Background: Neutrophil elastase (NE)-induced tumor cell proliferation requires endosomal entry of the proteinase.
Results: NE and CG bind to the surface of cancer cells and enter into tumor endosomes in a clathrin- and dynamin-dependent, caveolin- and flotillin-independent fashion.
Conclusion: Entry of NE and CG into tumor endosomes occurs via classic clathrin-pit mediated endocytosis, which requires the binding of the proteinase to a cell surface receptor.
Significance: This novel means of cell entry may grant tumor cells access to numerous, as yet unidentified bioactive molecules located in the extracellular environment.
Keywords: Cancer Biology, Endocytosis, Neutrophil, Receptor Endocytosis, Serine Protease, Cathepsin G, Neutrophil Elastase
Abstract
Neutrophil elastase (NE) is a neutrophil-derived serine proteinase with broad substrate specificity. We have recently demonstrated that NE is capable of entering tumor cell endosomes and processing novel intracellular substrates. In the current study, we sought to determine the mechanism by which NE enters tumor cells. Our results show that NE enters into early endosomal antigen-1+ endosomes in a dynamin- and clathrin-dependent but flotillin-1- and caveolin-1-independent fashion. Cathepsin G (but not proteinase-3) also enters tumor endosomes via the same mechanism. We utilized 125I-labeled NE to demonstrate that NE binds to the surface of cancer cells. Incubation of radiolabeled NE with lung cancer cells displays a dissociation constant (Kd) of 284 nm. Because NE is known to bind to heparan sulfate- and chondroitin sulfate-containing proteoglycans, we treated cells with glycanases to remove these confounding factors, which did not significantly diminish cell surface binding or endosomal entry. Thus, NE and CG bind to the surface of cancer cells, presumably to a cell surface receptor, and subsequently undergo clathrin pit-mediated endocytosis.
Introduction
Neutrophil elastase (NE),2 or human leukocyte elastase (encoded by Elane, formerly Ela2), is a neutrophil-derived serine proteinase with broad substrate specificity (1). The major physiologic role for NE resides in its ability to kill engulfed microorganisms within neutrophil phagolysosomes (2–5). However, most studies of NE have focused on its potentially destructive nature when released into the extracellular space (6). As is the case with most matrix-degrading enzymes, NE has been characterized by its ability to cleave extracellular matrix proteins, which include collagens (types I–III), fibronectin, laminin, entactin, type IV collagen, and the rather inert elastic fiber (7). The identification of nontraditional substrates for NE continues to emerge from the literature, with plasminogen, coagulation factors, immunoglobulins, C5a, and TGFα, among others (8, 9). Due to its broad substrate specificity and destructive properties when released freely into the extracellular matrix, NE has been implicated in the pathogenesis of several diseases, including acute lung injury, cystic fibrosis, and emphysema (10–12). However, two key aspects of NE release/function are often overlooked. First, NE is rarely dumped into the extracellular matrix, but rather is released by the neutrophil in quantum microbursts for the purpose of focused proteolysis (13). Second, NE frequently binds to heparan sulfate- and chondroitin sulfate-containing proteoglycans found on the surface of PMN, where it is catalytically active and resistant to its inhibitors (14, 15). Taken together, these findings suggest that the concentrations of NE commonly encountered in vivo are more likely to be modest and less likely to be destructive.
We have recently shown that these modest concentrations of NE promote lung cancer cell proliferation both in vitro and in vivo (16). NE accomplishes this by degrading a novel target substrate, insulin receptor substrate-1 (IRS-1), ultimately resulting in phosphoinositol 3-kinase (PI3K) hyperactivity and subsequent tumor cell proliferation (17). Surprisingly, we localized the site of NE:IRS-1 interaction within the tumor cell, and not in the extracellular space nor on the cell surface. Our report represents the first description of a secreted proteinase gaining access to another cell without killing it, as is the case with the granzymes, for example. Subsequently, NE has been shown to enter into breast cancer cells as well (18).
We were able to localize NE beyond the plasma membrane of tumor cells and within endosomal structures. Furthermore, we showed that entry of NE into tumor endosomes was required for its cell proliferative effects. These findings provide a novel means by which a secreted proteinase can impact the behavior of neighboring cells. Additionally, it increases the list of potential substrates for NE (and possibly for other secreted proteinases) to include novel targets that would previously have been considered restricted access. The purpose of the current study is to determine the means by which NE gains access to cancer cells. Here, we show that NE binds to the cancer cell surface with specificity and subsequently undergoes classic clathrin pit-mediated endocytosis.
EXPERIMENTAL PROCEDURES
Materials
Human neutrophil elastase (NE, also known as human leukocyte elastase), cathepsin G (CG), proteinase-3 (PR3), and myeloperoxidase (MPO), all purified from human sputum, were purchased from Elastin Products Co. (Owensville, MO). Transferrin, dynasore, nystatin, guanidine, sodium metabisulfite, cytochrome c, and phenylmethylsulfonyl fluoride (PMSF) were purchased from Sigma. Trypsin was purchased from Prospec (East Brunswick, NJ). Heparitinases I, II, and III (from Flavobacterium heparinum) and chondroitinase ABC (from Proteus vulgaris) were purchased from Seikagaku Corporation (Tokyo, Japan). Na-125I was obtained from PerkinElmer Life Sciences. Synthetic peptides were generated by the University of Pittsburgh Peptide Synthesis Core facility: P1 = ASEIVGGRRARPHAWPFMVSLQL; P2 = GGHFCGATLIAPNFVMSAAHC; P3 = LGAHNLSRREPTRQVFAVQRIFE; P4 = LNDIVILQLNGSATIN; P5 = CFGDSGSPLVCNGLIHGIASF; P6 = GGCASGLYPDAFAPVAQFVNWI; P7 = NVQNAQLPAQGRRLGNGVQC; P8 = MGWGLLGRNRGIASVLQEL.
Cells
Three human lung adenocarcinoma cell lines were utilized, A549, 343T, and H23. Each cell line was obtained from ATCC (Manassas, VA). Chinese hamster ovary (CHO) cells, xylosyltransferase I-deficient (proteoglycan-deficient) CHO cells (CHO-pgsa) (19), and CMP-sialic acid transporter mutant (sialic acid-deficient) CHO cells (CHO-Lec-2) (20) were also obtained from ATCC. All cells were cultured in T-75 flasks in DMEM plus 10% FBS and 1× penicillin/streptomycin. For the assays, cells were plated at 1 × 105 cells/well in 24-well plates and grown to confluence (∼4.0 × 105 cells/well) prior to changing to serum-free conditions.
Thymidine Incorporation Assay
H23 or A549 cells were plated at a concentration of 1 × 105 cells/well in 24-well plate before treatment with NE or CG at concentrations from 1 to 40 nm for 60 min in serum-free media. The cells were incubated with 1 μCi/ml thymidine (PerkinElmer Life Sciences) for 18 h. Assays were terminated by washing with PBS, fixing with 5% trichloroacetic acid, and washing with tap water. Cells were lysed with 200 mm NaOH neutralized with equimolar HCl and transferred to scintillation vials. Results are from a representative experiment in triplicate.
Radiolabeling of NE
NE was iodinated using the IODO-BEAD method according to the manufacturer's instructions (Pierce), as described previously (21). Briefly, two beads were washed in 500 μl of PBS for 5 min and dried on filter paper. The beads were then preincubated with 2 mCi of Na-125I in PBS for 5 min at which time the protein (NE or CG, in separate experiments) was added (200 μg) at a concentration of 10 μg/μl. After an 8-min incubation, the reaction was terminated with sodium metabisulfite. Cytochrome c was added as a marker, and the entire reaction mixture was run on a Sephadex G-25 column to separate bound 125I from free 125I. The column flow-through was collected in fractions and subjected to autoradiography to ensure that the collected radioligand was free of unlabeled 125I. Specific activity for 125I-NE was 29.4 cpm/fmol.
Radioligand Binding Assay
Cells were plated at 1 × 105 cells/well in a 24-well plate and grown to confluence prior to culturing under serum-free conditions. All binding assays were performed at 4 ºC in a walk-in cold room. The cells were washed in PBS buffer containing 1 mm CaCl2 and 1 mm MgCl2 before incubation with 125I-NE for 2 h. The incubation buffer consisted of PBS with 0.5% BSA and 0.01% v/v Tween 80. Initially, this experiment was performed using a fixed concentration of radiolabeled NE (25 nm) and increasing concentrations of unlabeled NE. At the conclusion of the experiment, the cells were washed in PBS buffer and lysed with 500 mm NaOH. The lysates were subject to γ-counting. Assays performed in triplicate and replicated in a separate experiment.
Determination of Dissociation Constant (Kd)
To determine the dissociation constant (Kd) of radiolabeled NE from the cancer cell surface, we repeated the binding assay in A549 cells described above using increasing concentrations of radiolabeled NE. A549 cells were incubated with the radiolabeled NE in both the absence and presence of 100-fold molar excess unlabeled NE. The results are presented as specific binding or total binding minus nonspecific (the amount remaining in the presence of 100-fold molar excess unlabeled ligand) binding. These data were analyzed using a nonlinear regression (curve fit) to determine the Kd and Bmax (maximum number of binding sites per cell). For context, we also performed a linear regression on transformed data to generate a Scatchard plot depicting bound NE versus bound/free NE. Assays were performed in triplicate and replicated in a separate experiment.
Determination of KI
To determine the half-maximal inhibitory concentration (IC50) and inhibition of binding affinity (KI) of unlabeled NE and CG as inhibitors, we treated A549 cells with a constant concentration of 125I-labeled NE (25 nm) in the presence of increasing concentrations of unlabeled NE and CG (in separate experiments) ranging from 1 nm to 10 μm. The data were analyzed using a nonlinear progression (Prism software). The “top” plateau was set as the cpm measurement using 25 nm 125I-NE in the absence of unlabeled ligand. The KI was calculated from this value using the formula KI = IC50/(1 + c/Kd), where c was the concentration of the radioligand, and Kd was its dissociation constant. Results are presented in logarithmic scale. Assays were performed in triplicate and replicated in a separate experiment.
Glycanase Treatment
NE has been previously reported to bind to the sulfate groups of heparan sulfate- and chondroitin sulfate-containing proteoglycans on the surface of PMNs (15). Therefore, we chose to perform the binding assays and NE trafficking experiments both before and after removing these potentially contaminating sulfate groups. For all glycanase experiments, cells were plated on type I collagen-coated plates (BD Biosciences) overnight and serum-starved for 1 h prior to treatment. Heparan sulfate and chondroitin sulfate groups were removed using 3.5 units/ml heparitinase I and 200 milliunits/ml chondroitinase ABC in a buffer composed of 50 mm NaCl, 4 mm CaCl2, 40 mm sodium acetate, 0.1% BSA in PBS for 1 h at 37C. Cleavage of heparan and chondroitin sulfate groups was verified using flow cytometry. Cell staining was performed using anti-δ-heparan sulfate (1:100, 3G10 clone; Seikagaku) and anti-chondroitin sulfate (1:100, CS-56 clone; Abcam) for 2 h at 4 ºC followed by secondary antibody stain (Alexa Fluor 488-anti-mouse IgG/IgM, 1:1000; Invitrogen). Verification of proteoglycan removal employed the above antibodies with the knowledge that the chondroitin sulfate antibody recognized shed proteoglycans, and the heparan sulfate antibody recognizes the exposed protein core on the cell surface once the heparan sulfate-containing proteoglycans have been shed (22). The samples were analyzed using a BD FacsCalibur instrument and CellQuest Pro software. Gates were set based on appropriate isotype controls.
Competition of Radiolabeled NE Binding
The above binding assays were repeated using a constant concentration of 125I-NE (25 nm) in the absence or presence of potentially competitive ligands in 100-fold molar excess. These ligands included proteins of neutrophil origin including CG, PR3, and MPO, as well as NE itself, both alone and in conjunction with the serine proteinase inhibitor PMSF. Transferrin was also used as an irrelevant protein control. Finally, this assay was repeated using synthetic peptides from different regions of NE as the competitor, all in 100-fold molar excess. Assays were performed in triplicate and replicated in a separate experiment.
Immunofluorescence Labeling and Confocal Microscopy
A549, H23, and 343T cells (5 × 104 cells) were plated on tissue culture-treated coverslips (Thermo Fisher) in 24-well plates, allowed to adhere overnight, and transferred to serum-free media for 24 h. NE was fluorescently labeled using the Alexa Fluor 488 protein labeling kit (Invitrogen) according to the manufacturer's instructions. Cells were treated with Alexa Fluor 488-NE (8 nm) for the times indicated, immediately fixed in 2% paraformaldehyde in PBS, and permeabilized with 0.1% Triton X-100. Cells were co-stained for phalloidin (Invitrogen), LAMP-1 (Developmental Studies Hybridoma Bank, Iowa City, IA), early endosomal antigen-1 (EEA-1; Abcam, Cambridge, MA), clathrin, caveolin-1, or flotillin-1 (all Cell Signaling, Danvers, MA). Donkey anti-rabbit 594 was used as the secondary antibody (Invitrogen). Cells were imaged using an Olympus Fluoview 100 upright confocal microscope.
Small Interfering RNA
To assess the dependence of NE endosomal entry on caveolin-1, flotillin-1 and clathrin, these genes were silenced using siRNA, in separate experiments. A549 cells were plated at a concentration of 1 × 104 cells/ml and transfected with siRNA for caveolin-1 (80 nm, Qiagen, Valencia, CA; Hs_CAV1_10), clathrin heavy chain (80 nm, Qiagen, Hs_CLTC_10), flotillin-1 (60 nm, Santa Cruz Biotechnology, Santa Cruz, CA), or scrambled (SCR) control (Qiagen) using Lipofectamine (Invitrogen) according to the manufacturers' instructions. Cells were transfected for 5 h on days 1 and 2. Adequacy of gene silencing was assessed using Western blotting. Immunofluorescence experiments (as above) were performed 48 h following initial transfection. NE entry into A549 cells in the presence of siRNA was quantified in at least 20 cells from four separate fields.
Immunoblotting
Proteins were separated by standard 10% SDS-PAGE followed by transfer of protein to nitrocellulose membranes. The following antibodies were used, all from Cell Signaling Technology: IRS-1 (1:250, 2382), pAkt (1:250, 9271), Akt (1:250, 9272), and β-actin (1:500, 4967).
Statistics
Assays were performed in duplicate or triplicate and replicated in a separate experiment. Data are expressed as the mean values ± S.E. Simple pairwise comparisons were analyzed using the Student's t test (two-tailed distribution with two-sample equal variance). For multiple comparisons, we used one-way ANOVA with Newman-Keuls post test. A p value of <0.05 was considered significant. Linear transformation of binding data were accomplished using Prism data analysis software.
RESULTS
Neutrophil Elastase Enters Tumor Cells
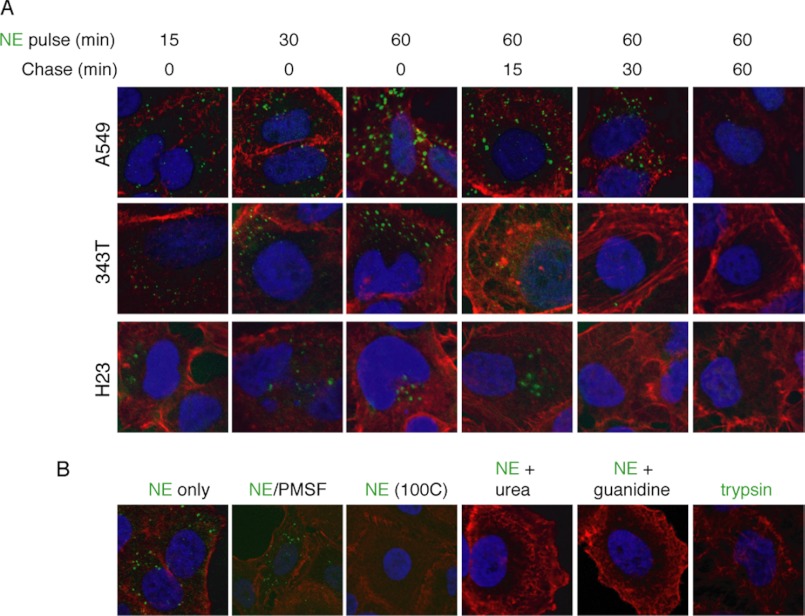
To determine the kinetics of NE entry into lung cancer cells, we treated three different lung cancer cell lines (A549, 343T, and H23) with Alexa Fluor 488-labeled NE over a time course. In each case, NE was located inside of the cell within 5 min, reached its maximal accumulation at ∼60 min, and disappeared from the cell during the subsequent 60 min (Fig. 1A).
FIGURE 1.
NE enters human lung adenocarcinoma cells using its tertiary structure. A, Alexa Fluor 488-labeled NE (8 nm) was incubated with three different human lung adenocarcinoma cell lines, A549, 343T, and H23, for 60 min, washed with PBS, and incubated for an additional 60 min. B, A549 cells were treated with Alexa Fluor-488-labeled NE (8 nm) in native form or denatured with PMSF (1:100 molar ratio), heat (100 ºC), urea (8 m), or guanidine-HCl (6 m); or the A549 cells were incubated with Alexa Fluor 488-labeled trypsin (8 nm). Cells were counterstained with phalloidin (red) and DAPI (blue). All images (magnification, ×1200) are representative data that were replicated in separate experiments.
We proceeded to determine whether NE required its catalytic domain or three-dimensional structure to enter tumor cells (Fig. 1B). Addition of the serine proteinase inhibitor PMSF does not prevent the entry of NE into tumor cells, thus demonstrating that the catalytic domain of NE is not required for cell entry. We subjected an aliquot of the NE/PMSF mixture to zymography, to prove that the catalytic activity of NE was indeed inhibited (data not shown). Next, we subjected NE to various means of denaturation, including heat (65 ºC and 100 ºC), guanidine, and urea. Whereas heating NE to 65 ºC had no effect on cell entry, heating to 100 ºC and both methods of chemical denaturation eliminated the ability of NE to enter tumor cells, suggesting that its three-dimensional structure is required for cell entry. As a relevant control, we incubated A549 cells with Alexa Fluor 488-labeled trypsin, a serine proteinase closely related to NE. The labeled trypsin did not enter A549 cells.
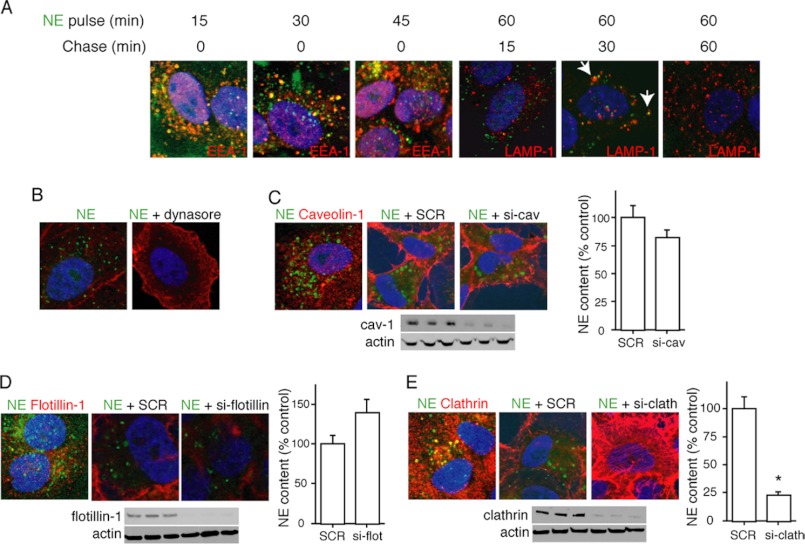
To determine the topographical location of NE within tumor cells, we repeated the NE exposure experiments in A549 cells using antibodies to markers of early to late endosomes. Within minutes of cell entry, NE can be located within early EEA-1+ (Fig. 2A), which are known to transport cargo directly after clathrin pit internalization to more mature endosomes (23). The majority of NE:EEA-1 co-localization occurs within the initial 10–15 of exposure, typical of clathrin pit-mediated endocytic processes. A modest proportion of the internalized NE reaches the lysosomes (LAMP-1+ endosomes), where it is presumably degraded (Fig. 2A). It remains unclear what proportion of NE reaches the lysosome and what proportion escapes prior to degradation. Because our data were consistent with a classic clathrin pit mode of entry into the cell, we repeated our assay in the presence of 40 μm dynasore, a dynamin inhibitor that prevents the formation of early endosomes (24). Accordingly, dynamin inhibition prevents the entry of NE into tumor cells (Fig. 2B), consistent with a clathrin pit to early endosome mechanism of cellular entry for NE.
FIGURE 2.
NE enters tumor endosomes via a dynamin- and clathrin-dependent, caveolin-1- and flotillin-independent mechanism. A, A549 cells were incubated with Alexa Fluor 488-labeled NE (8 nm) for 60 min, washed with PBS, and imaged over the following 60 min. Co-localization studies were performed using antibodies against EEA-1 and LAMP-1. B, A549 representative images show cells treated with Alexa Fluor 488-labeled NE (8 nm) alone or in combination with dynasore (40 nm). C, confocal image shows A549 cells treated with Alexa Fluor 488-labeled NE (8 nm) and stained with anti-caveolin-1 (red). A549 cells were also treated with scrambled (SCR) siRNA or siRNA against caveolin-1. Phalloidin (red) was the counterstain. Western blotting confirmed knockdown of caveolin-1. D, confocal image shows A549 cells treated with Alexa Fluor 488-labeled NE (8 nm) and stained with anti-flotillin-1 (red). A549 cells also treated with scrambled siRNA or siRNA against flotillin-1. Phalloidin (red) was the counterstain. Western blotting confirmed knockdown of flotillin-1. E, confocal image shows A549 cells treated with Alexa Fluor 488-labeled NE (8 nm) and stained with anti-clathrin (red). A549 cells were also treated with scrambled siRNA or siRNA against clathrin. Phalloidin (red) was the counterstain. Western blotting confirmed knockdown of clathrin.
We entertained two additional possible mechanisms for the entrance of NE into tumor vesicles. First, we considered the caveolar system, known to grant tumor cell access to extracellular proteins (25). We were unable to observe co-localization of Alexa Fluor 488-labeled NE with caveolin-1 over a time course (Fig. 2C). Additionally, the silencing caveolin-1 using siRNA did not impact the entrance of NE into A549 cells (Fig. 2C). Second, we evaluated the possibility that NE entered early endosomes in a dynamin-dependent, yet clathrin-independent fashion. Although this type of endocytosis is still poorly described, it is often flotillin-1 dependent, and does not necessarily utilize a cell surface receptor, as is typically the case for clathrin pit-mediated endocytosis (26, 27). Silencing of flotillin-1 did not inhibit the ability of NE to enter tumor endosomes (Fig. 2D). Furthermore, we were unable to co-localize Alexa Fluor 488-labeled NE with flotillin-1 using confocal microscopy (Fig. 2D). However, we were able to co-localize Alexa Fluor 488-labeled NE with clathrin using confocal microscopy and to inhibit the entry of NE into tumor endosomes by silencing the clathrin heavy chain using siRNA approaches (Fig. 2E). Thus, NE enters tumor endosomes in a dynamin- and clathrin-dependent, yet caveolin-1- and flotillin-1-independent fashion.
Endosomal Entry of NE Is Required for Tumor Cell Proliferation
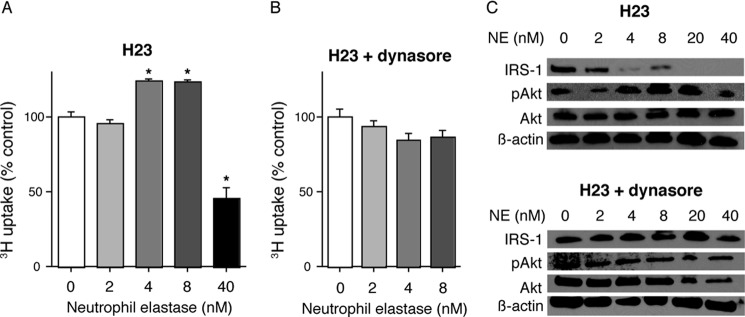
We treated H23 cells with NE both in the presence and absence of dynasore to demonstrate that NE must enter tumor endosomes to induce cellular proliferation (Fig. 3). Thymidine uptake experiments demonstrated a characteristic induction of cellular proliferation by NE in the 4–8 nm range. This finding was abolished by the addition of dynasore. Higher concentrations of NE were toxic, as previously described. Additionally, we utilized Western blotting to show that NE entry into endosomes is required for NE to access its target substrate, IRS-1, and to induce pAkt production (Fig. 3C). Thus, NE must enter tumor endosomes to degrade IRS-1, induce pAkt production, and cause cellular proliferation.
FIGURE 3.
NE endocytosis is required for cellular proliferation. A and B, 3H uptake for H23 cells treated with NE (0–40 nm) for 60 min in the absence (A) or presence (B) of dynasore. Results are shown as percentage of control. The experiment was done in triplicate. Error bars, S.E. *, p < 0.05. C, representative Western blots for IRS-1, pAkt, Akt, and β-actin.
NE Binds to the Surface of Tumor Cells
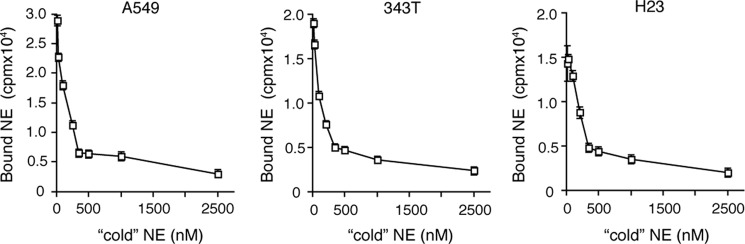
Classic clathrin pit-mediated endocytosis is generally facilitated by the existence of a specific cell surface receptor for the ligand in question. Similarly, a ligand may demonstrate the ability to bind a receptor with somewhat reduced affinity, for which it is not the primary ligand. Therefore, we assessed the ability of NE to bind to the cancer cell surface using a traditional radiolabeled ligand binding assay. Initially, we exposed a constant concentration of 125I-labeled NE to A549, 343T, and H23 lung cancer cells in the presence of increasing concentrations of unlabeled NE. These experiments were performed at 4 ºC to minimize endocytosis and focus on the binding of NE to the cell surface. The results indicate that NE bound to the surface of all three cell lines and displayed specificity, as increasing concentrations of unlabeled NE were capable of eliminating the binding in a dose-dependent manner (Fig. 4).
FIGURE 4.
NE binds to lung cancer cells. 125I-Labeled NE (25 nm) was incubated with A549, 343T, and H23 lung cancer cells (in separate experiments) for 2 h at 4 ºC. The cells were also treated with increasing concentrations of unlabeled NE for the same time period. Experiments were performed in triplicate. Error bars, S.E.
Cathepsin G Competes with NE for Cell Surface Binding Sites
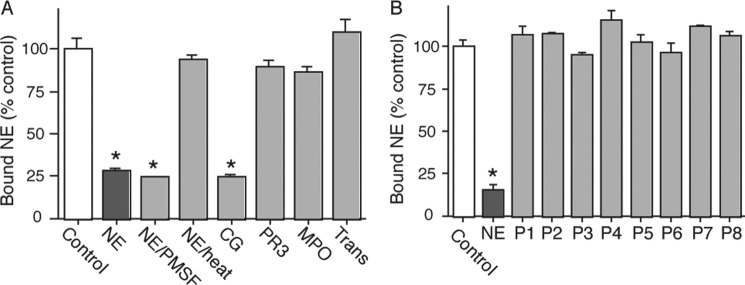
To demonstrate additional specificity for NE binding to cancer cells, we attempted to inhibit this binding with related proteins housed in neutrophils. The results show that the binding of NE to the surface of A549 cells can be inhibited by NE itself (both catalytically active and inactive) but not by heat-denatured NE, PR3, MPO, or transferrin (not neutrophil-derived but does undergo endocytosis). Interestingly, CG was able to inhibit the binding of NE to cancer cells (Fig. 5A). Because NE and CG share significant sequence identity, we investigated the possibility that a linear sequence common to both NE and CG was mediating cell binding. Therefore, we synthesized eight peptides (P1–P8) from regions of the NE sequence that shared sequence identity with CG based on sequence alignment (data not shown). However, none of these linear peptides was able to inhibit the binding of NE to cancer cells (Fig. 5B). These data are consistent with the prior observation that NE requires its tertiary structure to enter tumor endosomes and presumably to bind to the cell surface.
FIGURE 5.
Cathepsin G competes with NE for cell surface binding sites. A, A549 cells were incubated with 125I-labeled NE alone (control) or in combination with 100-fold molar excess unlabeled NE, NE/PMSF, NE + heat (100 ºC), CG, PR3, MPO, or transferrin for 2 h at 4 ºC. Experiments were performed in triplicate. Error bars, S.E. *, p < 0.05 from control. B, A549 cells were incubated with 125I-labeled NE alone (control) or in combination with 100-fold molar excess unlabeled NE, or one of eight linear peptides (P1–P8) derived from regions of the NE sequence that shared identity with CG.
CG Enters Tumor Endosomes
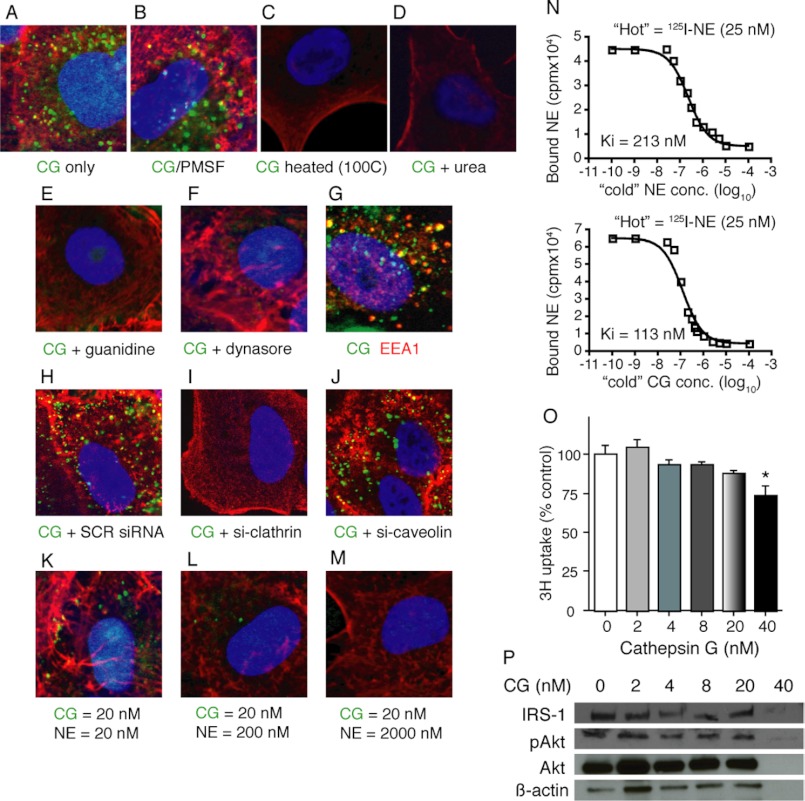
Because CG was able to inhibit the binding of NE to cancer cells, we hypothesized that CG may also utilize its ability to bind cancer cells ultimately to enter into tumor endosomes. Therefore, we labeled CG protein with Alexa Fluor 488 and treated A549 cells with the labeled protein. CG gains access to A549 cells beyond their plasma membrane over a time course similar to that described for NE (Fig. 6A). As is the case with NE, CG does not require its catalytic domain to enter cancer cells (Fig. 6B), but it does require its tertiary structure, as denaturation of the protein using heat, guanidine, or urea eliminates CG cellular entry (Fig. 6, C–E). As was the case for NE, CG enters into EEA-1+ endosomes in a dynamin- and clathrin-dependent but caveolin-1-indpendent fashion (Fig. 6, F–J), consistent with clathrin pit-mediated endocytosis. It appears that NE and CG utilize the same cell surface binding sites and mechanisms to gain access to tumor endosomes, as the addition of increasing concentrations of NE eliminated the entry of Alexa Fluor 488-labeled CG into cancer cells (Fig. 6, K–M). Interestingly, CG demonstrates a higher affinity for cell surface binding than NE. Determination of the KI, or the binding affinity of an inhibitor, demonstrated a KI of 113 nm for CG as opposed to a KI of 213 nm for NE (Fig. 6N). In other words, 113 nm CG inhibits the binding of radiolabeled NE to the cell surface to the same degree that it requires 213 nm NE to achieve.
FIGURE 6.
CG enters tumor endosomes. A549 cells were incubated with Alexa Fluor 488-labeled CG for 30 min either alone (A), or with the addition of PMSF (B), heat (100 ºC) (C), urea (D), guanidine (E), dynasore (F), scrambled siRNA (H), heavy chain clathrin siRNA (I), or caveolin-1 siRNA (J). G, EEA-1 antibody staining was employed to co-localize CG and EEA-1. K–M, increasing concentrations of unlabeled NE were added to Alexa Fluor 488-labeled CG-treated A549 cells. All images (magnification, ×1200) are representative data that were replicated in separate experiments. N, A549 cells were incubated with 125I-NE for 2 h at 4 ºC in the presence of increasing concentrations of unlabeled (“cold”) NE or CG to determine the KI values. Results are in logarithmic scale. Experiment was performed in triplicate. O, 3H uptake is shown for A549 cells treated with CG (0–40 nm) for 60 min. Results are shown as percentage of control. Experiment was performed in triplicate. Error bars, S.E. *, p < 0.05. P, representative Western blots for are shown for IRS-1, pAkt, Akt, and β-actin.
CG Does Not Induce Cellular Proliferation
Because CG enters tumor endosomes as does NE, we sought to determine whether CG was also capable of inducing cellular proliferation. Treatment of A549 cells with concentrations of CG from 2 to 40 nm did not induce cell proliferation, as determined by thymidine uptake assay (Fig. 6O). To explain this finding mechanistically, we subjected CG-treated A549 cell lysates to Western blotting for IRS-1, pAkt, and Akt. In contrast to NE, CG does not degrade IRS-1, nor does it induce pAkt, two events known to be essential for NE-induced cellular proliferation (Fig. 6P). Only the highest concentration of CG (40 nm) resulted in loss of IRS-1 signal; however, this concentration of CG was cell-toxic, as determined by reduced thymidine uptake and loss of β-actin expression.
NE Binds with Low Affinity to a Cell Surface Receptor
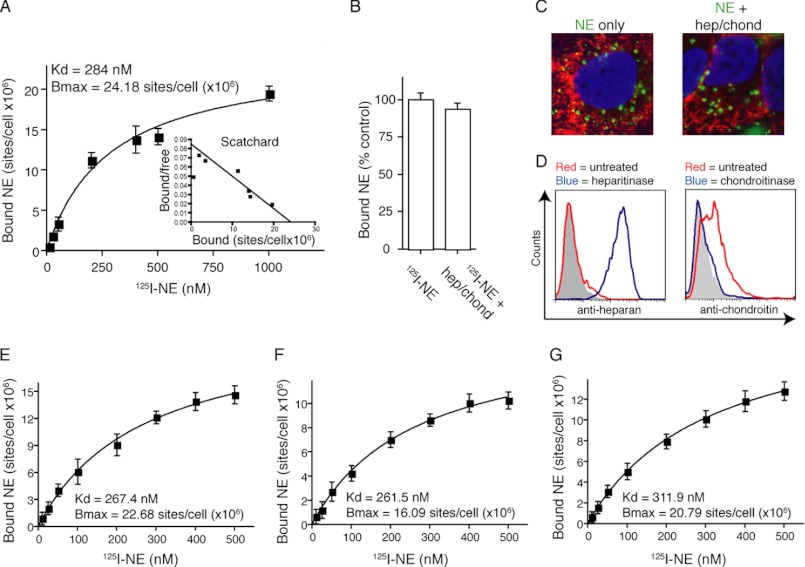
We performed standard ligand affinity binding curves for NE using A549 cells. 125I-Labeled NE was used in increasing concentrations from 10 to 1000 nm in both the presence and absence of a 100-fold molar excess of unlabeled NE. The results are expressed as specific binding, or the total binding minus the nonspecific binding (binding that remains in presence of 100-fold molar excess unlabeled ligand). The dissociation constant was calculated by performing a linear regression of the data using Prism software, which yielded a Kd of 284 nm (Fig. 7A). The maximum number of binding sites, or Bmax, was 24.18 × 106 sites/cell. We suspected that our data may be skewed by the presence of heparan sulfate- and chondroitin sulfate-containing proteoglycans, as NE has previously been shown to bind to these residues on the surface of PMN, albeit with a somewhat reduced affinity (Kd = 538 nm) (15). Suspecting that these low affinity sites may be masking our results, we repeated this assay following treatment of the cells with both heparitinase I and II and chondroitinase ABC. Removing these sulfate groups did not alter the Kd (276 nm; data not shown), nor did it reduce the number of binding sites (Fig. 7B). We also exposed heparitinase and chondroitinase treated A549 cells to Alexa Fluor 488-labeled NE to confirm that heparan and chondroitin sulfate-containing proteoglycans are not required for the entry of NE into tumor endosomes (Fig. 7C). We confirmed removal of the heparan sulfate- and chondroitin sulfate-containing proteoglycans using flow cytometry with the understanding that the antibody raised against chondroitin sulfate recognized shed proteoglycans and the heparan sulfate antibody recognized the exposed protein core after shedding of heparan sulfate-containing proteoglycan chain (Fig. 7D).
FIGURE 7.
NE binds cell surface receptor with low affinity. A, A549 cells incubated with 125I-labeled NE for 2 h at 4 ºC both with and without 100-fold molar excess of unlabeled NE. Specific binding (total − nonspecific) is depicted. Error bars, S.E. Curve represents linear transformation with nonlinear transformation (Scatchard) shown in inset. B, A549 cells incubated with 125I-labeled NE following treatment with heparitinase and chondroitinase. C, representative images (magnification, ×400) of Alexa Fluor 488-labeled NE incubated with A549 cells or A549 cells following heparitinase and chondroitinase treatment. D, flow diagram of A549 cells following treatment of heparitinase and chondroitinase, in separate experiments. 125I-labeled NE incubated with (E) CHO, (F) CHO-pgsa, and (G) CHO-Lec-2 cells for 2 h at 4 ºC both with and without 100-fold molar excess of unlabeled NE. Data represent specific binding. Experiment was performed in triplicate. Error bars, S.E.
Because the cells are capable of rapidly reconstituting their cell surface proteoglycan content, we chose to repeat the NE binding studies using genetically engineered proteoglycan-deficient cells. Xylosyltransferase-deficient CHO cells do not synthesize proteoglycans and were chosen for this purpose, using CHO cells as a control. We additionally employed CHO-Lec-2 cells, which contain a mutation in sialic acid transferase, which additionally reduces polyanionic residues from the cell surface. We initially confirmed that NE is capable of entering each of these cell types (data not shown). Standard ligand affinity binding curves were repeated for each cell line (Fig. 7, E–G). The Kd was not different for any of the three cell lines, nor were these values significantly different from the Kd measured for A549 cells. Proteoglycan-deficient CHO cells (CHO-pgsa) displayed a 29% reduction binding site number, suggesting that there is some binding of NE to these sites. However, the absence of these sites did not impact the binding curves or the entry of NE into endosomes.
DISCUSSION
We have recently shown that NE-deficient mice display reduced tumor burden, tumor cell proliferation, and mortality in the LSL-K-ras mouse model of lung tumorigenesis (16). The finding that NE actually entered tumor endosomes and altered cancer cell behavior represented a novel function of a secreted proteinase. Because endosomal entry was required for the cell proliferative effects of NE, we sought to determine the means by which NE gained cellular access, in an effort to identify a therapeutically relevant cell surface receptor for NE. Here, we have presented data that NE binds to the surface of cancer cells in a specific and saturable manner, indicative of a receptor-ligand interaction. Following cellular binding, NE enters tumor endosomes in a clathrin- and dynamin-dependent, flotillin-1 and caveolin-1-independent fashion, consistent with classic receptor-mediated clathrin pit endocytosis. Additionally, we have shown that the related proteinase CG binds to cancer cells and enters tumor endosomes in a similar manner.
The NE-cell surface receptor interaction displays relatively low affinity, illustrated by a Kd = 284 nm. There are a number of possible confounding factors that may have artificially reduced the calculated Kd. As mentioned, we initially suspected that heparan sulfate- and chondroitin sulfate-containing proteoglycans were binding NE, based on prior studies performed in neutrophils (15). We were able to detect small differences in specific binding using both enzyme treatment and genetically engineered cells deficient in proteoglycan synthesis (CHO-pgsa). Therefore, it appears that NE does bind to these moieties to some extent. However, we were not able to demonstrate a change in binding affinity (equivalent Kd) or a requirement of surface proteoglycan for endosomal entry of NE. These data additionally support the existence of a receptor-ligand interaction between NE and its putative receptor.
A more likely confounding factor is the presence of putative extracellular matrix secreted by cancer cells in culture. Many of the matrix proteins secreted by cancer cells, such as fibronectin, collagen, and laminin, all bind NE, as they are substrates for the enzyme. Thus, it is possible that the true Kd is somewhat lower. Although the binding affinity measured here would be quite low for a classic receptor-ligand interaction, we hypothesize that NE binds to a receptor for which it is not the primary ligand. In this model, NE would not serve as a receptor agonist, but simply utilize the low affinity interaction for the purpose of cell entry. This proposed mechanism is the major means by which viral pathogens enter host cells. For example, the measles virus enters host cells by binding the cell surface receptors CD46 and CD150 and subsequently undergoes endocytosis (28, 29).
The only cell surface receptor previously reported to bind NE is the serpin-enzyme complex receptor (30), which actually binds NE that has been irreversibly bound by its endogenous inhibitor, α1-antitrypsin. The serpin-enzyme complex receptor similarly displays a relatively low affinity for its ligand(s), ∼40 nm, which is within the same order of magnitude described here. It is difficult to imagine that a cell could tolerate a high affinity interaction with NE because this would put substantially more NE within the cell, likely in concentrations that would cause cell death. The identity of the receptor with which NE interacts is unknown and represents an ongoing area of investigation in our laboratory.
Following entry into EEA-1+ early endosomes, NE either traffics to LAMP-1+ lysosomes, or “leaks” into the cytosol. The presence of the latter is supported by two observations. First, Alexa Fluor 488-NE signal can be located within the cell but not in either vesicle. Second, preliminary observations using NE labeled with 5-nm gold particles can be detected within the cytosol of A549 cells using transmission electron microscopy (data not shown). The presence of NE within the cytosol would explain how it is capable of degrading IRS-1, a cytosolic protein that is endosome-associated (docks to the outside of endosomes), but not actually found within endosomes. The means by which NE escapes the endosomal compartment is not yet known. We would speculate that NE uses its catalytic domain to cleave through the endosomal wall, as PMSF-conjugated NE is found only within intact endosomes by transmission electron microscopy and not within the cytoplasm (not shown).
The finding that CG binds to the cancer cell surface and enters tumor endosomes is entirely novel. CG, like NE, was reported to bind to heparan sulfate- and chondroitin sulfate-containing proteoglycans on the surface of neutrophils. Of note, MPO and PR3, both neutrophil-derived, were capable of partially competing for these binding sites on PMN, which was not the case here. The fact that CG, but not PR3, was capable of competing for NE binding sites raises questions regarding the three-dimensional configuration of the enzymes in vivo. The degree of sequence homology among the three enzymes is similar. Linear peptide sequences displaying sequence homology between NE and CG failed to inhibit NE binding. Finally, denaturing NE with heat or chemicals eliminates binding. Therefore, there is likely a unique three-dimensional characteristic in NE and CG that has not been preserved in PR3. The only other notable difference is that NE and CG are both polycationic proteins, whereas PR3 is more neutral.
In contrast to NE, CG is unable to induce cellular proliferation. As illustrated here, NE-induced cell proliferation is dependent upon four events: (i) cell surface binding, (ii) entrance into tumor endosomes, (iii) degradation of IRS-1, and (iv) enhanced pAkt production. CG is capable of binding cancer cells and entering endosomes, but is incapable of degrading IRS-1 in this context, and therefore, subsequent pAkt production is not observed. It is likely that CG processes tumor-derived substrates that could impact tumor cell behavior. The nature of these events is currently under investigation in our laboratory.
In summary, we have provided evidence that NE and CG bind to the surface of cancer cells and subsequently undergo clathrin- and dynamin-dependent, and caveolin-1- and flotillin-1-independent endocytosis. These results strongly suggest the presence of a cell surface receptor capable of binding NE and CG with relatively low affinity, that allows for endosomal entry via clathrin pit-mediated endocytosis. The presence of such a receptor may provide endosomal access to numerous, as yet unidentified ligands, capable of altering cell behavior, as is clearly the case for NE, and may be for CG. Expression cloning approaches are under way in our laboratory to definitively identify the cell surface receptor in question. The identification of this receptor bears clinical significance. Because NE is a potent antimicrobial agent, inhibiting NE directly would theoretically carry some risk of immunocompromise. The ability to target the receptor that grants NE access to cancer cells would eliminate the tumor promoting effects of NE without compromising the enzymes ability to kill engulfed microorganisms within neutrophil phagolysosomes.
This work was supported, in whole or in part, by National Institutes of Health Grants R01HL108979 through the NHLBI (to A. M. H.) and 5T32HL007563-24 through the NHLBI (to A. D. G.). This work was also supported by an American Lung Association senior research award (to A. D. G.).
- NE
- neutrophil elastase
- CG
- cathepsin G
- EEA-1
- early endosomal antigen-1
- IRS-1
- insulin receptor substrate-1
- MPO
- myeloperoxidase
- PMN
- polymorphonuclear neutrophil
- PR3
- proteinase-3.
REFERENCES
- 1. Baugh R. J., Travis J. (1976) Human leukocyte granule elastase: rapid isolation and characterization. Biochemistry 15, 836–841 [DOI] [PubMed] [Google Scholar]
- 2. Belaaouaj A., McCarthy R., Baumann M., Gao Z., Ley T. J., Abraham S. N., Shapiro S. D. (1998) Mice lacking neutrophil elastase reveal impaired host defense against Gram-negative bacterial sepsis. Nat. Med. 4, 615–618 [DOI] [PubMed] [Google Scholar]
- 3. Belaaouaj A., Kim K. S., Shapiro S. D. (2000) Degradation of outer membrane protein A in Escherichia coli killing by neutrophil elastase. Science 289, 1185–1188 [DOI] [PubMed] [Google Scholar]
- 4. Weinrauch Y., Drujan D., Shapiro S. D., Weiss J., Zychlinsky A. (2002) Neutrophil elastase targets virulence factors of enterobacteria. Nature 417, 91–94 [DOI] [PubMed] [Google Scholar]
- 5. Gabay J. E., Scott R. W., Campanelli D., Griffith J., Wilde C., Marra M. N., Seeger M., Nathan C. F. (1989) Antibiotic proteins of human polymorphonuclear leukocytes. Proc. Natl. Acad. Sci. U.S.A. 86, 5610–5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee W. L., Downey G. P. (2001) Leukocyte elastase: physiological functions and role in acute lung injury. Am. J. Respir. Crit. Care Med. 164, 896–904 [DOI] [PubMed] [Google Scholar]
- 7. Hedstrom L. (2002) Serine protease mechanism and specificity. Chem. Rev. 102, 4501–4524 [DOI] [PubMed] [Google Scholar]
- 8. Lane A. A., Ley T. J. (2003) Neutrophil elastase cleaves PML-RARα and is important for the development of acute promyelocytic leukemia in mice. Cell 115, 305–318 [DOI] [PubMed] [Google Scholar]
- 9. Kohri K., Ueki I. F., Nadel J. A. (2002) Neutrophil elastase induces mucin production by ligand-dependent epidermal growth factor receptor activation. Am. J. Physiol. Lung Cell Mol. Physiol. 283, L531–540 [DOI] [PubMed] [Google Scholar]
- 10. Kaynar A. M., Houghton A. M., Lum E. H., Pitt B. R., Shapiro S. D. (2008) Neutrophil elastase is needed for neutrophil emigration into lungs in ventilator-induced lung injury. Am. J. Respir. Cell Mol. Biol. 39, 53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shapiro S. D., Goldstein N. M., Houghton A. M., Kobayashi D. K., Kelley D., Belaaouaj A. (2003) Neutrophil elastase contributes to cigarette smoke-induced emphysema in mice. Am. J. Pathol. 163, 2329–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Senior R. M., Tegner H., Kuhn C., Ohlsson K., Starcher B. C., Pierce J. A. (1977) The induction of pulmonary emphysema with human leukocyte elastase. Am. Rev. Respir. Dis. 116, 469–475 [DOI] [PubMed] [Google Scholar]
- 13. Liou T. G., Campbell E. J. (1996) Quantum proteolysis resulting from release of single granules by human neutrophils: a novel, nonoxidative mechanism of extracellular proteolytic activity. J. Immunol. 157, 2624–2631 [PubMed] [Google Scholar]
- 14. Owen C. A., Campbell M. A., Sannes P. L., Boukedes S. S., Campbell E. J. (1995) Cell surface-bound elastase and cathepsin G on human neutrophils: a novel, nonoxidative mechanism by which neutrophils focus and preserve catalytic activity of serine proteinases. J. Cell Biol. 131, 775–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Campbell E. J., Owen C. A. (2007) The sulfate groups of chondroitin sulfate- and heparan sulfate-containing proteoglycans in neutrophil plasma membranes are novel binding sites for human leukocyte elastase and cathepsin G. J. Biol. Chem. 282, 14645–14654 [DOI] [PubMed] [Google Scholar]
- 16. Houghton A. M., Rzymkiewicz D. M., Ji H., Gregory A. D., Egea E. E., Metz H. E., Stolz D. B., Land S. R., Marconcini L. A., Kliment C. R., Jenkins K. M., Beaulieu K. A., Mouded M., Frank S. J., Wong K. K., Shapiro S. D. (2010) Neutrophil elastase-mediated degradation of IRS-1 accelerates lung tumor growth. Nat. Med. 16, 219–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Metz H. E., Houghton A. M. (2011) Insulin receptor substrate regulation of phosphoinositide 3-kinase. Clin. Cancer Res. 17, 206–211 [DOI] [PubMed] [Google Scholar]
- 18. Mittendorf E. A., Alatrash G., Qiao N., Wu Y., Sukhumalchandra P., St John L. S., Philips A. V., Xiao H., Zhang M., Ruisaard K., Clise-Dwyer K., Lu S., Molldrem J. J. (2012) Breast cancer cell uptake of the inflammatory mediator neutrophil elastase triggers an anticancer adaptive immune response. Cancer Res. 72, 3153–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Esko J. D., Stewart T. E., Taylor W. H. (1985) Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 82, 3197–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eckhardt M., Gotza B., Gerardy-Schahn R. (1998) Mutants of the CMP-sialic acid transporter causing the Lec2 phenotype. J. Biol. Chem. 273, 20189–20195 [DOI] [PubMed] [Google Scholar]
- 21. Joslin G., Griffin G. L., August A. M., Adams S., Fallon R. J., Senior R. M., Perlmutter D. H. (1992) The serpin-enzyme complex (SEC) receptor mediates the neutrophil chemotactic effect of α1-antitrypsin-elastase complexes and amyloid-β peptide. J. Clin. Invest. 90, 1150–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. David G., Bai X. M., Van der Schueren B., Cassiman J. J., Van den Berghe H. (1992) Developmental changes in heparan sulfate expression: in situ detection with mAbs. J. Cell Biol. 119, 961–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rubino M., Miaczynska M., Lippé R., Zerial M. (2000) Selective membrane recruitment of EEA-1 suggests a role in directional transport of clathrin-coated vesicles to early endosomes. J. Biol. Chem. 275, 3745–3748 [DOI] [PubMed] [Google Scholar]
- 24. Macia E., Ehrlich M., Massol R., Boucrot E., Brunner C., Kirchhausen T. (2006) Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell 10, 839–850 [DOI] [PubMed] [Google Scholar]
- 25. Nabi I. R., Le P. U. (2003) Caveolae/raft-dependent endocytosis. J. Cell Biol. 161, 673–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McMahon H. T., Boucrot E. (2011) Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12, 517–533 [DOI] [PubMed] [Google Scholar]
- 27. Glebov O. O., Bright N. A., Nichols B. J. (2006) Flotillin-1 defines a clathrin-independent endocytic pathway in mammalian cells. Nat. Cell Biol. 8, 46–54 [DOI] [PubMed] [Google Scholar]
- 28. Buchholz C. J., Koller D., Devaux P., Mumenthaler C., Schneider-Schaulies J., Braun W., Gerlier D., Cattaneo R. (1997) Mapping of the primary binding site of measles virus to its receptor CD46. J. Biol. Chem. 272, 22072–22079 [DOI] [PubMed] [Google Scholar]
- 29. Erlenhoefer C., Wurzer W. J., Löffler S., Schneider-Schaulies S., ter Meulen V., Schneider-Schaulies J. (2001) CD150 (SLAM) is a receptor for measles virus but is not involved in viral contact-mediated proliferation inhibition. J. Virol. 75, 4499–4505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perlmutter D. H., Joslin G., Nelson P., Schasteen C., Adams S. P., Fallon R. J. (1990) Endocytosis and degradation of α1-antitrypsin-protease complexes is mediated by the serpin-enzyme complex (SEC) receptor. J. Biol. Chem. 265, 16713–16716 [PubMed] [Google Scholar]