Abstract
In the last years, there was an exponential increase in the number of publicly available genomes. Once finished, most genome projects lack financial support to review annotations. A few of these gene annotations are based on a combination of bioinformatics evidence, however, in most cases, annotations are based solely on sequence similarity to a previously known gene, which was most probably annotated in the same way. As a result, a large number of predicted genes remain unassigned to any functional category despite the fact that there is enough evidence in the literature to predict their function. We developed a classifier trained with term-frequency vectors automatically disclosed from text corpora of an ensemble of genes representative of each functional category of the J. Craig Venter Institute Comprehensive Microbial Resource (JCVI-CMR) ontology. The classifier achieved up to 84% precision with 68% recall (for confidence≥0.4), F-measure 0.76 (recall and precision equally weighted) in an independent set of 2,220 genes, from 13 bacterial species, previously classified by JCVI-CMR into unambiguous categories of its ontology. Finally, the classifier assigned (confidence≥0.7) to functional categories a total of 5,235 out of the ∼24 thousand genes previously in categories “Unknown function” or “Unclassified” for which there is literature in MEDLINE. Two biologists reviewed the literature of 100 of these genes, randomly picket, and assigned them to the same functional categories predicted by the automatic classifier. Our results confirmed the hypothesis that it is possible to confidently assign genes of a real world repository to functional categories, based exclusively on the automatic profiling of its associated literature. The LitProf - Gene Classifier web server is accessible at: www.cebio.org/litprofGC.
Introduction
In the last years, there was an exponential increase in the number of publicly available genomes. Once finished, most genome projects lack financial support to review annotations. Extracting knowledge from genome sequencing efforts requires the predicted genes to be functionally annotated. A few of these gene or genome annotations are based on a combination of computationally derived evidence, such as metabolic reconstruction, presence of candidate transcription factor binding sites or even the fact that functionally related genes tend to cluster on prokaryotic chromosomes [1]. However, in most cases, annotations are based solely on sequence similarity to a previously known gene, which was most probably annotated in the same way [2]. In addition, it is often difficult to find the genes that were experimentally validated to evaluate the reliability of these original annotations [3]. In cases where the reference sequence has no annotation or is annotated as “Unknown function”, or even is incorrectly annotated, then all sequences subsequently annotated based on their similarity to the references will inherit their inexactly assigned attributes. Current bioinformatics-based approaches cannot predict the function of up to one-third of sequenced genes. For prokaryote genomes deposited in the J. Craig Venter Institute Comprehensive Microbial Resource (JCVI-CMR) (http://cmr.jcvi.org/cgi-bin/CMR/shared/RoleList.cgi), genes classified as “Unknown function” account for ∼10% (26,390) of the total of deposited genes. Those assigned to the category “Unclassified” account for ∼18% (45,870). The “Unknown Function” and “Unclassified” genes represent ∼30% of all unique prokaryotic genes at JCVI-CMR. Furthermore, for some gene families, at least 60% of the gene predictions are wrong [4]. For many of these genes there is sufficient evidence in the literature to identify their function [5]. However, the costs involved in manually reviewing the literature to improve gene annotation in a whole genome sequencing project are prohibitive. Text-mining algorithms can help in this task.
We present, herein, LitProf – Gene classifier, a tool for automatically assigning prokaryote genes to functional categories of the JCVI-CMR ontology (File S1) based exclusively on the literature profiles extracted from their gene-specific collections of abstracts in MEDLINE database (http://www.pubmed.org). Using LitProf – Gene classifier we were able to propose functional categories to 5,235 out of the 69,088 genes previously assigned to categories “Unknown function” or “Unclassified” of the JCVI-CMR ontology.
Results
Disclosing the informative vocabulary required to describe each functional category
Starting from 2,201,517 loci from JCVI-CMR repository we filtered out genes that do not have a name (80%), and genes assigned to more than one functional category of JCVI-CMR ontology (12%). Were also genes assigned to the categories “unknown function”, “unclassified” and “disrupted reading frame”, which combined accounts for approximately 16% of all loci. For each group of homonymous genes we kept only one randomly chosen representative. The final dataset consisted of 59,830 unique canonical gene names from 117 genomes representing all prokaryote phylogenetic branches in JCVI-CMR (Table 1). After the redundancy reduction steps, the resultant gene set comprised 3,542 genes preserving the proportional contribution of each functional categories in JCVI-CMR (Table 2). LitProf retrieved 126,990 MEDLINE abstracts for the selected 3,542 genes (average = 35.8 abstracts per gene). From this text corpus LitProf disclosed the minimum informative vocabulary of 889 stemmed terms, and represented each gene as a term-frequency vector.
Table 1. Taxonomy distribution of genes in the training dataset.
| Phylum | # ofspecies |
| Acidobacteria | 1 |
| Actinobacteria | 5 |
| Aquificae | 1 |
| Bacteroidetes | 3 |
| Chlamydiae | 1 |
| Chlorobi | 1 |
| Chloroflexi | 4 |
| Crenarchaeota | 3 |
| Cyanobacteria | 9 |
| Deinococcus-Thermus | 2 |
| Euryarchaeota | 10 |
| Fibrobacteres | 1 |
| Firmicutes | 15 |
| Fusobacteria | 1 |
| Nanoarchaeota | 1 |
| Planctomycetes | 1 |
| Proteobacteria | 49 |
| Spirochaetes | 2 |
| Tenericutes | 3 |
| Thermotogae | 1 |
| Virus | 3 |
Table 2. Gene distribution in the functional categories of the JCVI-CMR ontology.
| Functional category | Dataset | ||
| Original (%) | Training (%) | Classified (%) | |
| Amino acid biosynthesis | 4102 (2.53) | 90 (2.54) | 115 (2.20) |
| Biosynthesis of cofactors, prosthetic groups, and carriers | 5482 (3,39) | 148 (4.18) | 179 (3.42) |
| Cell envelope | 227 (1.40) | 47 (1.33) | 41 (0.78) |
| Cellular processes | 17778 (10.99) | 353 (9.97) | 283 (5.41) |
| Central intermediar metabolism | 2517 (1.56) | 78 (2.20) | 92 (1.76) |
| DNA metabolism | 9238 (5.71) | 287 (8.10) | 344 (6.57) |
| Energy metabolism | 27132 (16.77) | 744 (21.01) | 1363 (26.04) |
| Fatty acid and phospholipid metabolism | 4823 (2.98) | 118 (3.33) | 253 (4.83) |
| Mobile and extrachromosomal element functions | 7716 (4.77) | 123 (3.47) | 119 (2.27) |
| Protein fate | 11611 (7.17) | 359 (10.14) | 550 (10.51) |
| Protein synthesis | 9044 (5.59) | 140 (3.95) | 149 (2.85) |
| Purines, pyrimidines, nucleosides, and nucleotides | 2678 (1.65) | 62 (1.75) | 77 (1.47) |
| Regulatory functions | 18817 (11.63) | 247 (6.97) | 327 (6.25) |
| Transcription | 2816 (1.74) | 86 (2.43) | 50 (0.96) |
| Transport and binding proteins | 25368 (15.68) | 363 (10.25) | 757 (14.46) |
| Mix category | 8297 (5.13) | 297 (8.39) | 536 (10.24) |
Only categories used to train the classifier are shown. Mix category regroups the noisy subcategories. The original column refers to the complete J. Craig Venter Institute Comprehensive Microbial Resource (JCVI-CMR). The training column refers to the dataset used to train the classifier. The classified column refers to the “Unknown function” and “Unclassified” genes that were classified by LitProf- Gene Classifier with confidence≥0.7. There is no significant difference between the original and training datasets (p>0.05 in paired t-test; confidence level of 95%).
SVM training
Genes represented by their term-frequency vectors together with the information about their respective functional categories, previously assigned to each gene by JCVI-CMR team, were used to train the SVM classifier. The “Signal transduction” category was underrepresented in our gene set, most probably because these genes are often assigned to more than one category in the JCVI-CMR. As a consequence, there were not enough genes assigned by JCVI-CMR only to the category “Signal transduction” making it impossible to train the SVM to recognize this category. For that reason this category was removed from the training dataset. During the cross-validation of the classifier we observed that the subcategories “Other” from categories “Cell envelope” and “Central intermediary metabolism” were frequently misclassified. For that reason, these subcategories were excluded. The subcategories “Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides” and “Biosynthesis of murein sacculus and peptidoglycan” from the category “Cell envelope”, “Biosynthesis and degradation of polysaccharides” from the category “Energy metabolism” and “Sugar-nucleotide biosynthesis and conversions” from category “Purines, pyrimidines, nucleosides, and nucleotides”, all related to polysaccharide biosynthesis added noise to the classifier and thus were merged into a new “Mix category”. In any case subcategories were exchanged between established JCVI-CMR categories. Thus, the category-subcategory hierarchy of the original JCVI-CMR ontology was not violated. After these adjustments, the rearranged ontology had 16 categories comprising 115 sub-categories.
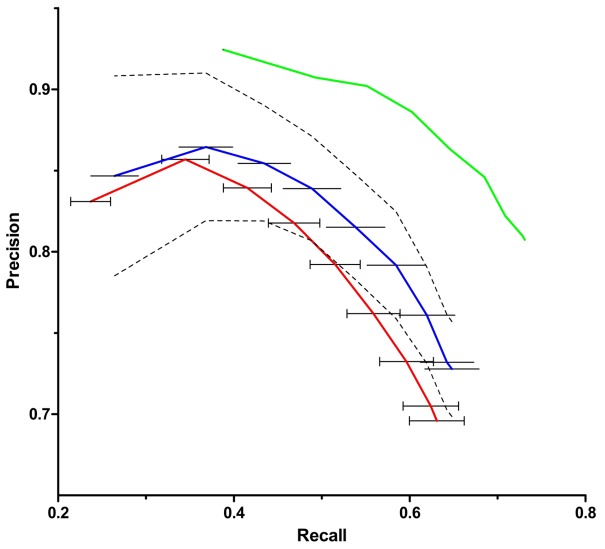
After the ontology rearrangement, the SVM classifier was retrained and cross-validation was performed again. The estimated average precision of the resultant classifier was 80±3%, and the average recall was 60±3% (confidence≥0.4); for equally weighted recall and precision, the F-measure was 0.7. Furthermore, for the independent gene set the classifier achieved 84% precision with 68% recall (F-measure = 0.76) for confidence≥0.4, and 90% precision with 55% recall (F-measure = 0.68) for confidence≥0.7, as shown in Figure 1.
Figure 1. Recall vs. precision of the classifier.
The red line represents the average performance of the initial classifier trained with the original categories of the JCVI-CMR ontology. The blue line, represents the average performance of the final classifier trained with a rearranged version of the ontology where noisy subcategories were merged together to create the Mix Category. For red and blue lines, the average was calculated from 100 replicates of 10-fold cross validation. The green line represents the performance of the final classifier in an independent gene set. Horizontal bars represent the standard deviations of recall. The dashed lines represent the standard deviation of precision for the blue curve.
It is important to highlight that the SVM classifier predicts categories, not subcategories. At present, there are not enough genes in JCVI-CMR assigned to only one functional subcategory so that we could have used them to compose a robust example gene set required to train the SVM with enough statistical support.
Classification of “Unknown function” and “Unclassified” genes
We used the classifier to tentatively classify the 69,088 genes previously assigned by JCVI-CMR to the categories “Unknown function” or “Unclassified”. However, only 34,033 genes had a name, and 23,973 of these genes had at least five abstracts in MEDLINE. Their text corpora retrieved by LitProf – Gene Classifier with the thresholds established in this experiment (min = 5 abstracts; max = 50) summed up 247,442 abstracts (average = 10.3 abstracts/gene). For a minimum confidence threshold of 0.7, LitProf – Gene Classifier unambiguously assigned 5,235 out of the ∼24 K genes with literature in MEDLINE to a functional category. Table 3 summarizes these results. For details on the classified genes see Table S3.
Table 3. Summary of the classification of genes previously assigned to categories “Unknown function” and “Unclassified” of the JCVI-CMR ontology.
| Filters | # of genes |
| Total number of “Unknown function” and “Unclassified” genes | 69,088 |
| Genes that have a name | 34,033 |
| Genes with at least five abstracts in MEDLINE | 23,973 |
| Classified genes (confidence threshold≥0.7) | 5,235 |
From the total number of “Unknown function” and “Unclassified” genes, nearly 50% have a name, with is crucial for text corpora retrieval. From those, ∼70% have enough literature (min = five abstracts; max = 50) for classification, and in this group, ∼22% could be assigned by LitProf - Gene Classifier to a functional category with high confidence.
JCVI-CMR = J. Craig Venter Institute Comprehensive Microbial Resource.
The classified genes span over all functional categories present in the training dataset. One-hundred randomly picked genes of this set were manually classified by two Biologists who reviewed their literature. Their manual classifications completely agreed with those of LitProf – Gene Classifier (see Table 4 for a sample of these results). No significant difference was found in the gene distribution in functional categories when comparing the classified gene set, the training gene set, and the original JCVI-CMR (Table 3).
Table 4. Examples of genes classified by LitProf- Gene Classifier and further validated by manually reviewing their literature.
| Name | JCVI-CMR Accession | Species | Predicted category | Confidence | PubmedIDs | GO Biological process (species with GO annotated ortholog) * |
| ArsR protein | NT01MC4786 | Magnetococcus sp. MC-1 | Regulatory functions | 0.98 | 20724137; 20586430 | GO:0006355: regulation of transcription, DNA-dependent (Pseudomonas aeruginosa PAO1) |
| Phosphatidylserine decarboxylase, putative | GSU_1908 | Geobacter sulfurreducens PCA | Fatty acid and phospholipids metabolism | 0.96 | 14651609; 16667073 | GO:0006660: phosphatidylserine catabolic process; GO:0004609: phosphatidylserine decarboxylase (Geobactersulfurreducens PCA) |
| UmuD protein [Contains: UmuD protein] | NT03PS1033 | Candidatus Protochlamydia amoebophila UWE25 | DNA metabolism | 0.97 | 14651609; 16667073 | GO:0009432: SOS response; GO:0009650: UV protection; GO:0008236: serine-type peptidase activity (Colwellia psychrerythraea 34H) |
| phage portal protein | NT03SP0558 | Streptococcus pyogenes MGAS8232 | Mobile and extrachromosomal element functions | 0.95 | 20467052; 19947526 | GO:0019068: virion assembly; GO:0019012: virion; GO:0005198: structural molecule activity (Clostridium perfringens ATCC13124) |
| Putative metalloprotease | pc0037 | Candidatus Protochlamydia amoebophila UWE25 | Protein fate | 0.94 | 20838651; 20812964 | GO:0006508: proteolysis; GO:0008233: peptidase activity (Methylococcus capsulatus str. Bath) |
| Lambda Kil | ECH74115_3562 | Escherichia coli O157:H7 str. EC4115 | Mobile and extrachromosomal element functions | 0.98 | 12441108; 11470529 | - |
| bacteriophage tail fiber assembly protein | NT06EC2684 | Erwinia carotovora atroseptica SCRI1043 | Mobile and extrachromosomal element functions | 0.95 | 20531477; 10051617 | - |
| staphylococcal respiratory response protein, SrrB | SAUSA300_1441 | Staphylococcus aureus subsp. aureus USA300-FPR3757 | Regulatory functions | 0.92 | 17697253; 17198402 | - |
| Clp amino terminal domain protein | NT01NFA0344 | Nocardia farcinica IFM10152 | Protein fate | 0.99 | 20014030; 19843523 | - |
| Putative malate dehydrogenase | nfa36620 | NocardiafarcinicaIFM10152 | Energy metabolism | 0.94 | 20127467; 19405028 | GO:0006108: malate metabolic process; GO:0016615: malate dehydrogenase activity (Bacillus anthracis) |
| (R)-2-hydroxyglutaryl-CoA dehydratase activator | NT01CA2639 | Clostridium acetobutylicumATCC824 | Regulatory functions | 0.76 | 11106419; 15374661 | GO:0006520: cellular amino acid metabolic process; GO:0008047: enzyme activator activity (Geobacter sulfurreducens PCA) |
| putative beta-lactamase II | NT05LB0990 | Leptospira biflexa serovar Patoc strain Patoc1 | Cellular processes | 0.58 | 19407375; 16452624 | GO:0017001: antibiotic catabolic process; GO:0008800: beta-lactamase activity (Bacillus anthracis) |
| Carbohydrate binding protein, cbp35C | CJA_0494 | Cellvibriojaponicus Ueda107 | Transport and binding proteins | 0.80 | 20816499; 20713592 | - |
| Serine acetyltransferase, putative | GFRORF1528 | Prevotella ruminicola 23 | Cellular processes | 0.58 | 20830571; 20189106 | GO:0006535: cysteine biosynthetic process from serine; GO:0009001:serine O-acetyltransferase (Campylobacter jejuni RM1221) |
| CoA ligase Family protein | NT01BT3039 | Bacteroides thetaiotaomicronVPI-5482 | Fatty acid and phospholipid metabolism | 0.81 | 20545743; 20534558 | GO:0008150: biological process; GO:00165878: acid-thiol ligase activity; GO:0016208: AMP binding (Colwellia psychrerythraea 34H) |
| Modification methylase SalI | NT09RC1177 | Roseiflexus castenholzii DSM 13941 | DNA metabolism | 0.81 | 9628360; 9130589 | - |
The GO terms from Biological Process, Molecular Function and Cellular Component ontologies associated with each gene in table 4 (or, in most cases, their prokaryotic orthologs) were retrieved from AmiGO (http://amigo.geneontology.org) by querying the database with their canonical gene names. In most cases the GO terms retrieved supported the functional categorization predicted by LitProf – Gene Classifier, although there is not an exact correspondence between GO and JCVI-CMR ontologies. Six gene names out 16 tested had no match in AmiGO.
Discussion
The current scenario of gene functional annotation (annotations based solely on sequence similarity to a previously known gene, which was most probably annotated in the same way [2]) condemns “Unknown function” and “Unclassified” genes to remain in this situation. Under this perspective this work brings significant contribution by being able to automatically capture reliable gene function categories to unknown genes. In practical terms it means that precision is a more important characteristic for a gene functional classifier than recall. Another important aspect that has to be considered is the classifier's ability to deal with real world datasets. In other words, the classifier has to present a good performance when any misclassified gene is submitted for functional assignment.
Other gene classifiers with good performance have already been proposed, but attention should be paid to the possible bias introduced in their models by the training and test datasets used. Artificial highly informative training datasets composed only of selected true positive examples (i.e., abstracts previously known to describe the gene function) will improve the performance of the classifier since the training process will not be disturbed by noise in the data. Classifiers created under such conditions tend to underperform in real-world situations when the text corpora associated to the test dataset contain informative and non-informative documents in unpredictable proportions.
In a previous work, Theodosiou and co-workers [6] developed a SVM classifier for assigning one of 12 selected GO categories [7] to a gene product by searching abstracts retrieved from MEDLINE for MeSH terms, previously associated to GO categories by the authors. That classifier achieved an average recall of 0.70 and precision of 0.68. For equally weighted recall and precision the F-measure was 0.69. However, both training and test datasets used in that study were composed of highly informative selected text corpora, retrieved based on MeSH terms used as proxy for GO categories. MeSH terms contribute to compose a highly informative training dataset that can bias the classifier.
Another work, that competed in BioCreAtIvE II [8], also presented a good performance achieving recall of 0.82 and precision of 0.67. With equally weighted recall and precision the F-measure was 0.74. The BioCreAtIvE II training dataset was mostly (64.3%) composed of true positive documents and the remaining 35.7% are known true negatives [8]. The classifier was trained with a percentage of noise documents that is unlikely to be observed in the text corpora of the “Unknown function” and “Unclassified” genes of any gene repository.
In the present work, the abstracts were retrieved solely by querying PubMed with the gene names. No strategy was used to enrich the text corpus with highly informative documents. The two datasets we used (one for training and cross-validation and one independent test dataset) were composed of text corpora from previously classified genes randomly picked from JCVI-CMR, covering 117 species. The informative vocabulary was then automatically disclosed by analyzing the frequency of terms in the text corpus of all genes. Importantly, the model was trained, tested and evaluated with datasets with a distribution of functional categories similar to that of the source (real life) repository of genes. Because we dramatically reduced the redundancy in the training dataset, when applied to an independent test dataset (2,220 previously classified genes randomly picked from JCVI-CMR), that represents the real world genes ensemble which contains a certain level of redundancy (orthologs, etc), the classifier performed even better than in the cross-validation step (Figure 1).
LitProf-Gene Classifier is species blind since we assumed that orthologs sharing the same name may also share similar functions. This assumption may be true in the majority of the cases and is also behind the popular strategy of gene annotation transfer based on sequence similarity. Our rationale in this work was that once a given gene “X” is named based on its sequence similarity to another gene “Y” which, at that time, was not yet assigned to any functional category, it will inherit this “misclassification”. Later on, new experimental studies can clarify the function of the ortholog “Y”, but its reviewed functional annotation will not be automatically transferred to gene “X” in the main public databases (we took JCVI-CMR as a case study). It is important to remark that the aim of the present work was to review gene functional annotation, not gene name assignment. Nevertheless, Litprof-Gene Classifier's performance depends on how informative is the text corpus of the gene to be classified. Incorrectly selecting documents due to gene name ambiguity has low impact in the training step of the SVM since the training gene set contains many examples of each functional category, the majority of them with unambiguous text corpora. However, individual genes with ambiguous text corpora will be classified by LitProf – Gene Classifier with low confidence. For example, querying PubMed with gene symbols mgtA and mgtB resulted in 69 and 39 abstracts respectively. Both genes are involved in magnesium transport in prokaryotes, but some abstracts indexed for gene symbol mgtB were clearly not related to this function (e.g. PMID: 21604018 – “male genital tuberculosis”). As a consequence, Litprof- Gene Classifier unambiguously assigned mgtA to functional category “Regulatory Functions” (confidence = 0.82), whereas mgtB could only be categorized with very low confidence (0.39), which is below the cutoff of 0.7 assumed to be sufficient for reliable classifications.
Differently from some of previous works, the main contribution of our initiative is that using the developed SVM model we could assign more than five thousand genes to functional categories, defined in a complete and well structured ontology. This represents more than 20% of the genes previously assigned to “Unknown function” or “Unclassified” categories despite having literature in MEDLINE. This number accounts for approximately two bacterial genomes completely annotated. These genes would probably never have their misannotation reviewed otherwise. From the total number of genes initially selected, only 49.3% have a name which is pre-requisite for text corpora retrieval. For genes with a name, 70% had enough abstracts in MEDLINE to be processed by LitProf. Table 5 shows the total number of genes classified by LitProf – Gene Classifier when no restrictions were set to confidence values. These numbers make evident the considerable amount of genes currently lacking functional annotation. Another relevant issue is the multiple category annotation of some genes. At present, only 12% of the genes in the JCVI-CMR are classified in more than one functional category. However, it is not surprising that some categories, such as the “Signal transduction”, are enriched in genes classified in more than one category. To better accommodate these cases, future work to improve LitProf – Gene Classifier will focus on predicting multiple classes instead of only the most probable class.
Table 5. Number of genes assigned to functional categories with different confidence thresholds.
| confidence | genes |
| ≥0.9 | 1804 |
| ≥0.8 | 3479 |
| ≥0.7 | 5235 |
| ≥0.6 | 7085 |
| ≥0.5 | 9055 |
| ≥0.4 | 11797 |
| ≥0.3 | 15584 |
| ≥0.2 | 23397 |
| ≥0.1 | 23973 |
As exemplified in table 4, the information granularity of the main categories of JCVI-CMR is similar to that of the upper levels of the GO “Biological Process” ontology, which is the only annotation available for many prokaryotic genes. As far as more genes classified in only one subcategory will be available in the JCVI-CMR database, it is conceivable that LitProf – Gene Classifier could be trained with these examples to also predict subcategories of the JCVI-CMR ontology. In principle, the same strategy could be used to train the classifier to predict functional categories of other complimentary ontologies, such as GO.
According to Blaby-Haas[2], the experimental characterization of the millions of genes sequenced is, to date, an impossible task. For that reason, automated methodologies for gene function annotation are essential in the post genomic era. Our results confirmed the hypothesis that it is possible to confidently assign genes of a real world repository to functional categories, based exclusively on the automatic profiling of its associated literature. The LitProf- Gene Classifier web server may be a valuable complimentary tool for the community involved in prokaryote gene annotation. A web server version of LitProf – Gene classifier is accessible at: www.cebio.org/litprofGC.
Methods
Disclosing the informative vocabulary required to describe each functional category
To compose the initial data set required to train the Support Vector Machine (SVM), canonical prokaryote gene names distributed in the 20 functional categories of the JCVI-CMR ontology (downloaded in 8th June 2010), were randomly selected from the genomes of 117 prokaryote taxa represented in the JCVI-CMR database. Each of these genes had been assigned by JCVI-CMR to only one functional category. We used as gene name, the content of the field “Putative identification” from JCVI-CMR. The initial gene set was then screened to eliminate homonymous genes and genes assigned to more than one functional category that could bias the gene set. Genes assigned to categories “Disrupted reading frame”, “Hypothetical proteins”, “Unclassified” or “Unknown function” of the JCVI-CMR ontology were also excluded. For details on the rearranged ontology, see File S2.
We used the software LitProf, a homemade customized implementation of the Chaussabel & Sher algorithm [9], to identify the minimum vocabulary required to describe the function of a given gene from a collection of its gene-specific abstracts in MEDLINE. LitProf was first used by Coimbra et al to cluster genes differentially expressed in infant rat brain in the course of experimental meningitis [10] and later, to estimate the ambiguity level of individual aliases in large gene terminologies based exclusively in their name-specific vocabulary fingerprints automatically extracted from abstracts in MEDLINE [11].
LitProf works in three fundamental steps. In the first step, it retrieves the abstracts from MEDLINE by sequentially querying the online version of PubMed with gene names in a list provided by the user. LitProf communicates with PubMed using the Entrez Programming Utilities (E-utilities), the structured interface to the Entrez system [http://eutils.ncbi.nlm.nih.gov/]. Searches are case insensitive and approximate matches are allowed. In this study we only included genes which had at least five abstracts in MEDLINE. For these genes, up to the 50 most recent abstracts were retrieved by LitProf. In the second step, for each gene a vector of stemmed terms, suffix stripped with the Porter stemming algorithm, and their relative frequencies is constructed. The term frequency is defined as the fraction of abstracts containing the term in the text corpus of a given gene. In the last step LitProf reduces the dimensionality of the vectors removing all the promiscuous or gene-specific terms. To this purpose, LitProf first determines the baseline frequencies of each term occurring in a set of 7,465 MEDLINE abstracts retrieved for a set of 230 human official gene symbols randomly picked. Terms with frequencies higher than a user-defined cut-off in the baseline (five percent in this study) are eliminated from the vectors of the experimental set of genes. Then, terms for which the difference between their frequencies in the text corpora of the experimental gene set and in baseline exceeds an optimized cut-off are excluded. The optimized cut-off is defined applying the equation: opt_cut-off = t+(k/n) where t is the minimum threshold (in this study t = 0.15), k is a constant (in this study k = 1.5) and n is the number of abstracts retrieved for a given gene. The optimized cut-off compensates for the differences in the number of abstracts retrieved for each gene.
To further decrease redundancy in the gene set, genes were grouped by Hierarchical Clustering algorithm implemented in GenePattern[12] (Pearson correlation as distance metric and average linkage as clustering method) using as input the term-frequency vectors produced by LitProf. For each cluster of highly correlated genes (≥0.99 correlation), one representative gene was randomly chosen. The resultant gene list was resubmitted to LitProf and the minimum informative vocabulary, now adjusted to avoid the bias of genes too closely related, was recreated. For details on the training dataset, see Table S1.
SVM training and evaluation
The term-frequency vectors together with the respective functional categories previously assigned to each gene by JCVI-CMR were used to train a gene classifier using the SVM implemented in GenePattern, designed to use a Radial Basis Function kernel function. We first assessed the classifier's performance using a 10-fold cross validation with 100 replicates. A homemade Perl script shuffled the original training dataset and divided it in training and test sets containing 90% and 10% of the genes, respectively. The SVM classifier from GenePattern was then automatically launched for each of the 100 replicas. After all the iterations, the standard deviations are calculated for recall and precision at each confidence level. We also assessed the recall and precision of classifier trained with the full training set against an independent set of 2,220 randomly picked genes, from 13 species not used in the training step, previously classified into unambiguous categories of the JCVI-CMR ontology. For details on the independent test dataset see Table S2.
LitProf - Gene Classifier
We developed LitProf - Gene Classifier, a web-based tool composed of a set of Perl scripts that integrate the validated SVM model and all the required steps for assign a gene to a functional category based solely on its available literature.
Given a list of genes names provided by the user, the tool retrieves a minimum of five and a maximum of 50 abstracts for each gene from MEDLINE repository. These abstracts compose the text corpora for each gene. From these text corpora LitProf – Gene Classifier calculates the frequency for each term of the minimum informative vocabulary used in the previous step to training the SVM. These term-frequency vectors for each gene to be classified are automatically loaded to the SVM (implemented in GenePattern) together with the predictive model produced in the training step. The output is a web page showing the gene name, the predicted category, and the confidence.
Statistical analysis
Gene distribution over functional categories in the classified gene set, the training gene set, and the original JCVI genome resource, were compared using a paired t-test, with a confidence level of 95%, using GraphPad Prism version 5.02 (GraphPad Software, Inc. CA, USA).
Supporting Information
Original JCVI-CMR ontology.
(DOC)
Rearranged JCVI-CMR ontology.
(DOC)
Training dataset – Used to train the SVM classifier and to perform the cross-validation.
(DOC)
Independent dataset – Used to test the SVM classifier.
(DOC)
Genes classified by LitProf - Gene Classifier which were previously assigned to ‘Unknown function’ and ‘Unclassified’ categories of JCVI-CMR.
(DOC)
Funding Statement
This work was supported by FAPEMIG (TCT 16.028/10; www.fapemig.br), NIH-FIC (TW007012; http://www.fic.nih.gov/Pages/Default.aspx), CNPq (564941/2010-7; www.cnpq.br), and FIOCRUZ-Minas (www.cpqrr.fiocruz.br). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, et al. (2005) The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33: 5691–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blaby-Haas CE, de Crécy-Lagard V (2011) Mining high-throughput experimental data to link gene and function. Trends Biotechnol 29: 174–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Poptsova MS, Gogarten JP (2010) Using comparative genome analysis to identify problems in annotated microbial genomes. Microbiology 156: 1909–1917. [DOI] [PubMed] [Google Scholar]
- 4. Schnoes AM, Brown SD, Dodevski I, Babbitt PC (2009) Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol 5: e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hunter L, Cohen KB (2006) Biomedical language processing: what's beyond PubMed? Mol Cell 21: 589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Theodosiou T, Angelis L, Vakali A, Thomopoulos GN (2007) Gene functional annotation by statistical analysis of biomedical articles. Int J Med Inform 76: 601–613. [DOI] [PubMed] [Google Scholar]
- 7. Raychaudhuri S, Chang JT, Sutphin PD, Altman RB (2002) Associating genes with gene ontology codes using a maximum entropy analysis of biomedical literature. Genome Res 12: 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lan M, Tan CL, Su J (2009) Feature generation and representations for protein-protein interaction classification. J Biomed Inform 42: 866–872. [DOI] [PubMed] [Google Scholar]
- 9. Chaussabel D, Sher A (2002) Mining microarray expression data by literature profiling. Genome Biol 3: RESEARCH0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coimbra RS, Voisin V, de Saizieu AB, Lindberg RL, Wittwer M, et al. (2006) Gene expression in cortex and hippocampus during acute pneumococcal meningitis. BMC Biol 4: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coimbra RS, Vanderwall DE, Oliveira GC (2010) Disclosing ambiguous gene aliases by automatic literature profiling. BMC Genomics 11 Suppl 5 S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, et al. (2006) GenePattern 2.0. Nat Genet 38: 500–501. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Original JCVI-CMR ontology.
(DOC)
Rearranged JCVI-CMR ontology.
(DOC)
Training dataset – Used to train the SVM classifier and to perform the cross-validation.
(DOC)
Independent dataset – Used to test the SVM classifier.
(DOC)
Genes classified by LitProf - Gene Classifier which were previously assigned to ‘Unknown function’ and ‘Unclassified’ categories of JCVI-CMR.
(DOC)