Abstract
INTRODUCTION
Paraplegia results in significant skeletal muscle atrophy through increases in skeletal muscle protein breakdown. Recent work has identified a novel SIRT1-p53 pathway that is capable of regulating autophagy and protein breakdown.
METHODS
Soleus muscle was collected from 6 male Sprague-Dawley rats 10 weeks following complete T(4)-T(5) spinal-cord transection (paraplegia) and 6 male sham-operated rats (control). We utilized immunoblotting methods to measure intracellular proteins and qRT-PCR to measure the expression of skeletal muscle microRNAs.
RESULTS
SIRT1 protein expression was 37% lower, and p53 acetylation (LYS379) was increased in the paraplegia rats (P<0.05). Atg7 and Beclin-1, markers of autophagy induction, were elevated in paraplegia compared to controls (P<0.05).
DISCUSSION
Severe muscle atrophy resulting from chronic paraplegia appears to increase skeletal muscle autophagy independent of SIRT1 signaling. We conclude that chronic paraplegia may cause an increase in autophagic cell-death and negatively impact skeletal muscle protein balance.
Keywords: SIRT1, atrophy, Atg7, Beclin-1, microRNA
INTRODUCTION
The ability of skeletal muscle to adapt to various loading and other environmental conditions is substantial. Muscle atrophy occurs at a fairly rapid rate in the initial period following a variety of disuse interventions, such as hindlimb suspension or spinal cord transection 1–6. Of the total soleus muscle weight loss that occurs following spinal cord transection, 60% is lost within the first 10 days, illustrating the rapid dysregulation of protein synthesis and breakdown 4. Considerable research has explored the regulation of muscle protein breakdown during muscle atrophy 7, and our lab recently illustrated a down regulation of the mTORC1 pathway governing protein synthesis in a model of chronic paraplegia 8.
Much of the work exploring proteolysis in atrophying muscle has focused on the ubiquitin proteasome system (UPS) and the contribution of the two primary muscle E3 ubiquitin ligases, MuRF1 (muscle RING finger 1) and MAFbx (Atrogin-1) to muscle protein breakdown during atrophy 9–11. Less is known regarding the involvement of autophagy in proteolysis and cellular homeostasis during skeletal muscle atrophy. Increased expression of the autophagy protein Beclin-1 was observed in the tibialis anterior (TA) muscle of rats following 7 days of denervation, and it was suggested that there was an increase in autophagic cell-death during muscle disuse 12.
Autophagy is a catabolic pathway capable of degrading cellular membranes, organelles and proteins in order to remove damaged or defective components in the cell 13, and through this process, the primary role of autophagy appears to be cellular housekeeping. In conjunction with the proteasome, autophagy is involved in protein turnover, which helps maintain the energy balance of the cell. However, autophagocytosis seems to be a double-edged sword which protects cells when it is moderately activated, but excessive activation can induce autophagic cell death 14–16.
The mammalian family of sirtuin proteins, SIRT1–7, are involved in the regulation of numerous cellular processes, including aging, metabolism, growth, survival and differentiation 17–20. A direct role for SIRT1 in the regulation of starvation-induced autophagy has been observed recently 21. SIRT1 deacetylates FoxO3a in the regulation of cellular stress, 22–23 and autophagic and proteasomal pathways are regulated in coordination via FoxO3a in atrophying muscle as deacetylated FoxO3a can induce expression of LC3 expression 24–25. Transcriptional activity of FoxO3a is also influenced via phosphorylation by Akt 26. On the other hand, p53 is also a known repressor of autophagy 27–29, and SIRT1 deacetylation of p53 inactivates its transcriptional capabilities 30. It is not known whether skeletal muscle SIRT1 expression and muscle autophagy are increased following prolonged inactivity (e.g., spinal cord injury).
Individuals with spinal cord injury are the most physically inactive members of society and are placed at the lowest end of the human fitness spectrum 31–32. The life expectancy of individuals with spinal cord injuries is increasing to near that of healthy bodied persons 33, thus interventions to improve cellular homeostasis and maintain skeletal muscle mass are clinically relevant. Improving muscle mass will help improve whole body metabolism and may aid in the rehabilitation of individuals with spinal cord injuries. Our purpose in this study was to assess the activation of skeletal muscle autophagy and its regulation via SIRT1 in a model of skeletal muscle induced atrophy. We hypothesized that chronic paraplegia would increase skeletal muscle autophagy by a SIRT1-dependent mechanism.
MATERIALS AND METHODS
Study design
All surgical procedures were reviewed and approved by the Institutional Animal Care and Use Committee at Wayne State University School of Medicine (Detroit, MI). Twelve male Sprague-Dawley rats (24 ± 0.2 wk of age at death) were separated into 2 groups: complete spinal cord transection between thoracic level 4 and 5 (T4–T5, paraplegia, n = 6) or sham spinal cord transection (control, n = 6). Following 10 wk of spinal cord transected-induced paralysis or sham-operated free living, the rats were deeply anesthetized by intraperitoneal injection of sodium pentobarbital (approximately 75 mg/kg body weight) at the same time of day (early morning), and the soleus muscle was isolated, removed and quickly freeze clamped, immediately frozen in liquid nitrogen, and stored at −80°C until analyzed. Subsequently, death was assured by excision of the heart. Soleus muscles were homogenized (1:9 wt/vol) in a buffer containing 50 mM Tris-HCl, 250 mM mannitol, 50 mM NaF, 5 mM Na pyrophosphate, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, pH 7.4, 1 mM DTT, 1 mM benzamadine, 0.1 mM PMSF, and 5 μg/ml of soybean trypsin inhibitor with protease inhibitors and reducing agents added just before use. Total protein concentrations were determined for all supernatants with the Bradford assay using a Smartspec spectrophotometer (Bio-Rad) on the day of homogenization.
Induced paraplegia
All surgical procedures were performed using aseptic techniques. Rats were anesthetized with pentobarbital sodium (45 mg/kg ip), and supplemental doses (10–20 mg/kg ip) were administered if rats regained the blink reflex or responded during the surgical procedure. After anesthesia, rats were positioned prone over a thoracic roll resulting in a moderately flexed spine. The T4 and T5 vertebrae were exposed via a midline dorsal incision. The underlying spinal cord between T4 and T5 was completely transected through the intervertebral space as described previously 34–35. The control animals (sham operated) had the spinal cord exposed in an identical procedure, however, the spinal cord was not transected. This approach allowed for minimal impact on the stability of the vertebral column. During the first week of recovery, all rats were handled several times daily. During the handling periods, visual inspection and physical manipulations were performed to prevent pressure ulcers. Additionally, bladder voiding was accomplished by manual compression, after which all animals were weighed. After the first week of recovery, handling was reduced to one time each day, and bladders no longer required manual compression for voiding. At day 7, rats underwent a motor activity score using previously described criteria 36. Briefly, the motor activity score was determined by placing the animal on a table and observing hindlimb movement for 1 min. Scores ranged from 0 to 5; a 5 indicated normal walking and a 0 indicated zero weight bearing or spontaneous movement specific to the hindlimbs. All paralyzed rats had a score of 0.
SDS-PAGE and immunoblotting
Details of the immunoblotting procedures have been published previously37. Briefly, aliquots from homogenates (described above) were boiled at 100°C for 3 min in 2X sample buffer (2XSB) containing 125 mM Tris, pH 6.8, 25% glycerol, 2.5% SDS, 2.5%-mercaptoethanol, and 0.002% bromophenol blue. Samples containing 50 μg of total protein per lane were loaded in duplicate and separated by SDS-PAGE for 1 hr at 150 V using 7.5 or 15% gels on Criterion electrophoresis cell. Following SDS-PAGE, proteins were transferred to polyvinylidene diflouride membranes (PVDF) (Hybond-P, Amersham Biosciences, Piscataway, NJ) at 50 V for 1 hr. Verification of loading and transfer uniformity for the western blots was confirmed by Ponceau S staining. Once transferred, PVDF membranes were placed in blocking buffer [5% nonfat dry milk (NFDM) in TBST (Tris-buffered saline and 0.1% Tween 20)] for 1 hr. Blots were then washed serially 2 times in deionized water and 2 more times in TBST and incubated with primary antibody in 5% NFDM in TBST overnight at 4°C with constant agitation. Blots were washed in TBST twice and incubated with secondary antibody for 1 hr in 5% NFDM in TBST at room temperature with constant agitation. Blots were washed for 15 min followed by three additional washes lasting 5 min with TBST. Blots were then incubated for 5 min with enhanced chemiluminescence reagent (ECL plus Western Blotting Detection System, Amersham Biosciences, Piscataway, NJ) to detect horseradish peroxidase activity. Optical density measurements were obtained with a charge coupled device camera mounted in a ChemiDoc XRS imaging system (Bio-Rad, Hercules, CA). Once the appropriate image was captured, Densitometric analysis was performed using Quantity One 1-D analysis Software (version 4.5.2, Bio-Rad). Data are expressed as the raw value of the band minus a representative background sample from the membrane, divided by an internal loading control (50 μg/lane) loaded on every gel for comparisons across membranes.
Antibodies
The primary antibodies used were all purchased from Cell Signaling (Beverly, MA): acetylated-p53 [Lys379 (catalog number 2570); 1:500]; p53 (catalog number 2527; 1:500); SIRT1 (catalog number 2493; 1:1000); Atg7 (catalog number 2631; 1:1000); Beclin-1 (catalog number 3738; 1:1000); LC3B (catalog number 4108; 1:1000); p-FoxO3a [Ser253 (catalog number 9466); 1:500]; FoxO3a (catalog number 2497; 1:500). Anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody was purchased from Amersham Bioscience (catalog number NA9340; 1:2000).
Real time PCR
RNA was isolated from 6 paraplegia and 6 control rats. Details of the RNA extraction, cDNA synthesis and semi-quantitative real-time PCR procedures have been published previously 38. Briefly, RNA concentration was assessed using the NanoDrop 2000 (Thermo Scientific, West Palm Beach, FL). The relative expression levels of miR-9, -30a and −34a were determined using Taqman MicroRNA Assay (Applied BioSystems, Foster City, CA) according to the manufacturer’s directions. To account for any starting difference in the amount of total RNA, gene expression levels were normalized to 4.5S RNA. Relative fold changes were determined from the Ct values using the 2−ΔΔCt method39.
Statistical analysis
All values are expressed as means ± SE. All comparisons between control and paraplegic rats were performed with an independent t-test using SigmaStat statistical software version 11.0 (Systat Software Inc, San Jose, CA, USA). Significance was set at P < 0.05.
RESULTS
Whole body and soleus muscle weight
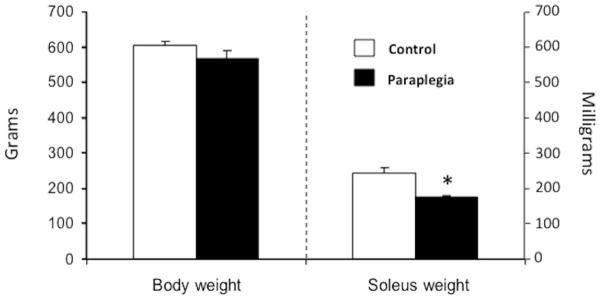
Initial body weight (~521 g) was not different between groups (data not shown; P = 0.8). Ten weeks later whole body weight was not different between paraplegic and control rats (568 ± 23 vs. 605 ± 11 g, respectively; P = 0.18; Fig. 1). Soleus muscle weight, however, was 28% lower in the paraplegic rats compared with control rats (175 ± 6 vs. 244 ± 17 mg, respectively; P = 0.003; Fig. 1).
Figure 1.
Whole body and soleus muscle weight. Whole body weight (g) and soleus muscle weight (mg) were measured in the Paraplegia group 10 weeks following complete spinal cord transection and in the Control group. Values are mean ± SE. *Significantly different from control (P < 0.05).
SIRT1 signaling
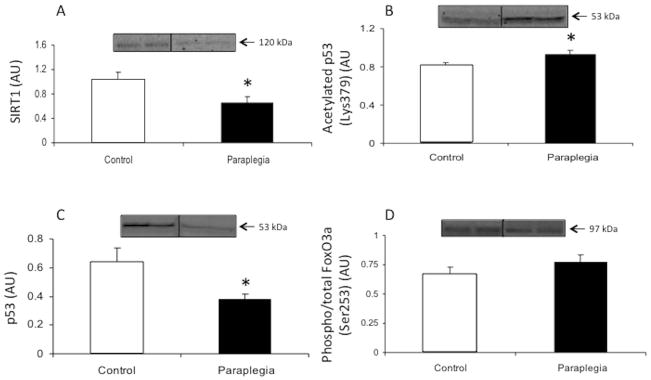
Total protein expression of SIRT1 was reduced 37% in the paraplegia group compared to the control rats (P = 0.05; Fig. 2A). Acetylation of p53 (Lys379) was increased in the paraplegia group compared to control rats (P = 0.03; Fig. 2B), however total p53 protein was lower in the paraplegia rats compared to the control group (P = 0.03; Fig. 2C). FoxO3a phosphorylation (Ser253) was not different between groups (P = 0.27; Fig. 2D).
Figure 2.
SIRT1 signaling in soleus muscle from Control and Paraplegic rats. Data represent SIRT1 total protein (A), acetylated p53 (Lys379) (B), p53 total protein (C) and phospho-FoxO3a (Ser253) (D). Values are mean ± SE.
*Significantly different from control (P < 0.05).
Markers of autophagy induction
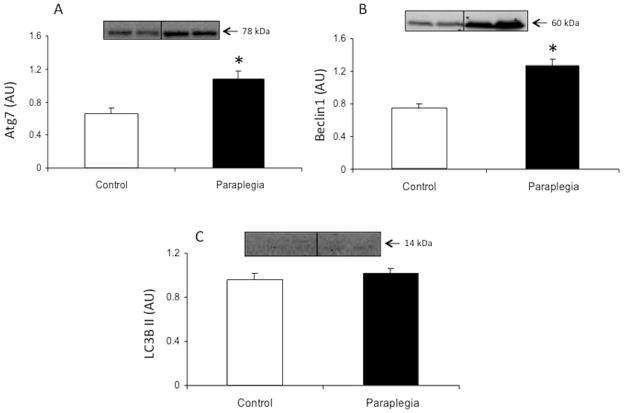
Total protein content of Atg7 was increased in the paraplegia rats compared to the control group (P = 0.01; Fig. 3A). Beclin-1 protein expression was also increased in the paraplegia rats compared to the control group (P = 0.001; Fig. 3B). Total protein levels of LC3B II were not different between groups (P = 0.41; Fig. 3C).
Figure 3.
Markers of autophagy induction in soleus muscle from Control and Paraplegic rats. Data represent Atg7 (A), Beclin-1 (B) and LC3B II (C) total protein. Values are mean ± SE.
*Significantly different from control (P < 0.05).
MicroRNA expression
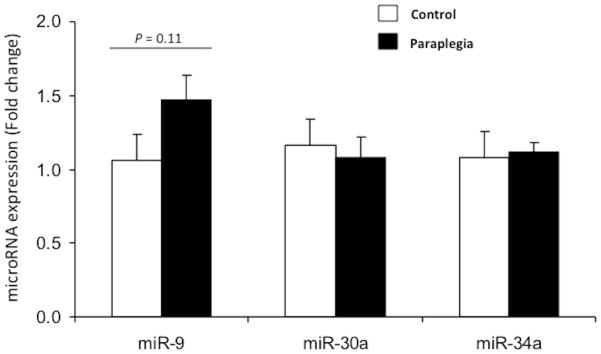
Expression levels of miR-30a and −34a were not different between groups (P = 0.79 and P = 0.85, respectively; Fig. 4). However, miR-9 expression was 47% higher in the paraplegia rats (P = 0.11).
Figure 4.
MicroRNA expression of miR-9, miR-30a and miR-34a in soleus muscle were measured in the Paraplegia group 10 weeks following complete spinal cord transection and in the Control group. Values are mean ± SE.
DISCUSSION
Following hindlimb suspension or spinal cord transection, there is a rapid onset of skeletal muscle wasting due to imbalances that occur in the rates of protein turnover 1–6. Our primary finding was an upregulation of markers of autophagy induction in the soleus muscle of rats 10 weeks following complete T(4)-T(5) spinal cord transection. Furthermore, we found a downregulation of SIRT1 protein expression and signaling, suggesting that the upregulation of autophagy in a chronic paraplegia model is independent of SIRT1.
Degradation of damaged cellular organelles and membranes is accomplished through autophagy 13, an evolutionarily conserved cellular housekeeping process. Autophagy, along with the proteasome, also contributes to protein breakdown in order to remove old and defective proteins and maintain cellular energy balance. Autophagy has also emerged as a promising target in longevity studies 40, and recent work has identified a direct role of SIRT1 in the regulation of autophagocytosis 21. In this study, we observed a decrease in the expression of SIRT1 and increased levels of acetylated p53 in the paraplegic rats. These findings illustrate decreased protein content and activity of SIRT1 in our model of chronic paraplegia. SIRT1, a member of the sirtuin family of histone deacetylases, is involved in numerous aspects of cellular metabolism, growth, differentiation and senescence 17–20. The study by Lee et al. demonstrated that SIRT1 is capable of forming complexes with and deacetylating several autophagy regulators, including Atg5, Atg7 and Atg8, and in the absence of SIRT1, fibroblasts were unable to stimulate autophagy during starvation 21. In contrast to our findings, Chabi et al used a short-term denervation model and found that 7, 14 and 21 days of denervation increased the expression of SIRT1 in skeletal muscle 41. It is likely that the increase in SIRT1 expression during the acute stages of denervation plays a role in both downregulation of mitochondrial biogenesis and induction of muscle autophagy. However, with prolonged inactivity (such as occurs with spinal cord injury in this study) the increase in muscle autophagy appears to be independent of SIRT1.
In addition to its direct ability to influence autophagocytosis, SIRT1 can also influence autophagy induction through its interaction with p53 and FoxO3a, known upstream effectors of autophagy 22–23,27–29. We were unable to measure the acetylation of FoxO3a, however, we used the phosphorylation of FoxO3a via Akt as an indicator of its transcriptional activity 26. We did not observe any differences in FoxO3a phosphorylation in the 2 groups. Acetylation of p53 is a marker for the deacetylase activity of SIRT1 30, and in this study we observed increased acetylation of p53 in the paraplegic rats, corresponding with lower total levels of SIRT1 protein. Acetylation of p53 increases its transcriptional activity, and p53 has been shown to be a negative regulator of autophagy. 29,28 In this study, the acetylation of p53 was greater in the paraplegic rats, and the paraplegic rats also had elevated expression of autophagy markers, suggesting dysregulation of the SIRT1-p53 mediated control of autophagy in chronic paraplegia 42. We have previously shown a downregulation of the mTOR signaling pathway following 10 weeks of complete T(4)-T(5) spinal cord transection 8, which contribute to the increase in autophagy induction in the current study, as the Akt-mTOR pathway is a negative regulator of autophagy 43.
While results from this study indicate reduced activation of SIRT1 signaling in our model of chronic paraplegia, the same rats had increased levels of autophagy-specific proteins (Atg7 and Beclin-1) compared to the control group. The increase in Atg7 and Beclin-1 expression may indicate an increase in autophagy mediated protein degradation. In healthy systems, increasing SIRT1 activity and autophagy can improve longevity and cellular homeostasis 44–46, but excessive activation can induce autophagic cell death 14–16. In a recent study, O’Leary et al. also found elevated levels of the autophagic protein Beclin-1 in skeletal muscle after 7 days of denervation in rats 12. The authors suggested that there was an increase in autophagic cell death that was contributing to muscle atrophy during denervation 12. As the majority of muscle loss occurs in the early period following spinal cord transection 4, it is unlikely that the increase in autophagy we observed 10 weeks after transection contributes greatly to loss of muscle due to autophagic cell death. However, our data provide intriguing evidence for an increased role of autophagy in a model of chronic paraplegia. Much of the rapid muscle atrophy immediately following spinal cord transection is attributed to increases in protein breakdown through the UPS 9–11, however, autophagy may play a more prominent role in regulating protein degradation during prolonged physical inactivity and/or paraplegia. Increased protein degradation through autophagic processes may contribute to decreased cellular size in paraplegia. Upregulation of autophagic proteins may also serve to improve cellular survival in the absence of neural input.
Upstream activity of SIRT1 and autophagy, specifically autophagic protein Beclin-1, can be regulated through various microRNAs, including miR-9, -30a and -34a 47–50. MicroRNAs are a class of endogenous, 22–24 nucleotide RNA molecules with the ability to induce mRNA degradation, translational repression, or both, via pairing with partially complementary sites in the 3' UTR of the targeted genes 51. The miRNAs measured in this study can negatively impact the expression of SIRT1 and Beclin-1 47–50. Although we found no significant differences in the expression of miR-30a and -34a between the control and paraplegia groups, miR-9 tended to be elevated (P=0.11) suggesting that this specific miRNA may play a modest role in the regulation of SIRT1 signaling and the induction of muscle autophagy 10 weeks following complete spinal cord transection.
Soleus muscle atrophy following 10 weeks of paraplegia induces dysregulation of SIRT1 signaling and increases skeletal muscle autophagy. We propose that the increase in autophagy plays a role in regulation of muscle size and metabolism during prolonged inactivity. Based on our data, we conclude that chronic paraplegia increases markers of autophagy in skeletal muscle independent of SIRT1, illustrating the potential for aberrant autophagy to contribute to muscle protein degradation during atrophy associated with spinal cord injury.
Acknowledgments
We would like to thank JunFang Hao for technical assistance. This study was supported by grants from NIAMS to BBR (AR049877), NHLBI to SED (HL088615) and NIH T32-HD07539.
ABREVIATIONS
- 2XSB
2X sample buffer
- Atg5
autophagy gene 5
- Atg7
autophagy gene 7
- Atg8
autophagy gene 8
- Ct
cycle threshold
- DTT
dithiothreitol
- ECL
enhanced chemiluminescence
- FoxO3a
forkhead box O3a
- hr
hour
- ip
intraperitoneal
- LC3B
light chain 3
- MAFbx
muscle atrophy factor 1; atrogin-1
- miRNA
microRNA
- min
minute
- mTORC1
mammalian target of rapamycin complex 1
- MuRF1
muscle RING finger 1
- NFDM
nonfat dry milk
- PMSF
phenylmethylsulfonylfluoride
- PVDF
polyvinylidene difluoride
- RT-PCR
real time polymerase chain reaction
- SDS-PAGE
sodium dodecylsulfate-polyacrymalide gel electrophoresis
- SE
standard error of the mean
- SIRT1
sirtuin 1
- T(4)-T(5)
thoracic level 4 and 5
- TA
tibialis anterior
- TBST
Tris buffered saline with 0.1% Tween-20
- UPS
ubiquitin proteasome system
- wk
week
References
- 1.Giger JM, Bodell PW, Zeng M, Baldwin KM, Haddad F. Rapid muscle atrophy response to unloading: pretranslational processes involving MHC and actin. J Appl Physiol. 2009;107(4):1204–1212. doi: 10.1152/japplphysiol.00344.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haddad F, Roy RR, Zhong H, Edgerton VR, Baldwin KM. Atrophy responses to muscle inactivity. I. Cellular markers of protein deficits. J Appl Physiol. 2003;95(2):781–790. doi: 10.1152/japplphysiol.00317.2003. [DOI] [PubMed] [Google Scholar]
- 3.Haddad F, Roy RR, Zhong H, Edgerton VR, Baldwin KM. Atrophy responses to muscle inactivity. II. Molecular markers of protein deficits. J Appl Physiol. 2003;95(2):791–802. doi: 10.1152/japplphysiol.01113.2002. [DOI] [PubMed] [Google Scholar]
- 4.Dupont-Versteegden EE, Houle JD, Gurley CM, Peterson CA. Early changes in muscle fiber size and gene expression in response to spinal cord transection and exercise. Am J Physiol Cell Physiol. 1998;275(4):C1124–C1133. doi: 10.1152/ajpcell.1998.275.4.C1124. [DOI] [PubMed] [Google Scholar]
- 5.Diffee GM, Caiozzo VJ, Herrick RE, Baldwin KM. Contractile and biochemical-properties of rat soleus and plataris after hindlimb suspension. Am J Physiol. 1991;260(3):C528–C534. doi: 10.1152/ajpcell.1991.260.3.C528. [DOI] [PubMed] [Google Scholar]
- 6.Diffee GM, Haddad F, Herrick RE, Baldwin KM. Ccontrol fo myosin heavy-chain expression - interaction of hypothyroidism and hindlimb suspension. Am J Physiol. 1991;261(6):C1099–C1106. doi: 10.1152/ajpcell.1991.261.6.C1099. [DOI] [PubMed] [Google Scholar]
- 7.Kandarian SC, Jackman RW. Intracellular signaling during skeletal muscle atrophy. Muscle Nerve. 2006;33(2):155–165. doi: 10.1002/mus.20442. [DOI] [PubMed] [Google Scholar]
- 8.Dreyer HC, Glynn EL, Lujan HL, Fry CS, DiCarlo SE, Rasmussen BB. Chronic paraplegia-induced muscle atrophy downregulates the mTOR/S6K1 signaling pathway. J Appl Physiol. 2008;104(1):27–33. doi: 10.1152/japplphysiol.00736.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bodine SC, Latres E, Baumhueter S, Lai VKM, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na EQ, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294(5547):1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 10.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci U S A. 2001;98(25):14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18(1):39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 12.O'Leary MFN, Hood DA. Effect of prior chronic contractile activity on mitochondrial function and apoptotic protein expression in denervated muscle. J Appl Physiol. 2008;105(1):114–120. doi: 10.1152/japplphysiol.00724.2007. [DOI] [PubMed] [Google Scholar]
- 13.Xie ZP, Klionsky DJ. Autophagosome formation: Core machinery and adaptations. Nat Cell Biol. 2007;9(10):1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 14.Song CW, Lee SJ, Jeen YT, Chun HJ, Um SH, Kim CD, Ryu HS, Hyun JH, Lee MS, Kahrilas PJ. Inconsistent association of esophageal symptoms, psychometric abnormalities and dysmotility. Am J Gastroenterol. 2001;96(8):2312–2316. doi: 10.1111/j.1572-0241.2001.04035.x. [DOI] [PubMed] [Google Scholar]
- 15.Kundu JK, Surh YJ. Cancer chemopreventive and therapeutic potential of resveratrol: Mechanistic perspectives. Cancer Lett. 2008;269(2):243–261. doi: 10.1016/j.canlet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- 16.Ferraro E, Cecconi F. Autophagic and apoptotic response to stress signals in mammalian cells. Arch Biochem Biophys. 2007;462(2):210–219. doi: 10.1016/j.abb.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature. 2006;444(7121):868–874. doi: 10.1038/nature05486. [DOI] [PubMed] [Google Scholar]
- 18.Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell. 2006;126(2):257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA. SIRT1 Redistribution on Chromatin Promotes Genomic Stability but Alters Gene Expression during Aging. Cell. 2008;135(5):907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105(9):3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berdichevsky A, Viswanathan M, Horvitz HR, Guarente L. C. elegans SIR-2. 1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125(6):1165–1177. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- 23.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin YX, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303(5666):2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 24.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6(6):458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the Autophagic/Lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6(6):472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 26.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a forkhead transcription factor. Cell. 1999;96(6):857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 27.Morselli E, Tasdemir E, Maiuri MC, Galluzzi L, Kepp O, Criollo A, Vicencio JM, Soussi T, Kroemer G. Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle. 2008;7(19):3056–3061. doi: 10.4161/cc.7.19.6751. [DOI] [PubMed] [Google Scholar]
- 28.Tasdemir E, Maiuri MC, Orhon I, Kepp O, Morselli E, Criollo A, Kroemer G. p53 represses autophagy in a cell cycle-dependent fashion. Cell Cycle. 2008;7(19):3006–3011. doi: 10.4161/cc.7.19.6702. [DOI] [PubMed] [Google Scholar]
- 29.Tavernarakis N, Pasparaki A, Tasdemir E, Maiuri MC, Kroemer G. The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy. 2008;4(7):870–873. doi: 10.4161/auto.6730. [DOI] [PubMed] [Google Scholar]
- 30.Vaziri H, Dessain SK, Eagon EN, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107(2):149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 31.Dearwater SR, Laporte RE, Robertson RJ, Brenes G, Adams LL, Becker D. Aactivity in the spinal cord-injured patient - an epidemiological analysis of metabolic parameters. Med Sci Sports Exerc. 1986;18(5):541–544. [PubMed] [Google Scholar]
- 32.Ginis KAM, Latimer AE, Buchholz AC, Bray SR, Craven BC, Hayes KC, Hicks AL, McColl MA, Potter PJ, Smith K, Wolfe DL. Establishing evidence-based physical activity guidelines: methods for the Study of Health and Activity in People with Spinal Cord Injury (SHAPE SCI) Spinal Cord. 2008;46(3):216–221. doi: 10.1038/sj.sc.3102103. [DOI] [PubMed] [Google Scholar]
- 33.Devivo MJ, Shewchuk RM, Stover SL, Black KJ, Go BK. A cross-sectional study of the relationship between age and current health-status for persons with spinal cord injuries. Paraplegia. 1992;30(12):820–827. doi: 10.1038/sc.1992.158. [DOI] [PubMed] [Google Scholar]
- 34.Collins HL, DiCarlo SE. Acute exercise reduces the response to colon distension in T-5 spinal rats. Am J Physiol Heart Circ Physiol. 2002;282(4):H1566–H1570. doi: 10.1152/ajpheart.00733.2001. [DOI] [PubMed] [Google Scholar]
- 35.Collins HL, DiCarlo SE. TENS attenuates response to colon distension in paraplegic and quadriplegic rats. Am J Physiol Heart Circ Physiol. 2002;283(4):H1734–H1739. doi: 10.1152/ajpheart.00253.2002. [DOI] [PubMed] [Google Scholar]
- 36.vonEuler M, Akesson E, Samuelsson EB, Seiger A, Sundstrom E. Motor performance score: A new algorithm for accurate behavioral testing of spinal cord injury in rats. Exp Neurol. 1996;137(2):242–254. doi: 10.1006/exnr.1996.0023. [DOI] [PubMed] [Google Scholar]
- 37.Dreyer HC, Fujita S, Cadenas JG, Chinkes DL, Volpi E, Rasmussen BB. Resistance exercise increases AMPK activity and reduces 4E-BP1 phosphorylation and protein synthesis in human skeletal muscle. J Physiol. 2006;576(2):613–624. doi: 10.1113/jphysiol.2006.113175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drummond MJ, McCarthy JJ, Fry CS, Esser KA, Rasmussen BB. Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Am J Physiol Endocrinol Metab. 2008;295(6):E1333–E1340. doi: 10.1152/ajpendo.90562.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(−Delta Delta C) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Yen WL, Klionsky DJ. How to Live Long and Prosper: Autophagy, Mitochondria, and Aging. Physiology. 2008;23(5):248–262. doi: 10.1152/physiol.00013.2008. [DOI] [PubMed] [Google Scholar]
- 41.Chabi B, Adhihetty PJ, O'Leary MFN, Menzies KJ, Hood DA. Relationship between Sirt1 expression and mitochondrial proteins during conditions of chronic muscle use and disuse. J Appl Physiol. 2009;107(6):1730–1735. doi: 10.1152/japplphysiol.91451.2008. [DOI] [PubMed] [Google Scholar]
- 42.Salminen A, Kaarniranta K. SIRT1: Regulation of longevity via autophagy. Cell Signal. 2009;21(9):1356–1360. doi: 10.1016/j.cellsig.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 43.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273(7):3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 44.Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C-elegans. Science. 2003;301(5638):1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 45.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410(6825):227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 46.Hars ES, Qi HY, Ryazanov AG, Jin SK, Cai L, Hu CC, Liu LF. Autophagy regulates ageing in C-elegans. Autophagy. 2007;3(2):93–95. doi: 10.4161/auto.3636. [DOI] [PubMed] [Google Scholar]
- 47.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105(36):13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mukhopadhyay P, Pacher P, Das DK. MicroRNA signatures of resveratrol in the ischemic heart. Ann N Y Acad Sci. 2011;1215:109–116. doi: 10.1111/j.1749-6632.2010.05866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu H, Wu H, Liu XP, Li BA, Chen Y, Ren XC, Liu CG, Yang JM. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy. 2009;5(6):816–823. doi: 10.4161/auto.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saunders LR, Sharma AD, Tawney J, Nakagawa M, Okita K, Yamanaka S, Willenbring H, Verdin E. miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging. 2010;2(7):415–431. doi: 10.18632/aging.100176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3' untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16(2):144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]