Abstract
Parvin is a putative F-actin binding protein important for integrin-mediated cell adhesion. Here we used overexpression of Drosophila Parvin to uncover its functions in different tissues in vivo. Parvin overexpression caused major defects reminiscent of metastatic cancer cells in developing epithelia, including apoptosis, alterations in cell shape, basal extrusion and invasion. These defects were closely correlated with abnormalities in the organization of F-actin at the basal epithelial surface and of integrin-matrix adhesion sites. In wing epithelium, overexpressed Parvin triggered increased Rho1 protein levels, predominantly at the basal side, whereas in the developing eye it caused a rough eye phenotype and severely disrupted F-actin filaments at the retina floor of pigment cells. We identified genes that suppressed these Parvin-induced dominant effects, depending on the cell type. Co-expression of both ILK and the apoptosis inhibitor DIAP1 blocked Parvin-induced lethality and apoptosis and partially ameliorated cell delamination in epithelia, but did not rescue the elevated Rho1 levels, the abnormal organization of F-actin in the wing and the assembly of integrin-matrix adhesion sites. The rough eye phenotype was suppressed by coexpression of either PTEN or Wech, or by knock-down of Xrp1. Two main conclusions can be drawn from our studies: (1), high levels of cytoplasmic Parvin are toxic in epithelial cells; (2) Parvin in a dose dependent manner affects the organization of actin cytoskeleton in both wing and eye epithelia, independently of its role as a structural component of the ILK-PINCH-Parvin complex that mediates the integrin-actin link. Thus, distinct genetic interactions of Parvin occur in different cell types and second site modifier screens are required to uncover such genetic circuits.
Introduction
Epithelial tissue morphogenesis involves cell shape changes that are induced by tightly regulated interactions between adhesion proteins and the associated actin cytoskeleton. Thus, proteins that modify either the adhesive properties of cells or the dynamics of actin organization have profound effects on epithelial patterning. In pathological situations including cancer, abnormal protein expression drives cells to acquire metastatic properties and break the epithelial integrity.
Integrins comprise a major cell surface protein family that mediate cell adhesion with the extracellular microenvironment and their function is essential for several tissue morphogenetic events during development [1]. Inside the cell, integrins organize the assembly of a large protein network, the adhesome, which mediates linkage with the actin cytoskeleton [2]. Parvin is a core component of the integrin adhesome and binds directly to integrin-linked kinase (ILK). Members of the highly conserved Parvin protein family contain two tandem unconventional Calponin-Homology (CH)-domains [3]. In contrast to mammalian α, β and γ-parvin, invertebrates have a single parvin homolog [4]. Genetic data in mice have demonstrated the important role of Parvin in integrin-mediated adhesion and our previous genetic analysis in Drosophila revealed that Parvin is also essential for adhesion in muscle and wing epithelia [5], [6]. In addition to these developmental functions, recent studies have linked β-Parvin expression to tumor suppressor effects during breast cancer formation in mice [7]. Misexpression studies and modifier screens aimed at identifying genetic circuits regulated by Parvin are of great importance to elucidate the tissue-specific molecular functions of Parvin in the context of a whole organism.
Here we took advantage of the Drosophila system to determine the effects of high levels of Parvin at the cellular level in several tissues and to investigate the tissue-specific suppression or enhancement of these defects by specific genes.
Materials and Methods
Genetics and Drosophila Stocks
All transgenic strains encoding UAS::Parvin and its mutated forms were previously described [6]. Recombinant lines of UAS::Parvin-GFP with longGMRGal4 were generated by standard meiotic recombination. In the eye modifier screen, virgin females of w;;longGMRGal4, UAS::Parvin-GFP were crossed with males of the tested strain from three different categories: (1) UAS lines expressing specific genes; (2) UAS::IR (RNAi-lines) derived either from the VDRC or the NIG collection; and (3) deficiencies included in the deficiency kit for the third chromosome derived from Bloomington. The following stocks were used: UAS::Wech-GFP (M. Hoch); UAS::PTEN (A. Manoukian and T. Xu); puc-laZ (S. Noselli); UAS::ABDMoesinRFP (T. Millard) and UAS::DIAP1 (Bloomington); UAS::ILK [8]; UAS::αPS1; UAS::αPS2; and UAS::βPS (N. Brown). Gal4 drivers were obtained from Bloomington. All crosses were performed at 25°C.
Immunohistochemistry and Confocal Microscopy
Eye and wing discs were dissected from third-instar larvae or 75% pupae and fixed according to standard protocols [6], [8]. Primary antibodies were against: active caspase-3 (1∶250, Cell Signaling); active JNK (1∶500, Cell Signaling); MMP1 (1∶50, mix in 1∶1∶1 of 5H7B11/3A6B4/3B8D12, DSHB); βPS-integrin (1∶10, CF.6G11, DSHB); Ena (1∶50, 5G2, DSHB); Cadherin (1∶50, DCAD2, DSHB); Rho1 (1∶50, p1D9, DSHB); LamininA (1∶500, [9]) and Dia (1∶250, provided by S. Wasserman, UCSD, USA). F-actin was labelled using either rhodamine or Alexa-Fluor-633 phalloidin (Molecular Probes). Secondary antibodies were used at a dilution of 1∶500 and were conjugated to Alexa-Fluor-488, -568, or -633 (Molecular Probes). Nuclei were labelled with DAPI. Images were obtained with a Leica SP5 confocal microscope, using the 20X/0.7NA objective or an oil 63X/1.4 NA objective. Leica SP5 software was used for quantitative analysis of the immunolabelled tissues. The compared images were acquired with identical settings of laser power, gain and iris while avoiding saturation of pixel intensity. Selected areas were outlined and the total intensity was measured and plotted using Excel. Images from adult eyes were obtained using either a Leica DFC500 cooled CCD camera or a Leica TCS LSI system. All images were assembled in Photoshop 7 and labelled in Corel Draw 12.
Results
Parvin Overexpression during Development Causes Morphogenetic Defects
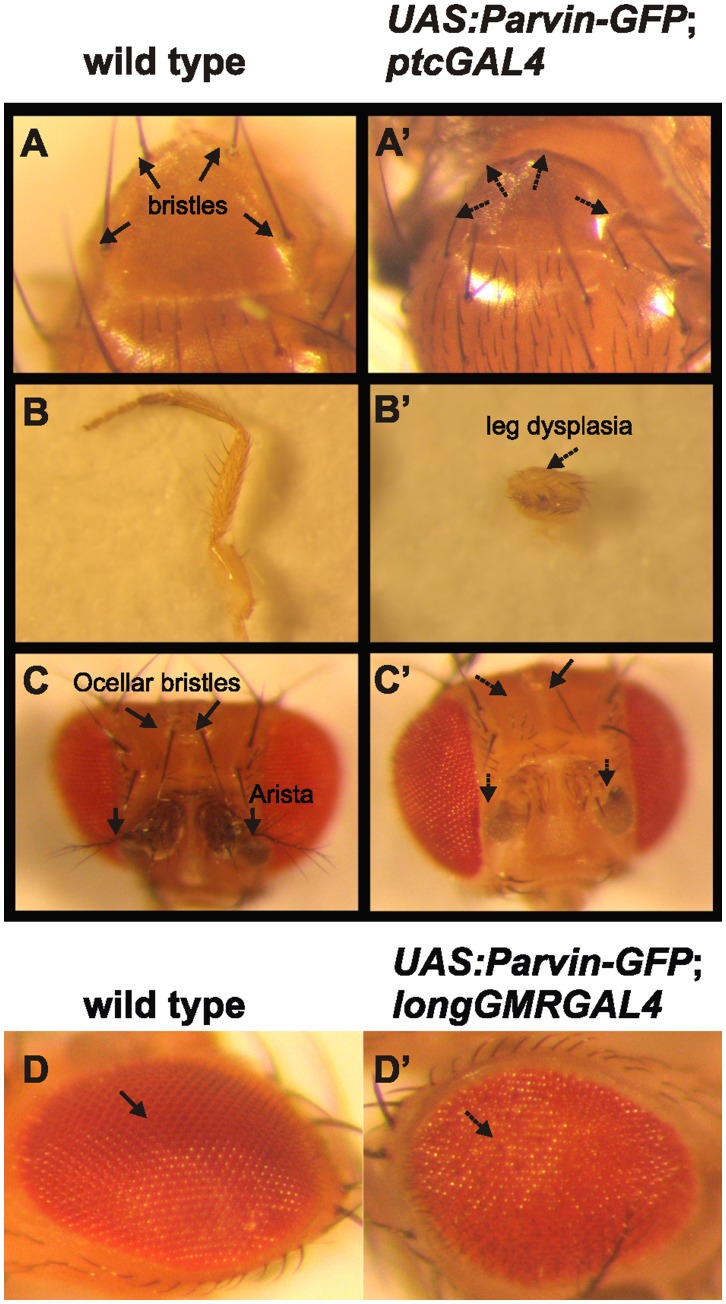
In mammalian cells, α-Parvin has an anti-apoptotic function whereas β-Parvin promotes apoptosis [10], [11]. We followed a gain-of function approach utilizing the UAS/Gal4 system [12] to overexpress Parvin in several tissues during development (Table 1). We focused mainly on the wing epithelium and the eye, using ptcGal4, enGal4 and longGMRGal4 drivers. Overexpression of Parvin by ptcGal4 resulted in several abnormal developmental defects including loss of thoracic bristles, dysplasia in legs, loss of arista and ocellar bristles in the head, whereas a fraction of flies died during pupae development (Figure 1A′–C′). Parvin overexpression driven by longGMRGal4 caused a rough eye phenotype (Figure 1D′). Finally, induction of Parvin expression with enGal4 mostly caused lethality, while the surviving flies had wing defects (Figure 2L2, L3). Fly morphogenesis was not interrupted by similar levels of overexpression of several domain deletion UAS::Parvin-GFP constructs (Table 2), suggesting that combinatorial interactions of Parvin domains are required to elicit a lethal effect and that only high levels of full-length Parvin are detrimental for the whole organism.
Table 1. Gal4 drivers used to direct expression of UAS::Parvin-GFP during Drosophila development.
| Gal4 driver | Tissue of expression | UAS::Parvin-GFP expression effect |
| mef2 | mesoderm | larval lethality |
| 24B | mesoderm and tendon cells | larval lethality |
| drm | gut and malpighian tubules | larval lethality |
| 48Y | endoderm | loss of maxillary palpus |
| segregation of cuticular vesicles | ||
| elav | nervous system | no phenotype |
| sim | ventral nerve cord | no phenotype |
| 69B | epidermis and imaginal discs | mild rough eye |
| missing posterior cross vein | ||
| sev | eye | no phenotype |
| en | epidermis and imaginal discs | pupae lethality |
| tissue loss in the wing discs | ||
| ptc | epidermis and imaginal discs | loss of arista |
| loss of ocellar bristles | ||
| loss of thoracic bristles | ||
| leg dysplasia | ||
| mild rough eye | ||
| longGMR | eye | mild rough eye |
Figure 1. Overexpression of Parvin results in morphogenetic defects at various tissues in the adult fly.
Images were collected with a cooled CCD camera for various adult structures. Thoracic bristles in wild type (A) were missing upon expression of UAS::Parvin-GFP under ptcGal4 (A′). Leg from a wild type adult fly (B) was malformed when UAS::Parvin-GFP was expressed under ptcGal4 (B′). Ocellar bristles and arista from the head of a wild type adult fly (C) were missing upon UAS::Parvin-GFP expression under ptcGal4 (C′). A compound eye from a wild type adult fly (D) took on a rough appearance when UAS::Parvin-GFP was expressed under longGMRGal4 (D′). Arrows depict a wild type tissue structure, whereas dashed arrows indicate defects.
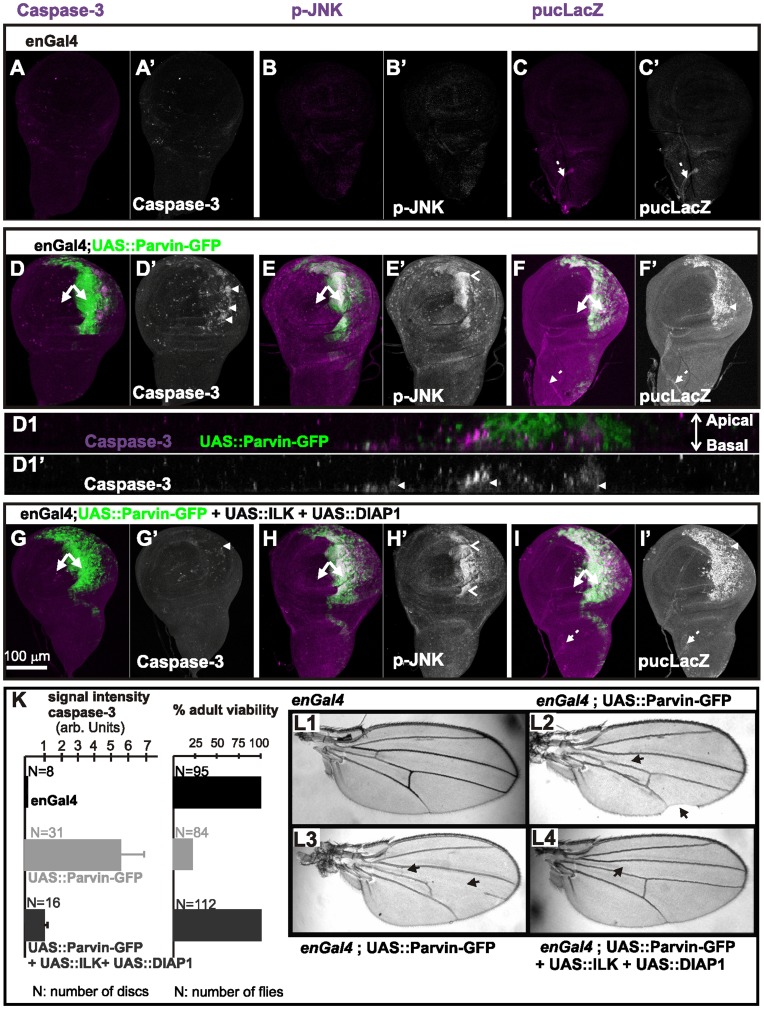
Figure 2. Parvin overexpression induces apoptosis and activation of JNK signaling.
(A–I) Wing imaginal discs of late third instar larvae from control enGal4 (A–C), enGal4/+;UAS::Parvin-GFP/+ (D–F), enGal4/UAS:ILK;UAS::Parvin-GFP/UAS:DIAP1 (G–I). Wing discs expressed either only Gal4 (A–C), or Parvin-GFP (green, D–F), or ILK, DIAP1 and Parvin-GFP together (green, G–I), and were probed for activated caspase-3 (magenta, A, D, G; white A′, D′, G′), active p-JNK (magenta, B, E, H; white B′, E′, H′), or lacZ expressed from the puc locus (magenta, C, F, I; white C′, F′, I′). (D1–D1′) A cross optical section taken in the middle of the wing poutch from the imaginal disc appearing in image D. Arrowheads indicate apoptotic cells; open arrowheads indicate areas of active JNK; arrows, closed areas in the posterior and anterior compartment of the wing pouch expressing (right) or not expressing (left) UAS::Parvin-GFP; dashed arrows indicate stalk cells in the wing notum area. The anterior part of the wing disc (where Parvin-GFP expression is not induced) serves as an internal control. (K) Quantification of caspase-3 signal intensity in wing imaginal discs and the percentage of adult viability. (L1–L4) Adult wings derived from flies expressing only enGal4 (L1), UAS::Parvin-GFP under enGal4 (L2, L3) and UAS:ILK together with UAS:DIAP1 and UAS::Parvin-GFP under enGal4 (L4). Arrows indicate vein defects and notched areas in the wing.
Table 2. Truncated forms of UAS::Parvin-GFP expressed with specific Gal4 drivers did not affect tissue morphogenesis.
| UAS Transgene | Gal4 drivers tested | Phenotypeinduced |
| UAS::ParvinΔCH2-GFP | mef2/24B/en/ptc/longGMR | no |
| UAS::ParvinΔCH1-GFP | mef2/24B/en/ptc/longGMR | no |
| UAS::ParvinCH1LCH2-GFP | mef2/24B/en/ptc/longGMR | no |
Parvin Overexpression in the Wing Epithelium Leads to Apoptosis and Activation of the JNK Pathway
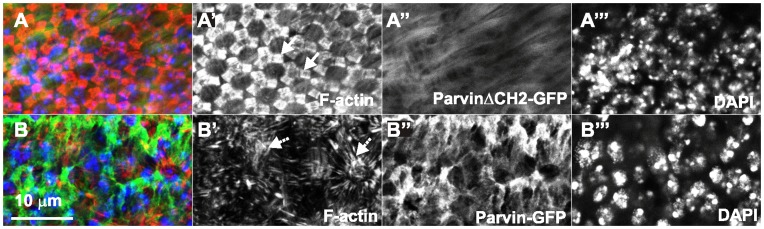
The morphogenetic defects caused by Parvin-GFP overexpression driven by enGal4 suggested a pro-apoptotic function for Parvin in Drosophila, similar to β-Parvin in mammalian cells [11]. To further verify if Parvin-GFP overexpression caused apoptosis, we examined the levels of active Caspase-3. Active Caspase-3 was undetectable in control enGal4 wing discs (Figure 2A, A′) or those expressing a CH2-domain deletion Parvin mutant fused to GFP (UAS::ParvinΔCH2-GFP) (Figure 3A). In contrast, Parvin-GFP overexpression induced a large increase in active Caspase-3, specifically in the posterior compartment of the wing disc, compared to only a few apoptotic cells in the anterior compartment which serves as an internal control (Figure 2D, D1′, K). We used a commercially available antibody against active Caspase-3 that was recently reported to recognize not only Caspase-3 but also additional substrates cleaved in a Drosophila Nedd2-like caspase (DRONC)-dependent manner [13]. Thus, we concluded that Parvin-GFP overexpression induced elevation of the Caspase-9-like initiator DRONC that resulted in apoptosis.
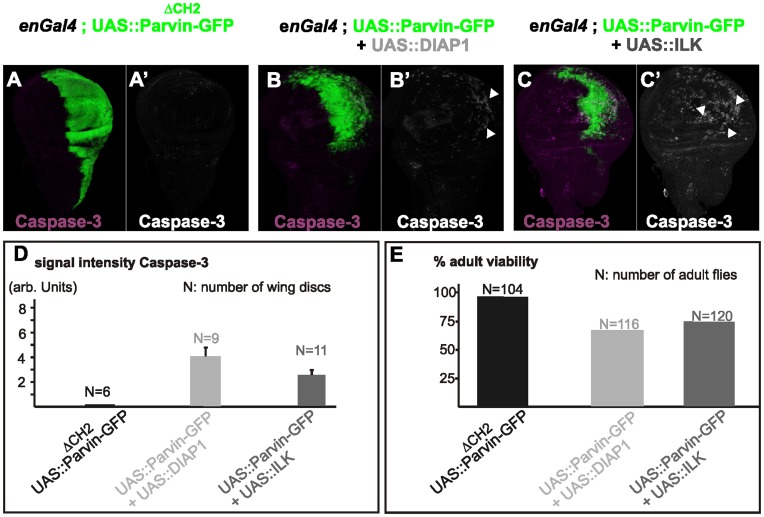
Figure 3. Expression of Parvin lacking the CH2-domain and Parvin coexpression with ILK or DIAP1.
Confocal optical sections acquired from wing imaginal discs of late third instar larvae expressing UAS::ParvinΔCH2-GFP (green, A), or coexpressing either UAS:DIAP1 and UAS::Parvin-GFP (green, B) or UAS:ILK and UAS::Parvin-GFP (green, C), under enGal4 in the posterior compartment of the disc, probed for activated caspase-3 (magenta, A–C; white A′–C′). Arrowheads indicate apoptotic cells. The anterior part of the wing disc (where Parvin-GFP expression is not induced) serves as an internal control. (D–E) Quantification of the caspase-3 signal intensity in wing imaginal discs (D) and the percentage of adult viability (E).
Apoptotic stimuli are known to activate JNK signaling at the imaginal discs [14]. We examined whether Parvin–induced apoptosis is mediated by the JNK pathway, by immunostaining for the phosphorylated active form of JNK. The Drosophila homolog of JNK, basket, was highly phosphorylated specifically at the posterior compartment of the wing disc (Figure 2E–E′), compared to low levels of active JNK in control discs (Figure 2B–B′). We used the downstream target, puckered, as another marker for activation of the JNK pathway [15]. Cells ectopically expressing Parvin-GFP strongly upregulated the puc-lacZ reporter in the posterior compartment (Figure 2F–F′), whereas in control discs, puc-lacZ was detected only in the stalk cells (Figure 2C–C′). Thus, JNK signaling was activated by increased levels of Parvin-GFP within the wing imaginal disc.
Increased Levels of Both ILK and DIAP1 Suppress Parvin-induced Apoptosis
Overexpression of Parvin-GFP driven by enGal4 resulted in lethality mainly during pupae development. Only 20% of the late pupae developed into adult flies (Figure 2K) which exhibited various developmental defects in the wings, including tissue loss and vein defects (Figure 2L2, L3). To dissect the molecular mechanism of UAS::Parvin-GFP-induced apoptosis, we coexpressed Parvin-GFP with either ILK, a binding partner of Parvin, or Drosophila Inhibitor of Apoptosis Protein (DIAP1) [16]. Coexpression of DIAP1 alone largely suppressed the Parvin-GFP-induced dominant lethality and apoptosis (73% rescue of adult viability, n = 116) (Figure 3B, D, E). Coexpression of ILK [8] was less efficient at reducing the activation levels of DRONC, but significantly rescued lethality (75% rescue of adult viability, n = 120), similarly to DIAP1 expression alone (Figure 3C–E). However, coexpression of both ILK and DIAP1 completely blocked UAS::Parvin-GFP-induced apoptosis (Figure 2G–G′) and lethality (100% rescue of adult viability, n = 104) (Figure 2K), and the adult flies displayed only loss of posterior cross-veins (Figure 2L4). DIAP1 coexpression appeared to block Parvin-GFP-induced apoptosis at the final step of the process, because JNK signaling remained active in cells overexpressing both Parvin-GFP and DIAP1 (Figure 2H, I).
Parvin Overexpression in the Wing Epithelium Leads to Cell Delamination, Cell Invasion and MMP1 Secretion
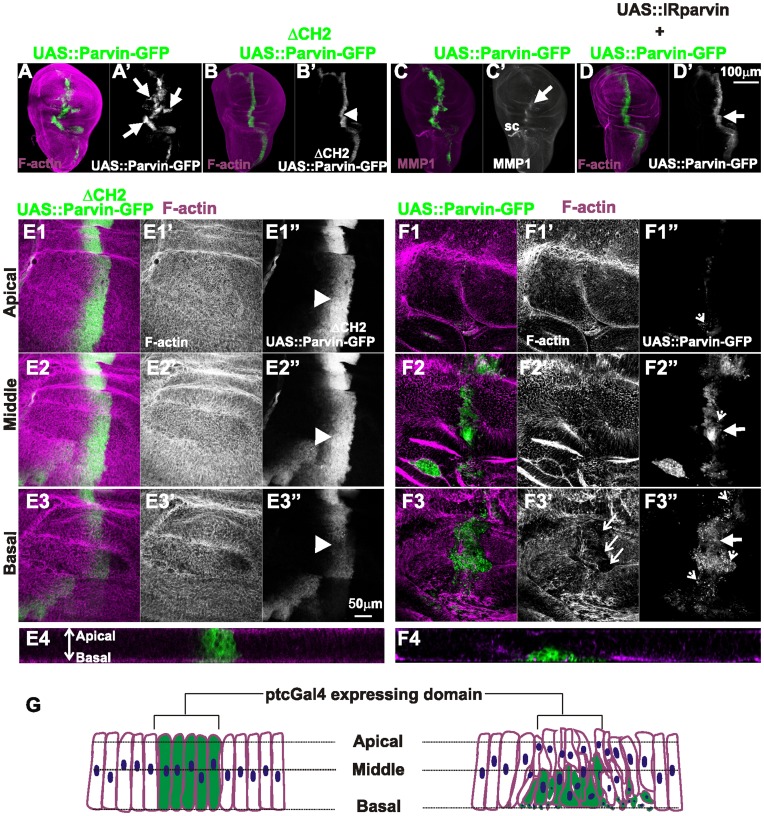
To further investigate the cellular consequences of Parvin-GFP overexpression, we used ptcGal4 to drive expression in a thin stripe of cells anterior to the anteroposterior (A/P) boundary of the wing disc. Overexpression of UAS::Parvin-GFP triggered cell invasion in areas proximal to ptcGal4 expression domain (Figure 4A). In contrast, overexpression of UAS::ParvinΔCH2-GFP that was expressed even at higher levels than full-length Parvin-GFP [6] did not cause epithelial morphogenetic defects, indicating that the invasive phenotype was not a consequence of protein overexpression in general (Figure 4B). High magnification optical sections along the apical/basal side of the wing pouch revealed a large reduction in Parvin-GFP expressing cells, whereas cells expressing UAS::ParvinΔCH2-GFP were maintained within the ptcGal4 domain (Figure 4E1–F1). The UAS::Parvin-GFP expressing cells were extruded toward the basal side of the epithelium, where they acquired invasive properties that render them capable of migrating to the basal side of the epithelium and spread distant from the ptcGal4 expression domain (Figure 4E2, E3, E4, F2, F3, F4, G). Several cells displayed small pyknotic nuclei indicative of apoptosis (Figure 4F3′′). Cell invasion was consistent with ectopic induction of matrix metalloproteinase-1 (MMP1) along the ptcGal4 expressing region (Figure 4C). MMP1 is a well established effector of cell invasion that is upregulated upon JNK activation and is normally expressed only in the stalk cells of the wing disc (Figure 4C) [17].
Figure 4. Parvin overexpression in the wing epithelium leads to cell extrusion, MMP1 secretion and invasion.
Confocal projections of wing imaginal discs of late third instar larvae (A–D), and optical sections acquired at different focal planes as indicated (E1, E2, E3, E4–F1, F2, F3, F4), with ptcGal4 driving expression of UAS::Parvin-GFP (A, C, F1–F4), or UAS::ParvinΔCH2-GFP (B, E1–E4), or UAS::Parvin-GFP together with UAS:IRParvin (D). Imaginal wing discs expressing Parvin-GFP (green, A–F3; white, A′–D′, E1′′–F3′′) were probed for rhodamine-phalloidin to visualize F-actin (magenta, A–B, D, E1–E4, F1–F4; white, E1′–E3′, F1′–F3′) or MMP1 (magenta, C; white C′). (E4–F4) Cross optical sections of the imaginal discs appearing in images E1–F1 taken in the middle of the wing poutch.Graphic cartoon based on the optical sections shown in images E4 and F4 (G). Arrowheads indicate the expression domain of ptcGal4 that drives expression of the transgene UAS::ParvinΔCH2-GFP; large arrows point to migrating cells that have invaded the proximal areas of UAS::Parvin-GFP expression; dashed arrowheads denote small cellular debris containing pyknotic nuclei; and open arrows show areas lacking F-actin staining in the basal side of discs expressing UAS::Parvin-GFP. sc; stalk cells.
To verify that a threshold level of UAS::Parvin-GFP is required to induce the invasive cell behavior, we coexpressed a UAS::RNAi construct known to knock down Parvin [6]. The moderate levels of Parvin-GFP expression along the ptcGal4 domain did not cause migration of these cells distant from their original position (Figure 4D).
Parvin Overexpression in the Wing Epithelium Results in Loss of Cell-matrix Adhesion and Extracellular Matrix Disassembly without Affecting Cadherin Levels
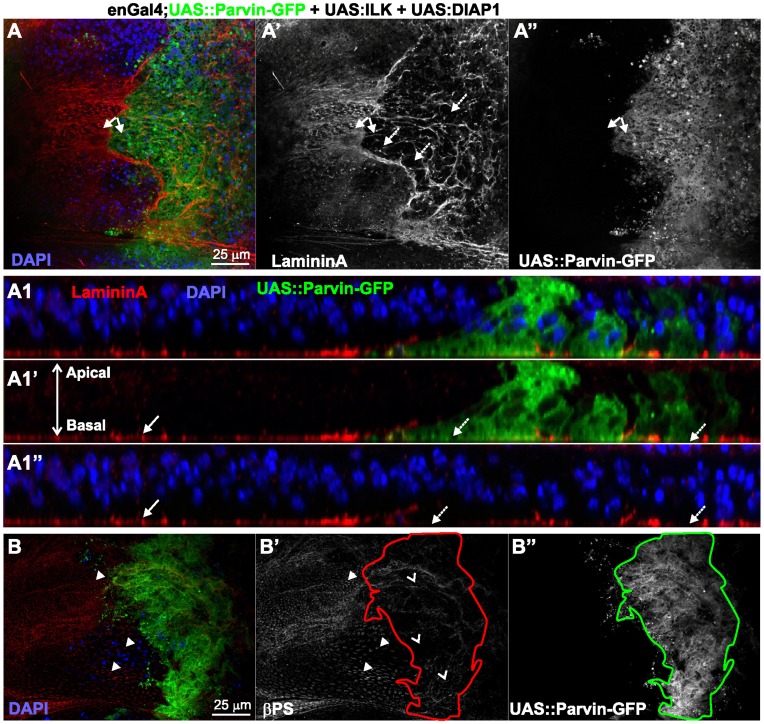
In the wing imaginal discs integrin localizes largely in clusters containing adhesome proteins on the basal side of the epithelium, resembling the focal adhesions of mammalian cells [6], [18]. The ectopic elevated levels of MMP1 upon UAS::Parvin-GFP overexpression, prompted us to further investigate cell-matrix adhesion organization. LamininA, is a major component of the extracellular matrix (ECM) and it has been shown to localize basaly in the wing disc, where it displays a fibrillar distribution [19]. We found that overexpression of Parvin caused disorganization of LamininA in the posterior compartment of the wing epithelium. LamininA was reduced in certain areas and accumulated in others, displaying a non-ordered pattern of distribution (Figure 5A, A1–A1′′). Similarly the typical punctuate integrin localization at the focal contact-like structures at the basal side of the wing epithelium was severely affected, specifically in the posterior compartment, whereas large areas of the basal epithelium lacked integrin deposition (Figure 5B). Enabled (Ena) plays a role in the elongation of F-actin barbed end filaments and recently it was shown that is expressed in the wing disc [20], [21]. We found that within the anterior compartment, Ena accumulated basally at the focal-contact like structures, similarly to integrins and other integrin adhesome proteins [6], [18], whereas in the posterior compartment expressing UAS::Parvin-GFP, Ena was largely diminished (Figure 5C, E–E′). In contrast, in the middle and apical areas of the disc, Ena distribution was not affected, suggesting that its basal reduction was most likely a consequence of disorganized cell-matrix adhesion sites (Figure 5D–D′). Thus, we concluded that in the basal wing epithelium high levels of Parvin-GFP disrupt integrin-matrix adhesion sites.
Figure 5. Parvin overexpression in the wing epithelium disrupts basal adhesion without affecting apicobasal polarity.
Confocal optical sections acquired from wing imaginal discs of late third instar larvae expressing UAS::Parvin-GFP (green, A–F; white, A′′–B′′) or UAS::ParvinΔCH2-GFP (green, G) under enGal4 in the posterior compartment of the disc, probed for laminin (red, A–A1′′; white A′), βPS integrin (red, B; white B′, B1′), Ena (magenta, C–E; white C′–E′), and Cadherin (magenta, F–G; white F′–G′). Panels C–D show two different focal planes: basal (C), and intermediate (D) sides of the epithelium. (A1–A1′′, E–E′) cross optical sections of the imaginal discs appearing in images A and C respectively taken in the middle of the wing poutch. Small arrows, closed areas in the posterior and anterior compartment of the wing pouch expressing (right) or not expressing (left) UAS::Parvin-GFP; big arrows, indicate areas with LamininA deposition; big dashed arrows indicate areas without LamininA deposition; open arrows, apical boundaries of wing epithelial cells; arrowheads, focal contact-like structures in the basal side of the anterior compartment; open arrowheads, areas lacking integrin-matrix adhesion sites. The anterior part of the wing disc (where Parvin-GFP expression is not induced) serves as an internal control.
Cadherin downregulation and initiation of the epithelial-mesenchymal transition (EMT) are typical features of cells acquiring invasive properties [22]. Although the majority of the cells expressing UAS::Parvin-GFP were extruded on the basal side of the posterior wing epithelium, the amount and pattern of cadherin distribution was unaffected in the remaining cells that maintained their plasma membrane in the apical side of the disc (Figure 5F–G). We therefore concluded that Parvin overexpression did not trigger EMT in the wing epithelium.
Features of the Wing Epithelial Cells Expressing High Levels of Parvin-GFP
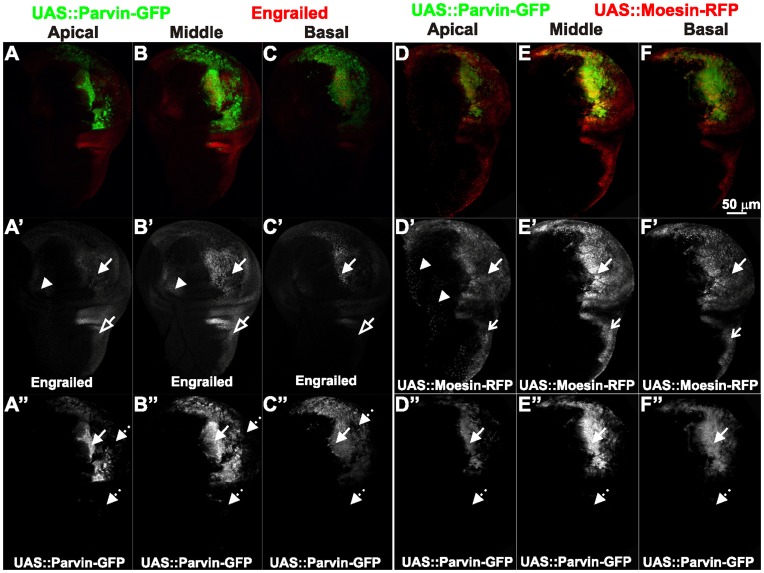
Upon Parvin-GFP overexpression in the posterior wing compartment, we noticed a mosaic expression of the transgene (Figures 2, 3, 5, 6). Certain areas within the enGal4 domain, notably in the hinge and notum, were not labelled for Parvin-GFP although they properly expressed Engrailed and retained their posterior compartment identity (Figure 6A–C). In these cells enGal4 was able to direct expression of a UAS::ABDMoesin-RFP transgene (Figure 6D–F), suggesting that the lack of Parvin-GFP labeling was not due to defective enGal4 activity. In aggrement with this, when we probed wing imaginal discs with an antibody against Parvin, we found that in certain areas of the epithelium, where Parvin-GFP was undetectable, high levels of the protein were present as expected due to overexpression (Fig. 7A–D). In some of these areas, we found apoptotic cells with basaly located pyknotic nuclei (Fig. 7C). We concluded that in these cells, GFP could be destabilized due to undergoing apoptosis. In other areas of the disc undetectable Parvin-GFP was correlated with high density of nuclei (Fig. 7B). That could reflect newly proliferating cells contributing in the regeneration of the damaged epithelium [23]. Therefore in these cells, GFP may have not matured yet to obtain fluorescent properties.
Figure 6. Posterior identity is retained in wing epithelial cells overexpressing Parvin.
Confocal optical sections were acquired at different focal planes (A, D) apically, (B, E) in the middle, (C, F) at the basal side of wing imaginal discs of late third instar larvae expressing UAS::Parvin-GFP (green, A–C; white, A′′–C′′) and probed with antibodies against Engrailed (red, A–C; white A′–C′) or coexpressing UAS:ABDmoesin-RFP (red, D–F; white D′–F′) and UAS::Parvin-GFP (green, D–F; white D′′–F′′) under enGal4 in the posterior compartment of the disc. Arrows: areas in the posterior compartment where expression of UAS::Parvin-GFP is retained; dashed arrows: areas in the posterior compartment that have lost expression of UAS::Parvin-GFP; arrowheads: cells in the anterior compartment expressing enGal4; open arrowheads: areas in the posterior compartment expressing Engrailed; open arrows: areas in the posterior compartment expressing UAS:ABDmoesin-RFP.
Figure 7. Comparison of anti-Parvin and Parvin-GFP detection in the wing imaginal discs.
Confocal optical sections acquired from wing imaginal discs of late third instar larvae expressing UAS::Parvin-GFP (green, A–D; white A′–D′) under enGal4 in the posterior compartment of the disc, probed with an antibody against Parvin (red, A–D; white A′′–D′′) and DAPI to visualize the nuclei (blue, A–D; white A′′′–C′′′). (D) cross optical section taken in the middle of the wing poutch from the imaginal disc appearing in images A–C. Arrows: cells expressing Parvin detected either by the antibody or by Parvin-GFP; dashed arrows: cells where Parvin-GFP is undetectable; open arrowheads:pyknotic nuclei; open arrow: an area in the posterior compartment with high density of nuclei outlined with a light blue dashed line.
High Levels of Parvin Induce Cell Delamination, Cell Shape Changes and Disorganization of F-actin on the Basal Side of the Wing Epithelium
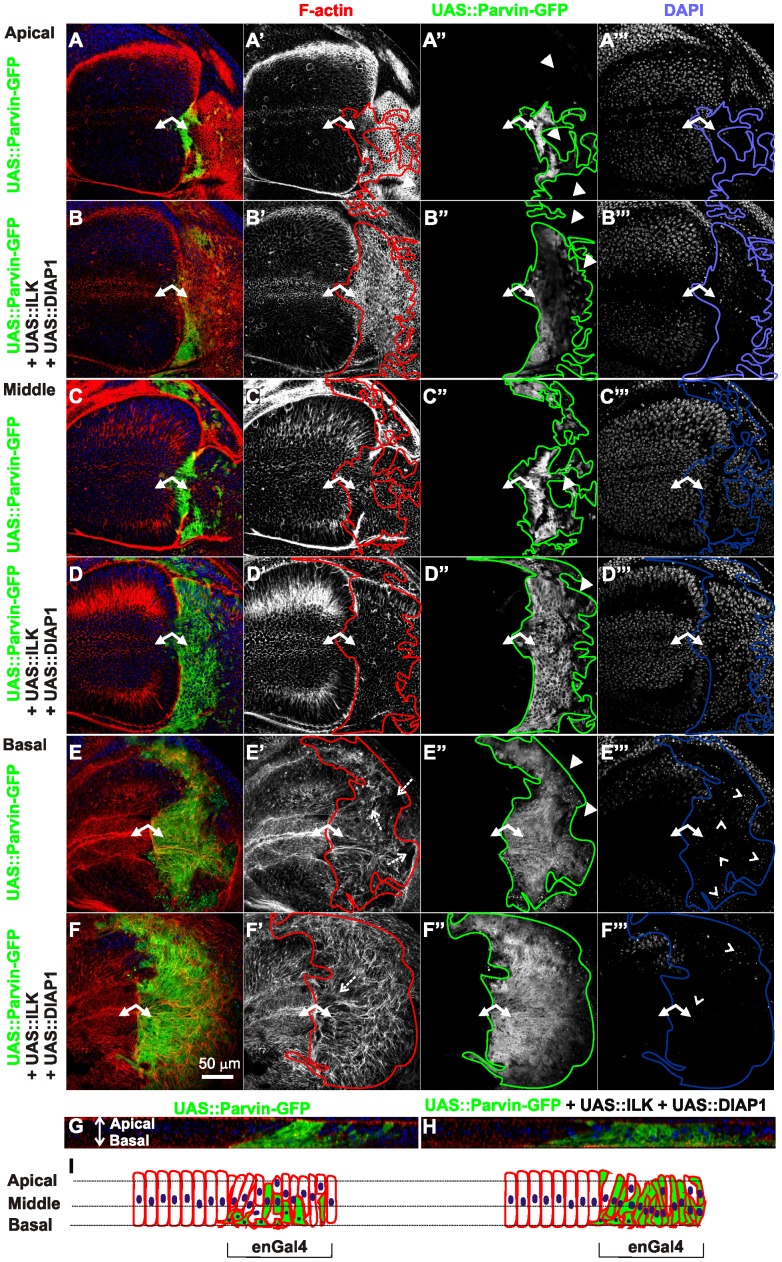
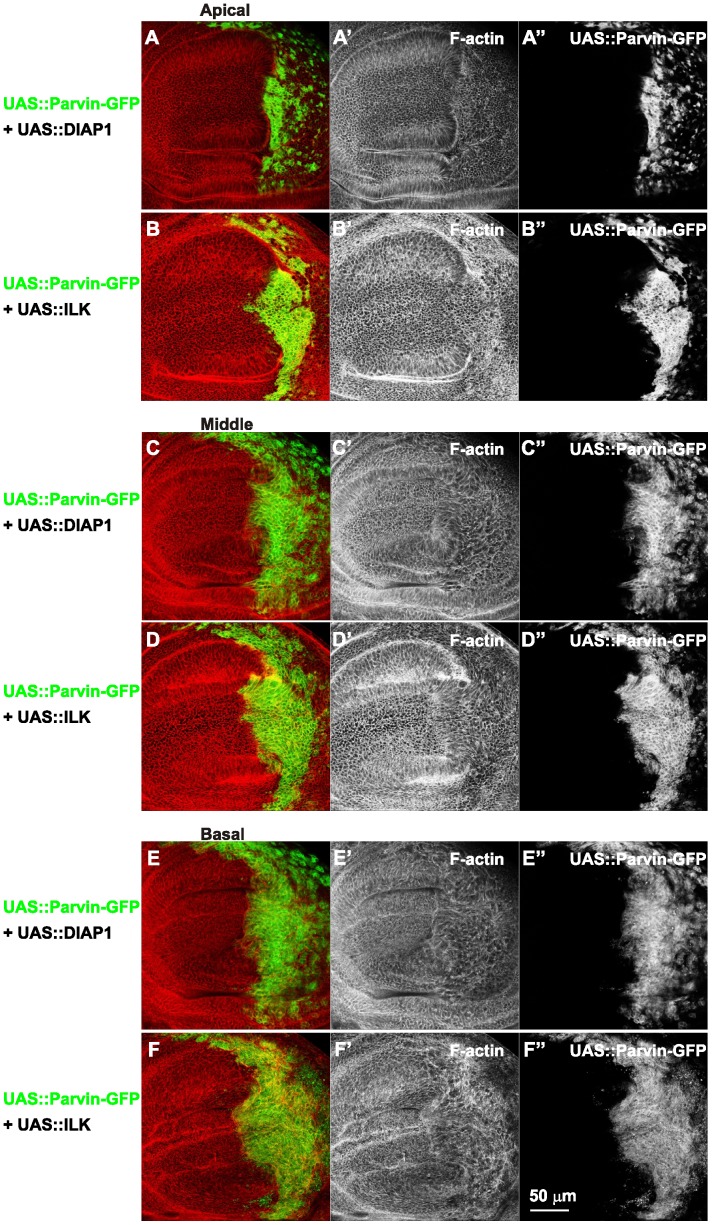
To address whether the mosaic expression of Parvin-GFP within the wing epithelium and cell delamination along the apicobasal axis of the blade were also accompanied by changes in cell shape, we examined the F-actin cytoskeleton organization. In the most apical area of the wing blade the tissue was folded and the posterior compartment appeared shrunken, while the amount and distribution of F-actin cortically appeared normal. From the location of the nuclei in optical cross-sections -obtained at the region between the dorsal-ventral boundary in the middle of the wing poutch- it was evident that cells were shorter (Figure 8A, G, I). In the middle area of the wing disc, cells expressing Parvin-GFP occupied a larger region of the wing blade whereas cell shape, as it was highlighted by F-actin, was similar to the flanking cells in the anterior compartment that did not express high levels of full-length Parvin-GFP (Figure 8C). On the basal side, Parvin-GFP expressing cells occupied almost the entire posterior wing blade, but they were missing from the regions flanking the wing margin (Fig. 8E). The organization of F-actin basaly was completely disrupted (Fig. 8E, E′). Actin filaments were accumulated ectopically in some areas of the wing poutch cells and were missing from others. The observed gaps containing pyknotic nuclei, indicating areas of dead delaminated cells (Fig. 8E, E′′), in accordance with previous studies describing the basal extrusion of dead cells in the wing epithelium [23], [24]. As consequence of the damaged epithelium, the basal cell periphery appeared enlarged and irregularly shaped (Figure 8E′). Coexpression of either ILK or DIAP1 with Parvin-GFP noticeably improved the cell delamination at the basal side (Figure 9), whereas simultaneous coexpression of both ILK and DIAP1 further improved cell extrusion, as was evident from the reduced number of pyknotic nuclei accumulated basaly (Figure 8B, D, F). However, in the wing blade F-actin organization was only modestly ameliorated by coexpression of both ILK and DIAP1. Actin filaments instead of decorating the outline of the cell, extended to the periphery and remained tangled resulting in a disordered meshwork pattern (Figure 8B–B′, D–D′, F–F′, H, I). To test whether the disorganised F-actin is correlating with abnormal cell-matrix adhesion mediated by increased levels of Parvin-GFP rather than being a consequence of apoptosis, we examined the distribution of integrins and lamininA in discs coexpressing ILK and DIAP1, where apoptosis was rescued (Fig. 2). No improvement in the abnormal organization of either integrin or lamininA basaly in the wing epithelium was observed (Figure 10A–B). Thus, defects in integrin-mediated adhesion in the basal side of the epithelium upon Parvin-GFP overexpression is not a consequence of Parvin-induced apoptosis, but rather a distinct effect that correlates with abnormalities in the organization of actin cytoskeleton.
Figure 8. Parvin overexpression in the wing imaginal disc epithelium disrupts the basal F-actin cytoskeleton.
Confocal optical sections acquired apically (A–B), in the middle (C–D) and at the basal side of the epithelium (E–F) from wing imaginal discs with enGal4 driving expression in the posterior compartment of UAS::Parvin-GFP alone (green, A, C, E; white, A′′, C′′, E′′) or coexpression with UAS:ILK and UAS:DIAP1 (green, B, D, F; white B′′, D′′, F′′). Imaginal discs were probed with rhodamine-labelled phalloidin to visualize F-actin (red, A–F; white A′–F′) and DAPI to visualize nuclei (blue, A–F; white A′′′–F′′′). (G) A cross optical section of the imaginal disc appearing in images A, C, and E. (H) a cross optical section of the imaginal disc appearing in images B, D, and F. (I) Graphic cartoon based on the cross optical sections G and H. Small arrows, closed areas in the posterior and anterior compartment of the wing pouch expressing (right) or not expressing (left) UAS::Parvin-GFP; Arrowheads, areas within the posterior compartment that do not express UAS::Parvin-GFP; dashed arrows, disrupted F-actin; open arrowheads, pyknotic nuclei. Lines encircle the region of the posterior compartment retained expression of UAS::Parvin-GFP. The anterior part of the wing disc (where Parvin-GFP expression is not induced) serves as an internal control.
Figure 9. ILK and DIAP1 individually coexpressed with Parvin partially ameliorate F-actin misorganization in the wing epithelium.
Confocal optical sections were acquired apically (A–B), in the middle (C–D) and on the basal side of the epithelium (E–F) from wing imaginal discs with coexpression driven by enGal4 in the posterior compartment of UAS::Parvin-GFP (green, A–F; white A–F′′) and UAS:DIAP1 (A, C, E) or UAS::Parvin-GFP and UAS:ILK (B, D, F). Imaginal discs were probed with rhodamine-labelled phalloidin to visualize F-actin (red, A–F; white A′–F′).
Figure 10. ILK and DIAP1 coexpressed with Parvin-GFP do not rescue the dissorganized integrin-matrix adhesion sites.
Confocal optical sections acquired from wing imaginal discs of late third instar larvae expressing UAS::Parvin-GFP (green, A–B, A1–A1′; white, A′′–B′′) with both UAS:ILK and UAS:DIAP1 driven by enGal4 in the posterior compartment and probed for LamininA (red, A, A1–A1′; white, A′) or βPS integrin (red, B; white, B′) and DAPI to visualize nuclei (blue, A, A1, A1′′). (A1) cross optical section of the imaginal disc appearing in image A taken in the middle of the wing poutch. Small arrows, closed areas in the posterior and anterior compartment of the wing pouch expressing (right) or not expressing (left) UAS::Parvin-GFP; big arrows, indicate areas with LamininA deposition; big dashed arrows indicate areas without LamininA deposition; arrowheads, focal contact-like structures in the basal side of the anterior compartment and open arrowheads indicate areas in the posterior compartment without integrin accumulation. The anterior part of the wing disc (where Parvin-GFP expression is not induced) serves as an internal control.
Parvin Overexpression Induces Up-regulation of Rho1 at the Basal Side of the Epithelium
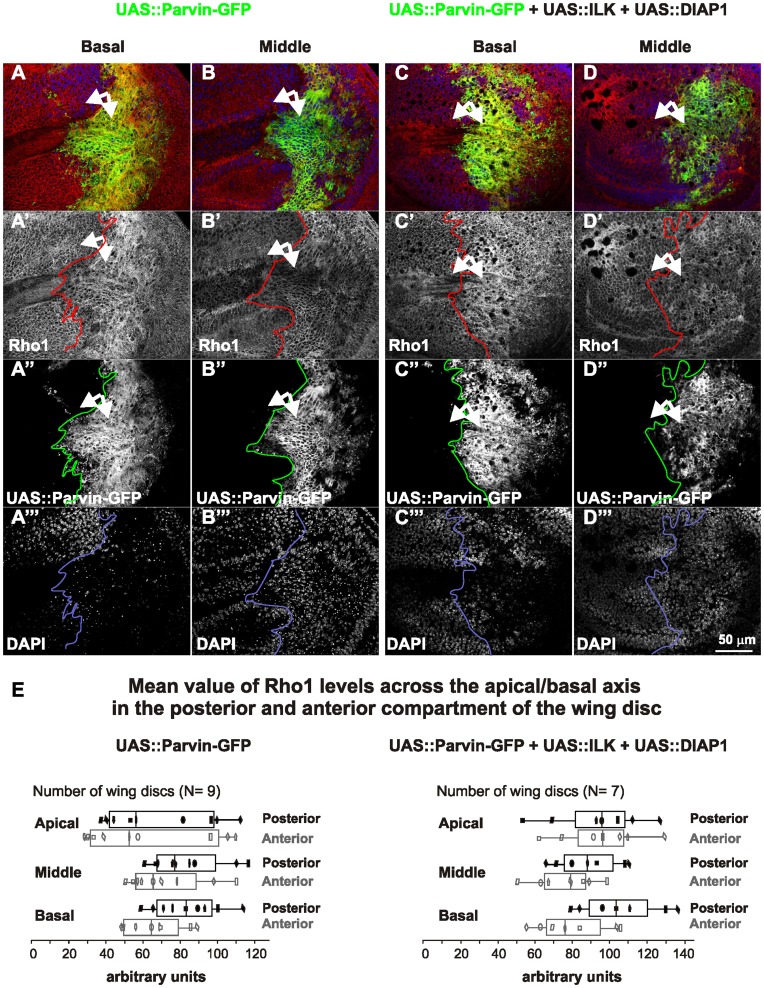
The Parvin-GFP induced alterations in the wing epithelium were highly reminiscent of those observed upon Rho1 overexpression [25], [26]. We found that cells overexpressing Parvin-GFP triggered a substantial increase in Rho1 protein levels, mostly on the basal side (Figure 11A, E), whereas Rho1 accumulation increased only modestly in the middle and in most apical areas of the epithelium (Figure 11B, E). However, the increase in Rho1 levels represented a distinct effect, different from Parvin-induced apoptosis, because elevated Rho1 levels were unaffected, even when both ILK and DIAP1 were coexpressed (Figure 11C–E). Diaphanus (Dia) is one of the main Rho1 downstream effectors. However, as previously found in wing discs [25], Rho1 elevation did not coincide with increased Dia levels upon Parvin-GFP overexpression (Figure 12).
Figure 11. Parvin overexpression in the wing epithelium leads to Rho1 elevation, mainly basally.
Confocal optical sections were acquired basally (A, C) or in the middle (B–D) from wing imaginal discs expressing UAS::Parvin-GFP alone (green, A, B; white, A′′, B′′) or with both UAS:ILK and UAS:DIAP1 (green, C, D; white C′′, D′′), as driven by enGal4 in the posterior compartment, probed for Rho1 (red, A–D; white A′–D′) and stained with DAPI to visualize nuclei (blue, A–D; white A′′′–D′′′). (E) Box-and-whisker plot of Rho1 levels indicating the means (vertical lines in the middle of the rectangular boxes) of measurements taken in apical, middle and basal focal planes. All individual measurements are superimposed on the box-and-whisker plots and are indicated by the same symbol in all focal planes to allow direct comparisons of the variation in pixel intensity. Arrows, closed areas in the posterior and anterior compartment of the wing pouch expressing (right) or not expressing (left) UAS::Parvin-GFP. The anterior part of the wing disc serves as an internal control.
Figure 12. Parvin overexpression does not affect the endogenous levels of Dia.
Confocal optical sections acquired basally (A), in the middle (B) or apically (C) of the wing imaginal discs with enGal4 driving expression in the posterior compartment of UAS::Parvin-GFP (green, A–C; white, A′–C′) probed for the Rho1 effector Dia (red, A–C; white A′′–C′′) or DAPI to visualize the nuclei (blue, A–C; white A′′–C′′). Small arrows, closed areas in the posterior and anterior compartment of the wing pouch expressing (right) or not expressing (left) UAS::Parvin-GFP. The anterior part of the wing disc (where Parvin-GFP expression is not induced) serves as an internal control.
Parvin Overexpression Disrupts F-actin Stress Fibers in the Pigment Cells of the Pupal Retina
Parvin overexpression by longGMRGal4 caused a rough eye phenotype (Table 1, Figure 1). This Gal4 driver is expressed in all cell types of the eye (pigment, cone and photoreceptor cells) [27]. The elavGal4 and sevGal4 drivers that limit expression of Parvin-GFP to only the photoreceptor [28], or specific photoreceptor and cone cells [29], respectively, did not cause any eye roughening (Table 1). Thus, the rough-eye phenotype is most likely caused by overexpression of Parvin in the pigment cells. Several morphogenetic defects during eye development could result in final eye roughening [30]. We therefore examined the organization of F-actin in both 3rd instar larvae and at 75% of pupal development (p.d). The later developmental stage was selected because in the retinal floor, F-actin displays a highly ordered structure of stress fibers within the pigment cells encircling the cone cells [31]. We did not find any defects in F-actin organization in the eye imaginal discs from 3rd instar larvae (data not shown). In contrast, when we examined retinas from late pupae, we found complete disorganization of actin stress fiber arrays in the retina floor, whereas retinas expressing the truncated UAS::ParvinΔCH2-GFP form appeared normal (Figure 13A, B). Thus, in the pupal retina Parvin-GFP overexpression severely disrupted F-actin stress fiber organization in the basal side of the pigment cells, similar to the wing epithelium phenotype.
Figure 13. Parvin overexpression in the eye pigment cells disrupts F-actin cytoskeleton basally.
Confocal optical sections acquired basally, in developing pupae eyes expressing control UAS::ParvinΔCH2-GFP (A, green; A′′, white), or UAS::Parvin-GFP (B, green; B′′, white) with longGMRGal4, probed with rhodamine-phalloidin to visualize F-actin (red, A–B; white A′–B′) and stained with DAPI to visualize the nuclei (blue, A–B; white A′′′–B′′′). The pattern of F-actin in the retina floor is illustrated with the arrow.
Genetic Interactors of Parvin in the Eye
The homozygous longGMRGal4 flies had wild type-like eye morphology when kept at 25°C (Figure 14A). In contrast, flies overexpressing Parvin-GFP under longGMRGal4 displayed distorted ommatidia and mild rough eyes (Figure 14B). This phenotype was sensitive to the copy number of both longGMRGal4 and UAS::Parvin-GFP transgenes (data not shown), and could therefore be exploited to identify genetic suppressors and enhancers of UAS::Parvin-GFP overexpression-induced rough-eye.
Figure 14. Rough eye phenotype and identification of enhancers and suppressors of UAS::Parvin-GFP, longGMRGal4.
Adult eye confocal optical projections from the longGMRGal4 stock served as control (A), the starting UAS::Parvin-GFP, longGMRGal4 genotype (B) and the recovered modifiers (C–L). The main suppressors of the modifier screen show morphology of the ommatidia that is reminiscent of the control: UAS:Wech (C), UAS:PTEN (E), Df(3L)Exel6119 (G), Df(3L)ED2 (I), and UAS:IRXrp1 (K). The main enhancers of the screen are Df(3R)e-GC3 (D), UAS:IREip93F (F), UAS:IRmys (H), UAS:IRzasp52 (J), and UAS:IRChd64 (L).
We investigated individual coexpression of a panel of selected available UAS genes in the eye. Loss of function mutations in both αPS1 and βPS integrin subunits affect the F-actin pattern at the basal surface of the eye retina in late pupae [31]. We hypothesized that if Parvin-GFP overexpression compromised integrin-containing adhesion sites, as we found in the wing epithelium, then coexpression of an integrin heterodimer (αPS1βPS or the αPS2βPS) would ameliorate the Parvin-induced phenotype [32]. In contrast, we found that elevated levels of either of the two coexpressed integrin heterodimers mildly enhanced the UAS::Parvin-GFP induced rough-eye phenotype (Table 3). However, because the levels of integrin expression are not accurately controlled in this experimental setting it is plausible that high levels of integrin expression could not reverse the Parvin-induced rough eye phenotype. Thus, we concluded that a tight balance of the intracellular amount of integrins appears to be rather crucial for the proper eye development. UAS::ILK weakly suppressed the Parvin induced rough-eye phenotype (Table 3). Surprisingly, coexpression of UAS::Wech-GFP, an ILK binding protein [33], completely suppressed the phenotype (Figure 14C). The morphology and organization of the ommatidia remained intact when UAS::Wech-GFP was overexpressed alone under longGMRGal4 (Table 3). A strong suppressive effect was also achieved by coexpression of UAS::PTEN [34] (Figure 14E). In contrast, the catalytically inactive PTENC124S mutant [35] was a poor suppressor, suggesting that enzymatically active PTEN is required to modulate Parvin effects.
Table 3. Summary of all identified modifiers of UAS::Parvin-GFP, longGMRGal4 in the developing eye, including information on the stock used, the cytogenetic map for the deficiencies and the effect of each modifier.
| Gene/Deficiency | Reference/stock | Cytogeneticmap | Effect |
| UAS::βPS;UAS::αPS1 | [31] | E/mr | |
| UAS::βPS;UAS::αPS1 | [32] | E | |
| UAS::Wech-GFP | [33] | S | |
| UAS::ILK | [8] | s | |
| UAS::dPTEN | [34] Line (2nd chr) | S | |
| [35] Line 31 | s | ||
| UAS::dPTENC124S | [35] | s | |
| UAS::IRβPS | mysKK100518 | 7D5 | E |
| UAS::IRZasp52 | Zasp52GD14816 | 52C4–52C7 | E |
| Zasp52KK101276 | E | ||
| UAS::IRparvin | ParvinGD3687 | 18E5–18F1 | S |
| ParvinKK102567 | S | ||
| UAS::IRChd64 | Chd64GD1212 | 64A6;A7 | E |
| UAS::IREip93F | Eip93FGD4449 | 93F14 | E/gl |
| UAS::IRXrp1 | Xrp1KK104477 | 91D3;D5 | S |
| Df(3R)ED2 | BL-6962 | 91A5;91F1 | S |
| Df(3L)Exel6119 | BL-7598 | 70B2;70C2 | S |
| Df(3R)e-GC3 | BL-5798 | 93C6;94A4 | E |
Table key. E: Enhancement; e: mild enhancement; S: Suppression; s: mild suppression. Modifiers phenotype when expressed under longGMR alone; mr: mild rough-eye, gl:glassy.
Next we conducted a dominant-modifier screen using chromosomal deficiency lines of the third chromosome (Bloomington kit) that covered almost 40% of the fly genome. In the first round we tested 111 deficiencies covering almost the entire 3rd chromosome and found 4 suppressors and 12 enhancers. We further narrowed down three genomic regions that were dominant modifiers (present as just one copy) of Parvin-induced rough-eye. The cytogenetic regions encompassing 70B2;70C2 {Df(3L)Exel6119} and 91A5;91F1 {Df(3R)ED2} were identified as strong suppressors (Figure 14G, I), whereas the region 93C6;94A4 {Df(3R) e-GC3} was an enhancer (Figure 14D). To identify candidate genes, we used individual knock-down of 391 specific genes in the eye utilizing UAS::IR lines [36] for the majority of the genes located in the identified genomic regions. No candidate gene was identified for the dominant suppressive effect of 70B2; 70C2. Knock-down of Xrp1 (CG17836, 91D3-D5) within Df(3R)ED2 was equally efficient at suppressing the removal of one copy of the genomic region 91A5; 91F1 (Figure 14K). In addition, knock-down of Eip93F (CG18389, 93F14) located in the genomic region 93C6; 94A4 enhanced the rough-eye, similarly to Df(3R) e-GC3 (Figure 14F). Lastly, knock-down of genes encoding βPS integrin, Zasp52, and the transgelin homolog Chd64 (CG14996) all enhanced the Parvin-induced rough-eye phenotype (Figure 14H, J, L).
Discussion
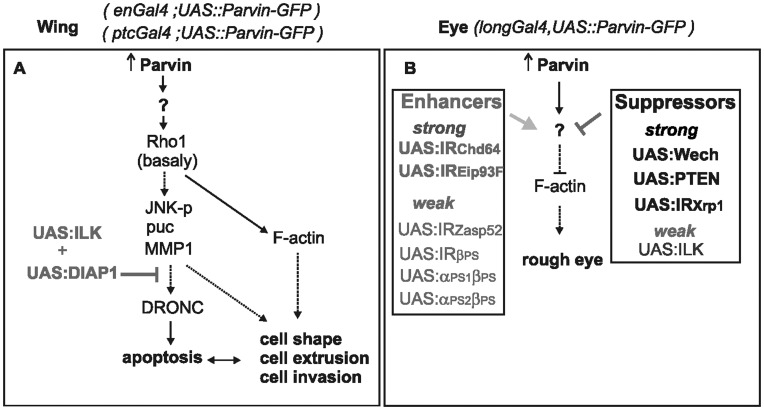
Parvin proteins are highly conserved and participate in the assembly and function of the integrin adhesome [3], [6]. Here we employed the UAS/Gal4 system to investigate additional functions of Parvin upon overexpression in a tissue specific manner and to identify novel genetic interactions in the wing and the eye (Figure 15).
Figure 15. Model of the genetic interactions of Parvin in the wing and eye.
(A) The consequences of elevated levels of Parvin in the wing epithelium based on data from this and previous studies [25], [26]. Parvin induces the upregulation of protein levels of Rho1 basally that leads to activation of the JNK pathway resulting in apoptosis. The abnormal levels of Rho1 could also affect the organization of F-actin leading to changes in cell shape, cell exclusion and cell invasion. Collectively these events can also enhance apoptosis and vice versa, as indicated by the double arrow. The coexpression of both ILK and DIAP1 suppresses the activation of DRONC and ameliorates cell exclusion. (B) The consequences of Parvin overexpression in the developing eye that lead to disruption of F-actin basally and the formation of a rough eye. This phenotype is suppressed or enhanced by overexpression or knockdown of specific genes.
We showed that Drosophila Parvin promoted apoptosis when overexpressed in vivo, similar to mammalian β-Parvin in HeLa cells [11]. Expression of β-Parvin in breast cancer cells was recently shown to inhibit tumor progression and cell proliferation [7] suggesting that our study of the cellular and molecular changes associated with Parvin overexpression in Drosophila may be relevant to cancer pathology. At the cellular level we demonstrated that overexpressed Parvin induced alterations in the organization of the actin cytoskeleton, disruption of cell-matrix adhesion, cell invasion and cell delamination. Mechanistically, we showed that overexpressed Parvin causes JNK activation and enhanced MMP1 levels. We also revealed a functional link between Parvin and subcellular distribution of Rho1. Interestingly, we showed that these Parvin-induced signaling effects are not dependent on its interaction with ILK.
Among the three counterparts of the ILK/PINCH/Parvin-complex, only overexpression of full-length Parvin induced ectopic apoptosis and excessive lethality in the larval and pupae developmental stages [6]. Nevertheless, in the wing imaginal discs overexpression of other components of the integrin adhesome such as tensin and paxillin also result in apoptosis and lethality, including activation of the JNK pathway and modulation of Rho1 activity, respectively [37], [38]. We showed that overexpression of Parvin increases Rho1 protein levels predominantly at the basal side of the wing epithelium, although loss of Parvin did not cause a reciprocal reduction of Rho1 levels [5], [6]. Given the previous reports that mammalian Parvins interact with two regulators of the small GPTases family, the GEF αpix and the CdGAP respectively [39], [40], one hypothesis would be that high levels of Parvin sequester these factors and interfere with their interaction with Rho1. As a consequence, Rho1 is released from the apicolateral side where normally is enriched [41]. The elevated Rho1 levels in the basal compartment of the epithelium could explain the formation of ectopic actin accumulation in accordance with previous studies [42]. As already described Rho1 is able to induce JNK-dependent apoptosis and F-actin organization defects in the wing epithelia cells [25], [26]. Therefore, it is plausible that the elevated JNK activity observed upon Parvin overexpression is caused by aberrant elevation of Rho1 basaly. Taken our findings together, we propose that Parvin-induced cellular defects in the wing epithelia are mediated by increased levels of Rho1, however, we cannot rule out a putative role of additional unidentified factors that are activated downstream of Parvin independently of Rho1.
We recently showed that coexpression of ILK together with Parvin-GFP in the mesoderm is sufficient to completely rescue Parvin-induced lethality and control Parvin subcellular localization [6], suggesting that coupling of Parvin to ILK could have a protective effect in epithelia viability. We performed rescue experiments to investigate whether Parvin function in the wing epithelium is mechanistically linked to its interaction with ILK, by coexpressing Parvin with ILK. Expression of ILK alone did not completely rescue the dominant effects of Parvin overexpression in the developing wing epithelia, had a mild suppressive effect on the rough eye phenotype and did not change the subcellular distribution of Parvin-GFP in the wing epithelial cells. Both the JNK activity and the increase in Rho1 protein levels were also not affected by ILK coexpression. Even when high levels of ILK are present, the putative interaction of Parvin with GTPase regulators is not disturbed and the imbalance of Rho1 subcellular distribution is maintained. That is not unexpected given that both αpix and CdGAP interact with the N-terminus region of Parvin, whereas ILK bind on the C-terminus. These findings demonstrate that the functional interplay between Parvin and ILK depends on the cell context and that Parvin interacts with other proteins and perform additional roles. In addition to functioning as a structural element of the integrin-actin link, it also acts as a dosage dependent modulator of actin cytoskeleton organization and cell homeostasis in the developing epithelia, via modulating the subcellular distribution of Rho1.
Because overexpression of Parvin caused extensive apoptosis in the wing epithelium, to mechanistically uncouple the Parvin-induced cellular defects from Parvin-induced apoptosis, we performed rescue experiments by coexpressing Parvin and DIAP1, which blocks apoptosis by inhibiting both the initiator caspase DRONC and the effector caspases DriCe and Dcp-1 [43]. DIAP1 alone did not efficiently suppress the cellular defects of Parvin in the wing. Both ILK and DIAP1 had to be coexpressed to completely rescue the lethality, presumably by coupling the reduction of excessive cytoplasmic Parvin by ILK and the inhibition of DRONC-mediated apoptosis by DIAP1. Coexpression of ILK and DIAP1 rescue both cell apoptosis and cell extrusion in the wing poutch cells, but not in the hinge and notum. These findings were not entirely unexpected, given previous documentation of regional differences within the wing imaginal disc regarding the differential requirement of actin regulators for epithelial integrity [44]. However, in consistence with our results from ILK rescue experiments, coexpression of DIAP1 or both ILK and DIAP1 did not ameliorate either the irregular organization of F-actin or the disorganized integrin-matrix adhesion sites and did not change the elevated levels of Rho1 in the basal side of the wing epithelium. These results demonstrate that the Parvin-induced cellular defects are not a simple consequence of apoptosis, but rather a distinct feature of Parvin function.
Overexpression of Parvin in the eye generated a rough eye phenotype. At the cellular level the basal actin cytoskeleton in the eye retina was severely disrupted, suggesting that a cause of the abnormal eye development could be initiated by abnormalities in the cell shape of pigment cells, as in the case of the wing epithelium. Because the Parvin-induced eye phenotype was sensitive to the copy number of Parvin transgenes and to temperature, we performed a modifier screen to uncover novel genetic interactors. We found that elevated levels of Wech and PTEN antagonized the Parvin-induced dominant effects in the developing eye and completely suppressed the rough eye phenotype, whereas high levels of ILK had only minimal suppression activity.
Wech is an ILK binding protein and it is not clear why it could suppress Parvin-induced dominant defects at elevated levels [33] rather than ILK itself, which directly binds to Parvin and rescues lethality completely in the mesoderm [6] and significantly in enGal4 expressing cells. The lack of data regarding Wech function in the eye, preclude further analysis at this point. The second surprising result of our study was the ability of high levels of PTEN to suppress the rough eye phenotype induced by Parvin overexpression. UAS::PTEN overexpression under GMRGal4 has been reported to induce a rough-eye phenotype by inhibiting cell-cycle progression in proliferating cells and inducing apoptosis in a cell-context dependent manner [35], [45]. In our experiments expression of the same UAS::PTEN lines obtained from two different donors [35], [45] did not result in eye roughening. One possible explanation could be the use of longGMRGal4 (Bloomington #8506) in our experiments, because previous studies drove expression of UAS::PTEN with GMRGal4 [46]. In addition, previous reports suggested that expression by longGMRGal4 driver in the developing eye follows a more strict pattern in the photoreceptor cells [47]. Taken together our data and previous reports, we speculate that Parvin and PTEN have antagonistic functions within the eye epithelium and coexpression of both proteins counterbalance their induced dominant effects upon overexpression. Currently we do not have sufficient data to point a specific pathway that could be modified by Parvin and PTEN and leads to rough eye phenotype. However, the recent report that Parvin is associated with PKB [48] together with previous data suggesting that Parvin may facilitates the recruitment of PKB at plasma membrane [10], suggests that Parvin could antagonized the negative effects of PTEN on PKB activation by reducing PIP3 levels [49].
The third suppressor gene we found was Xrp1. Xrp1 contains an AT-hook motif that is found in nuclear proteins with DNA binding activity. Currently, we lack sufficient knowledge to speculate on putative functional interaction between Parvin-induced signaling and nuclear activity. However, previous studies on Xrp1 point on its role as a p53-dependent negative regulator of cell proliferation following genotoxic stress [50]. Among the genes that enhanced the Parvin-induced rough eye were all of the integrin subunits known to be expressed in the eye, including αPS1, αPS2 and βPS [31], the cytoskeletal regulators ZASP52 [51] and the transgelin homolog encoded by CG14996 of unknown function.
In conclusion, our findings revealed novel cell context-dependent roles for Parvin in the whole organism. Besides its known function as a structural component of the IPP-complex that mediate the integrin-actin link, we demonstrated that Parvin can also affect cell-matrix adhesion, organization of actin cytoskeleton and cell homeostasis, by regulating Rho1 and JNK levels in an ILK-independent manner. These findings are relevant to situations where cell homeostasis is altered ranging from the physiological renewal of tissues to cancer pathology. In addition, our modifier genetic screen revealed novel interactors that affect Parvin function in a living organism. Our in vivo data provide the first insight into genetic circuits influenced by Parvin and offer a framework for additional detailed studies to elucidate how these genetic networks interact.
Acknowledgments
We thank Drs N. Brown, C. Delidakis, M. Hoch, A. Manoukian, T. Millard, M. Narasimha, S. Noselli, E. Skoulakis, M. Strigini, S. Wasserman, T. Xu for sharing fly stocks and antibodies. We thank Bloomington Drosophila Stock Center, VDRC (Vienna), NIG (Kyoto), Developmental Studies Hybridoma Bank at the University of Iowa (DSHB) for providing us with fly stocks and antibodies. We also thank Vasso Kostourou for critical reading of the manuscript, Stamatis Pagakis (Biological Imaging Unit of BRFAA) for imaging analysis suggestions, and Xiao Tan for his helpful comments on pupae dissection protocol.
Funding Statement
This research was supported by Greek Secretariat for Research and Technology (PENED-03ED777), Association Francaise contre les Myopathies (AFM;12641) and BRFAA intramural funds to CGZ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bokel C, Brown N (2002) Integrins in development: moving on, responding to, and sticking to the extracellular matrix. Dev Cell 3: 311–321. [DOI] [PubMed] [Google Scholar]
- 2. Geiger B, Yamada KM (2011) Molecular Architecture and Function of Matrix Adhesions Cold Spring Harb Perspect Biol. 3: a005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sepulveda JL, Wu C (2006) The parvins. Cell Mol Life Sci 63: 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wickstrom SA, Lange A, Montanez E, Fassler R (2010) The ILK/PINCH/parvin complex: the kinase is dead, long lived the pseudokinase! EMBO J. 29: 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Montanez E, Wickström SA, Altstätter J, Chu H, Fässler R (2009) Alpha-parvin controls vascular mural cell recruitment to vessel wall by regulating RhoA/ROCK signalling. EMBO J 28: 3132–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vakaloglou KM, Chountala M, Zervas CG (2012) Functional analysis of parvin and different modes of IPP-complex assembly at integrin sites during Drosophila development. J Cell Sci 125: 3221–3232. [DOI] [PubMed] [Google Scholar]
- 7. Johnstone CN, Mongroo PS, Rich AS, Schupp M, Bowser MJ, et al. (2008) Parvin-beta inhibits breast cancer tumorigenicity and promotes CDK9-mediated peroxisome proliferator-activated receptor gamma 1 phosphorylation. Mol Cell Biol 28: 687–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zervas CG, Gregory SL, Brown NH (2001) Drosophila integrin-linked kinase is required at sites of integrin adhesion to link the cytoskeleton to the plasma membrane. J Cell Biol 152: 1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gutzeit HO, Eberhardt W, Gratwohl E (1991) Laminin and basement membrane-associated microfilaments in wild-type and mutant Drosophila ovarian follicles. J Cell Sci 100: 781–788. [DOI] [PubMed] [Google Scholar]
- 10. Fukuda T, Guo L, Shi X, Wu C (2003) CH-ILKBP regulates cell survival by facilitating the membrane translocation of protein kinase B/Akt. J Cell Biol 160: 1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Y, Chen X, Tu Y, Wu C (2004) Distinct roles of two structurally closely related focal adhesion proteins, a-parvins and b-parvins, in regulation of cell morphology and survival. J Biol Chem 279: 41695–41705. [DOI] [PubMed] [Google Scholar]
- 12. Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- 13. Fan YaB, A. (2010 ) The cleaved-Caspase-3 antibody is a marker of Caspase-9-like DRONC activity in Drosophila. Cell Death Differ 17: 534–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Adachi-Yamada T, Fujimura-Kamada K, Nishida Y, Matsumoto K (1999) Distortion of proximodistal information causes JNK-dependent apoptosis in Drosophila wing. Nature 400: 166–169. [DOI] [PubMed] [Google Scholar]
- 15. Agnès F, Suzanne M, Noselli S (1999) The Drosophila JNK pathway controls the morphogenesis of imaginal discs during metamorphosis. Development 126: 5453–5462. [DOI] [PubMed] [Google Scholar]
- 16. Wang SL, Hawkins CJ, Yoo SJ, Müller HA, Hay BA (1999) The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell 98: 453–463. [DOI] [PubMed] [Google Scholar]
- 17. Page-McCaw A, Serano J, Sante JM, Rubin GM (2003) Drosophila matrix metalloproteinases are required for tissue remodeling, but not embryonic development. Dev Cell 4: 95–106. [DOI] [PubMed] [Google Scholar]
- 18. Brown NH, Gregory SL, Rickoll WL, Fessler LI, Prout M, et al. (2002) Talin is essential for integrin function in Drosophila. Dev Cell 3: 569–579. [DOI] [PubMed] [Google Scholar]
- 19. Domínguez-Giménez P, Brown N, Martín-Bermudo M (2007) Integrin-ECM interactions regulate the changes in cell shape driving the morphogenesis of the Drosophila wing epithelium. J Cell Sci 120: 1061–1071. [DOI] [PubMed] [Google Scholar]
- 20. Breitsprecher D, Kiesewetter AK, Linkner J, Vinzenz M, Stradal TE, et al. (2011) Molecular mechanism of Ena/VASP-mediated actin-filament elongation. EMBO J 30: 456–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Becam I, Rafel N, Hong X, Cohen SM, Milán M (2011) Notch-mediated repression of bantam miRNA contributes to boundary formation in the Drosophila wing. Development 138: 3781–3789. [DOI] [PubMed] [Google Scholar]
- 22. Thiery JP, Acloque H, Huang RY, Nieto MA (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139: 871–890. [DOI] [PubMed] [Google Scholar]
- 23. Bergantiños C, Corominas M, Serras F (2010) Cell death-induced regeneration in wing imaginal discs requires JNK signalling. Development 137: 1169–1179. [DOI] [PubMed] [Google Scholar]
- 24. Li W, Baker N (2007) Engulfment is required for cell competition. Cell 129: 1215–1225. [DOI] [PubMed] [Google Scholar]
- 25. Neisch AL, Speck O, Stronach B, Fehon RG (2010) Rho1 regulates apoptosis via activation of the JNK signaling pathway at the plasma membrane. J Cell Biol 189: 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vidal M, Larson DE, Cagan RL (2006) Csk-deficient boundary cells are eliminated from normal Drosophila epithelia by exclusion, migration, and apoptosis. Dev Cell 10: 33–44. [DOI] [PubMed] [Google Scholar]
- 27. Grzeschik NA, Knust E (2005) IrreC/rst-mediated cell sorting during Drosophila pupal eye development depends on proper localisation of DE-cadherin Development. 132: 2035–2045. [DOI] [PubMed] [Google Scholar]
- 28. Lin DMaG, C S (1994) Ectopic and increased expression of Fasciclin II alters motoneuron growth cone guidance. Neuron 13: 507–523. [DOI] [PubMed] [Google Scholar]
- 29. Tomlinson A, Bowtell DD, Hafen E, Rubin GM (1987) Localization of the sevenless protein, a putative receptor for positional information, in the eye imaginal disc of Drosophila. Cell 51: 143–150. [DOI] [PubMed] [Google Scholar]
- 30. Cagan R (2009) Principles of Drosophila eye differentiation. Curr Top Dev Biol 89: 115–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Longley RL, Ready DF (1995) Integrins and the development of three-dimensional structure in the Drosophila compound eye. Dev Biol 171: 415–433. [DOI] [PubMed] [Google Scholar]
- 32. Martin-Bermudo MD, Dunin-Borkowski OM, Brown NH (1997) Specificity of PS integrin function during embryogenesis resides in the a subunit extracellular domain. EMBO 16: 4184–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Löer B, Bauer R, Bornheim R, Grell J, Kremmer E, et al. (2008) The NHL-domain protein Wech is crucial for the integrin-cytoskeleton link. Nat Cell Biol 10: 422–428. [DOI] [PubMed] [Google Scholar]
- 34. Staveley BE, Ruel L, Jin J, Stambolic V, Mastronardi FG, et al. (1998) Genetic analysis of protein kinase B (AKT) in Drosophila . Current Biology 8: 599–602. [DOI] [PubMed] [Google Scholar]
- 35. Huang H, Potter CJ, Tao W, Li DM, Brogiolo W, et al. (1999) PTEN affects cell size, cell proliferation and apoptosis during Drosophila eye development. Development 126: 5365–5372. [DOI] [PubMed] [Google Scholar]
- 36. Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, et al. (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156. [DOI] [PubMed] [Google Scholar]
- 37. Lee SB, Cho KS, Kim E, Chung J (2003) blistery encodes Drosophila tensin protein and interacts with integrin and the JNK signaling pathway during wing development. Development 130: 4001–4010. [DOI] [PubMed] [Google Scholar]
- 38. Chen GC, Turano B, Ruest PJ, Hagel M, Settleman J, et al. (2005) Regulation of Rho and Rac signaling to the actin cytoskeleton by paxillin during Drosophila development. Mol Cell Biol 25: 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mishima W, Suzuki A, Yamaji S, Yoshimi R, Ueda A, et al. (2004) The first CH domain of affixin activates Cdc42 and Rac1 through alphaPIX, a Cdc42/Rac1-specific guanine nucleotide exchanging factor. Genes Cells 9: 193–204. [DOI] [PubMed] [Google Scholar]
- 40. LaLonde DP, Grubinger M, Lamarche-Vane N, Turner CE (2006) CdGAP Associates with Actopaxin to Regulate Integrin-Dependent Changes in Cell Morphology and Motility. Curr Biol 16: 1375–1385. [DOI] [PubMed] [Google Scholar]
- 41. Widmann TJ, Dahmann C (2009) Dpp signaling promotes the cuboidal-to-columnar shape transition of Drosophila wing disc epithelia by regulating Rho1. J Cell Sci 122: 1362–1373. [DOI] [PubMed] [Google Scholar]
- 42. Speck O, Hughes SC, Noren NK, Kulikauskas RM, Fehon RG (2003) Moesin functions antagonistically to the Rho pathway to maintain epithelial integrity. Nature 421: 83–87. [DOI] [PubMed] [Google Scholar]
- 43. Fan Y, Bergmann A (2008) Apoptosis-induced compensatory proliferation. The Cell is dead. Long live the Cell! Trends in Cell Biol 18: 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Janody F, Treisman JE (2006) Actin capping protein alpha maintains vestigial-expressing cells within the Drosophila wing disc epithelium. Development 133: 3349–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Scanga SE, Ruel L, Binari RC, Snow B, Stambolic V, et al. (2000) The conserved PI3′K/PTEN/Akt signaling pathway regulates both cell size and survival in Drosophila. Oncogene 19: 3971–3977. [DOI] [PubMed] [Google Scholar]
- 46. Kramer JM, Staveley BE (2003) GAL4 causes developmental defects and apoptosis when expressed in the developing eye of Drosophila melanogaster. Genet Mol Res 2: 43–47. [PubMed] [Google Scholar]
- 47. Ellis MC, O’Neill EM, Rubin GM (1993) Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development 119: 855–865. [DOI] [PubMed] [Google Scholar]
- 48. Kimura M, Murakami T, Kizaka-Kondoh S, Itoh M, Yamamoto K, et al. (2010) Functional molecular imaging of ILK-mediated Akt/PKB signaling cascades and the associated role of beta-parvin. J Cell Sci 123: 747–755. [DOI] [PubMed] [Google Scholar]
- 49. Radimerski T, Montagne J, Hemmings-Mieszczak M, Thomas G (2002) Lethality of Drosophila lacking TSC tumor suppressor function rescued by reducing dS6K signaling. Genes Dev 16: 2627–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Akdemir F, Christich A, Sogame N, Chapo J, Abrams JM (2007) p53 directs focused genomic responses in Drosophila. Oncogene 26: 5184–5193. [DOI] [PubMed] [Google Scholar]
- 51. Jani K, Schöck F (2007) Zasp is required for the assembly of functional integrin adhesion sites. J Cell Biol 179: 1583–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]