Background: Notch family transcriptional repressors explain the predominant production of IL-23 elicited by β-glucans.
Results: Association of SIRT1 with the il12a promoter and an enhanced production of the sirtuin co-substrate NAD+ is described.
Conclusion: SIRT1-mediated histone deacetylation regulates the accessibility of Rel family proteins to the il12a promoter.
Significance: Blocking the interactions of the transcriptional machinery with acetyl-histone might modulate the Th1/Th17 balance.
Keywords: Dendritic Cells, Histone Deacetylase, NAD, NF-κB (NF-KB), Sirtuins, Dectin-1, NF-κB, β-Glucans, Fungal Infection
Abstract
Stimulation of human dendritic cells with the fungal surrogate zymosan produces IL-23 and a low amount of IL-12 p70. Trans-repression of il12a transcription, which encodes IL-12 p35 chain, by proteins of the Notch family and lysine deacetylation reactions have been reported as the underlying mechanisms, but a number of questions remain to be addressed. Zymosan produced the location of sirtuin 1 (SIRT1) to the nucleus, enhanced its association with the il12a promoter, increased the nuclear concentration of the SIRT1 co-substrate NAD+, and decreased chromatin accessibility in the nucleosome-1 of il12a, which contains a κB-site. The involvement of deacetylation reactions in the inhibition of il12a transcription was supported by the absence of Ac-Lys-14-histone H3 in dendritic cells treated with zymosan upon coimmunoprecipitation of transducin-like enhancer of split. In contrast, we did not obtain evidence of a possible effect of SIRT1 through the deacetylation of c-Rel, the central element of the NF-κB family involved in il12a regulation. These data indicate that an enhancement of SIRT1 activity in response to phagocytic stimuli may reduce the accessibility of c-Rel to the il12a promoter and its transcriptional activation, thus regulating the IL-12 p70/IL-23 balance and modulating the ongoing immune response.
Introduction
The response of human monocyte-derived dendritic cells (DC)4 to the fungal surrogate zymosan shows a distinct pattern of production of IL-12 p70 and IL-23, which are involved in the development of the T-helper type 1 and type 17 responses, respectively. IL-12 p70 and IL-23 share a common chain IL-12 p40 (il12/23b) and differ by other chain, IL-12 p35 (il12a) in the case of IL-12 p70 and IL-23 p19 (il23a) in IL-23. Addressing the mechanism of IL-12 p70/IL-23 balance regulation is of clinical relevance, as IL-23 takes part in the defense against pathogens and in the development of autoimmunity (1–4). The production of IL-12 p70 and IL-23 is regulated at the transcriptional level and depends on chain pairing. il12/23b regulation depends on NF-κB activation (5, 6), whereas the regulation of il12a also requires a type I interferon autocrine-paracrine loop (7, 8). Stimulation of TLR4 induces both IL-12 p70 and IL-23, whereas the TLR2 and C-type lectin receptor routes mainly produce IL-23 (9, 10). Moreover, co-ligation of the β-glucan receptor dectin-1 and TLR2 enhances IL-23 and down-regulates IL-12 p70 (10, 11). Recent studies have shown that zymosan produces cross-inhibition through the transcriptional repressors hairy and enhancer of split 1 (HES1), hairy/enhancer-of-split related with YRPW motif 1 (HEY1), and the corepressor transducin-like enhancer of split (TLE). Zymosan also modulates the acetylation of lysines in histones, thereby modifying the accessibility of transcription factors to the il12a promoter (12). This molecular mechanism is of clinical relevance because inhibition with the synthetic acetyl-histone mimic i-BET of interactions between acetylated histones and the bromodomains of proteins involved in transcriptional activation is a promising therapy in bacteria-induced sepsis. In fact, i-BET has been found to produce a 6.8-fold reduction of il12a mRNA expression in bone marrow-derived macrophages stimulated with LPS (13).
The analysis of the IL-12 p70/IL-23 balance should focus on the activation of NF-κB, specially c-Rel (14–16), and take into account the different layers of regulation. In addition to the translocation of NF-κB proteins to the nucleus, post-translational modifications such as phosphorylation, acetylation, and ubiquitylation influence their transactivating activity, and changes in the structure of chromatin regulate the accessibility of these proteins to the promoters. In fact, remodeling of nucleosome-2 (Nuc-2) in the il12a promoter is an important factor in the regulation of this gene in DC (16, 17). Some studies have suggested that the regulation of il12a may depend on acetylation/deacetylation reactions involving class III Lys-deacetylases (sirtuins (SIRT)) (12, 18, 19). Sirtuins are highly conserved NAD+-dependent lysine deacetylases that could act on two different layers of regulation of κB-dependent transcription, i.e. regulation of NF-κB transactivating activity (20) and modulation of chromatin accessibility by promoting histone deacetylation (21, 22). SIRT1, the human ortholog of yeast Sir2, has been involved in the regulation of RelA/p65, and has marked anti-inflammatory effects in several systems (20, 23–25). The cellular NAD+ levels have been considered the primary mechanism regulating SIRT1 activity, although a recent report has stressed cyclic AMP-mediated phosphorylation of Ser-434 as a key event in the regulation of its activity independently of changes in NAD+ levels (26). In this study we have observed changes in the nuclear concentrations of NAD+, an increase of SIRT1 protein associated with the il12a promoter, and a correlation of SIRT1 activity with the inhibition of il12a transcription during the activation of DC by zymosan. After having analyzed acetylation/deacetylation reactions of NF-κB and histone proteins, we propose that the inhibition of il12a transcription elicited by zymosan is best explained by an increase of SIRT1 activity linked to an enhanced expression of the protein, an increased disposal of its co-substrate NAD+, and the ensuing deacetylation of histones.
EXPERIMENTAL PROCEDURES
Reagents, Cells, and Mice
Zymosan from Saccharomyces cerevisiae, LPS, and the catalytic subunit of protein kinase A (PKAc) were from Sigma. IL-12 p70 was assayed by ELISA (Thermo Scientific Pierce). A-769662, a reversible activator of AMPK, was from Tocris Bioscience (Bristol, UK). 3-Aminopyridine adenine dinucleotide (AAD), a NAD+ analog that increases the cellular concentration of NAD+ by inhibiting dehydrogenases (27), was synthesized from nicotinic acid adenine dinucleotide and sodium azide as described (28). Recombinant histone H3.3, BstXI, and PshAI were from New England BioLabs (Ipswich, MA). p300, pCAF, and SIRT1 recombinant proteins were from Active Motif (Carlsbad, CA). SRT1720, a selective activator of SIRT1, was from Cayman Chemical (Ann Arbor, MI). EX-527, a selective inhibitor of SIRT1, was from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). DCs were obtained from human monocytes as reported (29). Bone marrow-derived DC from mice were obtained by culture in the presence of murine recombinant GM-CSF. sirt1-Tg and heterozygous sirt1+/− mice in C57BL/6 background were provided by Dr. Manuel Serrano (Spanish National Cancer Centre, Madrid, Spain) (23).
Real-time RT-PCR
Purified RNA was depleted of genomic DNA by treatment with DNase and used for reverse transcription reactions. Cycling conditions were adapted to each set of primers. gapdh was used as a housekeeping gene to assess the relative abundance of the different mRNA using the comparative cycle threshold method. The sequences of the primers are shown in supplemental Methods.
Chromatin Accessibility Measured by Real-time PCR
To quantify Nuc remodeling at the il12a promoter, chromatin accessibility was measured by a real-time PCR (CHART-PCR) assay. About 5 × 106 DC were washed in ice-cold PBS, pelleted by centrifugation, resuspended in 1 ml of ice-cold lysis buffer (10 mm Tris-HCl, 15 mm NaCl, 3 mm MgCl2, 0.5% Nonidet P-40, 0.15 mm spermine, and 0.5 mm spermidine, pH 7.5), and incubated on ice for 5 min. The suspension was centrifuged to pellet the nuclei and resuspended in a buffer suitable for nuclease digestion according to the manufacturer's instructions (New England Biolabs). Nuclei were digested with 50 units of BstXI or 50 units PshAI for 1 h at 37 °C. After treatment with 0.1 mg/ml RNase A, reactions were stopped by treatment with 1 mg/ml proteinase K overnight at 37 °C. Genomic DNA was extracted with phenol/chloroform, and 150 ng of genomic DNA were used to perform SYBR Green quantitative PCR with triplicate samples. Nuc-1 and -2 of the il12a promoter have cleavage sites for restriction enzymes PshAI (located at nucleotide −69) and BstXI (located at nucleotide −298). Increased accessibility of the chromatin would result in an increased digestion of Nuc by the enzymes and thus a reduced amplification in the quantitative PCR. To normalize for DNA input amounts, each sample was analyzed with primers for cyclooxygenase-2 (ptgs2) promoter, a unique sequence in genome that is not digested by the restriction enzymes used in the assays. Results are expressed as a percentage of the undigested sample for each cell treatment using the formula (Ntno E − NtE/Ntno E) × 100%, with Nt = 2Ct(primer Pr ptgs) − Ct(primer Nuc). The sequence of the primers is shown in supplemental Methods.
NAD+ Extraction and Colorimetric Cycling Assay
Intracellular NAD+ concentrations were determined by an enzymatic cycling assay (30). 15–30 × 106 DC were pelleted, washed in PBS, and lysed in 400 μl of hypotonic medium. The remaining cells were eliminated by centrifugation, and the nuclei were pelleted and resuspended in hypertonic medium (20 mm Hepes, 25% glycerol, 0.5 m NaCl, 1.5 mm MgCl2, 0.5 mm EDTA, 50 mm NaF, 5 μg/ml leupeptin, and 0.5 mm PMSF, pH 7.9) for 10 min on ice. Chromatin was pelleted at 90,000 × g for 30 min. The cytosols, and nuclear extracts were treated with 250 mm HCl for 20 min to destroy NADH and then neutralized by adding 250 mm NaOH. Samples were mixed with cycling buffer containing 125 mm Tris-HCl, 1.25 mm phenazine methosulfate, 0.625 mm thiazolyl blue/methylthiazolyldiphenyl-tetrazolium bromide and 15 units alcohol dehydrogenase, pH 8.8. Optical density at 570 nm was measured as the blank, and then the cycling reaction was initiated by adding 20 μl of ethanol 6 m. The samples were incubated at 37 °C, and the optical density was measured at 570 nm after 5, 10, 15, and 20 min using an ELISA plate reader. The blank values were subtracted from all A570 nm for concentration analysis. Cellular NAD+ levels of unknown samples were calculated from the standard curve of serial known dilutions of NAD+. NAD+ values were normalized against protein levels and are presented as nmol NAD+/mg of protein compared with control.
Expression of Human Rel Family Proteins
c-Rel, the Rel-homology domain (RHD) of human c-Rel1–309, and the mutant Ser-267-Ala were constructed by PCR from a full-length c-Rel cDNA construct provided by Dr. Tse-Ha Tan (Baylor College of Medicine, Houston, TX) and fused in phase to the N-terminal histidine tag using SalI-NotI sites of a pET-28a vector (12). pCMV4, which encodes human RelA, was obtained from Addgene, and the coding sequence was fused in phase to the N-terminal histidine tag using HindIII-NotI sites of a pET-28a vector. pGEX-GSTp6512–317 and pGEX-GSTp6512–317(S276C) were from BCCM/LMBP collections (Gent, Belgium). Recombinant proteins were expressed in Escherichia coli strain BL21(DE3). His-tag proteins were purified with nickel beads and used for in vitro kinase assays. GST-tag proteins were purified with glutathione-SepharoseTM 4 Fast Flow GST-nickel beads (GE Healthcare) and the GST tag cleaved from the fusion protein by digestion with thrombin and separated from thrombin by incubation with benzamidine-SepharoseTM 4 beads (GE Healthcare).
In Vitro Kinase Assay
A 1-μg aliquot of substrate proteins was incubated at 30 °C for 20 min in reaction buffer: 12 mm MOPS/NaOH, 0.3 mm EDTA, 0.01% β-mercaptoethanol, 0.5% glycerol, 0.01 Brij-35, and 0.1 mg of BSA containing 15 ng of PKAc and supplemented with 10 mm Mg(Ac)2 and 20 μm ATP, pH 7. The reaction was stopped by the addition of Laemmli buffer, and the phosphorylated substrate proteins were separated by SDS/PAGE in 15% acrylamide. Phosphorylation of RelA/p65 and c-Rel was assayed by Western blot using anti-Ser(P)-276-RelA/p65-phosphospecific antibody. Ab reactive to the N-terminal domain of c-Rel and RelA/p65 (Cell Signaling #3034S) was used in assays where both proteins were used.
Acetylation and Deacetylation Assays
1 μg of recombinant histone H3, RelA/p65, or c-Rel was incubated with 200 ng of recombinant p300 or pCAF in lysine acetyltransferase buffer (50 mm Tris HCl, 0.1 mm EDTA, 10% glycerol, 1 mm DTT, and 200 μm acetyl-CoA, pH 8). Reaction mixtures were incubated at 30 °C for 1 h, stopped by the addition of Laemmli buffer and heating at 97 °C for 5 min, resolved by SDS-PAGE, and analyzed by Western blot. Deacetylase assays were performed after inhibition of the activities of lysine acetyltransferases with 30 μm garcinol. Thereafter, 1 μg of acetylated proteins was incubated with 100 ng of recombinant SIRT1 in deacetylase buffer (50 mm Tris-HCl, 137 mm NaCl, 2.7 mm KCl, 1 mm MgCl2, 1 mg/ml BSA, and 250 mm NAD+, pH 8) at 37 °C for 1 h. Reaction mixtures were subsequently analyzed by Western blot.
Chromatin Immunoprecipitation (ChIP) Assay, Nuc Reconstitution, and Sequential ChIP
ChIP assays were conducted with reagents and Ab against SIRT1 (Santa Cruz sc-15404) and anti-SIRT6 from Abcam plc (Cambridge, UK) as previously reported (31). Nuc-1 was reconstituted in vitro using the EpiMarkTM Nucleosome Assembly kit (New England Biolabs) according to the dilution assembly protocol. For this purpose, purified recombinant human histone H2A/H2B dimer and histone H3.1/H4 tetramer were mixed with a DNA substrate encoding Nuc-1 constructed by PCR with the primers used for the assay of chromatin accessibility of Nuc-1 of the il12a promoter. The salt concentration was diluted to allow each histone tetramer to associate with two histone dimers and form the histone octamer on the DNA. Reconstituted Nuc were subjected to phosphorylation and/or acetylation assays before performing the binding reactions between the Nuc and transcription factors. To assess the binding of NF-κB proteins to the Nuc, sequential ChIP was conducted using in the first step anti-histone H3 Ab. The immunoprecipitate was dissolved in 100 μl of 10 mm DTT to break apart the Ab chains, and then the final concentration of DTT diluted 40-fold. Ab reactive to the N-terminal domain of c-Rel and RelA/p65 was added and incubated overnight in a rotating shaker. After a new round of immunoprecipitation, a real time-PCR was carried with il12a promoter Nuc-1 primers.
Immunoblots, Immunoprecipitations, and Pulldown Assays
Proteins were separated by electrophoresis in SDS/PAGE and transferred to nitrocellulose membranes. The membranes were used for immunodetection of c-Rel (C-terminal, Santa Cruz sc-71), c-Rel (N-terminal, Santa Cruz sc-70), RelA/p65 (N-terminal, #06–418), histone H3 (#05-928), Ac-histone H3 (# 05-928), Ac-Lys-14-histone H3 (#07-353), and Ser(P)-10-histone H3 (#04-817) with Ab from Upstate Biotechnology. Ser(P)-276-RelA (#3031), Ac-Lys (#9441), Ac-Lys-310-RelA (#3045), AMPKα (#2532), and phospho-AMPKα (#2535) were assayed with Ab from Cell Signaling Technology. Immunoprecipitations were carried out as described (29). For immunoblots directed to assay nuclear proteins, the nuclear extracts were obtained by using a nuclear extract kit (Active Motif). Pulldown assays of the RHD of both RelA and c-Rel were carried out with biotinylated probes from the proximal κB site of human il12a promoter (supplemental Methods) and recombinant proteins, both acetylated in vitro and left untreated. The binding reactions were carried out with 5 μg of hybridized probe, 100 μl of streptavidin-agarose beads, 96 μl of acetylation buffer 5×, and different concentrations of recombinant RHD in a final volume of 500 μl. The incubation was carried out for 1 h at room temperature in a rotating shaker. Samples were centrifuged at 5000 × g in a microcentrifuge for 30 s, and the pellet was washed 4 times with PBS. The pulled down mixture was resuspended in 50 μl of Laemmli sample buffer and developed in a 12% SDS/PAGE. The proteins were transferred to a nitrocellulose membrane, and the proteins were identified with Ab reactive to the N-terminal domain of c-Rel and RelA/p65, anti-Ac-Lys-310-RelA, and anti-Ac-Lys.
RESULTS
SIRT1 and IL-12 p70 Production
Based on our previous findings suggesting that the sirtuin inhibitor EX-527 diminished the inhibitory effect of zymosan on the production of IL-12 p70 elicited by the combination of LPS and IFNγ, we tested whether activation of sirtuin activity with SRT1720 influenced IL-12 p70 in an opposite way. As shown in Fig. 1A, SRT1720 enhanced the inhibitory effect of zymosan on the production of IL-12 p70 elicited by LPS plus IFNγ when used at 10 μm, which most likely reflects the difficulty of obtaining a greater level of inhibition upon that elicited by zymosan. Because SIRT1 is a protein deacetylase dependent on NAD+, this nucleotide was assayed in DC. As shown in Fig. 1B, zymosan induced a robust increase of NAD+ levels in the nuclear fractions, whereas the concentration in the cytoplasm did not show significant change after stimulation. Because these results were consistent with an effect of SIRT1 on the regulation of IL-12 p70, we went on to address the expression of SIRT1 after stimulation of DC. As shown in Fig. 2A, zymosan induced a rapid appearance of SIRT1 protein in the nuclear fractions that persisted for at least 5 h, whereas only minor amounts of the protein were detected in the cytoplasm (not shown), thus agreeing with the preferential nuclear location of SIRT1. This was not observed after stimulation with LPS and IFNγ, which is in keeping with the reported inhibition of SIRT1 expression by these stimuli (33–36), nor were changes of the nuclear levels of the related protein SIRT6 detected (Fig. 2B). The accumulation of SIRT1 in the nucleus was not observed at 24 h, when similar amounts of the protein were detected in cytoplasm and nucleus (supplemental Fig. 1A, left panel). Similar to DC, zymosan induced the accumulation of SIRT1 in the nucleus of macrophages (supplemental Fig. 1A, right panel). Pure β-glucan particles also induced the accumulation of SIRT1 in the nucleus, and this was not inhibited by the soluble β-glucan laminarin (supplemental Fig. 1B), thus suggesting that particulate stimuli are most efficient to induce SIRT1. Parallel assays of the mRNA of sirt1 showed mild changes after zymosan stimulation (Fig. 2C), which probably indicates that the increased expression of SIRT1 protein induced by zymosan can be explained by mechanisms influencing mRNA translation rather than transcription rate. To link the changes in the levels of nuclear SIRT1 with il12a transcription, the presence of SIRT1 in the il12a promoter was assessed in ChIP assays. A shown in Fig. 2D, unlike LPS and IFNγ, zymosan enhanced the association of SIRT1 protein with the il12a promoter. In contrast, no significant changes of the association of SIRT6 with this promoter were observed, which agrees with the constitutive presence of SIRT6 in the nucleus. Because SIRT1 lacks a DNA binding domain, attempts to show the association of transcriptional repressors with the il12a promoter were carried out. TLE, which has been found in the nucleus after zymosan treatment (12), was detected in the il12a promoter in control DC and after zymosan stimulation (Fig. 2D), thus suggesting that transcriptional repressors that directly or indirectly bind SIRT1 could be operative during SIRT1 induction. We were not able to show the concomitant presence of HES1 in the il12a promoter, which might indicate the difficulty of the Ab to recognize epitopes in multimolecular inhibitory complexes.
FIGURE 1.
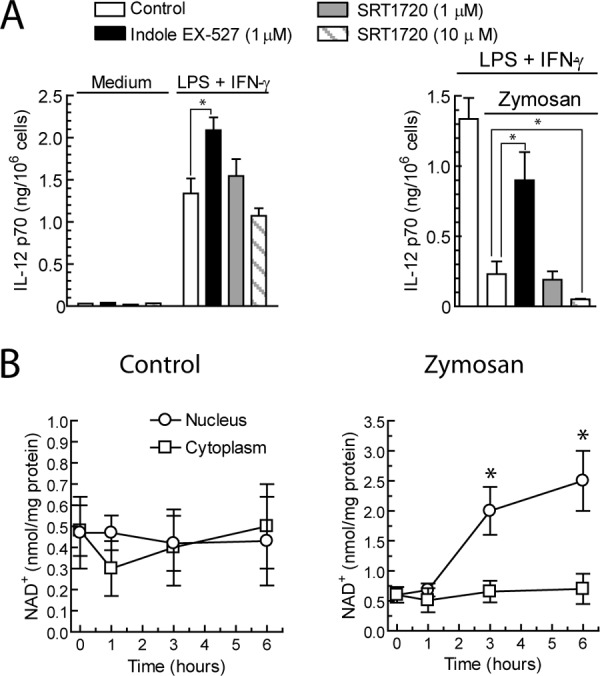
Effect of the pharmacological modulation of SIRT1 activity on IL-12 p70 production and assay of NAD+ production. A, DC were incubated with 10 μg/ml LPS plus 1000 units/ml IFNγ or 1 mg/ml zymosan in the presence of the indicated additions. 24 h after the addition of the stimuli, supernatants were collected for the assay of IL-12 p70 protein. B, NAD+ was assayed in the cytoplasm and nuclear fractions of DC stimulated with zymosan for the times indicated. Results represent the mean ± S.D. of four to six independent experiments. *, p < 0.05.
FIGURE 2.
SIRT protein expression in DC. A and B, DC were stimulated with zymosan and a combination of LPS and IFNγ at the concentrations described in the legend to Fig. 1. At the times indicated, nuclear fractions were collected for the assay of SIRT1 (A) and SIRT6 protein expression (B). TATA-box binding protein (TBP) was used as a load control. These are representative experiments of two with a similar trend. C, the mRNA expression of sirt1 was assayed at different times after zymosan stimulation. D, the binding of SIRT1 and SIRT6 to il12a promoter was assayed by ChIP assay in DC stimulated for 1 h with the indicated additions. This is a representative experiment of three. E, expression of the mRNA encoding Sirt1, Sirt2, and Sirt6 in bone marrow cells differentiated with murine GM-CSF from WT, sirt1-Tg, and sirt1+/− mice stimulated for 5 h with 1 mg/ml zymosan or left untreated. F and G, production of IL-12 p70 in these groups of mice is shown. The data represent the mean ± S.D. of 6–8 animals. H, shown is the effect of the pretreatment of EX-527 on the production of IL-12 p70 in a pool of cells from three mice. *, p < 0.05.
Further attempts to address the role of SIRT1 were carried out in sirt1-Tg and sirt1+/− mice, as the combined use of these animals provides a cogent appraisal of the phenotypical abnormalities resulting from controlled changes of Sirt1 function (18, 19). As expected, sirt1-Tg showed an enhanced expression of the mRNA encoding Sirt1. In contrast, a significant reduction of sirt1 mRNA was observed in the heterozygous mice. Zymosan induced a net increase of sirt1 mRNA, whereas the changes of sirt2 expression were less prominent, and sirt6 was barely detected (Fig. 2E). sirt1-Tg mice showed a reduced production of IL-12 p70 in response to the combination of LPS and IFNγ that was completely abrogated when the cells were preincubated with zymosan (Fig. 2F). On the other hand, sirt1+/− mice showed an enhanced production of IL-12 p70 (Fig. 2G) as well as some basal production in the absence of stimulus, but zymosan was also inhibitory, thus suggesting that the remaining expression of Sirt1 is sufficient to exert a blunting effect. In an experiment combining cells from three different sirt1+/− mice, preincubation with EX-527 showed an enhancing effect on the production of IL-12 p70 elicited by LPS and IFNγ as well as a reduction of the inhibition elicited by zymosan (Fig. 2H), whereas this could not be observed in the sirt1-Tg mice (not shown). Taken collectively, these results would suggest that SIRT1 may be the main sirtuin involved in the repression of il12a elicited by zymosan.
SIRT1 Regulation by Zymosan
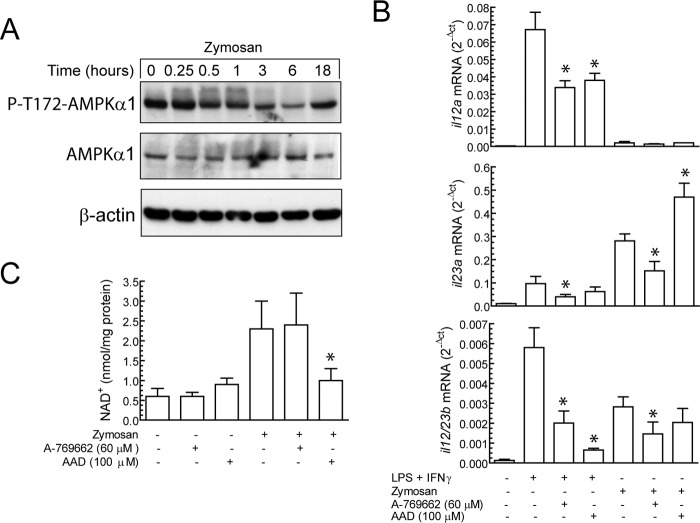
Because of the well known connection of AMPK with the activity of SIRT1 in metabolic systems, this topic deserves further analysis in view of the similar down-regulatory effect of SIRT1 (19) and AMPKα (37) on the production of proinflammatory cytokines. The first attempt to address this issue was the assay of the phosphorylation of Thr-172 of AMPKα1, which is the predominant isoform expressed in mononuclear phagocytes and undergoes dephosphorylation/inactivation in response to proinflammatory stimuli. Zymosan treatment did not change the level of Thr-172 phosphorylation of AMPKα1 up to 30 min, whereas a reduction was observed at 6 h and was followed by a recovery of the prestimulation level at 18 h (Fig. 3A). This is quite different from reports in human macrophages treated with LPS, where there is a decrease of Thr-172-AMPKα1 phosphorylation as soon as 10 min after stimulation and a return to prestimulation levels by 1 h (37). To further address the involvement of AMPK, pharmacological experiments were carried out with A-769662. As shown in Fig. 3B, activation of AMPK with A-769662 reduced the expression of il12a, il23a, and il12/23b transcripts. Unexpectedly, A-769662 did not influence the nuclear levels of NAD+ in DC treated with zymosan (Fig. 3C). This finding would suggest alternative mechanisms, for instance, inhibition by AMPK of NF-κB activation due to the recruitment of CREB-binding protein (CREB) by a concomitant enhanced activation of CREB and inhibition of IκB degradation (37). However, it seems likely that nicotinamide phosphoribosyltransferase expression might require not only phosphorylation of FOXO family transcription factors by AMPK but also a SIRT1-mediated deacetylation that is not induced by the sole addition of AMPK activators (38, 39). Treatment of DC with AAD, a compound that increases the cell level of NAD+ and whereby influences SIRT1 activity, reduced significantly the expression of il12a and il12/23b in response to the combination of LPS and IFNγ (Fig. 3B). In contrast, AAD increased il23a expression and did not influence il12a and il12/23b in DC treated with zymosan. Because we have been unable to show an increase of NAD+ by AAD in response to zymosan, but rather a decrease (Fig. 3C), it seems likely that AAD might influence NAD+ levels by NAD+/NADH ratio disequilibrium and product inhibition of NAD+ biosynthesis, whereas zymosan enhances nuclear NAD+ through NAD+ biosynthesis. Altogether, these results indicate that unlike LPS, which induces a rapid inactivation of AMPK activity, zymosan shows a more limited capacity to deactivate AMPK. The parallelism of the effects induced by modulating AMPK and SIRT1 activity could involve different mechanisms. The effect of AAD on the il23a mRNA expression elicited by zymosan might be explained by a concomitant action of high concentrations of NAD+ on poly(ADP-ribose) polymerase-dependent reactions (40, 41).
FIGURE 3.
AMPK activity, NAD+ production, and il12a expression. A, DC were stimulated with zymosan for the times indicated. At the end of these times cell lysates were collected for the assay of AMPKα1 phosphorylation/activation with a phosphospecific Ab reactive to P- Thr-172-AMPKα1. B, shown is the effect of the AMPK activator A-769662 and the NAD+ analog AAD on the expression of the mRNA encoding IL-12 p35 (il12a), IL-12 p19 (il23a), and IL-12 p40 (il12/23b). C, nuclear levels of NAD+ in DC treated with pharmacological agents acting on AMPK activity and NAD+ synthesis. The pharmacological agents were added 30 min before the stimuli, and the samples were collected 6 h after stimulation. Results represent the mean ± S.D. of four to five experiments with duplicate samples. *, p < 0.05.
Transcriptional Regulation of il12a and Acetylation Reactions
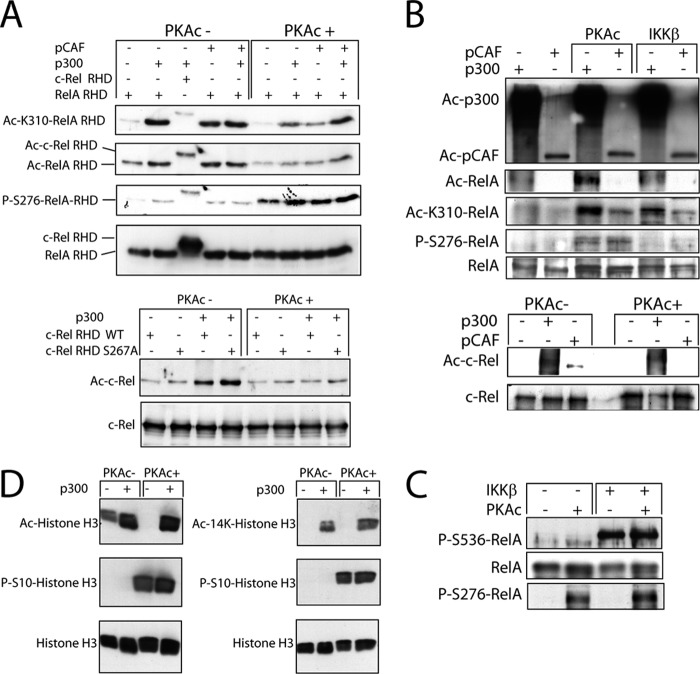
Because binding of transcription factors to cis-regulatory elements depends on post-translational modifications of the chromatin and the transcription factors and both can be influenced by SIRT1-catalyzed reactions, we went on to address the accessibility of the Nuc involved in the transcriptional regulation of il12a. As shown in Fig. 4, zymosan induced a significant decrease of Nuc-1 accessibility as compared with that observed in both control and LPS plus IFNγ-treated samples, which agrees with the presence of the κB-site in Nuc-1 and the relevance of NF-κB proteins in the transcriptional regulation of il12a. To assess whether post-translational modifications of either histones or NF-κB proteins are the targets of SIRT1, experiments were carried out using recombinant histone H3 and NF-κB proteins and the lysine acetyltransferases p300 and pCAF. As shown in Fig. 5A, both p300 and pCAF induced the acetylation of the RHD of both RelA/p65 and c-Rel. Notably, acetylation of the Lys-310 of RelA/p65, which enhances its transactivating ability (42), was clearly detected. By contrast, the anti-Ac-Lys-310-RelA/p65 Ab did not show any reactivity with the RHD of c-Rel, which agrees with the absence of homology of these proteins in the vicinity of the Lys-310 of RelA/p65. Mass spectrometric analysis of in vitro acetylated c-Rel RHD showed acetylations at Lys-116, -124, -193, -210, -261, -284, -295, and -302 (supplemental Fig. 2), as judged from the detection of peptides containing the acetylated immonium ion at m/z 126 that corresponds to acetylated lysine (43). Because it has been described that phosphorylation of Ser-267 of RelA/p65 may favor the acetylation of Lys-310 (44–46), additional experiments were conducted by carrying out in vitro phosphorylation of the RHD before acetylation. Under these conditions, no enhancement of the acetylation could be observed even though the occurrence of phosphorylation under these conditions was checked by immunoblotting with Ser(P)-276 phosphospecific anti-RelA Ab (Fig. 5A, right panels). Because RelA phosphorylation has been considered consistently as a post-translational modification that favors Lys-310-RelA/p65 acetylation, we carried out additional experiments using the whole protein as substrate. As shown in Fig. 5B, phosphorylation of RelA/p65 enhanced the acetylation of Lys-310, specially when p300 was utilized. This was observed both when the phosphorylation was carried out with PKAc and IKKβ. Because IKKβ only phosphorylates Ser-536 (Fig. 5C), these results indicate that acetylation of RelA/p65 is not only enhanced by Ser-276 phosphorylation but also facilitated by the presence of Ser and/or Thr residues out of the RHD, that the effect of IKKβ is observed in the absence of Ser-276 phosphorylation, and that the whole protein is needed to observe enhancement of acetylation by phosphorylation. When WT c-Rel and the Ser-267-Ala mutant were used in the assay, the acetylation was even lower in samples treated with PKAc (Fig. 5A, right lanes on the lower panels), thus suggesting that phosphorylation of c-Rel is not a requisite for acetylation by both p300 and pCAF. This was confirmed in assays with whole c-Rel protein (Fig. 5B, lower panels). When histone H3 was used as substrate, no difference was observed as regards the extent of acetylation of Lys-14 in previously phosphorylated samples as compared with non-phosphorylated samples (Fig. 5D). We next assessed the effect of SIRT1 on acetylated NF-κB proteins and histone H3. As shown on the right lanes of the upper panel of Fig. 6A, SIRT1 was only active on Ac-Lys-310 of RelA/p65, as judged from the disappearance of the Ac-Lys-310-RelA-RHD band in samples previously acetylated with p300, whereas blotting with the anti-Ac-Lys Ab did not show any significant change on both c-Rel-RHD and RelA-RHD (Fig. 6A, middle panel). Similar results were obtained when the RHD domains were acetylated with pCAF (Fig. 6B). In contrast, when acetylated histone H3 was treated with SIRT1, a significant reduction of Ac-histone H3 was observed (Fig. 6C). Overall, these results suggest that both pCAF and, to a greater extent p300, can acetylate NF-κB proteins, but only Ac-Lys-310 of RelA/p65 and Ac-histone H3 are significantly deacetylated by SIRT1. Taken collectively, these results agree with previous studies showing that p300/CBP is more active than pCAF in RelA acetylation (42, 44–46), whereas pCAF/Gcn5 is more active to acetylate histones (47).
FIGURE 4.
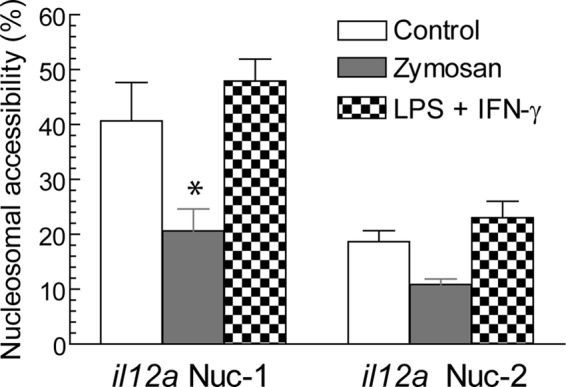
Accessibility of Nuc-1 and Nuc-2 of the il12a promoter. Accessibility was assayed by CHART-PCR. Nuclei were digested with 50 units BstXI or 50 units PshAI for 1 h at 37 °C, and genomic DNA was used to perform SYBR Green quantitative PCR. Amplification with primers for the Nuc-1-encompassing PshAI site is sensitive to remodeling of Nuc-1, and amplification with primers for Nuc-2 encompassing BstXI is sensitive to remodeling of Nuc-2. Results are expressed as a percentage of the undigested sample for each cell treatment as described under “Experimental Procedures.” Results represent the mean ± S.D. of five experiments with triplicate samples. *, p < 0.05.
FIGURE 5.
Acetylation of NF-κB proteins. A, RelA/p65 and c-Rel RHD were phosphorylated with PKAc or left untreated and then acetylated with pCAF and p300. Reaction mixtures were incubated at 30 °C for 1 h, resolved by SDS-PAGE, and analyzed by Western blot to assess Ac-Lys-310-RelA/p65, Ac-Lys, and Ser(P)-276-RelA/p65. c-Rel acetylation was also assayed in the WT and Ser-267-Ala-c-Rel RHD to address the effect of the Ser homologous of Ser-276-RelA on the acetylation of c-Rel RHD. B, the effect of RelA/p65 and c-Rel phosphorylation on the acetylation by pCAF and p300 was studied using the recombinant whole proteins as substrate and PKAc and IKKβ as kinases. The autoacetylation of pCAF and p300 is shown in the upper panel. C, shown is the effect of PKAc and IKKβ on Ser-536-RelA/p65 phosphorylation. D, shown is the effect of the phosphorylation of histone H3 by PKAc on the acetylation reaction. These are representative experiments of three showing a similar trend.
FIGURE 6.

Deacetylation of NF-κB family proteins and histone H3 by SIRT1. A, c-Rel and RelA/p65 RHD were acetylated with p300 and then deacetylated with SIRT1. The reaction was analyzed by Western blot to assess Lys-310-RelA/p65 (upper panel), Ac-c-Rel, and Ac-RelA/p65 (middle panel). The bottom panel shows the load control of c-Rel and RelA/p65 RHD. B, a similar experiment was conducted to compare the effect of SIRT1 on the acetylation of Lys-310-RelA carried out by pCAF and p300. C, shown is the effect of SIRT1 on histone H3 acetylated by p300 and pCAF.
NF-κB Protein Acetylation and DNA Binding
To address whether acetylation of RHD domains influences binding to the κB site of the il12a promoter, pulldown assays were carried out using RHD. As shown in Fig. 7, acetylation of both RelA/p65 RHD and c-Rel RHD did not modify the extent of binding to the DNA probe. To address the effect of zymosan on acetylation reactions in DC, coimmunoprecipitation assays were carried out in nuclear fractions using anti-TLE Ab. As shown in Fig. 8A, zymosan-treated cells showed a decreased amount of Ac-Lys-14-histone H3. In contrast, levels of Ac-Lys-14-histone H3 similar to those detected in control cells were observed after treatment with the lysine deacetylase inhibitors trichostatin and EX-527, thus suggesting that zymosan enhances deacetylation of histone H3 in a TLE-dependent manner. To address NF-κB protein acetylation, immunoprecipitations were conducted on nuclear fractions as lysine acetylation cannot be assessed with certainty in cell lysates. As shown in Fig. 8B, nuclear translocation of both RelA/p65 and c-Rel was observed 1 and 5 h after zymosan stimulation. RelA/p65 showed some degree of Lys-310 acetylation at 1 h and to a lesser extent, if any, at 5 h, which could indicate that deacetylation by SIRT1 could have occurred at Lys-310. A low level of c-Rel acetylation was observed 1 h after stimulation with both zymosan and LPS plus IFNγ, but not at 5 h. To address the effect of Nuc-1 acetylation on transcription factor binding, phosphorylation and acetylation reactions were carried out on Nuc reconstituted with the DNA-containing Nuc-1 sequence. The most prominent changes were observed on c-Rel binding. Notably, phosphorylation of the nucleosome induced a 5-fold increase of c-Rel binding, and acetylation induced an additive increase. In contrast, the increase of RelA/p65 binding did not reach significance (Fig. 8, C and D). Overall, these results suggest that phosphorylation and acetylation reactions influence DNA accessibility of Nuc-1 in the il12a promoter.
FIGURE 7.

Effect of the acetylation of RelA/p65 and c-Rel RHD on the binding to the proximal κB site of the il12a promoter. Pulldown assays were carried out with biotinylated probes and different concentrations of the recombinant proteins both acetylated in vitro and left untreated. The incubation was carried out for 1 h at room temperature in a rotating shaker. The pulled-down mixture was developed in a 12% SDS/PAGE. The proteins transferred to a nitrocellulose membrane were identified with Ab reactive to the N-terminal domain of c-Rel and RelA/p65, anti Ac-Lys-310-RelA, and anti-Ac-Lys. This is an experiment representative of three with an identical trend.
FIGURE 8.
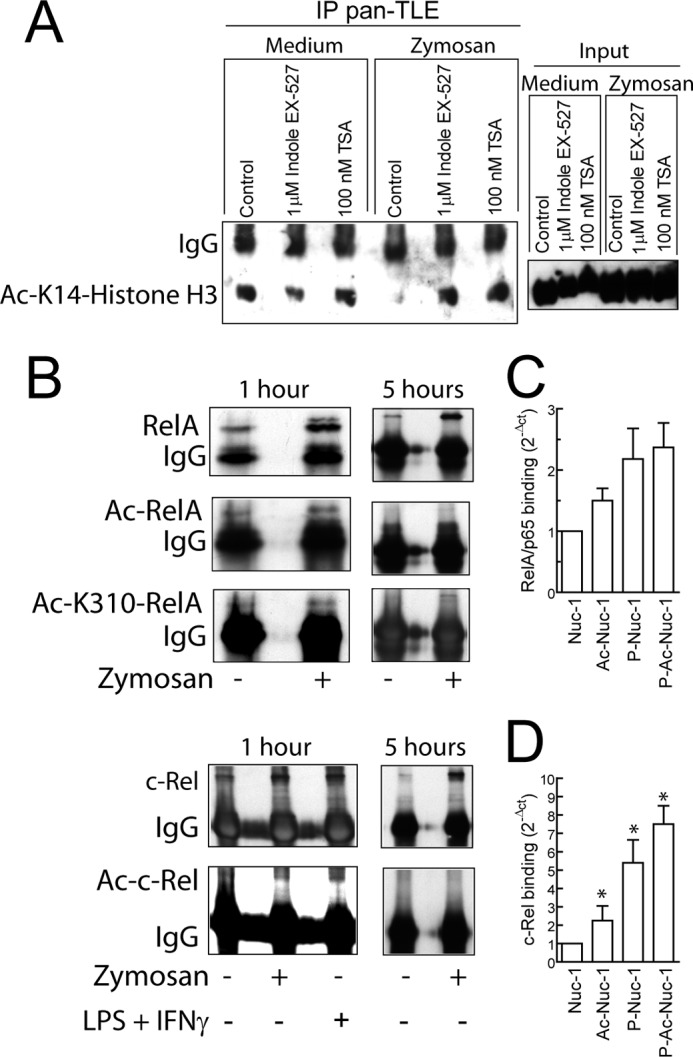
Immunoprecipitation of Ac- Lys-14-histone H3 and NF-κB proteins. A, DC were lysed, and the proteins of the nuclear extracts were obtained by using a nuclear extract kit. The clarified lysates were preabsorbed on protein G-Sepharose and then incubated overnight with precipitating Ab followed by a 2-h incubation with protein G-Sepharose beads. Immune complexes were extensively washed and subjected to SDS/PAGE. To address the presence of deacetylated histones, nuclear proteins were immunoprecipitated (IP) with anti-pan-TLE Ab, and after SDS/PAGE and membrane transfer, Ac-Lys-14-histone H3 was assayed. TSA indicates trichostatin. B, nuclear extracts from DC stimulated with zymosan or the combination LPS and IFNγ were immunoprecipitated with anti-RelA/p65 and anti-c-Rel Ab. The presence of Ac-RelA/p65 and Ac-c-Rel was assayed by immunoblotting. The blots are representative of three independent experiments with a similar trend. C and D, binding of RelA/p65 and c-Rel to in vitro reconstituted Nuc-1 is shown. 0.5 μg of either RelA/p65 or c-Rel was incubated with phosphorylated and/or acetylated reconstituted Nuc-1 containing 0.5 μg of histones for 1 h at 37 °C. At the end of this time sequential ChIP was carried out using anti-histone H3 Ab in the first step and after breaking apart the Ab chains with DTT, Ab reactive to the N-terminal domain of RelA/p65, and C-terminal domain of c-Rel in the second step. Results represent mean ± S.D. of five experiments. *, p < 0.05.
DISCUSSION
These data show that modulation of SIRT1 activity by pharmacological mechanisms is accompanied by changes in the production of IL-12 p70 by DC in response to the optimal stimuli and also by a reduction of the inhibitory effect on IL-12 p70 production observed when the fungal homolog zymosan is used before stimulation. These data are of clinical significance, as β-glucans are a central pathogen-associated molecular pattern during the yeast (conidia) to hyphal (filamentous) transition, which occurs when dimorphic fungi invade target organs and is characterized by changes in the expression of cell wall proteins and carbohydrates, including surface-exposed β-glucans in swelling conidia and early germlings, as disclosed during Aspergillus fumigatus infection (48), but this may also apply to Candida, Cryptococcus, and Pneumocystis jirovecii. On this basis, Th17 responses can be predominant during β-glucan accessibility, but Th1 responses can also occur as a result of the distinct time-frame of pathogen-associated molecular pattern accessibility.
Our results agree with previous findings suggesting that the balance IL-12 p70/IL-23 elicited by zymosan is best explained by an activation of class III lysine deacetylases coincidental with the activation of transcriptional repressors (12). Several mechanisms can regulate the expression of SIRT1 protein. On the one hand, SIRT1 expression is inhibited by miR-34a (49) and miR-200a (50) and by Jun N-terminal kinase-2-mediated regulation of SIRT1 stability (51). These mechanisms would agree with our findings and with previous reports on an increase of SIRT1 protein levels in the absence of an attendant increase of sirt1 mRNA (52, 53). On the other hand, a recent report has shown CREB-dependent transcriptional regulation of sirt1 mRNA in response to calorie restriction, norepinephrine, and glucagon (54), thus stressing the existence of cell-specific mechanisms to control SIRT1 protein levels. Our data are consistent with all of these views as we have found an increase of sirt1 mRNA after zymosan stimulation of mouse DC and an increase of SIRT1 protein after zymosan challenge in human DC. The regulation of sirt1 mRNA by CREB might be of particular importance in this system because CREB is activated after zymosan challenge (31, 55) and is involved in the transcriptional regulation of hes1 (56). Notably, one of the best established mechanisms whereby mammal Hes proteins repress transcription is by their ability to recruit the Groucho homologs TLE-1–4 to generate a transcriptional repressor complex (57). TLE interacts with the N-terminal residues 1–46 of histone H3 and attracts further corepressors like class I and III histone deacetylases and members of the Sin3 complex (58). An example of this mechanism of transcriptional repression involving HES1, TLE, and Sirt1 similar to our findings is the inhibition by mild oxidation of the pro-neural transcription factor Mash 1 during the differentiation of neural progenitors (59). Unlike SIRT6, zymosan induced a net increase of SIRT1 binding to the il12a promoter, thus pointing to a central role of SIRT1 on IL-12 p70 production regulation. These data agree with current views on the role of SIRT6 on the regulation of κB-dependent transcription, as SIRT6 interacts with RelA/p65 but not with other elements of the NF-κB family and exerts its effect via histone H3 Lys-9 deacetylation (60). This would explain a lack of direct effect of SIRT6 on c-Rel but does not rule out a combinatorial effect of both sirtuins and even a cooperation of other sirtuins (61). Interestingly, we have observed an ∼5-fold increase of sirt1 mRNA in dendritic cells of WT mouse stimulated with zymosan as well as in the engineered mice, thus suggesting that cell and species differences may exist and that results obtained in mouse models cannot be straightforwardly applied to the human. Our results showing that Sirt1 and Sirt2 are the sirtuins expressed at a higher extent in mouse DC are in keeping with a recent report (62) and indicate a difference with human DC where SIRT6 was expressed constitutively in the nuclei. As regards the results obtained in sirt1-Tg and sirt1+/− mice, the data are consistent with the involvement of Sirt1 in the modulation of IL-12 p70 levels by showing a reduction of IL-12 p70 production in the sirt1-Tg animals and an increase in the sirt1+/−, although the reduction of the inhibitory effect of zymosan did not reach statistical significance, most likely reflecting that the remaining Sirt1 is sufficient to exert inhibition. In keeping with this interpretation, an enhancement of IL-12 p70 production was elicited by EX-527 in a pool of DC from three sirt−/+ mice.
In agreement with the requirement of NAD+ for SIRT1 activity, we have observed that zymosan increases the nuclear levels of NAD+. This could be explained mechanistically through the ability of SIRT1 to bind nicotinamide nucleotide adenylyltransferase-1, a nuclear isoform of a central enzyme of NAD+ synthesis that creates nuclear microdomains of high concentration of NAD+ and allows SIRT1 activity without significantly altering the total cellular levels of NAD+, which rarely fluctuate more than 2-fold (63). SIRT1 activity has been related to AMPK activity through the NAD+ biosynthetic enzymes nicotinamide phosphoribosyltransferase and through the combined action of AMPK and SIRT1 on FOXO transcription factors. Regardless of which could be the ultimate mechanism, AMPK activation may be a relevant event in view of its modulatory effect on the inflammatory reaction (37) and its purported activation during phagocytic challenge due to the energetic challenge that leads to ATP consumption and AMP accumulation. Our data, showing the effect of an AMPK activator and the maintained activity of AMPK during the early phase of zymosan stimulation would point out to the involvement of AMPK in the IL-12 p70/IL-23 balance. However, we have not been able to disclose elevation of NAD+ levels after A-769662, thus suggesting that the connection between AMPK and NAD+ might be due to their concomitant activation and functional cooperation.
As regards the effect of zymosan on the regulation of il12a transcription, our data agree with previous reports stressing the relevance of mechanisms involving accessibility of transcription factors to the promoter (16, 17). Our study has disclosed more remarkable changes in Nuc-1 than in Nuc-2 accessibility. This can be explained by the central role of the κB site of Nuc-1 in DC, whereas Sp1 binding sites might play this role in THP-1 monocytic cell line (17). Acetylation of RelA/p65 at Lys-218, -221, and -310 plays a central role in the function of the protein (42). Acetylation of Lys-218 and -221 enhances DNA binding and reduces IκBα binding, whereas Lys-310 acetylation is required for full transcriptional activity. Notably, SIRT1 treatment induced a selective decrease of the acetylation of Lys-310-RelA/p65 as well as histone H3 acetylation, whereas it did not modify noticeably the overall acetylation of both RelA/p65 and c-Rel, thus suggesting a selective effect of SIRT1 on histone H3 and Lys-310-RelA/p65 acetylation. Attempts to address whether c-Rel and RelA/p65 acetylation influences DNA binding were carried out by pulldown experiments, but we found that binding occurred at the same extent in both p300-treated and untreated samples, thus indicating that c-Rel acetylation does not influence DNA binding. This agrees with current views on the function of c-Rel, as c-Rel homodimers bind with substantially higher affinity than RelA/p65 homodimers to all κB sites (64), although they do not to interact with CBP (65).
To address whether NF-κB proteins and/or histones are acetylated in vivo after stimulation by zymosan, nuclear extract proteins were immunoprecipitated with anti-TLE Ab to assess the acetylation of the coimmunoprecipitated histone H3. The finding of significant deacetylation of histone H3 associated with TLE and the lack of evidence of a SIRT1-sensitive acetylation of c-Rel homologous to Ac-Lys-310 in RelA/p65 would suggest that histone H3 deacetylation elicited by zymosan could play a major role on the accessibility of c-Rel to the Nuc-1 of il12a promoter. The low degree of RelA/p65 and c-Rel acetylation that could be observed at 1 h in the nuclear extracts can be related to other lysines like Lys-218 in RelA and Lys-210 in c-Rel that seem to be insensitive to SIRT1 but may be the target of other lysine deacetylases.
Attempts to show by sequential ChIP whether posttranslational modifications of Nuc-1 histones influenced NF-κB protein binding showed a prominent effect of histone phosphorylation on c-Rel binding, whereas acetylation increased slightly the extent of binding. This result agrees with previous reports on the relevance of histone phosphorylation for IL-12 p70 production (12) and also suggests that the effect of histone acetylation on transcriptional activation might involve the cooperation of other proteins that are not present in the in vitro assay. The analysis of these results indicates that the addition of phosphate groups to serine residues provokes repulsion forces with the DNA backbone phosphates, whereas acetylation of histones neutralizes the positive charge of lysines and only loosens the interactions with DNA phosphate groups. On this basis, il12a promoter might become most accessible to transcription factors by phosphorylation but only slightly by acetylation. The most likely consequence of acetylation could be the recruitment of proteins with bromodomains, like the coactivator SMARCA4 subunit of the remodeling complex SWI/SNF (a human ortholog of yeast Swi2/Snf2) (47) and RSC (remodels structure of chromatin) complex (66) that are involved in the displacement of DNA from histones, and BET (bromodomain and extra terminal domain) proteins, the function of which is the recruitment of the transcription machinery. In fact, the i-BET compound, which selectively inhibits the binding of BET proteins BRD2, BRD3, and BRD4 to acetylated histones, blunts the transcription of secondary response genes in macrophages like il12a (13). An alternative mechanism could be the inhibition of the interaction with proteins involved in the down-regulation of NF-κB activity, for instance IκBα, the binding of which to RelA/p65 is diminished by Lys-218 acetylation (42), and most likely by the acetylation of the homologous Lys-210 we detected in c-Rel (supplemental Fig. 2). The abrogation of IL-12 p70 production and the maintenance of IL-23 during phagocytic stress induced by a fungal cell wall mimic could be envisioned as an attempt to enhance the phagocytic potential in view of the ability of IL-23 to maintain the expansion and function of Th17 cells and the role of IL-17 family cytokines in the induction of neutrophil-activating factors and acute phase proteins in fungal defense. On this basis, pharmacological modulation of SIRT1 activity could be a good approach to tailor the cytokine response for the activation of phagocytes. In summary, our data show an enhanced activity of SIRT1 after zymosan challenge as judged from its increased location in the il12a promoter, the evidence of histone H3 deacetylation in areas in close contact with the transcriptional repressor TLE, and the increased nuclear concentration of the co-substrate NAD+ (see the schematic diagram in Fig. 9).
FIGURE 9.
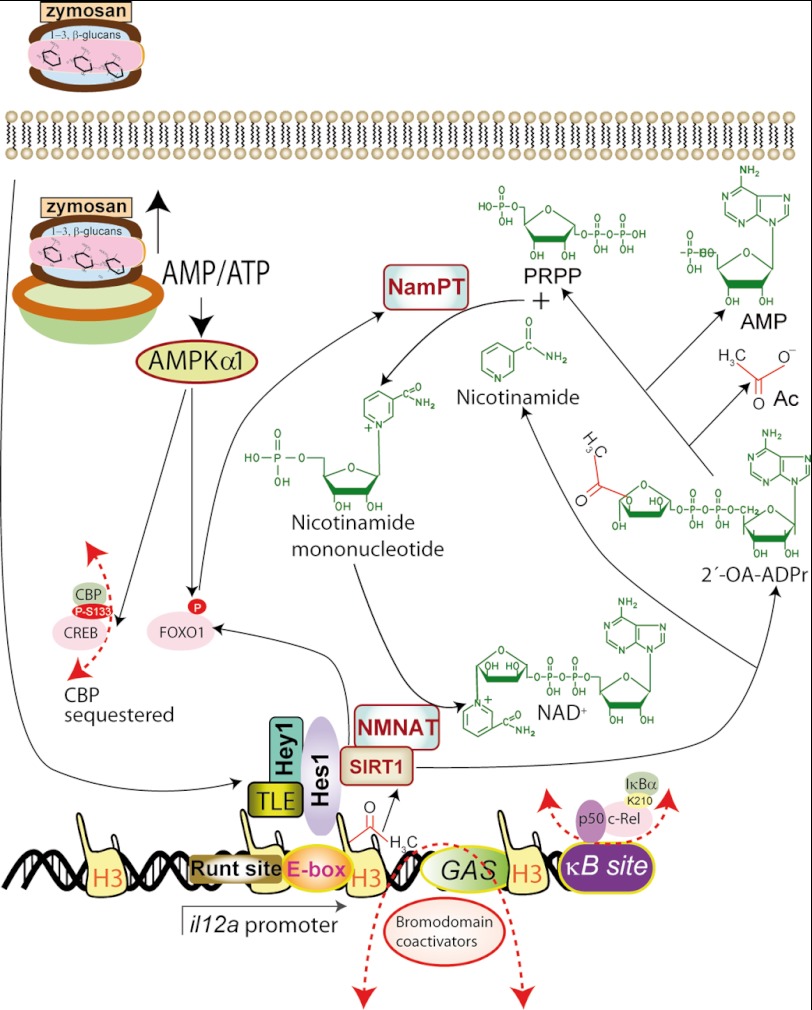
Schematic representation of the effect of zymosan on the repression of il12a. Phagocytosis of zymosan is an energetic challenge that can lead to ATP depletion and increase of the AMP/ATP ratio. This may increase SIRT1 activity by a mechanism dependent on AMPK but that may also require additional steps to induce NamPT expression (32, 38, 39). NAD+ and histone acetyl-Lys are substrates for SIRT1 leading to the production of nicotinamide and 2′-O-acetyl-ADP-ribose (2′-OA-ADPr). In addition to the possible effect on AMPK1α, which may affect gene expression by different mechanisms involving the function of CREB, FOXO, and PGC-1α, zymosan also elicits the formation of a supramolecular complex of transcriptional repressors, in which nicotinamide nucleotide adenylyltransferase 1 (NMNAT1) could be included because of its capacity to bind to SIRT1 (63). The overall functional consequence of this cascade of signaling events is the blockade of the binding of bromodomain-containing transcriptional coactivators and/or shuttling of c-Rel from the κB-site by IκBα.
Supplementary Material
Acknowledgments
We thank Dr. Manuel Serrano from Spanish National Cancer Centre (CNIO) for providing engineered mice and critical review of the manuscript. We thank the staff from Centro de Hemoterapia y Hemodonación de Castilla y León for help with blood cell purification. We thank Tse-Hua Tan and Kelly M. Stehling for the c-Rel cDNA. We also thank Andrés Alonso for help with protein expression.
This work was supported by Plan Nacional de Salud y Farmacia Grants SAF2007-60446 and SAF2010-15070, Fundación Ramón Areces, Red Temática de Investigación Cardiovascular, and Junta de Castilla y León Grant CSI003A11-2.

This article contains supplemental Methods, Figs. 1 and 2, and references.
- DC
- dendritic cells
- AMPK
- AMP-activated kinase
- CREB
- CRE binding protein
- CBP
- CREB-binding protein
- Nuc
- nucleosome
- RHD
- Rel-homology domain
- SIRT
- sirtuin
- Tg
- transgenic
- TLE
- transducin-like enhancer of split
- PKAc
- catalytic subunit of PKA
- AAD
- 3-aminopyridine adenine dinucleotide
- Ab
- antibody.
REFERENCES
- 1. Abraham C., Cho J. H. (2009) IL-23 and autoimmunity: New insights into the pathogenesis of inflammatory bowel disease. Annu. Rev. Med. 60, 97–110 [DOI] [PubMed] [Google Scholar]
- 2. Cua D. J., Sherlock J., Chen Y., Murphy C. A., Joyce B., Seymour B., Lucian L., To W., Kwan S., Churakova T., Zurawski S., Wiekowski M., Lira S. A., Gorman D., Kastelein R. A., Sedgwick J. D. (2003) Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature 421, 744–748 [DOI] [PubMed] [Google Scholar]
- 3. Lee E., Trepicchio W. L., Oestreicher J. L., Pittman D., Wang F., Chamian F., Dhodapkar M., Krueger J. G. (2004) Increased expression of interleukin 23 p19 and p40 in lesional skin of patients with psoriasis vulgaris. J. Exp. Med. 199, 125–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Murphy C. A., Langrish C. L., Chen Y., Blumenschein W., McClanahan T., Kastelein R. A., Sedgwick J. D., Cua D. J. (2003) Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J. Exp. Med. 198, 1951–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gautier G., Humbert M., Deauvieau F., Scuiller M., Hiscott J., Bates E. E., Trinchieri G., Caux C., Garrone P. (2005) A type I interferon autocrine-paracrine loop is involved in Toll-like receptor-induced interleukin-12 p70 secretion by dendritic cells. J. Exp. Med. 201, 1435–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu J., Cao S., Herman L. M., Ma X. (2003) Differential regulation of interleukin (IL)-12 p35 and p40 gene expression and interferon (IFN)-γ-primed IL-12 production by IFN regulatory factor 1. J. Exp. Med. 198, 1265–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goriely S., Molle C., Nguyen M., Albarani V., Haddou N. O., Lin R., De Wit D., Flamand V., Willems F., Goldman M. (2006) Interferon regulatory factor 3 is involved in Toll-like receptor 4 (TLR4)- and TLR3-induced IL-12p35 gene activation. Blood 107, 1078–1084 [DOI] [PubMed] [Google Scholar]
- 8. Kollet J. I., Petro T. M. (2006) IRF-1 and NF-κB p50/cRel bind to distinct regions of the proximal murine IL-12 p35 promoter during costimulation with IFN-γ and LPS. Mol. Immunol. 43, 623–633 [DOI] [PubMed] [Google Scholar]
- 9. Gerosa F., Baldani-Guerra B., Lyakh L. A., Batoni G., Esin S., Winkler-Pickett R. T., Consolaro M. R., De Marchi M., Giachino D., Robbiano A., Astegiano M., Sambataro A., Kastelein R. A., Carra G., Trinchieri G. (2008) Differential regulation of interleukin 12 and interleukin 23 production in human dendritic cells. J. Exp. Med. 205, 1447–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dennehy K. M., Willment J. A., Williams D. L., Brown G. D. (2009) Reciprocal regulation of IL-23 and IL-12 following co-activation of Dectin-1 and TLR signaling pathways. Eur. J. Immunol. 39, 1379–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang H., Ostroff G. R., Lee C. K., Wang J. P., Specht C. A., Levitz S. M. (2009) Distinct patterns of dendritic cell cytokine release stimulated by fungal β-glucans and toll-like receptor agonists. Infect. Immun. 77, 1774–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alvarez Y., Municio C., Hugo E., Zhu J., Alonso S., Hu X., Fernández N., Sánchez Crespo M. (2011) Notch- and transducin-like enhancer of split (TLE)-dependent histone deacetylation explain interleukin 12 (IL-12) p70 inhibition by zymosan. J. Biol. Chem. 286, 16583–16595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nicodeme E., Jeffrey K. L., Schaefer U., Beinke S., Dewell S., Chung C. W., Chandwani R., Marazzi I., Wilson P., Coste H., White J., Kirilovsky J., Rice C. M., Lora J. M., Prinjha R. K., Lee K., Tarakhovsky A. (2010) Suppression of inflammation by a synthetic histone mimic. Nature 468, 1119–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carmody R. J., Ruan Q., Liou H. C., Chen Y. H. (2007) Essential roles of c-Rel in TLR-induced IL-23 p19 gene expression in dendritic cells. J. Immunol. 178, 186–191 [DOI] [PubMed] [Google Scholar]
- 15. Grumont R., Hochrein H., O'Keeffe M., Gugasyan R., White C., Caminschi I., Cook W., Gerondakis S. (2001) c-Rel regulates interleukin 12 p70 expression in CD8+ dendritic cells by specifically inducing p35 gene transcription. J. Exp. Med. 194, 1021–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hovius J. W., de Jong M. A., den Dunnen J., Litjens M., Fikrig E., van der Poll T., Gringhuis S. I., Geijtenbeek T. B. (2008) Salp15 binding to DC-SIGN inhibits cytokine expression by impairing both nucleosome remodeling and mRNA stabilization. PLoS Pathog. 4, e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goriely S., Demonté D., Nizet S., De Wit D., Willems F., Goldman M., Van Lint C. (2003) Human IL-12 p35 gene activation involves selective remodeling of a single nucleosome within a region of the promoter containing critical Sp1-binding sites. Blood 101, 4894–4902 [DOI] [PubMed] [Google Scholar]
- 18. Purushotham A., Xu Q., Li X. (2012) Systemic SIRT1 insufficiency results in disruption of energy homeostasis and steroid hormone metabolism upon high fat diet feeding. FASEB J. 26, 656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schug T. T., Xu Q., Gao H., Peres-da-Silva A., Draper D. W., Fessler M. B., Purushotham A., Li X. (2010) Myeloid deletion of SIRT1 induces inflammatory signaling in response to environmental stress. Mol. Cell. Biol. 30, 4712–4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yeung F., Hoberg J. E., Ramsey C. S., Keller M. D., Jones D. R., Frye R. A., Mayo M. W. (2004) Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23, 2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Imai S., Armstrong C. M., Kaeberlein M., Guarente L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 [DOI] [PubMed] [Google Scholar]
- 22. Landry J., Sutton A., Tafrov S. T., Heller R. C., Stebbins J., Pillus L., Sternglanz R. (2000) The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. U.S.A. 97, 5807–5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pfluger P. T., Herranz D., Velasco-Miguel S., Serrano M., Tschöp M. H. (2008) SIRT1 protects against high fat diet-induced metabolic damage. Proc. Natl. Acad. Sci. U.S.A. 105, 9793–9798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yoshizaki T., Milne J. C., Imamura T., Schenk S., Sonoda N., Babendure J. L., Lu J. C., Smith J. J., Jirousek M. R., Olefsky J. M. (2009) SIRT1 exerts anti-inflammatory effects and improves insulin sensitivity in adipocytes. Mol. Cell. Biol. 29, 1363–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yoshizaki T., Schenk S., Imamura T., Babendure J. L., Sonoda N., Bae E. J., Oh D. Y., Lu M., Milne J. C., Westphal C., Bandyopadhyay G., Olefsky J. M. (2010) SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity. Am. J. Physiol. Endocrinol. Metab. 298, E419–EE428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gerhart-Hines Z., Dominy J. E., Jr., Blättler S. M., Jedrychowski M. P., Banks A. S., Lim J. H., Chim H., Gygi S. P., Puigserver P. (2011) The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD+. Mol. Cell 44, 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fisher T. L., Vercellotti S. V., Anderson B. M. (1973) Interaction of 3-aminopyridine adenine dinucleotide with dehydrogenases. J. Biol. Chem. 248, 4293–4299 [PubMed] [Google Scholar]
- 28. Palmere R. M., Conley R. T. (1970) The Schmidt reaction of α-aralkyl-substituted carboxylic acids. J. Org. Chem. 35, 2703–2707 [Google Scholar]
- 29. Valera I., Fernández N., Trinidad A. G., Alonso S., Brown G. D., Alonso A., Crespo M. S. (2008) Co-stimulation of dectin-1 and DC-SIGN triggers the arachidonic acid cascade in human monocyte-derived dendritic cells. J. Immunol. 180, 5727–5736 [DOI] [PubMed] [Google Scholar]
- 30. Van Gool F., Gallí M., Gueydan C., Kruys V., Prevot P. P., Bedalov A., Mostoslavsky R., Alt F. W., De Smedt T., Leo O. (2009) Intracellular NAD levels regulate tumor necrosis factor protein synthesis in a sirtuin-dependent manner. Nat. Med. 15, 206–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alvarez Y., Municio C., Alonso S., Sánchez Crespo M., Fernández N. (2009) The induction of IL-10 by fungi in dendritic cells depends on CREB activation by the coactivators CBP and TORC2 and autocrine PGE2. J. Immunol. 183, 1471–1479 [DOI] [PubMed] [Google Scholar]
- 32. Fulco M., Cen Y., Zhao P., Hoffman E. P., McBurney M. W., Sauve A. A., Sartorelli V. (2008) Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev. Cell 14, 661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li P., Zhao Y., Wu X., Xia M., Fang M., Iwasaki Y., Sha J., Chen Q., Xu Y., Shen A. (2012) Interferon-γ (IFN-γ) disrupts energy expenditure and metabolic homeostasis by suppressing SIRT1 transcription. Nucleic Acids Res. 40, 1609–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shen Z., Ajmo J. M., Rogers C. Q., Liang X., Le L., Murr M. M., Peng Y., You M. (2009) Role of SIRT1 in regulation of LPS- or two ethanol metabolites-induced TNF-α production in cultured macrophage cell lines. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G1047–G1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang Z., Kahn B. B., Shi H., Xue B. Z. (2010) Macrophage α1 AMP-activated protein kinase (α1AMPK) antagonizes fatty acid-induced inflammation through SIRT1. J. Biol. Chem. 285, 19051–19059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Z., Lowry S. F., Guarente L., Haimovich B. (2010) Roles of SIRT1 in the acute and restorative phases following induction of inflammation. J. Biol. Chem. 285, 41391–41401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sag D., Carling D., Stout R. D., Suttles J. (2008) Adenosine 5′-monophosphate-activated protein kinase promotes macrophage polarization to an anti-inflammatory functional phenotype. J. Immunol. 181, 8633–8641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cantó C., Gerhart-Hines Z., Feige J. N., Lagouge M., Noriega L., Milne J. C., Elliott P. J., Puigserver P., Auwerx J. (2009) AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458, 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tao R., Wei D., Gao H., Liu Y., DePinho R. A., Dong X. C. (2011) Hepatic FoxOs regulate lipid metabolism via modulation of expression of the nicotinamide phosphoribosyltransferase gene. J. Biol. Chem. 286, 14681–14690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim M. Y., Zhang T., Kraus W. L. (2005) Poly(ADP-ribosyl)ation by PARP-1: “PAR-laying” NAD+ into a nuclear signal. Genes Dev. 19, 1951–1967 [DOI] [PubMed] [Google Scholar]
- 41. Schreiber V., Dantzer F., Ame J. C., de Murcia G. (2006) Poly(ADP-ribose): Novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 7, 517–528 [DOI] [PubMed] [Google Scholar]
- 42. Chen L. F., Mu Y., Greene W. C. (2002) Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. EMBO J. 21, 6539–6548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang K., Yau P. M., Chandrasekhar B., New R., Kondrat R., Imai B. S., Bradbury M. E. (2004) Differentiation between peptides containing acetylated or tri-methylated lysines by mass spectrometry: An application for determining lysine 9 acetylation and methylation of histone H3. Proteomics 4, 1–10 [DOI] [PubMed] [Google Scholar]
- 44. Chen L. F., Williams S. A., Mu Y., Nakano H., Duerr J. M., Buckbinder L., Greene W. C. (2005) NF-κB RelA phosphorylation regulates RelA acetylation. Mol. Cell. Biol. 25, 7966–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhong H., May M. J., Jimi E., Ghosh S. (2002) The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol. Cell 9, 625–636 [DOI] [PubMed] [Google Scholar]
- 46. Zhong H., SuYang H., Erdjument-Bromage H., Tempst P., Ghosh S. (1997) The transcriptional activity of NF-κB is regulated by the IκB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell 89, 413–424 [DOI] [PubMed] [Google Scholar]
- 47. Hassan A. H., Awad S., Prochasson P. (2006) The Swi2/Snf2 bromodomain is required for the displacement of SAGA and the octamer transfer of SAGA-acetylated nucleosomes. J. Biol. Chem. 281, 18126–18134 [DOI] [PubMed] [Google Scholar]
- 48. Steele C., Rapaka R. R., Metz A., Pop S. M., Williams D. L., Gordon S., Kolls J. K., Brown G. D. (2005) The β-glucan receptor dectin-1 recognizes specific morphologies of Aspergillus fumigatus. PLos Pathog. 1, e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yamakuchi M., Ferlito M., Lowenstein C. J. (2008) miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. U.S.A. 105, 13421–13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eades G., Yao Y., Yang M., Zhang Y., Chumsri S., Zhou Q. (2011) miR-200a regulates SIRT1 expression and epithelial to mesenchymal transition (EMT)-like transformation in mammary epithelial cells. J. Biol. Chem. 286, 25992–26002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ford J., Ahmed S., Allison S., Jiang M., Milner J. (2008) JNK2-dependent regulation of SIRT1 protein stability. Cell Cycle 7, 3091–3097 [DOI] [PubMed] [Google Scholar]
- 52. Kanfi Y., Peshti V., Gozlan Y. M., Rathaus M., Gil R., Cohen H. Y. (2008) Regulation of SIRT1 protein levels by nutrient availability. FEBS Lett. 582, 2417–2423 [DOI] [PubMed] [Google Scholar]
- 53. Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005) Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 54. Noriega L. G., Feige J. N., Canto C., Yamamoto H., Yu J., Herman M. A., Mataki C., Kahn B. B., Auwerx J. (2011) CREB and ChREBP oppositely regulate SIRT1 expression in response to energy availability. EMBO Rep. 12, 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kelly E. K., Wang L., Ivashkiv L. B. (2010) Calcium-activated pathways and oxidative burst mediate zymosan-induced signaling and IL-10 production in human macrophages. J. Immunol. 184, 5545–5552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Herzig S., Hedrick S., Morantte I., Koo S. H., Galimi F., Montminy M. (2003) CREB controls hepatic lipid metabolism through nuclear hormone receptor PPAR-γ. Nature 426, 190–193 [DOI] [PubMed] [Google Scholar]
- 57. Grbavec D., Stifani S. (1996) Molecular interaction between TLE1 and the C-terminal domain of HES-1 containing the WRPW motif. Biochem. Biophys. Res. Commun. 223, 701–705 [DOI] [PubMed] [Google Scholar]
- 58. Palaparti A., Baratz A., Stifani S. (1997) The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined N-terminal silencing domain of histone H3. J. Biol. Chem. 272, 26604–26610 [DOI] [PubMed] [Google Scholar]
- 59. Prozorovski T., Schulze-Topphoff U., Glumm R., Baumgart J., Schröter F., Ninnemann O., Siegert E., Bendix I., Brüstle O., Nitsch R., Zipp F., Aktas O. (2008) Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat. Cell Biol. 10, 385–394 [DOI] [PubMed] [Google Scholar]
- 60. Kawahara T. L., Michishita E., Adler A. S., Damian M., Berber E., Lin M., McCord R. A., Ongaigui K. C., Boxer L. D., Chang H. Y., Chua K. F. (2009) SIRT6 links histone H3 lysine 9 deacetylation to NF-κB-dependent gene expression and organismal life span. Cell 136, 62–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rothgiesser K. M., Erener S., Waibel S., Lüscher B., Hottiger M. O. (2010) SIRT2 regulates NF-κB dependent gene expression through deacetylation of p65 Lys310. J. Cell Sci. 123, 4251–4258 [DOI] [PubMed] [Google Scholar]
- 62. Legutko A., Marichal T., Fiévez L., Bedoret D., Mayer A., de Vries H., Klotz L., Drion P. V., Heirman C., Cataldo D., Louis R., Thielemans K., Andris F., Leo O., Lekeux P., Desmet C. J., Bureau F. (2011) Sirtuin 1 promotes Th2 responses and airway allergy by repressing peroxisome proliferator-activated receptor-γ activity in dendritic cells. J. Immunol. 187, 4517–4529 [DOI] [PubMed] [Google Scholar]
- 63. Zhang T., Berrocal J. G., Frizzell K. M., Gamble M. J., DuMond M. E., Krishnakumar R., Yang T., Sauve A. A., Kraus W. L. (2009) Enzymes in the NAD+ salvage pathway regulate SIRT1 activity at target gene promoters. J. Biol. Chem. 284, 20408–20417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Siggers T., Chang A. B., Teixeira A., Wong D., Williams K. J., Ahmed B., Ragoussis J., Udalova I. A., Smale S. T., Bulyk M. L. (2012) Principles of dimer-specific gene regulation revealed by a comprehensive characterization of NF-κB family DNA binding. Nat. Immunol. 13, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang J., Wang X., Hussain S., Zheng Y., Sanjabi S., Ouaaz F., Beg A. A. (2007) Distinct roles of different NF-κB subunits in regulating inflammatory and T cell stimulatory gene expression in dendritic cells. J. Immunol. 178, 6777–6788 [DOI] [PubMed] [Google Scholar]
- 66. Chatterjee N., Sinha D., Lemma-Dechassa M., Tan S., Shogren-Knaak M. A., Bartholomew B. (2011) Histone H3 tail acetylation modulates ATP-dependent remodeling through multiple mechanisms. Nucleic Acids Res. 39, 8378–8391 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.