Background: Mia40 is regenerated by the sulfhydryl oxidase Erv1 in the disulfide relay system.
Results: Crystal structures of the Erv1 core domain and full length of Erv1 were determined.
Conclusion: The Erv1 N-terminal amphipathic helix is critical for electron transfer from Mia40 to the core redox center of Erv1.
Significance: This is the first structural snapshot of the electron transfer process in Mia40-Erv1 disulfide relay system.
Keywords: Cysteine-mediated Cross-linking, Electron Transfer, Oxidation-Reduction, Sulfhydryl, X-ray Crystallography, Yeast, Disulfide Relay System
Abstract
The disulfide relay system in the mitochondrial intermembrane space drives the import of proteins with twin CX9C or twin CX3C motifs by an oxidative folding mechanism. This process requires disulfide bond transfer from oxidized Mia40 to a substrate protein. Reduced Mia40 is reoxidized/regenerated by the FAD-linked sulfhydryl oxidase Erv1 (EC 1.8.3.2). Full-length Erv1 consists of a flexible N-terminal shuttle domain (NTD) and a conserved C-terminal core domain (CTD). Here, we present crystal structures at 2.0 Å resolution of the CTD and at 3.0 Å resolution of a C30S/C133S double mutant of full-length Erv1 (Erv1FL). Similar to previous homologous structures, the CTD exists as a homodimer, with each subunit consisting of a conserved four-helix bundle that accommodates the isoalloxazine ring of FAD and an additional single-turn helix. The structure of Erv1FL enabled us to identify, for the first time, the three-dimensional structure of the Erv1NTD, which is an amphipathic helix flanked by two flexible loops. This structure also represents an intermediate state of electron transfer from the NTD to the CTD of another subunit. Comparative structural analysis revealed that the four-helix bundle of the CTD forms a wide platform for the electron donor NTD. Moreover, computational simulation combined with multiple-sequence alignment suggested that the amphipathic helix close to the shuttle redox enter is critical for the recognition of Mia40, the upstream electron donor. These findings provide structural insights into electron transfer from Mia40 via the shuttle domain of one subunit of Erv1 to the CTD of another Erv1 subunit.
Introduction
Formation of correct disulfide bonds is important for the structure and function of most proteins. In eukaryotic cells, the intermembrane spaces (IMS)2 of mitochondria and endoplasmic reticulum are the two major locations for the introduction of disulfide bonds (1). The IMS has a dedicated disulfide relay system to introduce disulfide bonds into the small cysteine-rich substrate proteins (2), such as small Tim proteins and copper chaperone Cox17, which are nuclearly encoded and cytosolically synthesized (3, 4). These substrate proteins are characterized by a relatively low molecular mass in the range of 8–17 kDa and a conserved motif of cysteine pairs. These are twin CX3C or twin CX9C motifs that are crucial for the import of preprotein and accumulation of mature proteins in the IMS (5–7). Newly synthesized unfolded substrate proteins would pass through the translocase of the outer membrane and form a mixed disulfide bonded intermediate with Mia40 (mitochondrial intermembrane space import and assay/oxidoreductase 40) in the IMS (8–10). Mia40 is a conserved oxidoreductase that is soluble in mammals and plants but membrane-anchored in fungi (9, 11). It harbors an conserved redox-active motif of -CPC-CX9C-CX9C- (8, 11), using a CPC site to form an intermolecular disulfide bond with substrate proteins (12, 13). In the disulfide exchange reaction, a disulfide bond is introduced into the substrate protein, accompanied by the release of the reduced Mia40, which is reoxidized/regenerated to a functional state by the sulfhydryl oxidase Erv1 (essential for respiration and viability/FAD-linked sulfhydryl oxidase 1) (2, 14). Thereafter, the reduced Erv1 passes the electron to either cytochrome c or molecular oxygen (15–18). Together, Mia40 and Erv1 are two essential components of the disulfide relay system that is of crucial importance for mitochondrial biogenesis (2, 19, 20).
The FAD-linked sulfhydryl oxidase Erv1 (EC 1.8.3.2) is essential for the respiration and vegetative growth of the yeast (21, 22). A number of Erv1 homologs have been characterized in plants (23), mammals (24, 25), and double-stranded DNA viruses (26, 27). The mammalian homologs are called augmenters of liver regeneration (ALRs). All Erv1/ALR family members share a conserved core domain harboring a CXXC motif (the core redox center), juxtaposed with FAD and involved in redox reactions. To date, the structures of core domains have been determined for human ALR (28, 29), Arabidopsis thaliana Erv1 (30), Rattus norvegicus ALR (31), and Saccharomyces cerevisiae Erv2 (32). All exist as a homodimer, with each subunit composed of a four-helical bundle that accommodates the isoalloxazine ring of FAD with an additional single-turn helix.
In addition to the conserved core redox center, Erv1/ALR proteins, except for the viral homologs, possess another cysteine pair. This is at the N-terminal domain (NTD) in fungi and mammals and at the C-terminal segment in plants (33). Genetic studies demonstrated that the N-terminal CXXC motif of yeast Erv1 was required for in vivo functions (34). In fact, the NTD of yeast Erv1 is necessary and sufficient for interaction with Mia40. Moreover the N-terminal cysteine pair is required for the formation of a mixed disulfide intermediate with Mia40 (35). Because of its role in forwarding electrons from Mia40 to the C-terminal core domain (CTD), the NTD is termed the shuttle domain, and the CXXC motif at the NTD is termed the shuttle redox center (16, 20, 33, 35).
To gain insights into the structural basis of this electron transfer process, we determined the structure of the CTD at 2.0 Å resolution and the structure of the C30S/C133S double mutant of full-length Erv1 (Erv1FL) at 3.0 Å resolution. The structure of the N-terminal shuttle domain and its interactions with the CTD led us to propose a putative model of electron transfer from Mia40 via the shuttle domain of one subunit to the CTD of another subunit of Erv1.
EXPERIMENTAL PROCEDURES
Overexpression and Purification of Erv1 and Mutants
The coding sequences of the intact yeast ERV1/YGR029W and the C-terminal core domain (Asp86–Glu189, designated as Erv1CTD) were amplified by PCR using S. cerevisiae S288c genomic DNA as the template and cloned into a pET28a-derived vector, respectively. The constructs add a hexahistidine tag to the N terminus of the recombinant protein, which were overexpressed in Escherichia coli BL21 (DE3) (Novagen, Madison, WI) strain using 2× YT culture medium. Expression was started by adding 0.2 mm isopropyl-β-d-thiogalactoside, and the cells continued growing for another 20 h at 16 °C before harvesting. The cells were harvested by centrifugation at 8000 × g for 10 min and resuspended in lysis buffer (20 mm Tris-HCl, pH 8.0, 200 mm NaCl). After 5 min of sonication (power-on for each 1 s with an interval of 3 s in a total time of 20 min) and centrifugation at 12,000 × g for 25 min, the supernatant containing the soluble target protein was collected and loaded to a HiTrap nickel-chelating column (GE Healthcare) equilibrated with binding buffer (20 mm Tris-HCl, pH 8.0, 200 mm NaCl). The target protein was eluted with 300 mm imidazole buffer and further loaded onto a Superdex 75 column (GE Healthcare) equilibrated with 20 mm Tris-HCl, pH 8.0, 200 mm NaCl (20 mm sodium citrate, pH 5.38, 50 mm NaCl for Erv1CTD). Fractions containing the target protein were pooled and concentrated to 10 mg/ml by ultrafiltration (Millipore; 10-kDa cut-off). The purity of proteins was estimated by SDS-PAGE, and the proteins were stored at −80 °C. For expression of the N-terminal domain of Erv1 (Met1–Asp83, designated as Erv1NTD), the nucleotide sequence was PCR-amplified and cloned in pGEX-4T-2 expression vector. The Erv1NTD was expressed and purified as previously described (36). The mutant proteins were expressed, purified, and stored in the same manner as the wild type.
Analysis of the Complexes between Erv1NTD-C30S and Erv1CTD Mutant
Erv1NTD-C30S, Erv1CTD-C130S, and Erv1CTD-C133S were purified and reduced by DTT, respectively. After desalting, five samples were prepared (sample A, NTD-C30S; sample B, CTD-C130S; sample C, CTD-C133S; sample D, NTD-C30S + CTD-C130S; and sample E, NTD-C30S + CTD-C133S) and incubated at 25 °C for 30 min. Each sample was divided into two parts, with or without 5 mm DTT, and subjected to SDS-PAGE to test the quantity of the complex.
Crystallization, Data Collection, and Processing
Before crystallization, Erv1FL was incubated with 5 mm GSSG:GSH at a molar ratio of 3:1 for 1 h at 4 °C. Crystals of Erv1CTD and Erv1FL were grown by hanging drop vapor diffusion at 30 and 16 °C, respectively, with the initial condition by mixing 1 μl of the 10 mg/ml protein sample with equal volume of reservoir solution (Erv1CTD: 25% (w/v) polyethylene glycol 3350, 0.2 m ammonium acetate, 0.1 m Tris-HCl, pH 8.5; Erv1FL: 15% (w/v) polyethylene glycol 6000, 0.1 m HEPES, pH 8.0). The crystals of Erv1CTD appeared in 3 days, whereas that of the Erv1FL appeared in ∼2 days. The crystals were transferred to cryoprotectant (reservoir solution supplemented with 25% (v/v) glycerol) and flash-cooled at 100 K in liquid nitrogen. Both data were collected at a radiation wavelength of 0.979 Å at the Shanghai Synchrotron Radiation Facility (Shanghai Institute of Applied Physics, Chinese Academy of Sciences), using the Beamline BL17U at 100 K with a MX-225 CCD (Marresearch). The data sets of Erv1CTD was processed using the HKL2000 package (37), and that of Erv1FL was processed with iMosflm.
Structure Determination and Refinement
Both crystal structures were determined by the molecular replacement method with MOLREP (38) in the CCP4 suite (39) using the coordinates of R. norvegicus FAD-dependent sulfhydryl oxidase (Protein Data Bank code 1OQC) (31) as the search model. Refinement was carried out using REFMAC5 (40) and COOT (41). The overall assessment of model quality was performed using MOLPROBITY (42). The final atomic coordinates and structure factors were deposited in the Protein Data Bank under the accession codes 4E0H and 4E0I. The crystallographic parameters of the structures are listed in Table 1. All of the structure figures were prepared with PyMOL (43).
TABLE 1.
Data collection and refinement statistics
| Erv1CTD | Erv1FL | |
|---|---|---|
| Data collection | ||
| Space group | P22121 | P21221 |
| Unit cell (90°, Å) | 38.55, 46.60, 58.88 | 63.28, 77.68, 116.23 |
| Resolution range (Å)a | 50.00–2.00 (2.07–2.00) | 49.06–3.00 (3.16–3.00) |
| Unique reflections | 7,532 (727) | 11,892 (1,688) |
| Completeness (%) | 99.7 (100.0) | 99.2 (98.6) |
| <I/σ(I)> | 30.88 (10.68) | 11.00 (3.80) |
| Rmerge (%)b | 6.4 (23.8) | 12.6 (41.1) |
| Average redundancy | 10.4 | 5.7 |
| Structure refinement | ||
| Resolution range (Å)a | 50.00–2.00 (2.06–2.00) | 46.53–3.00 (3.08–3.00) |
| R factorc/Rfreed (%) | 19.2/22.9 | 25.5/30.4 |
| Number of protein atoms | 890 | 3250 |
| Number of water atoms | 21 | 0 |
| RMSD bond lengths (Å)e | 0.017 | 0.005 |
| RMSD bond angles (°) | 1.753 | 0.902 |
| Mean B factors (Å2) | 35.15 | 50.73 |
| Ramachandran plotf | ||
| Most favored (%) | 98.1 | 95.8 |
| Additional allowed (%) | 1.9 | 3.7 |
| Outliers (%) | 0 | 0.5 |
| Protein Data Bank entry | 4E0H | 4E0I |
a The values in parentheses are for the highest resolution shell.
b Rmerge = ΣhklΣi|Ii(hkl)−<I(hkl)>|/ΣhklΣi|Ii(hkl)|, where Ii(hkl) is the intensity of an observation, and <I(hkl)> is the mean value for its unique reflection. Summations are over all reflections.
c R factor = Σh‖Fo(h)| − |Fc(h)‖/Σh|Fo(h)|, where Fo and Fc are the observed and calculated structure-factor amplitudes, respectively.
d Rfree was calculated with 5% of the data excluded from the refinement.
e Root mean square deviation from ideal values.
f Categories as defined by MolProbity.
RESULTS AND DISCUSSION
Overall Structure of the Highly Conserved Erv1CTD
The yeast Erv1 has two domains. The highly conserved CTD follows a flexible N-terminal domain (20). We first determined the structure of Erv1CTD, which is from Asp86 to Asp188. Each asymmetric unit consists of one Erv1CTD molecule, which adopts an all-α overall structure (Fig. 1A), similar to previous core domain structures. For example, the root mean square deviation (RMSD) between Erv1CTD and human ALR is only 0.69 Å over 104 Cα atoms. Erv1CTD consists of a four-helix bundle (helices α1–α4) and an additional single-turn helix α5 perpendicularly packed against the bundle. The C terminus of α4 extends out from the N terminus of α3, and the FAD binds at the center of the four-helix bundle. The core redox center (Cys130–Cys133) is in proximity to the isoalloxazine ring of FAD, and the structural disulfide pair Cys159–Cys176 is proximal to the adenine ring of FAD (32, 34).
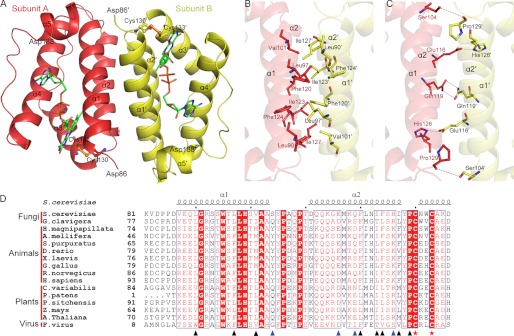
FIGURE 1.
Overall structure of Erv1CTD. A, schematic representation of the Erv1CTD dimer. The active sites are shown with ball and stick models, and the bound FAD molecules are shown as green sticks. B and C, hydrophobic (B) and hydrophilic (C) residues at the dimeric interface are shown as sticks. D, sequence comparison of the residues involved in dimeric interface. Sequences of Erv1/ALR proteins are from S. cerevisiae (NP_011543.2), Grosmannia clavigera kw1407 (EFX00923.1), Hydra magnipapillata (XP_002163122.1), Apis mellifera (XP_001120016.1), Strongylocentrotus purpuratus (XP_786637.1), Danio rerio (NP_001082855.1), Xenopus laevis (BC_097922.1), Gallus gallus (XP_414848.2), R. norvegicus (NP_037354.2), Homo sapiens (NP_005253.3), Chlorella variabilis (EFN55272.1), Physcomitrella patens subsp. Patens (XP_001774132.1), Picea sitchensis (ADE75626.1) Zea mays (NP_001148317.1), A. thaliana (NP_564557.1), and Feldmannia species virus (YP_002154679.1). Secondary structure elements of Erv1 (Protein Data Bank code 4E0H) are at the top. Residues involved in hydrophobic and hydrophilic interactions are marked with black and blue triangles, respectively. Cysteines of the redox center are marked with red stars. Alignments were performed with ClustalW and ESPript.
Erv1CTD form a noncovalently linked homodimer along a crystallographic 2-fold axis (Fig. 1A). Gel filtration chromatography also suggests that Erv1CTD most likely exists as a dimer in solution with an estimated mass of ∼27 kDa (Data not shown). Helices α1 and α2 (Asp86–Ser104 and Asp111–Ile127) from each subunit form a bundle of four helices with a buried interface of 1830 Å2. Residues Leu90, Leu97, Val101, Phe120, Ile123, Phe124, Ile127, and Pro129 from each subunit form hydrophobic patches at the center of the dimeric interface (Fig. 1B). In addition, residues Ser104, Glu116, Gln119, His126, and Pro129 contribute to the intermolecular hydrogen bonds and salt bridges (Fig. 1C). Multiple sequence alignment showed that most hydrophobic and polar residues involved in the dimeric interface are conserved (Fig. 1D). This conserved dimer interface provides the structural basis for the intersubunit electron transfer, in agreement with previous results that the dimerization promotes the functionally essential intersubunit disulfide exchange reaction (20, 30, 32).
Favored Disulfide Bond between NTD and CTD
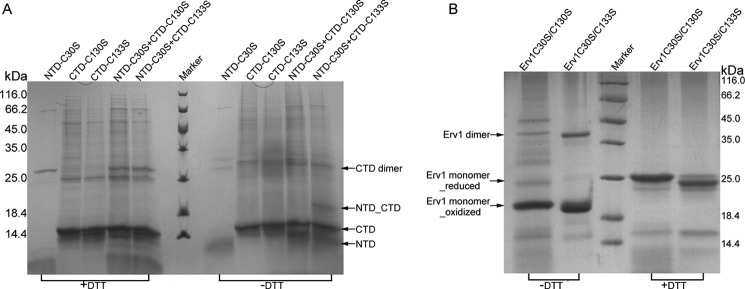
Erv1 contains two redox centers (Cys30–Cys33 at the NTD and Cys130–Cys133 at the CTD), both of which are indispensable for the disulfide relay/oxidation activity (17). The importance of the four cysteine residues of the two CXXC motifs has been assessed, and a mutation of Cys30 to Ser30 was not harmful to the in vivo function of Erv1 (20). This indicates that Cys33 of NTD might form a mixed disulfide with the core redox center in the electron transfer process. However, the cysteine of the core redox center (Cys130 or Cys133) that is favored by Cys33 remains unknown. To assign this cysteine, we overexpressed and purified three mutants of the NTD and CTD: NTD-C30S, CTD-C130S, and CTD-C133S. The reaction efficiency of the two CTD mutants toward NTD-C30S was semiquantatively compared using SDS-PAGE. A complex of NTD-C30S with CTD-C133S (Fig. 2A, eleventh lane), but not CTD-C130S (Fig. 2A, tenth lane), was observed. Moreover, the oxidized Erv1C30S/C133S mutant formed more mixed disulfide dimers than the Erv1C30S/C130S mutant (Fig. 2B). Thus, we concluded that Cys130 is the favored cysteine for the disulfide intermediate with the N-terminal Cys33. These results are in agreement with the somewhat buried position of Cys133, whereas Cys130 is relatively exposed to the solvent, as shown in the CTD structure (Fig. 1A). Thus, it should be easier for the NTD to access Cys130 than Cys133.
FIGURE 2.
Electrophoresis of the complexes between NTD and CTD mutants. A, the Coomassie-stained gel shows the formation of intermolecular disulfide bonds between NTD-C30S and CTD mutants after incubation under nonreducing conditions (see “Experimental Procedures”). First through fifth lanes, NTD-C30S, CTD-C130S, CTD-C133S, NTD-C30S + CTD-C130S, and NTD-C30S + CTD-C133S, with 5 mm DTT; sixth lane, protein marker; seventh through eleventh lanes, samples corresponding to first through fifth lanes, respectively, without DTT. B, the Coomassie-stained gel shows the formation of mixed disulfide dimer after incubation under nonreducing conditions. First and second lanes, Erv1C30S/C130S and Erv1C30S/C133S, without DTT; third lane, protein marker; fourth and fifth lanes, samples corresponding to first and second lanes, respectively, with 5 mm DTT.
Overall Structure of Erv1FL
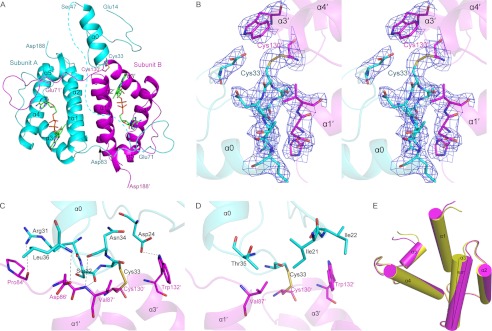
The failure of our initial attempt to crystallize the full-length Erv1 might be due to the high flexibility of the NTD. Because the NTD might be fixed by the core domain via a mixed disulfide between Cys33 and Cys130, the C30S/C133S double mutant of full-length Erv1 was incubated with 5 mm GSSG:GSH at a molar ratio of 3:1. This provides an oxidation environment for forming a mixed disulfide bond between the favored cysteine residue Cys33 and Cys130 from a neighboring subunits. The sample was crystallized, and the structure was determined at 3.0 Å resolution. Each asymmetric unit contains three molecules of Erv1 (subunit A, B, and C). Subunits A and B could form a homodimer along a noncrystallographic 2-fold axis with an interface of 1870 Å2, mainly contributed by the conserved core domain, and the interface is the same as the dimer interface in the Erv1CTD structure. Subunits A and C has a buried interface of 640 Å2 that might be due to the crystal packing. Moreover, subunit C forms a dimer with a subunit related by a crystallographic 2-fold axis. Residues Glu71–Asp188 could be traced in all three molecules, whereas the segment with residues Glu14–Ser47 could be fitted to the electron density map of only a single molecule. This segment of the NTD is folded into an α helix (α0, Leu36–Ser47) following a long defined loop (Glu14–Thr35), and locates proximally to the core redox center of subunit B (Fig. 3A). The shuttle redox center (Cys30–Cys33) is on a loop very close to first turn of helix α0. Although residues Glu48–Asp83 form a flexible linker between the CTD and helix α0, residues Met1–Gln13 and Gln48–Ser70 are not visible in the final 2Fo − Fc electron density map, presumably because of their high flexibility. Based on cross-linking results shown in Fig. 2B and previous reports on electron transfer from the reduced shuttle redox center to the core redox center of another subunit in the dimer (20, 30), we assign the visible NTD to subunit A (Fig. 3A). Helix α0 of subunit A is linked to the core domain of subunit B via an intermolecular disulfide bond (Cys33–Cys130′).
FIGURE 3.
Overall structure of Erv1FL. A, cartoon representation of Erv1FL dimer (subunit A, cyan; subunit B, magenta). The broken lines indicate the residues between NTD and CTD that could not be modeled into the electron density map. Cys33 of NTD and Cys130′ of CTD′ form a disulfide bond. B, stereo view of the 2Fo − Fc electron density map contoured at 1.00′ around the mixed disulfide bond between Cys33 of subunit A and Cys130′ of subunit B. The Cys33 and Cys130′ are shown as sticks and sulfur atoms colored yellow. C and D, hydrogen bonds (C) and hydrophobic interactions (D) between NTD of subunit A and CTD′ of subunit B. The backbone of protein is presented as a semitransparent cartoon. E, superposition of the individual core domain (yellow) and that of Erv1FL subunit B (magenta).
Structural Basis for the Intersubunit Electron Transfer
The structure of Erv1FL captures an intermediate state of electron transfer from one NTD to the CTD of another subunit (designated CTD′) via a mixed disulfide bond between Cys33 and Cys130′ (Fig. 3A). The 2Fo − Fc electron density map clearly displays a disulfide bond-linked complex in the asymmetric unit (Fig. 3B). NTD binds to CTD′ at the surface adjacent to the core redox center with a total interface area of 880 Å2 (450 Å2 for NTD and 430 Å2 for CTD′). This interface has a typical area for redox protein complexes because of their short-lived interactions (44). In addition to the intersubunit disulfide bond, five hydrogen bonds are involved in stabilizing the conformation of NTD (Fig. 3C). In particular, the carbonyl oxygen of Ser32 and Asn34 forms hydrogen bonds with the amide nitrogen of Val87′, respectively. The amide nitrogen of Leu36 forms a hydrogen bond with Asp86′-Oδ2, whereas Arg31-Nη1 makes a hydrogen bond with the carbonyl oxygen of Pro84′. These four hydrogen bonds close to the shuttle redox center form a network of seven residues, five of which contribute with the main chain atoms, whereas residues Arg31 and Asp86′ donate their side chain atoms. Sequence alignment showed that Arg31 and Asp86 are conserved in Erv1/ALR and homologs (data not shown). In addition, Asp24-Oδ2 forms a hydrogen bond with Trp132′-Nϵ1 of the CTD′. Moreover, hydrophobic contacts between a hydrophobic patch (Val87′ and Trp132′) of the CTD′ and the complementary side (Ile21, Ile22, and Thr35) of the NTD also contribute a part to the interface (Fig. 3D).
The overall structure of CTD′ of Erv1FL subunit B is quite similar to that of subunit A and the isolated CTD, with RMSD of 0.32 and 0.69 Å over 104 Cα atoms, respectively. However, when approaching the NTD, helix α3 (Cys130–Glu143) at the core redox center rotates outwards against the isoalloxazine ring of FAD at an angle of ∼6.0° along its C terminus (Fig. 3E). In addition, the other three helices of the four-helix bundle also shift slightly outwards. These conformational changes lead to a wider bundle mouth to interact with the approaching NTD. The segments that locate on the top of the four helices of CTD′ constitute a platform that plays a crucial role in the NTD interaction.
Although the NTD of subunit A is cross-linked to the CTD of subunit B, the traceable segment Glu14–Ser47 exhibits relatively high B factors. This structural flexibility is considered necessary for recognition of both Erv1CTD and Mia40 (29). Moreover, the linker between the shuttle redox center and the core domain displays a much higher flexibility, and most of the linker could not be traced in the electron density map. This highly flexible linker enables the shuttle redox center to easily flip between the Erv1CTD and Mia40.
A Putative Binding Model between Erv1 and Mia40
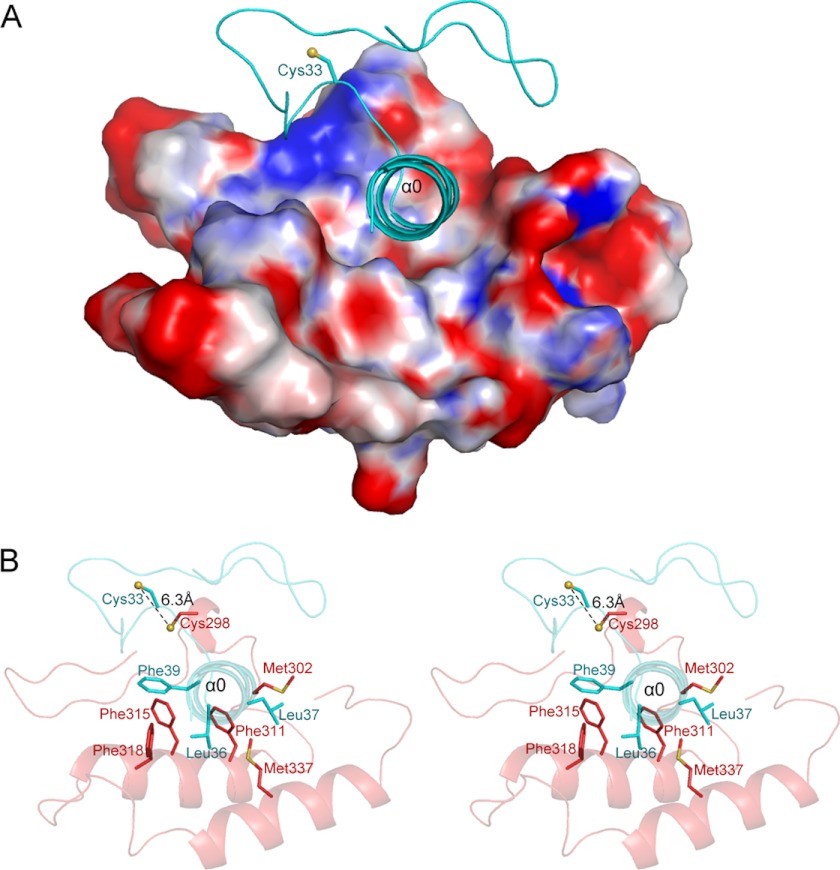
After determining the structure of Erv1NTD, we attempted but failed to obtain a crystal of Erv1NTD in complex with Mia40 by a similar cross-linking strategy. Alternatively, we simulated a model of the Mia40-Erv1NTD complex using the program HADDOCK (45), based on our Erv1FL structure and the previously reported structure of Mia40 (Protein Data Bank code 2ZXT, without the maltose-binding protein tag). This was driven by interaction restraints between the active site residues of Erv1 and Mia40, as defined by the program WHISCY (46). Among the 15 output clusters, the cluster of lowest energy with eight members satisfied the best interaction restraints and had the largest buried solvent-accessible interface area of ∼1050 Å2 (595 Å2 for Erv1NTD and 455 Å2 for Mia40). The overall backbone RMSD of 0.5 ± 0.3 Å for the eight members indicated that the model of Mia40-Erv1NTD was somewhat reliable. In the model, helix α0 makes extensive interactions with the hydrophobic cleft of Mia40 (Fig. 4A). This cleft is composed of a cluster of highly conserved hydrophobic residues Met302, Phe311, Phe315, Phe318, and Met337 (Fig. 4B). These residues are also involved in recognizing the CX9C and CX3C substrates (10), suggesting that Mia40 uses the same site to bind both protein substrates and the electron acceptor Erv1. Notably, this model is in accordance with the previous report that Erv1 competitively binds to the substrate-binding site on Mia40 (29).
FIGURE 4.
A putative binding pattern between Mia40 and Erv1. A, a docking model of Mia40-Erv1NTD complex. Erv1NTD is shown as cartoon, whereas Mia40 is shown as surface potential (contoured at ± 8.0 kT/e). B, stereo view of the hydrophobic residues at the interface between Erv1NTD and Mia40.
The docked interface on Erv1NTD that comprises hydrophobic residues Leu36, Leu37, and Phe39 (Fig. 4B) is in agreement with the results of Banci et al. (29). They used NMR titration to determine that the CRACVDFKTWM segment of ALR (homologous to the Erv1 CRSCNTL36L37DF39Q segment) is critical for recognition of Mia40. Using mutagenesis in combination with complementation assays, they confirmed that the hydrophobic residues downstream of the Erv1 shuttle redox center (Leu36, Leu37, and Phe39) play a vital role in complex formation with Mia40 in vitro and in vivo. These residues on the amphipathic helix α0, as shown in the structure of Erv1FL, are solvent-exposed before being recognized by Mia40. Once fitted into the hydrophobic cleft of Mia40, helix α0 will enable Cys33 of Erv1NTD to come as close as ∼6.3 Å to Cys298 of Mia40. With slight conformational changes, these two cysteine residues can form a transient mixed disulfide bond.
Universal Mode of Electron Transfer from Mia40 to Erv1 Shuttle Domain in Animals and Fungi
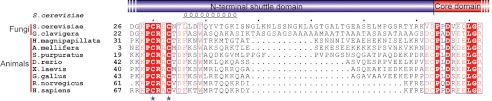
Erv1 homologs have a highly conserved core domain but variable shuttle domains (33). To find the probable original shuttle domain and its distribution, the sequence of yeast Erv1 was used in a BLAST search against the nonredundant protein sequences database. We chose 10 representatives of various species and compared the residues around the shuttle redox centers (Fig. 5). Both fungi and animals have an N-terminal shuttle domain with a highly conserved redox center (CXXC) but variable linkers between the shuttle and the CTD. Similar to the residues that constitute the amphipathic helix α0 in the yeast Erv1NTD, the corresponding residues in the animal homologs were also predicted to have a high propensity to form a helix (residues FKTWM in human ALR) (29). In yeast and human Erv1/ALR, the hydrophobic residues are somewhat aligned on one side of the amphipathic helix α0, indicating a universal hydrophobic interaction pattern between Mia40 and Erv1/ALR from fungi and animals.
FIGURE 5.
Multiple-sequence alignment of the shuttle domains of Erv1/ALR proteins from S. cerevisiae (NP_011543.2), G. clavigera kw1407 (EFX00923.1), H. magnipapillata (XP_002163122.1), A. mellifera (XP_001120016.1), S. purpuratus (XP_786637.1), D. rerio (NP_001082855.1), X. laevis (BC_097922.1), G. gallus (XP_414848.2), R. norvegicus (NP_037354.2), and H. sapiens (NP_005253.3). Cysteines of the shuttle redox center are marked with blue stars. Alignments were performed with ClustalW and ESPript.
In fungi and animals, the N-terminal shuttle redox center of Erv1/ALR functions as an antenna stretched from the core domain. This antenna is held by the CTD with a flexible linker of varied lengths. Of note, the linker is gradually truncated during evolution from lower to higher organisms (Fig. 5). The linker of yeast Erv1 is composed of 38 amino acids, whereas the linker of human ALR is of only 14 amino acids. A shorter linker might be helpful in enhancing electron transfer efficiency from the shuttle to the core redox center.
A Working Model of Electron Transfer Process in the Mia40-Erv1 Disulfide Relay System
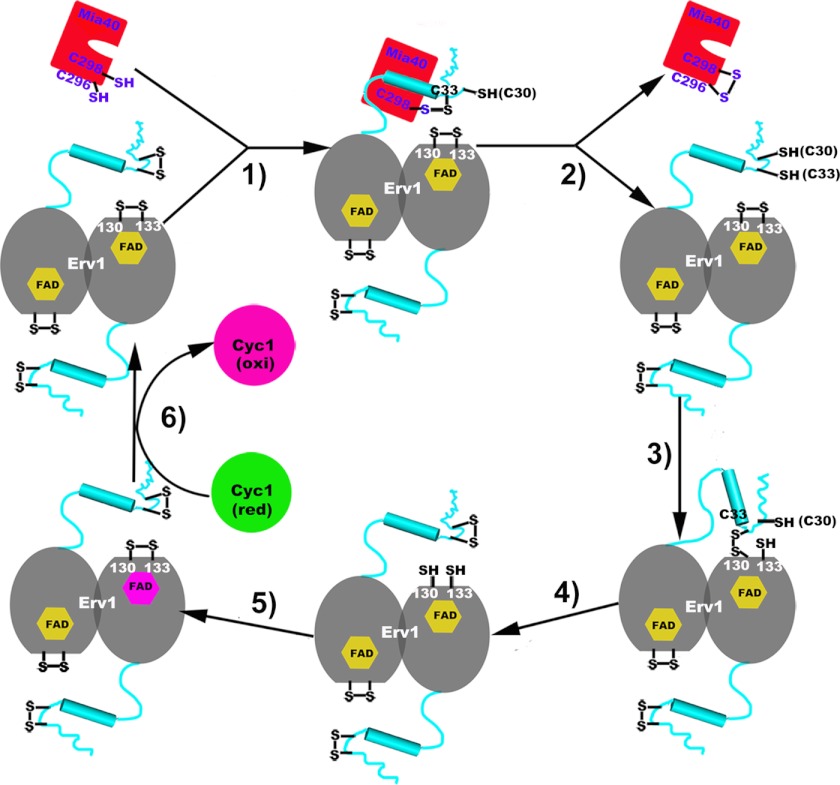
Based on our structural analyses and previous reports, we propose a working model of electron transfer in the Mia40-Erv1 disulfide relay system (Fig. 6). To simplify the illustration, we show that only one of two shuttle domains of the Erv1 dimer is working with a molecule of Mia40 during the catalysis cycle. In our model, 1) after the oxidation and release of a given substrate protein, the hydrophobic cleft of the reduced Mia40 is exposed (12). Meanwhile, the amphipathic helix α0 at the shuttle domain of the oxidized Erv1 is recognized by the hydrophobic cleft of Mia40. 2) The shuttle redox center forms an intermolecular disulfide bond with Mia40 (between Mia40-Cys298 and Erv1-Cys33) (12, 20). Consequently, this transient disulfide bond is exchanged upon the attack of Mia40-Cys296 to release oxidized Mia40. Regenerated Mia40 is ready to oxidatively refold another substrate protein. The electron transfer from Mia40 to the shuttle domain of Erv1 requires mechanisms to overcome the thermodynamically unfavorable redox gradient. This might be driven by conformational changes of Erv1NTD at different redox states (17). 3) The reduced shuttle domain of Erv1 swings back and lands on a platform on the core redox center of a neighboring subunit in the same dimer. The conformational changes of the core domain facilitate the formation of an intersubunit disulfide bond between Cys33 and Cys130′. 4) The intersubunit disulfide bond is subsequently attacked by Cys30 at the shuttle domain to regenerate an oxidized shuttle redox center. Simultaneously, the core redox center of Erv1 is reduced. This process is spontaneously driven by differences in redox potential between the shuttle and core redox centers (17, 20). 5) Electron transfer from the core redox center to FAD does not require a conformational change (the distance from the flavin C4α to the thiol group of Cys133 is ∼3.3Å) but must overcome the unfavorable redox gradient (17). This process might be driven by coupling to the efficient downstream electron flow to cytochrome c (47). 6) The reduced FAD efficiently transfers the electron to the most favorable physiological electron acceptor, cytochrome c, which passes the electron to the respiratory chain (15, 17, 18). Thus, Erv1 is ready for another electron transfer cycle.
FIGURE 6.
A schematic electron transfer cycle of the Mia40-Erv1 disulfide relay system.
Conclusions
This work captured an intermediate structure of the N-terminal shuttle domain cross-linked to the core domain of Erv1 via an introduced disulfide bond between Cys33 of one subunit and Cys130′ of another subunit. The frozen position of the highly flexible shuttle domain enabled us to determine a model of electron transfer from the upstream Mia40 to the downstream cytochrome c via a recognition pattern similar to the interaction between Mia40 and its substrate protein. These findings provide for the first time structural insights into the overall Mia40-Erv1 disulfide relay system.
Acknowledgments
We thank the staff at the Shanghai Synchrotron Radiation Facility for the data collection. We are grateful to all the developers of the CCP4 Suite, ESPript, MOLPROBITY, and PyMOL.
This work was supported by Ministry of Science and Technology of China Project 2012CB911000.
The atomic coordinates and structure factors (codes 4E0H and 4E0I) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- IMS
- intermembrane space
- NTD
- N-terminal shuttle domain
- CTD
- C-terminal core domain
- Erv1FL
- C30S/C133S double mutant of the full-length Erv1
- RMSD
- root mean square deviation
- ALR
- augmenter of liver regeneration.
REFERENCES
- 1. Riemer J., Bulleid N., Herrmann J. M. (2009) Disulfide formation in the ER and mitochondria. Two solutions to a common process. Science 324, 1284–1287 [DOI] [PubMed] [Google Scholar]
- 2. Mesecke N., Terziyska N., Kozany C., Baumann F., Neupert W., Hell K., Herrmann J. M. (2005) A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell 121, 1059–1069 [DOI] [PubMed] [Google Scholar]
- 3. Beers J., Glerum D. M., Tzagoloff A. (1997) Purification, characterization, and localization of yeast Cox17p, a mitochondrial copper shuttle. J. Biol. Chem. 272, 33191–33196 [DOI] [PubMed] [Google Scholar]
- 4. Koehler C. M. (2004) The small Tim proteins and the twin Cx3C motif. Trends Biochem. Sci 29, 1–4 [DOI] [PubMed] [Google Scholar]
- 5. Lutz T., Neupert W., Herrmann J. M. (2003) Import of small Tim proteins into the mitochondrial intermembrane space. EMBO J. 22, 4400–4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heaton D., Nittis T., Srinivasan C., Winge D. R. (2000) Mutational analysis of the mitochondrial copper metallochaperone Cox17. J. Biol. Chem. 275, 37582–37587 [DOI] [PubMed] [Google Scholar]
- 7. Lu H., Allen S., Wardleworth L., Savory P., Tokatlidis K. (2004) Functional TIM10 chaperone assembly is redox-regulated in vivo. J. Biol. Chem. 279, 18952–18958 [DOI] [PubMed] [Google Scholar]
- 8. Terziyska N., Lutz T., Kozany C., Mokranjac D., Mesecke N., Neupert W., Herrmann J. M., Hell K. (2005) Mia40, a novel factor for protein import into the intermembrane space of mitochondria is able to bind metal ions. FEBS Lett. 579, 179–184 [DOI] [PubMed] [Google Scholar]
- 9. Chacinska A., Pfannschmidt S., Wiedemann N., Kozjak V., SanjuánSzklarz L. K., Schulze-Specking A., Truscott K. N., Guiard B., Meisinger C., Pfanner N. (2004) Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 23, 3735–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Banci L., Bertini I., Cefaro C., Cenacchi L., Ciofi-Baffoni S., Felli I. C., Gallo A., Gonnelli L., Luchinat E., Sideris D., Tokatlidis K. (2010) Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import. Proc. Natl. Acad. Sci. U.S.A. 107, 20190–20195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Naoé M., Ohwa Y., Ishikawa D., Ohshima C., Nishikawa S., Yamamoto H., Endo T. (2004) Identification of Tim40 that mediates protein sorting to the mitochondrial intermembrane space. J. Biol. Chem. 279, 47815–47821 [DOI] [PubMed] [Google Scholar]
- 12. Banci L., Bertini I., Cefaro C., Ciofi-Baffoni S., Gallo A., Martinelli M., Sideris D. P., Katrakili N., Tokatlidis K. (2009) MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat. Struct. Mol. Biol. 16, 198–206 [DOI] [PubMed] [Google Scholar]
- 13. Kawano S., Yamano K., Naoé M., Momose T., Terao K., Nishikawa S., Watanabe N., Endo T. (2009) Structural basis of yeast Tim40/Mia40 as an oxidative translocator in the mitochondrial intermembrane space. Proc. Natl. Acad. Sci. U.S.A. 106, 14403–14407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Daithankar V. N., Farrell S. R., Thorpe C. (2009) Augmenter of liver regeneration. Substrate specificity of a flavin-dependent oxidoreductase from the mitochondrial intermembrane space. Biochemistry 48, 4828–4837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bihlmaier K., Mesecke N., Terziyska N., Bien M., Hell K., Herrmann J. M. (2007) The disulfide relay system of mitochondria is connected to the respiratory chain. J. Cell Biol. 179, 389–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ang S. K., Lu H. (2009) Deciphering structural and functional roles of individual disulfide bonds of the mitochondrial sulfhydryl oxidase Erv1p. J. Biol. Chem. 284, 28754–28761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dabir D. V., Leverich E. P., Kim S. K., Tsai F. D., Hirasawa M., Knaff D. B., Koehler C. M. (2007) A role for cytochrome c and cytochrome c peroxidase in electron shuttling from Erv1. EMBO J. 26, 4801–4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Allen S., Balabanidou V., Sideris D. P., Lisowsky T., Tokatlidis K. (2005) Erv1 mediates the Mia40-dependent protein import pathway and provides a functional link to the respiratory chain by shuttling electrons to cytochrome c. J. Mol. Biol. 353, 937–944 [DOI] [PubMed] [Google Scholar]
- 19. Rissler M., Wiedemann N., Pfannschmidt S., Gabriel K., Guiard B., Pfanner N., Chacinska A. (2005) The essential mitochondrial protein Erv1 cooperates with Mia40 in biogenesis of intermembrane space proteins. J. Mol. Biol. 353, 485–492 [DOI] [PubMed] [Google Scholar]
- 20. Bien M., Longen S., Wagener N., Chwalla I., Herrmann J. M., Riemer J. (2010) Mitochondrial disulfide bond formation is driven by intersubunit electron transfer in Erv1 and proofread by glutathione. Mol. Cell 37, 516–528 [DOI] [PubMed] [Google Scholar]
- 21. Lisowsky T. (1994) ERV1 is involved in the cell-division cycle and the maintenance of mitochondrial genomes in Saccharomyces cerevisiae. Curr. Genet. 26, 15–20 [DOI] [PubMed] [Google Scholar]
- 22. Lee J., Hofhaus G., Lisowsky T. (2000) Erv1p from Saccharomyces cerevisiae is a FAD-linked sulfhydryl oxidase. Febs. Letters 477, 62–66 [DOI] [PubMed] [Google Scholar]
- 23. Levitan A., Danon A., Lisowsky T. (2004) Unique features of plant mitochondrial sulfhydryl oxidase. J. Biol. Chem. 279, 20002–20008 [DOI] [PubMed] [Google Scholar]
- 24. Lisowsky T., Lee J. E., Polimeno L., Francavilla A., Hofhaus G. (2001) Mammalian augmenter of liver regeneration protein is a sulfhydryl oxidase. Dig. Liver Dis. 33, 173–180 [DOI] [PubMed] [Google Scholar]
- 25. Pagani M., Fabbri M., Benedetti C., Fassio A., Pilati S., Bulleid N. J., Cabibbo A., Sitia R. (2000) Endoplasmic reticulum oxidoreductin 1-Lβ (ERO1-Lβ), a human gene induced in the course of the unfolded protein response. J. Biol. Chem. 275, 23685–23692 [DOI] [PubMed] [Google Scholar]
- 26. Senkevich T. G., White C. L., Koonin E. V., Moss B. (2000) A viral member of the ERV1/ALR protein family participates in a cytoplasmic pathway of disulfide bond formation. Proc. Natl. Acad. Sci. U.S.A. 97, 12068–12073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rodríguez I., Redrejo-Rodríguez M., Rodríguez J. M., Alejo A., Salas J., Salas M. L. (2006) African swine fever virus pB119L protein is a flavin adenine dinucleotide-linked sulfhydryl oxidase. J. Virol. 80, 3157–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Daithankar V. N., Schaefer S. A., Dong M., Bahnson B. J., Thorpe C. (2010) Structure of the human sulfhydryl oxidase augmenter of liver regeneration and characterization of a human mutation causing an autosomal recessive myopathy. Biochemistry 49, 6737–6745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Banci L., Bertini I., Calderone V., Cefaro C., Ciofi-Baffoni S., Gallo A., Kallergi E., Lionaki E., Pozidis C., Tokatlidis K. (2011) Molecular recognition and substrate mimicry drive the electron-transfer process between MIA40 and ALR. Proc. Natl. Acad Sci. U.S.A. 108, 4811–4816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vitu E., Bentzur M., Lisowsky T., Kaiser C. A., Fass D. (2006) Gain of function in an ERV/ALR sulfhydryl oxidase by molecular engineering of the shuttle disulfide. J. Mol. Biol. 362, 89–101 [DOI] [PubMed] [Google Scholar]
- 31. Wu C. K., Dailey T. A., Dailey H. A., Wang B. C., Rose J. P. (2003) The crystal structure of augmenter of liver regeneration. A mammalian FAD-dependent sulfhydryl oxidase. Protein Sci. 12, 1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gross E., Sevier C. S., Vala A., Kaiser C. A., Fass D. (2002) A new FAD-binding fold and intersubunit disulfide shuttle in the thiol oxidase Erv2p. Nat. Struct. Biol. 9, 61–67 [DOI] [PubMed] [Google Scholar]
- 33. Fass D. (2008) The Erv family of sulfhydryl oxidases. Biochim. Biophys. Acta 1783, 557–566 [DOI] [PubMed] [Google Scholar]
- 34. Hofhaus G., Lee J. E., Tews I., Rosenberg B., Lisowsky T. (2003) The N-terminal cysteine pair of yeast sulfhydryl oxidase Erv1p is essential for in vivo activity and interacts with the primary redox centre. Eur. J. Biochem. 270, 1528–1535 [DOI] [PubMed] [Google Scholar]
- 35. Lionaki E., Aivaliotis M., Pozidis C., Tokatlidis K. (2010) The N-terminal shuttle domain of Erv1 determines the affinity for Mia40 and mediates electron transfer to the catalytic Erv1 core in yeast mitochondria. Antioxid. Redox Signal. 13, 1327–1339 [DOI] [PubMed] [Google Scholar]
- 36. Sideris D. P., Petrakis N., Katrakili N., Mikropoulou D., Gallo A., Ciofi-Baffoni S., Banci L., Bertini I., Tokatlidis K. (2009) A novel intermembrane space-targeting signal docks cysteines onto Mia40 during mitochondrial oxidative folding. J. Cell Biol. 187, 1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Macromol. Crystallogr. A 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 38. Vagin A., Teplyakov A. (1997) MOLREP. An automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 39. Bailey S. (1994) CCP4. The Ccp4 Suite. Programs for Protein Crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 40. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 41. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 42. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Structure validation by Cα geometry. φ, ψ and Cβ deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 43. DeLano W. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 44. Janin J., Bahadur R. P., Chakrabarti P. (2008) Protein-protein interaction and quaternary structure. Q. Rev. Biophys. 41, 133–180 [DOI] [PubMed] [Google Scholar]
- 45. de Vries S. J., van Dijk M., Bonvin A. M. (2010) The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 5, 883–897 [DOI] [PubMed] [Google Scholar]
- 46. de Vries S. J., van Dijk A. D., Bonvin A. M. (2006) WHISCY. What information does surface conservation yield? Application to data-driven docking. Proteins Struct. Funct. Bioinformat. 63, 479–489 [DOI] [PubMed] [Google Scholar]
- 47. Endo T., Yamano K., Kawano S. (2010) Structural basis for the disulfide relay system in the mitochondrial intermembrane space. Antioxid. Redox Signal. 13, 1359–1373 [DOI] [PubMed] [Google Scholar]