It is likely that the radial peak at 5 Å is a more prominent characteristic of this interaction, although the angular dependence could be more dependent of the chemical surroundings of each specific motif. Collectively, these results highlight the radial dependence of the Met sulfur-aromatic interaction and, to a lesser extent, its angular dependence. Moreover, the MD trajectory is sampling a range of biological sulfur-aromatic interactions consistent with a range of different protein structures, and these structures can be used to more accurately estimate the interaction energy in an ensemble of relevant conformations.
Thus, to probe the sulfur-aromatic interaction energy in a range of biological conformations, we randomly sampled 128 structures from the MD simulation trajectories. For each conformation, we computed the QM interaction energy and plotted the resulting histograms for the TRAIL-DR5 complex (Fig. 6G) and the LTα-TNFR1 complex (Fig. 6I). As expected, a wide distribution of interaction energies is sampled in both systems. The mean interaction energy can be bracketed between 1.4 and 1.9 kcal/mol (M06-2X and MP2 results, respectively) for Met-Tyr in TRAIL-DR5 and between 1.9 and 2.8 kcal/mol for Met-Trp in LTα-TNFR1. The calculated energies, in the 1–2 kcal/mol range, agree with experimentally determined results for this type of interaction (1, 6). The observation that in the TRAIL-DR5 complex an arginine residue (Fig. 6A, see Arg-191) is also in the proximity of the side chain of Met-99 made us wonder if this could provide the means for an additional stabilization. Indeed, as shown in Fig. 6H, the presence of Arg-191 strengthens the interaction, bringing the total interaction energy to 5.2 kcal/mol, which is in rough agreement with the importance of this arginine residue shown experimentally in which mutation to alanine results in a 3.9-fold increase in the dissociation constant as measured by surface plasmon resonance (11). The arginine residue in the TRAIL-DR5 interaction thus provides a means for additional stabilization, which may have evolved as a result of the lower interaction energy between Met and tyrosine in this complex, whereas the tryptophan residue in the LTα-TNFR1 complex is sufficient alone for complex formation.
DISCUSSION
The biophysical and biochemical roles of nearly all the amino acid residues have been well described. However, the precise role of the sulfur-containing Met residue remains poorly understood. Hydrophobic contacts via the Met residue, including methylene and methyl groups, are widely considered to be its major contribution to protein stability. If this is the case, what is the biological advantage of having a sulfur atom, rather than a methylene group, at the δ position? We present here the first case for a selective advantage, ∼1.5 kcal/mol, of the sulfur-aromatic interaction in conferring conformational specificity at distances up to ∼5–6 Å.
How does the energy associated with the sulfur-aromatic interaction motif compare with other stabilizing interactions found within protein complexes? The energy associated with a single sulfur-aromatic interaction is comparable with the interaction energy of a single salt bridge, a common interaction found in protein complexes with an interaction energy ranging from less than 1 to ∼3.2 kcal/mol (29–32) and greater depending on the local environment (33, 34). However, the sulfur-aromatic interaction also occurs at a larger distance (∼5–6 Å) than a salt bridge (typically <4 Å) and may be less sensitive to local environmental changes, including pH values of the solvent and the solvent itself. Additionally, although salt bridges have a significant energy penalty associated with side chain desolvation, which may in fact exceed the energy of salt bridge formation (35), the nonpolar nature of the Met and aromatic residues minimizes the desolvation energy penalty. The combination of these attributes in a sulfur-aromatic interaction yields a potentially more robust, and certainly unique, interaction than either purely hydrophobic contacts or salt bridges and an interaction that occurs at longer distances.
Given the prevalence of the Met sulfur-aromatic interaction in known protein structures, it is conceivable that mutations of these residues may play a role in the protein structure of disease-related proteins. We found several examples of known disease-related Met mutations in which the structure of the protein of interest shows the Met in proximity to one or more aromatic residues. In von Willebrand disease, a single amino acid substitution (valine replacing the normal methionine residue) results in an overactive glycoprotein Ibα, associated with a bleeding disorder (36, 37). Upon examination of this structure, we found the Met residue at the center of three aromatic residues (<5 Å separation), with a total of five aromatic residues within 7 Å (see PDB code 1sq0), and mutation to valine was considered simply a nonspecific loss of hydrophobic contact (38). In a second example, mutation of prion protein Met-129 is an allelic risk factor for the development of neurodegenerative diseases, such as Alzheimer (39) as well as fatal familial insomnia and familial Creutzfield-Jacob (40). This Met residue in the wild type prion protein (PDB code 2kun) has three aromatic tyrosine residues within 10 Å, the closest at 5.5 Å separation (41). Another example, a known mutation of the RET proto-oncogene resulting in an M918T mutation within RET protein, is associated with the development of thyroid carcinoma (42, 43), and Met-918 in the wild type structure (PDB code 2x2k) has six aromatic groups within 10 Å, with the nearest at 5.4 Å (44). In addition to these examples, there are a number of disease-related Met mutations with no associated protein structure, including one structure where Met substitution results in thermal instability and improper protein folding (45). It is conceivable that other pathological Met mutations are a direct result of lost or gained interactions with aromatic residues, thereby changing protein stability, structure, and function.
Another interesting implication in the nature of the Met sulfur-aromatic interaction is the oxidation of certain amino acid side chains by reactive oxygen species, with Met being the most readily oxidized. Oxidized proteins accumulate during aging (46) and are associated with a number of age-associated pathologies, including Parkinson disease, Alzheimer disease, cataracts, emphysema, and bronchitis (47–51). The oxidation of methionine to methionine sulfoxide, MetO, occurs via the addition of an oxygen atom to the δ-sulfur atom in the methionine. Furthermore, irreversible oxidation via the addition of a second oxygen atom results in methionine sulfone, MetO2. Although the oxidation of Met residues is known to decrease protein conformational stability (52), it is unknown whether this loss of stability is associated with energetic changes in the Met sulfur-aromatic interactions. Moreover, in the case where sulfur- and aromatic-containing residues are important in complex formation, shown here in the TRAIL-DR5 and LTα-TNFR1, it is unknown how the oxidation of Met sulfur may influence binding, complex formation, and subsequent function. However, it is conceivable that the addition of an oxygen atom (or atoms) to the δ-sulfur would disrupt both dispersion and electrostatic interactions present in the sulfur-aromatic motif, as described here, or at least reduce them in view of the subtle distance and angular requirements for the interactions to be effective.
Given the potential impact of the Met sulfur-aromatic motif in the stabilization of protein structures and complexes, and its role in disease, this motif presents a potentially enormous opportunity for rational drug design. Interestingly, certain TNF ligand-targeted peptide antagonists may already be taking advantage of this motif. A peptide designed to mimic the Met-containing loop within the TNFR1, known as WP9QY (amino acid sequence YCWSQYLCY, with the Trp-107 analog underlined), has been found to bind both RANKL and LTα (53, 54), and it has been shown to reduce inflammation and bone resorption in mouse arthritis models (55, 56). The peptide is thought to antagonize these TNF ligands by binding the Met-containing surface of LTα, thus preventing the LTα-TNFR1 interaction. Although the role of the sulfur-aromatic interaction in this peptide-protein interaction has not been discussed, the sequence homology between the peptide antagonist and TNFR1, including the tryptophan residue in TNFR1 involved in the Met-aromatic motif, demonstrates that the sulfur-aromatic interaction may be exploited in the rational design of therapeutics.
To conclude, the methionine sulfur-aromatic interaction is a unique motif that is prevalent in protein structures and provides a specific role in stabilization of protein structures. The prevalence of the sulfur-aromatic interaction and its relevance in a number of pathologies, including known disease-associated mutations of Met as well as age-associated diseases related to Met oxidation, presents a unique opportunity to utilize this motif in the rational design of therapeutics.
Acknowledgment
We thank the Minnesota Supercomputing Institute for computational resources.
This work was supported, in whole or in part, by National Institutes of Health Grant R21 CA152668. This work was also supported by American Cancer Society Grant IRG-58-001-49 IRG49-28.
- DMS
- dimethyl sulfide
- TRAIL
- tumor necrosis factor-related apoptosis-inducing ligand
- PDB
- Protein Data Bank
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- DME
- dimethyl ether
- MD
- molecular dynamics
- QM
- quantum mechanical
- HF
- Hartree-Fock.
REFERENCES
- 1. Morgan R. S., Tatsch C. E., Gushard R. H., McAdon J., Warme P. K. (1978) Chains of alternating sulfur and π-bonded atoms in eight small proteins. Int. J. Pept. Protein Res. 11, 209–217 [DOI] [PubMed] [Google Scholar]
- 2. Morgan R. S., McAdon J. M. (1980) Predictor for sulfur-aromatic interactions in globular proteins. Int. J. Pept. Protein Res. 15, 177–180 [DOI] [PubMed] [Google Scholar]
- 3. Rosenfield R. E., Parthasarathy R., Dunitz J. D. (1977) Directional preferences of nonbonded atomic contacts with divalent sulfur. 1. Electrophiles and nucleophiles. J. Am. Chem. Soc. 99, 4860–4862 [Google Scholar]
- 4. Reid K. S., Lindley P. F., Thornton J. M. (1985) Sulfur-aromatic interactions in proteins. FEBS Lett. 190, 209–213 [Google Scholar]
- 5. Zauhar R. J., Colbert C. L., Morgan R. S., Welsh W. J. (2000) Evidence for a strong sulfur-aromatic interaction derived from crystallographic data. Biopolymers 53, 233–248 [DOI] [PubMed] [Google Scholar]
- 6. Bodner B. L., Jackman L. M., Morgan R. S. (1980) NMR study of 1:1 complexes between divalent sulfur and aromatic compounds. A model for interactions in globular proteins. Biochem. Biophys. Res. Commun. 94, 807–813 [DOI] [PubMed] [Google Scholar]
- 7. Viguera A. R., Serrano L. (1995) Side-chain interactions between sulfur-containing amino acids and phenylalanine in α-helices. Biochemistry 34, 8771–8779 [DOI] [PubMed] [Google Scholar]
- 8. Cregut D., Serrano L. (1999) Molecular dynamics as a tool to detect protein foldability. A mutant of domain B1 of protein G with non-native secondary structure propensities. Protein Sci. 8, 271–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hymowitz S. G., Christinger H. W., Fuh G., Ultsch M., O'Connell M., Kelley R. F., Ashkenazi A., de Vos A. M. (1999) Triggering cell death. The crystal structure of Apo2L/TRAIL in a complex with death receptor 5. Mol. Cell 4, 563–571 [DOI] [PubMed] [Google Scholar]
- 10. Banner D. W., D'Arcy A., Janes W., Gentz R., Schoenfeld H. J., Broger C., Loetscher H., Lesslauer W. (1993) Crystal structure of the soluble human 55-kDa TNF receptor-human TNFβ complex. Implications for TNF receptor activation. Cell 73, 431–445 [DOI] [PubMed] [Google Scholar]
- 11. Hymowitz S. G., O'Connell M. P., Ultsch M. H., Hurst A., Totpal K., Ashkenazi A., de Vos A. M., Kelley R. F. (2000) A unique zinc-binding site revealed by a high resolution x-ray structure of homotrimeric Apo2L/TRAIL. Biochemistry 39, 633–640 [DOI] [PubMed] [Google Scholar]
- 12. Cha S. S., Kim M. S., Choi Y. H., Sung B. J., Shin N. K., Shin H. C., Sung Y. C., Oh B. H. (1999) 2.8 Å resolution crystal structure of human TRAIL, a cytokine with selective antitumor activity. Immunity 11, 253–261 [DOI] [PubMed] [Google Scholar]
- 13. Mongkolsapaya J., Grimes J. M., Chen N., Xu X. N., Stuart D. I., Jones E. Y., Screaton G. R. (1999) Structure of the TRAIL-DR5 complex reveals mechanisms conferring specificity in apoptotic initiation. Nat. Struct. Biol. 6, 1048–1053 [DOI] [PubMed] [Google Scholar]
- 14. Bin L., Thorburn J., Thomas L. R., Clark P. E., Humphreys R., Thorburn A. (2007) Tumor-derived mutations in the TRAIL receptor DR5 inhibit TRAIL signaling through the DR4 receptor by competing for ligand binding. J. Biol. Chem. 282, 28189–28194 [DOI] [PubMed] [Google Scholar]
- 15. Schneider P. (2000) Production of recombinant TRAIL and TRAIL receptor. Fc chimeric proteins. Methods Enzymol. 322, 325–345 [DOI] [PubMed] [Google Scholar]
- 16. Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) The Protein Data Bank. Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Berman H., Henrick K., Nakamura H. (2003) Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 10, 980. [DOI] [PubMed] [Google Scholar]
- 18. Zhao Y., Schultz N. E., Truhlar D. G. (2006) Design of density functionals by combining the method of constraint satisfaction with parameterization for thermochemistry, thermochemical kinetics, and noncovalent interactions. J. Chem. Theory Comput. 2, 364–382 [DOI] [PubMed] [Google Scholar]
- 19. Zhao Y., Truhlar D. G. (2006)A new local density functional for main-group thermochemistry, transition metal bonding, thermochemical kinetics, and noncovalent interaction. J. Chem. Phys. 125, 194101. [DOI] [PubMed] [Google Scholar]
- 20. Sherrill C. D., Takatani T., Hohenstein E. G. (2009) An assessment of theoretical methods for nonbonded interactions. Comparison with complete basis set limit coupled-cluster potential energy curves for the benzene dimer, the methane dimer, benzene-methane, and benzene-H2S. J. Phys. Chem. A 113, 10146–10159 [DOI] [PubMed] [Google Scholar]
- 21. Boys S. F., Bernardi F. (1970) The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors. Mol. Physics 19, 553–566 [Google Scholar]
- 22. Cock P. J., Antao T., Chang J. T., Chapman B. A., Cox C. J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B., de Hoon M. J. (2009) Biopython. Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pal D., Chakrabarti P. (2001) Non-hydrogen bond interactions involving the methionine sulfur atom. J. Biomol. Struct. Dyn. 19, 115–128 [DOI] [PubMed] [Google Scholar]
- 24. Imai Y. N., Inoue Y., Yamamoto Y. (2007) Propensities of polar and aromatic amino acids in noncanonical interactions. Nonbonded contacts analysis of protein-ligand complexes in crystal structures. J. Med. Chem. 50, 1189–1196 [DOI] [PubMed] [Google Scholar]
- 25. Ringer A. L., Senenko A., Sherrill C. D. (2007) Models of S/π interactions in protein structures. Comparison of the H2S benzene complex with PDB data. Protein Sci. 16, 2216–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cabaleiro-Lago E. M., Carrazana-García J. A., Rodríguez-Otero J. (2009) Study of the interaction between water and hydrogen sulfide with polycyclic aromatic hydrocarbons. J. Chem. Phys. 130, 234307. [DOI] [PubMed] [Google Scholar]
- 27. Némethy G., Scheraga H. A. (1981) Strong interaction between disulfide derivatives and aromatic groups in peptides and proteins. Biochem. Biophys. Res. Commun. 98, 482–487 [DOI] [PubMed] [Google Scholar]
- 28. Duan G., Smith V. H., Jr., Weaver D. F. (2001) Characterization of aromatic-thiol π-type hydrogen bonding and phenylalanine-cysteine side chain interactions through ab initio calculations and protein database analyses. Mol. Physics 99, 1689–1699 [Google Scholar]
- 29. Strop P., Mayo S. L. (2000) Contribution of surface salt bridges to protein stability. Biochemistry 39, 1251–1255 [DOI] [PubMed] [Google Scholar]
- 30. Iqbalsyah T. M., Doig A. J. (2005) Anticooperativity in a Glu-Lys-Glu salt bridge triplet in an isolated α-helical peptide. Biochemistry 44, 10449–10456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tsai J., Levitt M. (2002) Evidence of turn and salt bridge contributions to β-hairpin stability. MD simulations of C-terminal fragment from the B1 domain of protein G. Biophys. Chem. 101, 187–201 [DOI] [PubMed] [Google Scholar]
- 32. Singh U. C. (1988) Probing the salt bridge in the dihydrofolate reductase-methotrexate complex by using the coordinate-coupled free-energy perturbation method. Proc. Natl. Acad. Sci. U.S.A. 85, 4280–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wimley W. C., Gawrisch K., Creamer T. P., White S. H. (1996) Direct measurement of salt-bridge solvation energies using a peptide model system. Implications for protein stability. Proc. Natl. Acad. Sci. U.S.A. 93, 2985–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Waldburger C. D., Schildbach J. F., Sauer R. T. (1995) Are buried salt bridges important for protein stability and conformational specificity? Nat. Struct. Biol. 2, 122–128 [DOI] [PubMed] [Google Scholar]
- 35. Bosshard H. R., Marti D. N., Jelesarov I. (2004) Protein stabilization by salt bridges. Concepts, experimental approaches, and clarification of some misunderstandings. J. Mol. Recognit. 17, 1–16 [DOI] [PubMed] [Google Scholar]
- 36. Russell S. D., Roth G. J. (1993) Pseudo-von Willebrand disease. A mutation in the platelet glycoprotein Ibα gene associated with a hyperactive surface receptor. Blood 81, 1787–1791 [PubMed] [Google Scholar]
- 37. Moriki T., Murata M., Kitaguchi T., Anbo H., Handa M., Watanabe K., Takahashi H., Ikeda Y. (1997) Expression and functional characterization of an abnormal platelet membrane glycoprotein Ibα (Met-239 → Val) reported in patients with platelet-type von Willebrand disease. Blood 90, 698–705 [PubMed] [Google Scholar]
- 38. Dumas J. J., Kumar R., McDonagh T., Sullivan F., Stahl M. L., Somers W. S., Mosyak L. (2004) Crystal structure of the wild type von Willebrand factor A1-glycoprotein Ibα complex reveals conformation differences with a complex bearing von Willebrand disease mutations. J. Biol. Chem. 279, 23327–23334 [DOI] [PubMed] [Google Scholar]
- 39. Gacia M., Safranow K., Styczyńska M., Jakubowska K., Pepłońska B., Chodakowska-Zebrowska M., Przekop I., Słowik A., Golańska E., Hułas-Bigoszewska K., Chlubek D., Religa D., Zekanowski C., Barcikowska M. (2006) Prion protein gene M129 allele is a risk factor for Alzheimer disease. J. Neural Transm. 113, 1747–1751 [DOI] [PubMed] [Google Scholar]
- 40. Goldfarb L. G., Petersen R. B., Tabaton M., Brown P., LeBlanc A. C., Montagna P., Cortelli P., Julien J., Vital C., Pendelbury W. W. (1992) Fatal familial insomnia and familial Creutzfeldt-Jakob disease. Disease phenotype determined by a DNA polymorphism. Science 258, 806–808 [DOI] [PubMed] [Google Scholar]
- 41. Ilc G., Giachin G., Jaremko M., Jaremko Ł., Benetti F., Plavec J., Zhukov I., Legname G. (2010) NMR structure of the human prion protein with the pathological Q212P mutation reveals unique structural features. PloS One 5, e11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gimm O., Neuberg D. S., Marsh D. J., Dahia P. L., Hoang-Vu C., Raue F., Hinze R., Dralle H., Eng C. (1999) Over-representation of a germline RET sequence variant in patients with sporadic medullary thyroid carcinoma and somatic RET codon 918 mutation. Oncogene 18, 1369–1373 [DOI] [PubMed] [Google Scholar]
- 43. Eng C., Smith D. P., Mulligan L. M., Nagai M. A., Healey C. S., Ponder M. A., Gardner E., Scheumann G. F., Jackson C. E., Tunnacliffe A. (1994) Point mutation within the tyrosine kinase domain of the RET proto-oncogene in multiple endocrine neoplasia type 2B and related sporadic tumors. Hum. Mol. Genet. 3, 237–241 [DOI] [PubMed] [Google Scholar]
- 44. Mologni L., Rostagno R., Brussolo S., Knowles P. P., Kjaer S., Murray-Rust J., Rosso E., Zambon A., Scapozza L., McDonald N. Q., Lucchini V., Gambacorti-Passerini C. (2010) Synthesis, structure-activity relationship, and crystallographic studies of 3-substituted indolin-2-one RET inhibitors. Bioorg. Med. Chem. 18, 1482–1496 [DOI] [PubMed] [Google Scholar]
- 45. Li T., Sandberg M. A., Pawlyk B. S., Rosner B., Hayes K. C., Dryja T. P., Berson E. L. (1998) Effect of vitamin A supplementation on rhodopsin mutants threonine 17 → methionine and proline 347 → serine in transgenic mice and in cell cultures. Proc. Natl. Acad. Sci. U.S.A. 95, 11933–11938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stadtman E. R., Van Remmen H., Richardson A., Wehr N. B., Levine R. L. (2005) Methionine oxidation and aging. Biochim. Biophys. Acta 1703, 135–140 [DOI] [PubMed] [Google Scholar]
- 47. Glaser C. B., Yamin G., Uversky V. N., Fink A. L. (2005) Methionine oxidation, α-synuclein and Parkinson disease. Biochim. Biophys. Acta 1703, 157–169 [DOI] [PubMed] [Google Scholar]
- 48. Schöneich C. (2005) Methionine oxidation by reactive oxygen species. Reaction mechanisms and relevance to Alzheimer disease. Biochim. Biophys. Acta 1703, 111–119 [DOI] [PubMed] [Google Scholar]
- 49. Garner M. H., Spector A. (1980) Selective oxidation of cysteine and methionine in normal and senile cataractous lenses. Proc. Natl. Acad. Sci. U.S.A. 77, 1274–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vogt W. (1995) Oxidation of methionyl residues in proteins. Tools, targets, and reversal. Free Radic. Biol. Med. 18, 93–105 [DOI] [PubMed] [Google Scholar]
- 51. Maier K., Leuschel L., Costabel U. (1991) Increased levels of oxidized methionine residues in bronchoalveolar lavage fluid proteins from patients with idiopathic pulmonary fibrosis. Am. Rev. Respir. Dis. 143, 271–274 [DOI] [PubMed] [Google Scholar]
- 52. Gao J., Yin D. H., Yao Y., Sun H., Qin Z., Schöneich C., Williams T. D., Squier T. C. (1998) Loss of conformational stability in calmodulin upon methionine oxidation. Biophys. J. 74, 1115–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ta H. M., Nguyen G. T., Jin H. M., Choi J., Park H., Kim N., Hwang H. Y., Kim K. K. (2010) Structure-based development of a receptor activator of nuclear factor-κB ligand (RANKL) inhibitor peptide and molecular basis for osteopetrosis. Proc. Natl. Acad. Sci. U.S.A. 107, 20281–20286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Saito H., Kojima T., Takahashi M., Horne W. C., Baron R., Amagasa T., Ohya K., Aoki K. (2007) A tumor necrosis factor receptor loop peptide mimic inhibits bone destruction to the same extent as anti-tumor necrosis factor monoclonal antibody in murine collagen-induced arthritis. Arthritis Rheum. 56, 1164–1174 [DOI] [PubMed] [Google Scholar]
- 55. Aoki K., Saito H., Itzstein C., Ishiguro M., Shibata T., Blanque R., Mian A. H., Takahashi M., Suzuki Y., Yoshimatsu M., Yamaguchi A., Deprez P., Mollat P., Murali R., Ohya K., Horne W. C., Baron R. (2006) A TNF receptor loop peptide mimic blocks RANK ligand-induced signaling, bone resorption, and bone loss. J. Clin. Invest. 116, 1525–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Suzuki Y., Aoki K., Saito H., Umeda M., Nitta H., Baron R., Ohya K. (2006) A tumor necrosis factor-α antagonist inhibits inflammatory bone resorption induced by Porphyromonas gingivalis infection in mice. J. Periodontal Res. 41, 81–91 [DOI] [PubMed] [Google Scholar]