Abstract
According to the tRNA punctuation model, the mitochondrial genome (mtDNA) of mammals and arthropods is transcribed as large polycistronic precursors that are maturated by endonucleolytic cleavage at tRNA borders and RNA polyadenylation. Starting from the newly sequenced mtDNA of Ixodes ricinus and using a combination of mitogenomics and transcriptional analyses, we found that in all currently-sequenced tick lineages (Prostriata, Metastriata and Argasidae) the 3′-end of the polyadenylated nad1 and rrnL transcripts does not follow the tRNA punctuation model and is located upstream of a degenerate 17-bp DNA motif. A slightly different motif is also present downstream the 3′-end of nad1 transcripts in the primitive chelicerate Limulus polyphemus and in Drosophila species, indicating the ancient origin and the evolutionary conservation of this motif in arthropods. The transcriptional analyses suggest that this motif directs the 3′-end formation of the nad1/rrnL mature RNAs, likely working as a transcription termination signal or a processing signal of precursor transcripts. Moreover, as most regulatory elements, this motif is characterized by a taxon-specific evolution. Although this signal is not exclusive of ticks, making a play on words it has been named “Tick-Box”, since it is a check mark that has to be verified for the 3′-end formation of some mt transcripts, and its consensus sequence has been here carefully characterized in ticks. Indeed, in the whole mtDNA of all ticks, the Tick-Box is always present downstream of nad1 and rrnL, mainly in non-coding regions (NCRs) and occasionally within trnL(CUN). However, some metastriates present a third Tick-Box at an intriguing site - inside the small NCR located at one end of a 3.4 kb translocated region, the other end of which exhibits the nad1 Tick-Box - hinting that this motif could have been involved in metastriate gene order rearrangements.
Introduction
Chelicerates constitute a major lineage within Arthropoda and encompass taxa of evolutionary interest, such as the deep-branching lineage Xiphosura (including the living fossil Limulus polyphemus), and species of medical relevance, such as the Arachnida (e.g. ticks, mites, scorpions, spiders). Mitogenomic studies of chelicerates are thus conducted both to elucidate their evolutionary biology and to derive mitochondrial sequences for use in species identification. Ticks (Ixodida) are obligate blood-sucking ectoparasites that originated in early/middle Permian (300–260 million years ago, Mya) [1], [2], [3] and now parasitize a variety of terrestrial vertebrates [4], [5]. The approximately 870 described species of ticks are subdivided into three families: Argasidae, Ixodidae and Nuttalliellidae [6]. Ixodidae (hard ticks) can be divided in two morphological groups: Prostriata, including only the genus Ixodes, and Metastriata, including the remaining 12 genera [7]. Ticks can transmit a variety of pathogenic agents to humans and animals [4]. In particular, the sheep tick Ixodes ricinus (Linnaeus 1758), the most common blood-feeding ectoparasite in Europe, is the vector of Lyme disease and other bacteria, protozoa and viruses [8]. I. ricinus is also of particular interest in that it harbours a symbiont, “Candidatus Midichloria mitochondrii” [9], that resides in the intermembrane space of mitochondria. It can thus be considered a model of a three-levels relationship: the vertebrate host, the tick ectoparasite, and the intra-mitochondrial bacterium “Candidatus M. mitochondrii”.
Currently the complete mitochondrial genome (mtDNA) has been sequenced in 55 chelicerate species, including the living fossil L. polyphemus, whose gene order is considered to be ancestral for all arthropods [10], [11]. Chelicerate mitogenomes show several distinctive features compared to other arthropods: bizarre tRNA structures [12], [13], [14], [15]; unusual rRNAs [16], [17]; fast nucleotide substitution rate [18]; and extensive gene order rearrangements even between closely related species [17], [19], [20], [21], [22], [23], [24]. Indeed, among the 55 complete mtDNAs of chelicerates, the primitive gene order of L. polyphemus is shared only by two whip spiders from the order Amblypygi (Phrynus sp. and Damon diadema,), the mesothele spider Heptathela hangzhouensis, the scorpion Uroctonus mordax, and several tick species (suborder Ixodida) (see http://www.caspur.it/mitozoa).
Given the general interest in the Ixodida, we sequenced the complete mtDNA of the sheep tick I. ricinus. The comparison to other tick mtDNAs highlighted several oddities in the nad1 and rrnL genes that prompted us to investigate the transcription of these genes in all major tick lineages (Prostriata, Metastriata and Argasidae). Therefore, we carried out 3′ RACE experiments in I. ricinus, and mapped the exact 3′-end of these transcripts in several other ticks, using thousands of available tick EST sequences and according to the strategy described in Gissi et al. [25].
In this paper, after a brief summary of the main features of the I. ricinus mtDNA, we describe the identification of a degenerate 17 bp sequence motif directing the 3′-end formation of nad1 and rrnL transcripts in all major tick lineages. This motif represents an exception to the tRNA punctuation model, which predicts that arthropod mtDNA is transcribed in large polycistronic RNA precursors maturated through endonucleolytic cleavages and polyadenylation at sites immediately adjacent to tRNA genes [26], [27], [28]. Using genomics and transcriptional data, we also demonstrate the presence of a similar sequence motif, playing a similar function, downstream of the only nad1 gene in the basal chelicerate L. polyphemus and in the model hexapod Drosophila melanogaster. Finally, we illustrate a possible evolutionary scenario of this motif from chelicerates to hexapods. Making a play on word, we have named this motif “Tick-Box”, since it is a “check mark” that has to be verified for the 3′-end formation of nad1 and sometimes also rrnL transcripts, and its consensus sequence has been carefully characterized here, for the first time, in the “tick” group.
Methods
I. ricinus mtDNA Annotation and Analyses
The amplification and sequencing of the complete mtDNA of I. ricinus is described in Text S1. The mt sequence was deposited at EMBL database under accession number JN248424.
Protein coding genes (PCG) of I. ricinus were annotated by sequence similarity to the orthologous PCGs of other ticks. Partial stop codons were assumed only to avoid overlap with a downstream gene located on the same strand, while the 3′-end of nad1 was experimentally identified by 3′ RACE and EST analyses (see below). Overlaps between genes located on the same strand were kept as short as possible. tRNA annotation was performed comparing the predictions of tRNAscan-SE [29] and ARWEN [30] to the tRNAs annotated in other ticks (LocARNA multi-alignment [31]). Small (rrnS) and large (rrnL) ribosomal subunit rRNAs were identified by sequence similarity and their boundaries were settled as adjacent to those of the flanking genes. As an exception, the 3′-end of rrnL was experimentally determined by 3′ RACE and EST analyses (see below).
In the I. ricinus mtDNA analyses, the gene boundaries of the 10 previously published mtDNAs of Ixodida (Table 1) were revised based on sequence multi-alignment, transcriptional data, and the criterion of “minimum gene overlap”. Using this approach, we optimized the annotation of a total of 93 genes (i.e., 60 tRNAs and 33 PCGs) in 10 species, with up to 14 gene boundaries modified in I. hexagonus (data available on request).
Table 1. Completely sequenced mitochondrial genomes of Ixodida.
| Family | Group | Subfamily | Species | mtDNA |
| Ixodidae | Prostriata | Ixodinae | Ixodes ricinus | This study |
| “ | “ | “ | Ixodes hexagonus | NC_002010 |
| “ | “ | “ | Ixodes persulcatus | NC_004370 |
| “ | Australasian Prostriata | “ | Ixodes holocyclus | NC_005293 |
| “ | “ | “ | Ixodes uriae | NC_006078 |
| “ | Metastriata | Amblyomminae | Amblyomma triguttatum | NC_005963 |
| “ | “ | Haemaphysalinae | Haemaphysalis flava | NC_005292 |
| “ | “ | Rhipicephalinae | Rhipicephalus sanguineus | NC_002074 |
| Argasidae | Ornithodorinae | Carios capensis | NC_005291 | |
| “ | “ | Ornithodoros moubata | NC_004357 | |
| “ | “ | Ornithodoros porcinus | NC_005820 |
Secondary structures of the major non-coding region (the control region; CR) were predicted with Mfold [32].
Exact direct repeats longer than 9 bp were searched in the mtDNA sequences with RepFind [33], setting the P-value cut-off at 0.01 and with no filter for low-complexity sequences.
The Tick-Box motif was searched in complete and partial mt sequences using PatSearch [34], [35]. The Tick-Box consensus sequence (ttgyrtchwwwtwwgda) was defined as the sequence with the highest sensitivity in PatSearch analyses against all analysed Ixodida species. Tick-Box searches in the whole mtDNA sequences of two Xiphosura and 14 Drosophila species were carried out allowing mismatches and/or indels to the original consensus sequence. Tick-Box sequence logos [36] were generated by WebLogo [37] using all occurrences of the Tick-Box in the analysed species (Table S1). The possible presence of conserved secondary structure around the Tick-Box was verified by LocARNA [31].
Gene order, non-coding regions, and gene sequences of all mtDNAs analysed in this study were retrieved from MitoZoa Rel. 9.1 [38], [39] (http://www.caspur.it/mitozoa), a database collecting one representative and manually-curated mtDNA entry for each metazoan species. Therefore, the 474 complete nad1 sequences of arthropods analysed in this study were retrieved from MitoZoa Rel. 9.1.
3′ RACE of rrnL and nad1 Genes
Since both rrnL and nad1 transcripts are polyadenylated in Drosophila melanogaster [26], [27], [40], the 3′-end of these transcripts was identified by 3′ RACE (Random Amplification of cDNA End) or by identification of the start site of the polyA tail in mitochondrial ESTs, according to the method used in [25]. The 3′ RACE of nad1 and rrnL transcripts of I. ricinus was carried out using gene-specific inner (nad1-620pr and rrnL-1050pr) and outer (nad1-173pr and rrnL-850pr) primers (see Text S1).
One partially engorged adult female of I. ricinus was collected in Monte Bollettone (Como, Italy) and the total RNA was extracted following the total RNA isolation procedure of the mirVana™ miRNA Isolation Kit (Ambion). RNA was retrotranscribed to cDNA using an adaptor-ligated oligo (dT)-primer (FirstChoice RML-RACE Kit, Invitrogen) and the reverse transcriptase of the QuantiTect Reverse Transcription Kit (Qiagen). The first PCR reaction was assembled coupling the 3′ RACE outer adaptor primer (FirstChoice RML-RACE Kit, Invitrogen) with the nad1-173pr or rrnL-850pr primer. The second nested PCR reaction was assembled coupling the 3′ RACE inner adaptor primer (FirstChoice RML-RACE Kit, Invitrogen) with the nad1-620pr or rrnL-1050pr primer. All PCR reactions were performed in a total volume of 25 µl with 1.25 units of GoTaq (Promega), according to the manufacturer’s protocol. A single band of approximately the expected size was observed as product of the inner and outer PCRs in both the nad1 and rrnL 3′ RACE. In order to identify possible alternative polyadenylation sites located few nucleotides apart, nested PCR products were cloned (CloneJET PCR Cloning Kit, Fermentas) and a total of six positive clones were sequenced for each fragment. The partial RNA sequences of the I. ricinus rrnL and nad1 transcripts were deposited at EMBL database under accession numbers HE798553, HE798554 and HE798555.
EST Analyses of rrnL and nad1 Genes
EST sequences highly similar to the rrnL and nad1 genes of a given tick species were identified by Blast search [41] using as a probe the mt gene sequence of the same or of a congeneric species. Blast searches were carried out against the “Est_other” database that, at February 2012, included 297,856 ESTs of 20 Ixodida species. ESTs with statistically significant matches were assembled together with the corresponding mitogenomic sequence using Geneious [42]. The polyA start site was identified by visual inspection of the assembly. In particular, “A” or “T” stretches >10 bp located at the end of EST sequences were considered equivalent to the polyA tail of a mature transcript. In some cases, the lack of EST quality data and/or the presence of A stretches on the genomic mtDNA does not allowed mapping this site with single-nucleotide resolution, but only in a range of 2–5 nucleotides. The rrnL polyA site of Boophilus microplus and Dermacentor andersoni, and the nad1 polyA site of L. polyphemus were determined by analysis of the original untrimmed ESTs, kindly provided by the authors (see Acknowledgements).
Phylogenetic Analyses
Phylogenetic analyses were performed on the 13 PCGs of the 10 complete mtDNA of Ixodida (Table 1), using Argasidae as outgroup species. PCGs were aligned at the amino acid level with Muscle [43], and the equivalent nucleotide alignments were generated by “back-translation”. Ambiguous alignment regions were trimmed with Gblocks [44] using default parameters. The single PCG alignments were then concatenated with SEAVIEW [45].
Bayesian phylogenetic analyses were carried out on both amino acid and nucleotide alignments. The evolutionary models best fitting to the analyzed datasets were selected with ProtTest 1.4 [46] for amino acid, and ModelTest [47] for nucleotide datasets, according to the Akaike Information Criterion (AIC). The selected substitution model was the MtArt [48] with a proportion of invariant sites (I) and a gamma distribution for rate heterogeneity across sites (Γ) for the amino acid dataset, and the GTR+I+Γ for the nucleotide dataset [49]. Bayesian trees were calculated using MrBayes 3.1.2 [50]. Due to the absence of MtArt, the more general GTR and MtRev [51] model were applied in the amino acid analyses. Two different partitions based on the 13 genes and on the 3 codon positions were used in the nucleotide dataset analysis. One partition based on the 13 different proteins was used for the amino acid dataset. Two parallel analyses, each composed of one cold and three incrementally heated chains, were run for 2.5 million generations. Trees were sampled every 100 generations and burn-in fraction was calculated as 25% of total sampled trees, according to lnL stationary analyses.
Results and Discussion
Ixodes ricinus Genome Organization and Phylogeny
The mtDNA of I. ricinus is 14,566 bp long and encodes the 37 mt genes typical of other metazoans. The general features of this genome, together with peculiarities of the protein-coding genes (PCGs), the tRNA genes, the control region, and the small non-coding regions (NCRs) are illustrated in the Text S1, Figure S1 and Figure S2.
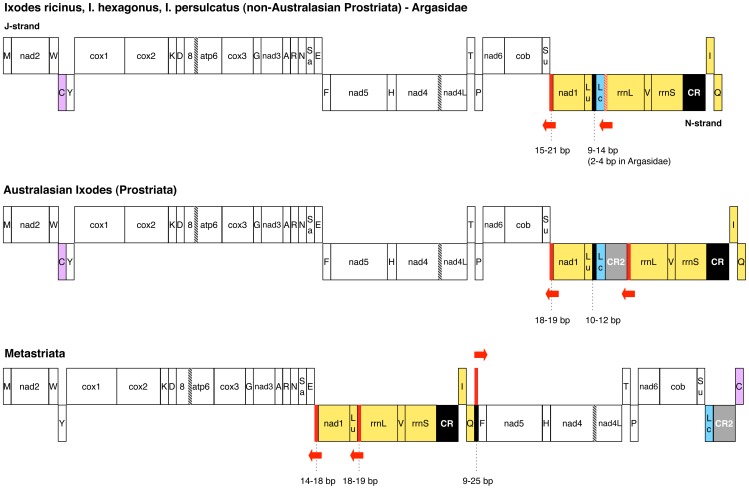
Figure 1 compares the genome organization of all available complete mtDNAs of ticks, taking also into account the location of the control region (CR), which contains the regulatory elements of mt transcription and replication. The genome organization of I. ricinus is identical to that found in all other available non-Australasian Ixodes species and in Argasidae (Figure 1). Since it is also shared with L. polyphemus, this organization is considered to be ancestral to all arthropods [10], [11], [52]. Australasian Ixodes species (I. uriae and I. holocyclus) have a genome organization almost identical to that of other Ixodes and Argasidae, except for the presence of a duplicate control region (CR2) between trnL(CUN) and rrnL (Figure 1), suggesting possible differences in mtDNA replication/transcription mechanisms [23]. With respect to the I. ricinus genome organization, the Metastriata exhibit: (1) the translocation of a large genomic block comprising 7 genes and the CR (yellow block in Figure 1); (2) the translocation plus inversion of trnC (violet blocks in Figure 1); (3) the presence of a duplicate CR2 between trnL(CUN) and trnC (grey blocks in Figure 1) [24], [53]. As already observed, the duplicate CR2s of both Metastriata and Australasian Ixodes exhibit concerted evolution and probably originated, together with the identified genome rearrangements, through two distinct events of tandem duplication and random gene loss [23], [53].
Figure 1. Mitochondrial gene arrangement of Ixodes ricinus and 10 other Ixodida species.
Translocated genes are reported in the same colour. Black block: non-coding regions ≥9 bp in all species of a taxonomic group, with bp range indicated by dashed lines; red block: Tick-Box within a non-coding region; red-hatched block: Tick-Box overlapped to trnL(CUN); red arrow: direction of the Tick-Box; black-hatched block: overlaps between genes; grey block: duplicated control region. The majority (J) and the minority (N) DNA strands, defined by the number of encoded genes, are also indicated. Gene abbreviations: 8, atp6: subunits 8 and 6 of the F0 ATPase; cox1-3: cytochrome c oxidase subunits 1–3; cob: cytochrome b; nad1–6 and nad4L: NADH dehydrogenase subunits 1–6 and 4L; rrnS and rrnL: small and large subunit rRNAs. tRNA genes are indicated by the one-letter code of the transported amino acid, with Lu: trnL(UUR); Lc: trnL(CUN); Sa: trnS(AGN); Su: trnS(UCN). Analysed mtDNAs are listed in Table 1.
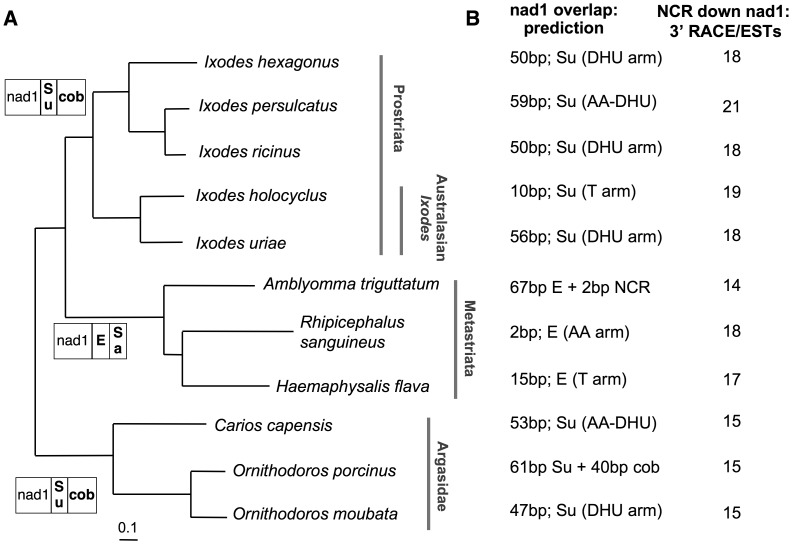
All Bayesian phylogenetic analyses of Ixodida, carried out on the 13 PCGs at both nucleotide and amino acid level, give congruent results and support the monophyly of the major Ixodida lineages. In particular, all phylogenetic reconstructions unambiguously identifies I. ricinus as sister taxon to I. persulcatus, with non-Australasian Ixodes positioned in a distinct highly supported clade (see nucleotide Bayesian tree in Figure 2A). This topology is in agreement with previous phylogenies based only on molecular data [2], [3], [54] or based on both morphological characters and nucleotide sequences (18S and 28S nuclear rRNAs; 16S mt rRNA) [55].
Figure 2. Features of the nad1 3′-end and the downstream non-coding region, mapped on the Ixodida phylogeny.
(A) Ixodida Bayesian tree calculated on the nucleotide sequence of the 13 mt protein-coding genes, and gene order downstream of nad1. Bayesian tree was calculated according to the GTR+I+gamma model, using 13 partitions, and all branches have a posterior probability value equal to 1. In gene order scheme, the genes encoded by the strand opposite to that of nad1 are reported in bold. Gene abbreviations as in Figure 1. (B) Predicted overlap between nad1 and the downstream gene, and length of the non-coding region experimentally identified downstream of nad1 by transcriptional data. tRNA regions containing the nad1 complete stop codon are indicated in brackets, with the following abbreviations: DHU: DHU arm; AA: amino acid acceptor arm; AA-DHU: spacer between the AA and DHU arms. Gene abbreviations as in Figure 1.
Partial Stop Codons and nad1 Annotation
In the mtDNA, partial stop codons are completed by polyadenylation of mature transcripts that are produced by endonucleolytic cleavages of precursor RNAs at sites immediately adjacent to tRNA genes [26], [27], [28]. It should be also noted that the usage of a partial stop codon eliminates the overlap between two consecutive genes (a PCG and a tRNA) encoded by the same strand, allowing the production of two full-length transcripts by cleavage of the same polycistronic RNA precursor. Thus, partial stop codons are commonly predicted according to the presence of an abutted tRNA gene and to the rule of “minimum overlap” between genes encoded by the same strand. In I. ricinus, the partial stop codons of five PCGs can be predicted according to the above-described rules (partial “T” stop codon in cox2, cox3, nad5, and cob; “TA” in nad2). On the contrary, the identification of the correct stop codon of nad1 is quite tricky because the 3′ end of this gene has unique peculiarities that do not fit to the known transcript maturation process and the derived annotation rules. In particular, nad1 is the only PCG followed by a gene encoded on the opposite strand (Figure 1). Therefore, based on the punctuation model of transcript maturation, the annotation of a partial stop codon is not strictly required in this case, since nad1 and the downstream gene are transcribed by two different strands. Moreover, the complete stop codon of the nad1 ORF is surprisingly located well inside the opposite strand-encoded trnS(UCN) gene, producing a large gene overlap of 50 bp.
Strikingly, even the 3′-ends of the nad1 genes/proteins currently annotated in all other ticks present similar unusual features.
Firstly, in almost all published tick mtDNA [23], [24], [53], the currently annotated 3′-end of nad1 has a complete stop codon located inside the first or even the second downstream, opposite strand-encoded, gene. This annotation gives rise to a gene overlap whose size is highly variable between species and ranges from 2 to 101 bp (Figure 2B). The most extreme case is in the argasid tick Ornithodoros porcinus, where the annotated nad1 contains the reverse complement of the entire downstream trnS(UCN), and the complete nad1 stop codon is located inside the following cob gene. Similarly, the nad1 ORF of the metastriate A. triguttatum contains the entire trnE gene. It is noteworthy that the predicted nad1 overlap size is not related to species phylogeny or gene order around nad1 (Figure 2), and that the currently annotated nad1 complete stop codons fall in different regions of the downstream tRNA gene, depending on the species (Figure 2B).
Secondly, assuming the veracity of these complete stop codons, the nad1 protein of Ixodida should have an extra C-terminal tail compared to the nad1 of D. melanogaster, ranging from 6 to 38 amino acids (20 amino acids in I. ricinus). The analysis of a multi-alignment of 474 nad1 proteins belonging to different arthropod species (see Materials and Methods) shows that this putative C-terminal tail is Ixodida-specific, being absent in all other available chelicerates (45 species) and in 96% of the whole arthropod dataset. Finally, this putative C-terminal tail has a low amino acid sequence similarity even within Ixodida (data not shown).
All these peculiarities prompted us to experimentally determine the actual nad1 stop codon of Ixodida by: (a) 3′ RACE in I. ricinus; (b) identification of the polyA start site of nad1 ESTs in all other tick species for which EST data are available.
In I. ricinus, the 3′ RACE analysis shows that the nad1 mRNA ends with a TAA stop codon created by the polyA tail and located exactly at the same position of the complete DNA-encoded stop codon of D. melanogaster (Figure 3). nad1 ESTs confirm this site in I. ricinus and in seven additional tick species (Ixodes scapularis, three Argasidae and three Metastriata species; see Table 2 and Figure 3). These data unambiguously demonstrate that, in all major Ixodida lineages, the putative C-terminal tail and the gene overlap of nad1, predicted in silico, result from the misannotation of the actual nad1 stop codon. Most importantly, the accurate annotation of the nad1 3′-end by transcriptional data identifies an unexpected NCR between nad1 and the downstream tRNA encoded by the opposite J-strand (i.e., trnS(UCN) in Argasidae and Prostriata, and trnE in Metastriata). This NCR has been identified in all the 11 analysed complete mtDNAs (Figure 2B) and in all the partial mt sequences available for other 17 tick species (Figure 3 and Table S1). We can thus conclude that the NCR downstream of nad1 is a common and ancestral character of the Ixodida mtDNA.
Figure 3. Non-coding region between nad1 and trnS(UCN)/trnE, and the Tick-Box degenerate consensus sequence.
Bold face: species listed in Table 2, for which the 3′-end of the nad1 transcript was experimentally determined by ESTs or 3′ RACE. Bold face only for the genus name: when the DNA sequence of a given species was unknown, the 3′-end of nad1 reconstructed by ESTs was mapped on the sequence of a congeneric species. Bold and underlined genus name: the nad1 3′-end of the argasid Argas monolakensis (Table 2) was mapped on the sequence of the argasid Carios capensis. Red colour: last DNA-encoded nucleotide preceding the nad1 polyA tail. Underlined nucleotides: complete stop codons predicted in silico; bold lower case nucleotides with grey background: differences to the Tick-Box consensus sequence; rpt: presence of a repeated sequence containing the Tick-Box (see main text). Degenerate nucleotide symbols according to the IUPAC code. Analyses species and sequence accession numbers are listed in Table S1. Gene abbreviations as in Figure 1.
Table 2. ESTs matching to the nad1 gene, and nad1 ESTs with a polyA stretch corresponding to the polyA tail of the mature nad1 transcript.
| Taxon | Species | AvailableESTsa | nad1 ESTs | mtDNAc | |
| tot | polyAb | ||||
| Prostriata | Ixodes ricinus | 1,969 | 2 | 2 | This study |
| Prostriata | Ixodes scapularis | 193,773 | 42 | 3 | congeneric |
| Argasidae | Ornithodoros coriaceus | 923 | 1 | 1 | congeneric |
| Argasidae | Ornithodoros parkeri | 1,563 | 1 | 1 | congeneric |
| Argasidae | Argas monolakensis | 2,914 | 7 | 6 | – |
| Metastriata | Amblyomma rotundatum | 1,230 | 2 | 1 | congeneric |
| Metastriata | Dermacentor variabilis | 2,090 | 8 | 7 | AY059254s1 |
| Metastriata | Boophilus microplus | 52,901 | 12 | 4 | AF110621 |
| Xiphosura | Limulus polyphemus | 8,488 | 12 | 7 | NC_003057 |
ESTs publicly available at 20 Feb, 2012.
ESTs with a terminal polyA stretch >10 bp, mapping to the end of nad1. Polyadenylated ESTs mapping well inside the nad1 gene have not been considered, as they are mostly cDNA artefacts originated from the annealing of the oligo-dT primer to an A-rich inner gene region during the cDNA first strand synthesis.
accession number of the mt genomic sequence, if available. “Congeneric” means that only the sequence of a congeneric species is available, as reported in Table S1.
As shown in Figure 3, this NCR is AT-rich (mean AT% = 76%), ranges from 14 to 30 bp in length, and is characterized by the presence of a degenerate 17 bp motif that includes the two last conserved nucleotides of nad1. Moreover, it can be observed that:
this degenerate motif is associated with the 3′-end of nad1 even when nad1 is translocated in Metastriata (Figure 1);
this motif is located at the boundaries between two large blocks of genes encoded by opposing genomic strands (Figure 1);
the polyA start site of the nad1 mRNA does not map at the boundary of the downstream tRNA gene in any analysed tick (Figure 3), thus excluding a nad1 transcript maturation according to the tRNA punctuation model [27], [28];
this motif is absent in the nad1 mature transcript, thus its sequence is either un-transcribed or quickly removed from the nad1 precursor transcript.
All these data suggest that this motif, that we have named the “Tick-Box”, directs the 3′-end formation of the polyadenylated nad1 transcripts in Ixodida, and likely works as a maturation signal for the cleavage of a large precursor transcript, or as a transcription termination signal.
We need to stress that this motif has been originally included inside the nad1 gene, and its identification has been made possible starting from the observations of: i) unusual position of the complete stop codon of nad1; ii) unusually large overlap between genes encoded by opposite strand; iii) an extra not-conserved C-terminal tail in the nad1 proteins of ticks. Thus, far from being a simple case of nad1 misannotation, this is an emblematic case that emphasizes how detailed analyses of unusual gene features can help to identify hidden functional element, and how gene misannotations can hamper the recognition of conserved regulatory elements.
The Tick-Box Downstream of rrnL
Sequences similar to the Tick-Box motif were sought along the entire mt sequences of all 11 ticks (Table 1) using pattern matching software, and were found to be present in only two or three fixed genomic positions (red blocks in Figure 1):
downstream of nad1;
near the 3′-end of rrnL;
inside a small NCR located between trnQ and trnF in some Metastriata.
Available partial mt sequences of 41 additional prostriates and metastriates (Table S1) contain Tick-Box motifs only in these genomic positions.
The exact location of Tick-Box motif near the 3′-end of rrnL depends on the taxa, indeed this Tick-Box falls:
in the DHU and anticodon arms of trnL(CUN) in Argasidae and non-Australasian Ixodes lineages (Figures 1);
at the end of CR2 in Australasian Ixodes (Figure 1);
a few bp upstream of the 3′-end of the currently annotated rrnL in Metastriata (Figure 1).
In order to study the potential functional role of the rrnL associated Tick-Box, we experimentally determined the 3′-end of rrnL transcripts through 3′ RACE in I. ricinus, and by using EST data in 10 other species (Table 3).
Table 3. ESTs matching to the rrnL gene, and rrnL ESTs with a polyA stretch corresponding to the polyA tail of the mature rrnL transcript.
| Taxon | Species | Available ESTsa | rrnL ESTs | polyA ESTsb | polyA start at: | mtDNAc | |
| 5′-end of Tick-Box | Other | ||||||
| Prostriata | Ixodes ricinus | 1,969 | 23 | 13 | 13 | 0 | This study |
| Prostriata | Ixodes scapularis | 193,773 | 33 | 8 | 8 | 0 | AB161439 |
| Argasidae | Ornithodoros coriaceus | 923 | 20 | 5 | 4 | 1 | congeneric |
| Metastriata | Amblyomma americanum | 6,480 | 852 | 30 | 30 | 0 | congeneric |
| Metastriata | Amblyomma rotundatum | 1,230 | 18 | 14 | 12 | 2 | congeneric |
| Metastriata | Amblyomma tuberculatum | 387 | 17 | 2 | 2 | 0 | congeneric |
| Metastriata | Hyalomma anatolicum | 736 | 5 | 5 | 2 | 3 | congeneric |
| Metastriata | Hyalomma marginatum | 2,110 | 27 | 24 | 18 | 6 | congeneric |
| Metastriata | Dermacentor andersoni | 1,387 | 67 | 25 | 16 | 9 | congeneric |
| Metastriata | Boophilus microplus | 52,901 | 296 | 130 | 115 | 15 | AF110619 |
| Metastriata | Rhipicephalus sanguineus | 2,899 | 428 | 57 | 50 | 7 | NC_002074 |
| Xiphosura | Limulus polyphemus | 8,488 | 2 | 2 | 0 | 2 | NC_003057 |
ESTs publicly available at 20 Feb, 2012.
ESTs with a terminal polyA stretch >10 bp, mapping to the end of rrnL. Polyadenylated ESTs mapping well inside the rrnL gene have not been considered, as they are mostly cDNA artefacts originated from the annealing of the oligo-dT primer to an A-rich inner gene region during the cDNA first strand synthesis.
accession number of the mt genomic sequence, if available. “Congeneric” means that only the sequence of a congeneric species is available, as reported in Table S1.
In I. ricinus, the rrnL polyadenylated transcript ends at two alternative sites, separated by 1 bp and located inside trnL(CUN), immediately before the 5′-end of the Tick-Box motif (red sites in Figure 4). Indeed, most rrnL 3′ RACE clones stop 14–19 bp inside trnL(CUN), while only one clone stops 11–12 bp inside trnL(CUN): the presence of one/multiple “A” nucleotides on the mitogenomic sequence prevents precisely mapping these polyA start sites. Even rrnL ESTs of I. ricinus confirm these two alternative 3′-ends of rrnL. Moreover, these ESTs do not provide support for the existence of rrnL transcripts terminating at the 5′-end of trnL(CUN), as predicted by the tRNA punctuation model.
Figure 4. The Tick-Box motif located downstream of rrnL.
Bold face: species listed in Table 3, for which the 3′-end of the rrnL transcript was experimentally determined by ESTs or 3′ RACE. Bold face only for the genus name: when the DNA sequence of a given species was unknown, the 3′-end of nad1 reconstructed by ESTs was mapped on the sequence of a congeneric species. The rrnL 3′-end of Hyalomma anatolicum and marginatum (Table 3) were both mapped on the sequence of Hyalomma aegyptium (Hyalomma_ae). Red colour: last DNA-encoded nucleotide preceding the rrnL polyA tail. Dashed line: overlap between rrnL and trnL(CUN). Genus names were abbreviated to a single letter for Ixodes (I), Haemaphysalis (H), Rhipicephalus (R) and Drosophila (D). Underlined nucleotides: tRNA anticodon; bold lower case nucleotide with grey background: differences to the Tick-Box consensus sequence; blue lines: original annotation of the rrnL 3′-end, with a dot indicating the presence of additional nucleotides; “repeat”: 71 bp-long inverted repeat located in the CR2 and rrnL gene of I. uriae (position 12431–12501 and 12606–12676, respectively, of NC_006078). Degenerate nucleotide symbols according to the IUPAC code. Analyses species and sequence accession numbers are listed in Table S1. Gene abbreviations as in Figure 1.
In I. scapularis, EST data identify the 3′-end of rrnL at two sites corresponding exactly to those found in I. ricinus (Table 3 and red sites in Figure 4). In Ornithodoros (Argasidae) and in all analysed metastriates, rrnL terminates always at the beginning of the Tick-Box. Moreover, in Ornithodoros an additional rrnL 3′-end site is located at the 5′-end of trnL(CUN), and in some metastriate species additional rrnL 3′-end sites can be observed very close to the 5′-end of the nearby trnL(UUR) gene, as predicted by the tRNA punctuation model (Table 3, and red sites in Figure 4). However, in each analysed species the majority of ESTs support the positioning of the rrnL 3′-end at the beginning of the Tick-Box motif (Table 3), suggesting that this site could be used more frequently than the other (given the different nature of the original cDNA libraries, definitive quantitative data cannot be inferred. Moreover, in some species the lack of EST quality data and/or of the mitogenomic sequence does not allow mapping of the rrnL polyA start site at single-nucleotide resolution). The lack of ESTs for Australasian Ixodes precludes validation of the 3′-end of rrnL in this lineage. However, based on sequence similarity to other Prostriata and on the lack of a tRNA abutted to rrnL, we hypothesize that in Australasian Ixodes species the 3′-end of rrnL occurs immediately before the identified Tick-Box motif (Figure 4).
In conclusion, as for nad1, transcriptional data are consistent with a functional role of the Tick-Box sequence in the 3′-end formation of polyadenylated rrnL transcripts. Indeed, in all analysed species the rrnL polyA tail starts immediately before or within the first 5 nt of the Tick-Box motif, independently of the gene/NCR downstream of rrnL. All additional rrnL polyadenylation sites, observed mainly in Metastriata, conform to the predictions of the tRNA punctuation model (i.e., they fall at the 5′-end of the downstream tRNA gene, considering the ambiguities due to EST quality) and appear infrequently used, as roughly estimated by the number of supporting ESTs (Table 3).
The presence of a Tick-Box near the 3′-end of rrnL is intriguing since a transcription termination signal has been functionally identified downstream of rrnL in Mammalia: this signal is a tridecamer sequence entirely contained in the trnL(UUR) gene [56], [57] and functions as a binding site for the mitochondrial transcription termination factor (mTERF) [58], [59]. Based only on sequence similarity to this mammalian tridecamer sequence, Valverde et al. [60] identified a “TGGCAGA” heptamer conserved downstream of rrnL from mammals to insects and protozoans, and hypothesized its function as an “rRNA termination box”. However, later functional studies have not validated the Valverde’s “rRNA termination box” as a binding site to the mTERF homologs of sea urchin and D. melanogaster [61], [62], [63], [64]. We need to stress that our Tick-Box does not coincide with the Valverde’s rRNA termination box either in sequence or exact genomic position. Moreover, unlike the rRNA termination box, our motif has been defined using both sequence similarity and transcriptional data. Finally, it should be noted that in Argasidae and non-Australasian Ixodes the exact location of the rrnL Tick-Box generates an overlap between rrnL and trnL(CUN) (dashed line in Figure 4). This situation recalls the overlap between rrnL and trnL(UUR) found in mammals because of the presence of the rrnL transcription termination signal inside trnL(UUR) [65].
As in the case of nad1, the determination of the rrnL 3′-end by transcriptional data has allowed the discovery of: i) an unexpected NCR downstream of rrnL in Metastriata (11–22 bp long); ii) an overlap between rrnL and trnL(CUN) in Argasidae and non-Australasian Ixodes (12–19 bp long; see dashed line in Figure 4); iii) the misannotations of rrnL in most ticks (Figure 4). However, we need to emphasize that the determination of the exact boundaries of rrnL only by comparative analyses is complicated by difficulties in the prediction of the rRNA secondary structure and by the low sequence conservation at both ends of this gene.
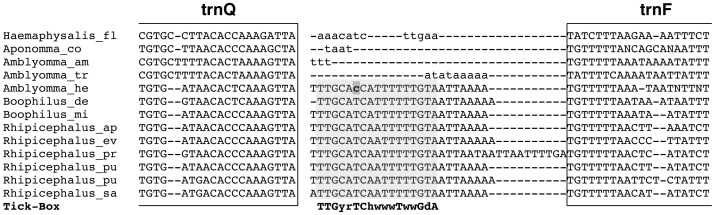
The Tick-Boxes of Metastriata
As shown in Figure 5, a third Tick-Box motif is located in the NCR between trnQ and trnF in 9 out of 13 analysed metastriates (complete and partial mtDNAs, see Table S1). In the remaining 4 metastriates, the trnQ-trnF NCR is always shorter than 12 bp, and does not contain an even partial Tick-Box sequence.
Figure 5. Tick-Box motif located in the NCR between trnQ and trnF of Metastriata.
Bold lower case with grey background: differences to the Tick-Box consensus sequence. Analysed species and sequence accession numbers are listed in Table S1.
This third Tick-Box is characterized by several oddities:
It is always on the opposite strand compared to the two Tick-Boxes situated downstream of nad1 and rrnL in the same genome (red arrows in Figure 1);
It is located in a NCR shared only by Metastriata, since the trnQ-trnF gene adjacency is specific of the metastriate gene rearrangement. Thus, if present, this third Tick-Box sequence gives rise to an inverted repeat (21 bp-long) that flanks the large translocated mt region of Metastriata ranging from nad1 to trnQ (yellow block in Metastriata of Figure 1). Even more surprisingly, in B. microplus [66] this large translocated mt region is preceded by a fivefold tandem repeat (126 bp unit) composed of trnE+Tick-Box+3′-end of nad1, and is followed by a single inverted copy of the Tick-Box sequence;
The phylogenetic distribution of this third Tick-Box is quite erratic, since it is absent in Haemaphysalinae, present in Rhipicephalinae, and present/absent even in congeneric species of Amblyomminae (Figure 5, and Table S1). Thus, it is difficult to discriminate between ancient or recent origins of this third Tick-Box.
As further peculiarity, the Tick-Box sequences present in the same mtDNA of metastriates are almost identical (maximum of 2 nt differences, observed only in one among the 13 analysed species), while the Tick-Boxes present in the same mtDNA of Argasidae and Prostriata species differ for 3–6 nucleotides. More interestingly, in the three complete mtDNAs of metastriates, the Tick-Boxes downstream of nad1 and rrnL are located inside a perfect direct repeat of 28–30 bp. On the contrary, perfect direct repeats of the same size are absent in Argasidae and Prostriata. These data suggest that the Metastriata Tick-Box motifs likely undergo to concerted evolution, as the duplicated CR2 of these taxa [24], [53]. It should be noted that this observation does not hold for Australasian Ixodes, where the intra-genome Tick-Boxes differ for 4–5 nucleotides and the identified duplicated CR2s also evolve by concerted evolution [23]. Although we have no convincing explanations for this observation, we hypothesize that the strong intra-genome Tick-Box conservation in Metastriata is related to the peculiar mt gene arrangement of this taxon.
The functional role of this third Tick-Box is enigmatic, and the absence of EST data for the trnQ-trnF region complicates the verification of possible functional hypotheses. However, since the sequence of this third Tick-Box is almost identical to that of functional Tick-Boxes identified in the same genome, we suggest that even the third Tick-Box is functional. We could tentatively hypothesize that this third Tick-Box motif plays the role of terminating the transcription of the J-strand, started at the CR, downstream of trnI. Indeed, in metastriates the movement of trnI far away from the cluster of other J-encoded genes makes J-strand transcription after trnI pointless (compare the J-strand gene distribution of Prostriata/Argasidae to Metastriata in Figure 1). Such a role in the rearranged mtDNAs might have represented a selective constraint for the conservation of the third Tick-Box in Metastriata. Finally, the presence of the Tick-Box motif at both ends of the large translocated mt regions of Metastriata (yellow blocks in Figure 1) might suggest its involvement in recombination events responsible for genome rearrangements. Indeed, signs of recombination have been found in several chelicerates based on the observation of concerted evolution, gene conversion, and translocation of genes to the opposite strand [17], [20], [23], [67].
Origin and Evolution of Tick-Box
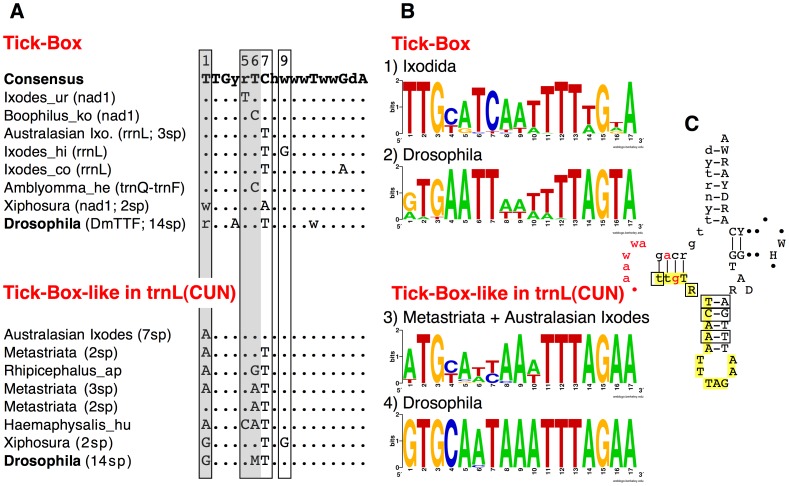
Figure 6A shows the consensus sequence of the Tick-Box motif and few rare variants, differing only in 1 or 2 positions. Noteworthy, the Tick-Box consensus sequence is quite degenerate, showing nucleotide ambiguity codes in almost half of the 17 sites (Figure 6A). This relatively high degeneration of the Tick-Box consensus is in accordance with its nature of regulatory element, and can be related to its possible functioning through interactions with one or more nuclear-encoded proteins. Thus, as usual for regulatory elements, the precise sequence of the Tick-Box is quite different from one species to the other, and we expect this element to be subject to a taxon-specific evolution. In this respect, the Tick-Box shows an evolutionary pattern very similar to the CR, a mt region also known to evolve in a taxon-specific way [68]. Remarkably, in addition to the control region, the Tick-Box is the only NCR conserved in all Ixodida species (Figure 1), while all other NCRs of ticks are unalignable (even those located at the same relative position in different species) and mainly shorter than 9 bp (see Text S1).
Figure 6. Tick-Box and Tick-Box-like sequences inside trnL(CUN) of Chelicerata and Drosophila species.
(A) Consensus and variants Tick-Box motifs of Ixodida, Xiphosura, and Drosophila species, together with non-functional Tick-Box-like sequences overlapping trnL(CUN). Boxes: positions with nucleotide differences between Tick-Box and Tick-Box-like sequences; grey background: crucial positions discriminating functional Tick-Box from non-functional Tick-Box-like sequences (see main text). In brackets is reported the genomic position of the sequence (i.e., downstream nad1, downstream rrnL, or between trnQ-trnF) and the number of species (sp) showing that sequence. DmTTF: consensus binding sites of DmTTF in Drosophila, considering sequences located downstream nad1 and between trnE-trnF. Analysed species and sequences are listed in Table S1. (B) Sequence logo for: (1) Tick-Box sequences of Ixodida; (2) DmTTF binding site of 14 Drosophila species, including both the sequences downstream of nad1 and between trnE-trnF; (3) Tick-Box-like sequences inside trnL(CUN) of Metastriata and Australasian Ixodes; (4) Tick-Box-like sequences inside trnL(CUN) of 14 Drosophila species. Sequence logos were generated as described in Materials and Methods, using sequences listed in Table S1. (C) Consensus sequence and secondary structure of the trnL(CUN) genes of Argasidae and non-Australasian Ixodes containing a functional Tick-Box. Boxes: positions with nucleotide differences between Tick-Box and Tick-Box-like sequences; yellow background: Tick-Box motif; red colour: polyA starts sites determined by 3′ RACE or ESTs in Argasidae and non-Australasian Ixodes; lower case: overlap region between rrnL and trnL(CUN); dot symbol: indels. Degenerate nucleotide symbols according to the IUPAC code. Analysed sequences are listed in Table S1.
The analysis of the single Tick-Box motifs indicates that the Tick-Box does not form a secondary structure, neither alone nor including surrounding sequences. The only exceptions are the few Tick-Boxes located downstream rrnL in non-Autralasian Ixodes and Argasidae, that are characterized by the overlapping with trnL(CUN) (Figure 6C). In these cases, the identified secondary structure has been evolutionary preserved because of the functional constraints of the tRNA gene rather than of the presence of the Tick-Box regulatory element (see also below). Therefore, the Tick-Box appears very different from the other hypothesized mt transcript processing sites. Indeed, in some cases, the absence of a tRNA punctuation mark has been supposed to be compensated by stem-loop structures resembling a tRNA portion [28].
In order to define the evolutionary origin of the Tick-Box, we have carefully investigated the presence of the Tick-Box in the basal chelicerate Xiphosura and in Drosophila, a highly derived insect genus belonging to the relatively recent Diptera lineage (divergence 228–245 Mya [69], [70]). These taxa have been selected due to their peculiar phylogenetic position and also because of the availability of a large amount of ESTs, useful for mt transcripts analyses. Moreover, there are several functional studies on the mt transcription of D. melanogaster [26], [27], [40], [64], [71], and the complete mtDNA is available for 14 congeneric Drosophila species (Table S1).
As for Xiphosura, we have considered the horseshoe crabs L. polyphemus (for which mtDNA and ESTs are available) and Tachypleus tridentatus (for which only the mtDNA is available). Our Xiphosura analyses show that:
Both species have a Tick-Box sequence (not perfectly matching to the Ixodida consensus) near the 3′-end of nad1. This Tick-Box includes the predicted nad1 complete stop codon and a short downstream NCR (Figure 3). The ESTs of L. polyphemus show that the mRNA of nad1 terminates immediately upstream of the Tick-Box sequence with a partial stop codon located at the same position of that of Ixodida (Table 2 and Figure 3). Thus, in Xiphosura the existence of a functional Tick-Box motif downstream of nad1 is supported by both transcriptional and sequence data.
A divergent Tick-Box sequence (3 mismatches compared to the Ixodida consensus) can be identified near the 3′-end of rrnL, exactly inside trnL(CUN), in both horseshoe crabs (Figure 4). However, ESTs of L. polyphemus show that the 3′-end of rrnL transcript is not located at the beginning of the Tick-Box sequence but just at the 5′-end of trnL(CUN), i.e., at the site predicted by the tRNA punctuation model (Figure 4 and Table 3). In conclusion, in Xiphosura a functional Tick-Box motif is absent downstream of rrnL, and the similar sequence identified inside trnL(CUN) probably results from the functional constraints of trnL(CUN).
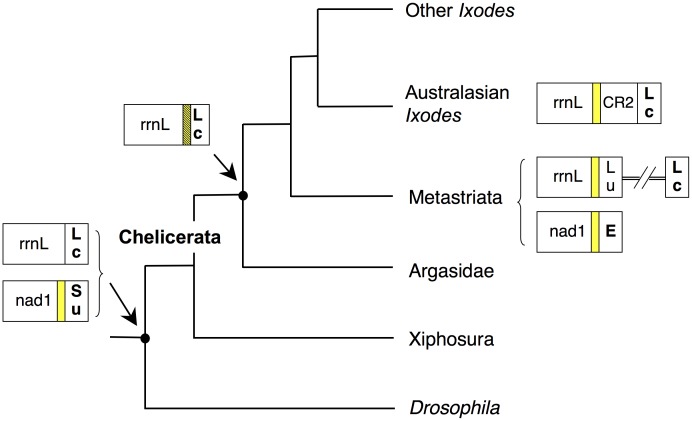
Based on these data, we suggest that the Tick-Box downstream of nad1 is an ancient signal that has been functionally conserved, in spite of the sequence changes, at least over the time separating Xiphosura from Ixodida (about 400 million years), while the Tick-Box downstream of rrnL is a specific invention of Ixodida (Figure 7). We hypothesize that the Tick-Box downstream of rrnL has evolved from a portion of trnL(CUN), through acquisition of a new function related to post-transcriptional regulation (Figure 7). After this gain-of-function, the trnL(CUN) and the Tick-Box have become overlapped elements and have coevolved in Ixodida for long time, until genome rearrangement events have disrupted the adjacency rrnL-trnL(CUN) (two independent events: one in Metastriata and the other in Australasian Ixodes). We suggest that in these rearranged mtDNAs, the sequence including the two overlapped Tick-Box and trnL(CUN) elements has been duplicated, and then the two copies have started diverging. In particular, due to the need to regulate the rrnL 3′-end formation, the Tick-Box function has been preserved at the position immediately downstream of rrnL, where the trnL(CUN) function has been instead lost. On the contrary, in these rearranged mtDNAs, the Tick-Box function has been disrupted in the position actually preserving the trnL(CUN) function (Figure 7). Based on the proposed evolutionary scenario, the Tick-Box sequence downstream of rrnL in Metastriata and Australasian Ixodes should be the only remnant of a duplicated trnL(CUN)/Tick-Box sequence that has lost all but the essential rrnL post-transcriptional regulatory motif.
Figure 7. Evolutionary scenario of the Tick-Box motif in Ixodida and other arthropods.
Tree topology according to [80]. Yellow block: Tick-Box motif; hatched yellow background: Tick-Box overlapped to trnL(CUN); bold case: genes encoded by the J-strand.
As for Drosophila, no sequence identical to the Tick-Box consensus motif is present in the whole mtDNA of D. melanogaster and congeneric species. However, the D. melanogaster nad1 gene is followed by a 17 bp-long NCR that is one of the two binding sites of the DmTTF transcription termination factor, the other site being an almost identical sequence located between trnE and trnF [64], [71]. The consensus of the DmTTF binding site for the 14 available Drosophila species matches to the Tick-Box degenerate consensus of Ixodida in all but 3–4 positions (Figure 6A; logos n° 1 and n° 2 in Figure 6B). Noteworthy, the D. melanogaster nad1 transcript is not 3′-processed at the site predicted by the tRNA punctuation model [27] but it terminates 16 bp upstream of the 5′-end of trnS(UCN) and 1 bp downstream the complete nad1 stop codon (red colour in Figure 3). It should be also noted that nad1 ends with a partial stop codon in 6 out of the 13 additional Drosophila mtDNAs, and that in all 14 available Drosophila species nad1 is followed by a NCR ranging from 15 to 25 bp [72] and having a 41–65% identity to the I. ricinus NCR downstream of nad1. We conclude that the Drosophila has a Tick-Box signal downstream of nad1 but this Tick-Box has a sequence quite divergent from the Ixodida consensus (Figure 7). This sequence variability between taxa follows the expected evolutionary pattern for a regulatory element, thus it is likely that the Tick-Box signal downstream of nad1 is also present in other arthropod lineages with even more divergent sequences. We need also to emphasize that the Tick-Box of Drosophila functions as a binding site of DmTTF [64], [71].
In D. melanogaster the 3′-end of the rrnL polyadenylated transcript falls exactly at the site predicted by the tRNA punctuation model [27] (i.e., at the 5′-end of trnL(CUN); see Figure 4), and no DmTTF binding site is present immediately downstream of rrnL [64]. However, we identified a sequence similar to the Tick-Box inside the trnL(CUN) gene, which is located in all 14 Drosophila species just downstream of rrnL. This sequence shows 3 mismatches to the Tick-Box consensus sequence of Ixodida (Figure 4, Figure 6A, logo n° 4 of Figure 6B) and 76% identity to the I. ricinus Tick-Box inside trnL(CUN). As for horseshoe crabs, we conclude that in Drosophila there is no functional Tick-Box downstream of rrnL (Figure 7) and that the observed sequence conservation is due to the functional constraints of trnL(CUN). It should be also noted that, in the comparison D. melanogaster - I. ricinus, the identity percentage is higher in the Tick-Box-like sequences overlapping trnL(CUN) than in the functional Tick-Boxes located in the NCR downstream nad1 (76% and 65%, respectively). This indicates that the overlapping of Tick-Box to trnL(CUN), and the degenerate nature of this Tick-Box regulatory signal can lead to misinterpretation of the Tick-Box presence/absence especially in taxa phylogenetically distant from Ixodida, and especially when only sequence similarity data are taken into account.
Coevolution of Tick-Box and trnL(CUN)
To better investigate the coevolution of Tick-Box and trnL(CUN), we have compared the identified Tick-Box motif to the similar sequences found inside the trnL(CUN) genes that lack a functional Tick-Box (Figure 6A). This comparison can assist in the identification of nucleotide positions discriminating functional Tick-Boxes from non-functional Tick-Box-like sequences overlapping trnL(CUN). The Tick-Box-like sequences overlapping trnL(CUN) were defined as non-functional based on EST data and mismatches to the Tick-Box consensus, and are present in the trnL(CUN) of Metastriata, Australasian Ixodes, Xiphosura, and all Drosophila species (see also Figures 3, 4). The logo of the Tick-Box-like sequences located inside trnL(CUN) is shown in Figure 6B, separately for ticks (logo n° 3) and Drosophila species (logo n° 4). Moreover, Figure 6C illustrates the consensus of functional Tick-Boxes located inside trnL(CUN) in non-Australasian Ixodes and Argasidae: as already discussed, these are the only Tick-Box elements showing a conserved secondary structure, since they are superimposed to a functional tRNA gene (see above).
As shown in Figure 6C (yellow background), the Tick-Box sequence inside trnL(CUN) is superimposed to half of the DHU and anticodon stems, plus the entire anticodon loop. According to the typical tRNA substitution pattern, the nucleotide substitutions observed in the trnL(CUN) with Tick-Box (boxed positions in Figure 6C) are mainly compensatory substitutions falling in the stem regions, while substitutions carefully avoid the anticodon loop. As reported in Figure 6A, the Tick-Box-like sequences inside trnL(CUN) differ in 2–4 positions from the consensus Tick-Box. As an exception, the Tick-Box-like sequence of Australasian Ixodes differs from the consensus Tick-Box only for a single substitution (T−>A) at position 1. Among positions with differences, we observe that positions 7 and 9 share the same substitution types in both Tick-Box variants and Tick-Box-like sequences (Figure 6A), thus these positions seem not to be crucial for the Tick-Box functionality. On the contrary, positions 5 and 6 (grey background in Figure 6A) have different substitution types in Tick-Box and Tick-Box-like sequences, indicating that they can be discriminating positions for the Tick-Box functionality. Finally, nucleotide substitutions at the first position of the consensus seem to inactivate the Tick-Box depending on the substitution type, the additional substitutions co-occurring in other positions, and the taxon (i.e., compare Xiphosura and Drosophila Tick-Box and Tick-Box-like sequences in Figure 6A). Thus, from the comparison between functional Tick-Boxes and Tick-Box like sequences, we can conclude that positions 1, 5 and 6 are the most important sites for the functionality of Tick-Box.
Overall, these data further support the hypothesis that Tick-Box is a highly dynamic and degenerate signal whose sequence variability is due to its specific regulatory function (i.e., the possible interaction with regulatory proteins encoded by the nuclear genome) and also to the overlap with coding sequences.
Conclusions
In this study we describe the identification of the Tick-Box, a degenerate 17-bp DNA motif involved in post-transcriptional processes. In particular, Tick-Box directs the 3′-end formation of nad1 and rrnL transcripts in all Ixodida lineages, as well as the 3′-end formation of the single nad1 transcript in basal chelicerates of the Xiphosura order and in Diptera insects of the Drosophila genus. Although this motif is not restricted to tick species, it has been named “Tick-Box” because its consensus sequence has been here carefully characterized in Ixodida and because it is a “tick box” necessary for the 3′-end formation of some mt transcripts. We have not investigated in details the phylogenetic distribution of this motif in Chelicerata and Arthropoda, however its presence in Drosophila and Limulus suggest that it could be a quite ubiquitous signal, whose existence has been obscured by its taxon-specific evolutionary pattern and by its nature of post-transcriptional regulatory element. Indeed, as most regulatory elements, Tick-Box is a short and degenerate motif, showing a low sequence similarity within Ixodida and even lower sequence conservation in the more distant species of Limulus and Drosophila. Therefore, additional studies combining sequence similarity and transcriptional analyses are needed to define the Tick-Box consensus sequence in other arthropods and to clarify its phylogenetic distribution in the main arthropod groups.
With regard to the exact Tick-Box function, this element is associated to the 3′-end of the nad1 and rrnL genes independently of the downstream gene/NCR. Moreover, it is absent in the mature transcripts. Therefore, we suggest that Tick-Box is either un-transcribed or quickly removed from the primary precursor transcripts of nad1 and rrnL. According to this observation, Tick-Box might be one of the few exceptions to the tRNA punctuation model of mt transcript maturation [28] or a transcription termination signal, whose existence was originally hypothesized in D. melanogaster by Berthier [26]. Remarkably, the Tick-Box downstream of nad1 found in D. melanogaster has been functionally described some time ago as one of the two binding sites of the DmTTF transcription termination factor [71]. Far from reducing the novelty of this study, the similarity between the DmTTF binding site of D. melanogaster and the Tick-Box downstream nad1 of Ixodida supports the functional role of Tick-Box as a transcription termination site. Moreover, it testifies the poor link between functional and evolutionary studies on the mtDNA, and the difficulties of mere mt comparative analyses in the detection of regulatory elements. Indeed, to our knowledge, after its functional characterization, the binding site of DmTTF has not been further investigated at level of taxonomic distribution, consensus sequence or exact mitogenomic location(s) within the numerous available mtDNA sequences of arthropods.
The discrimination between the two hypothesized Tick-Box functions, precursor transcript maturation or transcription termination, can be experimentally tested in Prostriata and Metastriata by qualitative and quantitative analyses of the whole mt transcriptome and/or experiments aimed at demonstrating the binding of this motif by mt regulatory proteins, such as members of the MTERF protein family [73], [74]. The availability of cell lines for both these tick taxa can also help these analyses [75], [76].
Finally, we would like to emphasize that the small Tick-Box and the large mt control region are the only non-coding regions conserved in all mtDNAs of ticks. To our knowledge, there is only one other small NCR conserved in all mtDNAs of a large metazoan group, i.e. the L-strand replication origin (oriL) of vertebrates [77], [78]. The oriL is a 20–30 bp sequence embedded in a tRNA cluster and forms a stable stem-loop structure partially overlapped to trnC. On the contrary, the Tick-Box is a degenerate DNA motif that does not show a conserved secondary structure and, like the control region, is characterized by a taxon-specific evolution. Moreover, based on the presence of a third Tick-Box in Metastriata and of a second DmTTF binding site in D. melanogaster, we anticipate the presence of Tick-Box in different mitogenomic positions depending on the overall genome organization and on the details of the transcriptional process (i.e., number and type of transcriptional units).
Supporting Information
Putative secondary structure of the 22 tRNAs of I. ricinus .
(PDF)
Conserved motifs and secondary structures of the tick control region, mapped on the I. ricinus sequence.
(PDF)
Accession numbers of 98 mitochondrial sequences belonging to 68 species analysed in this study.
(XLS)
Amplification strategy and general features of tRNAs, control region, and small non-coding regions of the I. ricinus mtDNA.
(DOC)
Acknowledgments
We thanks Thorsten Burmester, Karen Meusemann, Bernhard Misof, Francisco J. Alarcon-Chaidez, and Felix D. Guerrero for kindly providing untrimmed EST sequences of several tick species and of Limulus polyphemus; Adriana Giumbo, David S. Horner, Giulio Pavesi, Graziano Pesole, Paola Loguercio-Polosa and Marina Roberti for helpful discussion and comments on the manuscript.
Funding Statement
This work was supported by Ministero dell’Istruzione, dell’Università e della Ricerca, Italy (MIUR: PRIN-2009 to CB and PRIN-2009 to CG); European Cooperation in Science and Technology (Cost Action FA0701 to CB) and Università degli Studi di Milano (PUR project to CG). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dunlop JA, Selden PA (2009) Calibrating the chelicerate clock: a paleontological reply to Jeyaprakash and Hoy. Exp Appl Acarol 48: 183–197. [DOI] [PubMed] [Google Scholar]
- 2. Jeyaprakash A, Hoy MA (2009) First divergence time estimate of spiders, scorpions, mites and ticks (subphylum: Chelicerata) inferred from mitochondrial phylogeny. Exp Appl Acarol 47: 1–18. [DOI] [PubMed] [Google Scholar]
- 3. Mans BJ, de Klerk D, Pienaar R, Latif AA (2011) Nuttalliella namaqua: a living fossil and closest relative to the ancestral tick lineage: implications for the evolution of blood-feeding in ticks. PLoS One 6: e23675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonenshine DE (1991) Biology of ticks. Vol. 1. New York: Oxford Univ. Press. 447 p.
- 5.Sonenshine DE (1993) Biology of ticks. Vol. 2 New York: Oxford Univ. Press. 465 p.
- 6. Horak IG, Camicas JL, Keirans JE (2002) The Argasidae, Ixodidae and Nuttalliellidae (Acari: Ixodida): a world list of valid tick names. Exp Appl Acarol 28: 27–54. [DOI] [PubMed] [Google Scholar]
- 8.Keirans JE, Needham GR, Oliver JH (1999) The Ixodes (Ixodes) ricinus complex worldwide: Diagnosis of species in the complex, host and distribution. In: Glen R, Needham, Mitchell R, Horn DJ, Welbourn WC, editors. Acarology IX. Columbus, Ohio: The Ohio Biological Survey. 344.
- 9. Sassera D, Beninati T, Bandi C, Bouman EA, Sacchi L, et al. (2006) Candidatus Midichloria mitochondrii, an endosymbiont of the tick Ixodes ricinus with a unique intramitochondrial lifestyle. Int J Syst Evol Microbiol 56: 2535–2540. [DOI] [PubMed] [Google Scholar]
- 10. Boore JL, Collins TM, Stanton D, Daehler LL, Brown WM (1995) Deducing the pattern of arthropod phylogeny from mitochondrial DNA rearrangements. Nature 376: 163–165. [DOI] [PubMed] [Google Scholar]
- 11. Boore JL, Lavrov DV, Brown WM (1998) Gene translocation links insects and crustaceans. Nature 392: 667–668. [DOI] [PubMed] [Google Scholar]
- 12. Klimov PB, Oconnor BM (2009) Improved tRNA prediction in the American house dust mite reveals widespread occurrence of extremely short minimal tRNAs in acariform mites. BMC Genomics 10: 598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Masta SE (2000) Mitochondrial sequence evolution in spiders: intraspecific variation in tRNAs lacking the TPsiC Arm. Mol Biol Evol 17: 1091–1100. [DOI] [PubMed] [Google Scholar]
- 14. Masta SE, Boore JL (2004) The complete mitochondrial genome sequence of the spider Habronattus oregonensis reveals rearranged and extremely truncated tRNAs. Mol Biol Evol 21: 893–902. [DOI] [PubMed] [Google Scholar]
- 15. Masta SE, Boore JL (2008) Parallel evolution of truncated transfer RNA genes in arachnid mitochondrial genomes. Mol Biol Evol 25: 949–959. [DOI] [PubMed] [Google Scholar]
- 16. Klimov PB, Knowles LL (2011) Repeated parallel evolution of minimal rRNAs revealed from detailed comparative analysis. J Hered 102: 283–293. [DOI] [PubMed] [Google Scholar]
- 17. Masta SE (2010) Mitochondrial rRNA secondary structures and genome arrangements distinguish chelicerates: comparisons with a harvestman (Arachnida: Opiliones: Phalangium opilio). Gene 449: 9–21. [DOI] [PubMed] [Google Scholar]
- 18. Park SJ, Lee YS, Hwang UW (2007) The complete mitochondrial genome of the sea spider Achelia bituberculata (Pycnogonida, Ammotheidae): arthropod ground pattern of gene arrangement. BMC Genomics 8: 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gissi C, Iannelli F, Pesole G (2008) Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity 101: 301–320. [DOI] [PubMed] [Google Scholar]
- 20. Shao R, Barker SC, Mitani H, Takahashi M, Fukunaga M (2006) Molecular mechanisms for the variation of mitochondrial gene content and gene arrangement among chigger mites of the genus Leptotrombidium (Acari: Acariformes). J Mol Evol 63: 251–261. [DOI] [PubMed] [Google Scholar]
- 21. Jones M, Gantenbein B, Fet V, Blaxter M (2007) The effect of model choice on phylogenetic inference using mitochondrial sequence data: lessons from the scorpions. Mol Phylogenet Evol 43: 583–595. [DOI] [PubMed] [Google Scholar]
- 22. Choi EH, Park SJ, Jang KH, Hwang W (2007) Complete mitochondrial genome of a Chinese scorpion Mesobuthus martensii (Chelicerata, Scorpiones, Buthidae). DNA Seq 18: 461–473. [DOI] [PubMed] [Google Scholar]
- 23. Shao R, Barker SC, Mitani H, Aoki Y, Fukunaga M (2005) Evolution of duplicate control regions in the mitochondrial genomes of metazoa: a case study with Australasian Ixodes ticks. Mol Biol Evol 22: 620–629. [DOI] [PubMed] [Google Scholar]
- 24. Black WC, Roehrdanz RL (1998) Mitochondrial gene order is not conserved in arthropods: prostriate and metastriate tick mitochondrial genomes. Mol Biol Evol 15: 1772–1785. [DOI] [PubMed] [Google Scholar]
- 25. Gissi C, Pesole G (2003) Transcript mapping and genome annotation of ascidian mtDNA using EST data. Genome Res 13: 2203–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Berthier F, Renaud M, Alziari S, Durand R (1986) RNA mapping on Drosophila mitochondrial DNA: precursors and template strands. Nucleic Acids Res 14: 4519–4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stewart JB, Beckenbach AT (2009) Characterization of mature mitochondrial transcripts in Drosophila, and the implications for the tRNA punctuation model in arthropods. Gene 445: 49–57. [DOI] [PubMed] [Google Scholar]
- 28. Ojala D, Montoya J, Attardi G (1981) tRNA punctuation model of RNA processing in human mitochondria. Nature 290: 470–474. [DOI] [PubMed] [Google Scholar]
- 29. Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33: W686–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Laslett D, Canback B (2008) ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 24: 172–175. [DOI] [PubMed] [Google Scholar]
- 31. Smith C, Heyne S, Richter AS, Will S, Backofen R (2010) Freiburg RNA Tools: a web server integrating INTARNA, EXPARNA and LOCARNA. Nucleic Acids Res 38: W373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Betley JN, Frith MC, Graber JH, Choo S, Deshler JO (2002) A ubiquitous and conserved signal for RNA localization in chordates. Curr Biol 12: 1756–1761. [DOI] [PubMed] [Google Scholar]
- 34. Grillo G, Licciulli F, Liuni S, Sbisa E, Pesole G (2003) PatSearch: A program for the detection of patterns and structural motifs in nucleotide sequences. Nucleic Acids Res 31: 3608–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pesole G, Liuni S, D’Souza M (2000) PatSearch: a pattern matcher software that finds functional elements in nucleotide and protein sequences and assesses their statistical significance. Bioinformatics 16: 439–450. [DOI] [PubMed] [Google Scholar]
- 36. Schneider T, Stephens R (1990) Sequence logos: A new way to display consensus sequences. Nucleic Acids Res 18: 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Crooks G, Hon G, Chandonia J, Brenner S (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. D’Onorio de Meo P, D’Antonio M, Griggio F, Lupi R, Borsani M, et al. (2012) MitoZoa 2.0: a database resource and search tools for comparative and evolutionary analyses of mitochondrial genomes in Metazoa. Nucleic Acids Res 40: D1168–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lupi R, D’Onorio De Meo P, Picardi E, D’Antonio M, Paoletti D, et al. (2010) MitoZoa: a curated mitochondrial genome database of metazoans for comparative genomics studies. Mitochondrion 10: 192–199. [DOI] [PubMed] [Google Scholar]
- 40. Benkel BF, Duschesnay P, Boer PH, Genest Y, Hickey DA (1988) Mitochondrial large ribosomal RNA: an abundant polyadenylated sequence in Drosophila . Nucleic Acids Res 16: 9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 42.Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, et al. (2010) Geneious v5.5.7 created by Biomatters. Available: http://www.geneious.com. Accessed 2010.
- 43. Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552. [DOI] [PubMed] [Google Scholar]
- 45. Galtier N, Gouy M, Gautier C (1996) SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci 12: 543–548. [DOI] [PubMed] [Google Scholar]
- 46. Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105. [DOI] [PubMed] [Google Scholar]
- 47. Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- 48. Abascal F, Posada D, Zardoya R (2007) MtArt: a new model of amino acid replacement for Arthropoda. Mol Biol Evol 24: 1–5. [DOI] [PubMed] [Google Scholar]
- 50. Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- 51. Adachi J, Hasegawa M (1996) Model of amino acid substitution in proteins encoded by mitochondrial DNA. J Mol Evol 42: 459–468. [DOI] [PubMed] [Google Scholar]
- 52. Lavrov DV, Boore JL, Brown WM (2000) The complete mitochondrial DNA sequence of the horseshoe crab Limulus polyphemus . Mol Biol Evol 17: 813–824. [DOI] [PubMed] [Google Scholar]
- 53. Shao R, Aoki Y, Mitani H, Tabuchi N, Barker SC, et al. (2004) The mitochondrial genomes of soft ticks have an arrangement of genes that has remained unchanged for over 400 million years. Insect Mol Biol 13: 219–224. [DOI] [PubMed] [Google Scholar]
- 54. Xu G, Fang QQ, Keirans JE, Durden LA (2003) Molecular phylogenetic analyses indicate that the Ixodes ricinus complex is a paraphyletic group. J Parasitol 89: 452–457. [DOI] [PubMed] [Google Scholar]
- 55. Klompen JSH, Black IV WC, Keirans JE, Norris DE (2000) Systematics and biogeography of hard ticks: a total evidence approach. Cladistics 16: 79–102. [DOI] [PubMed] [Google Scholar]
- 56. Christianson TW, Clayton DA (1986) In vitro transcription of human mitochondrial DNA: accurate termination requires a region of DNA sequence that can function bidirectionally. Proc Natl Acad Sci USA 83: 6277–6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Christianson TW, Clayton DA (1988) A tridecamer DNA sequence supports human mitochondrial RNA 3′-end formation in vitro. Mol Cell Biol 8: 4502–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fernandez-Silva P, Martinez-Azorin F, Micol V, Attardi G (1997) The human mitochondrial transcription termination factor (mTERF) is a multizipper protein but binds to DNA as a monomer, with evidence pointing to intramolecular leucine zipper interactions. Embo J 16: 1066–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kruse B, Narasimhan N, Attardi G (1989) Termination of transcription in human mitochondria: identification and purification of a DNA binding protein factor that promotes termination. Cell 58: 391–397. [DOI] [PubMed] [Google Scholar]
- 60. Valverde JR, Marco R, Garesse R (1994) A conserved heptamer motif for ribosomal RNA transcription termination in animal mitochondria. Proc Natl Acad Sci U S A 91: 5368–5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fernandez-Silva P, Loguercio Polosa P, Roberti M, Di Ponzio B, Gadaleta MN, et al. (2001) Sea urchin mtDBP is a two-faced transcription termination factor with a biased polarity depending on the RNA polymerase. Nucleic Acids Res 29: 4736–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Loguercio Polosa P, Roberti M, Musicco C, Gadaleta MN, Quagliariello E, et al. (1999) Cloning and characterisation of mtDBP, a DNA-binding protein which binds two distinct regions of sea urchin mitochondrial DNA. Nucleic Acids Res 27: 1890–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Roberti M, Mustich A, Gadaleta MN, Cantatore P (1991) Identification of two homologous mitochondrial DNA sequences, which bind strongly and specifically to a mitochondrial protein of Paracentrotus lividus . Nucleic Acids Res 19: 6249–6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Roberti M, Loguercio Polosa P, Bruni F, Musicco C, Gadaleta MN, et al. (2003) DmTTF, a novel mitochondrial transcription termination factor that recognises two sequences of Drosophila melanogaster mitochondrial DNA. Nucleic Acids Res 31: 1597–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Van Etten RA, Bird JW, Clayton DA (1983) Identification of the 3′-ends of the two mouse mitochondrial ribosomal RNAs. The 3′-end of 16S ribosomal RNA contains nucleotides encoded by the gene for transfer RNALeuUUR. J Biol Chem 258: 10104–10110. [PubMed] [Google Scholar]
- 66. Campbell NJH, Barker SC (1999) The novel mitochondrial gene arrangement of the cattle tick, Boophilus microplus: fivefold tandem repetition of a coding region. Mol Biol Evol 16: 732–740. [DOI] [PubMed] [Google Scholar]
- 67. Shao R, Mitani H, Barker SC, Takahashi M, Fukunaga M (2005) Novel mitochondrial gene content and gene arrangement indicate illegitimate inter-mtDNA recombination in the chigger mite, Leptotrombidium pallidum . J Mol Evol 60: 764–773. [DOI] [PubMed] [Google Scholar]
- 68. Pesole G, Gissi C, De Chirico A, Saccone C (1999) Nucleotide substitution rate of mammalian mitochondrial genomes. J Mol Evol 48: 427–434. [DOI] [PubMed] [Google Scholar]
- 69. Friedrich M, Tautz D (1997) Evolution and phylogeny of the Diptera: a molecular phylogenetic analysis using 28S rDNA sequences. Systematic Biology 46: 674–698. [DOI] [PubMed] [Google Scholar]
- 70. Krzeminski W, Krzeminska E (2003) Triassic Diptera: description, revisions, and phylogenetic relations. Acta Zoologica Cracoviensia 46 Supp: 153–184 [Google Scholar]
- 71. Roberti M, Bruni F, Loguercio Polosa P, Gadaleta MN, Cantatore P (2006) The Drosophila termination factor DmTTF regulates in vivo mitochondrial transcription. Nucleic Acids Res 34: 2109–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Montooth KL, Abt DN, Hofmann JW, Rand DM (2009) Comparative genomics of Drosophila mtDNA: Novel features of conservation and change across functional domains and lineages. J Mol Evol 69: 94–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Roberti M, Loguercio Polosa P, Bruni F, Manzari C, Deceglie S, et al. (2009) The MTERF family proteins: mitochondrial transcription regulators and beyond. Biochim Biophys Acta 1787: 303–311. [DOI] [PubMed] [Google Scholar]
- 74. Linder T, Park CB, Asin-Cayuela J, Pellegrini M, Larsson NG, et al. (2005) A family of putative transcription termination factors shared amongst metazoans and plants. Curr Genet 48: 265–269. [DOI] [PubMed] [Google Scholar]
- 75. Munderloh UG, Liu Y, Wang M, Chen C, Kurtti TJ (1994) Establishment, maintenance and description of cell lines from the tick Ixodes scapularis . J Parasitol 80: 533–543. [PubMed] [Google Scholar]
- 76. Najm N-A, Silaghi C, Bell-Sakyi L, Pfister K, Passos L (2012) Detection of bacteria related to "Candidatus" Midichloria mitochondrii in tick cell lines. Parasitology Research 110: 437–442. [DOI] [PubMed] [Google Scholar]
- 77. Hixson JE, Wong TW, Clayton DA (1986) Both the conserved stem-loop and divergent 5′-flanking sequences are required for initiation at the human mitochondrial origin of light-strand DNA replication. J Biol Chem 261: 2384–2390. [PubMed] [Google Scholar]
- 78. Macey JR, Larson A, Ananjeva NB, Fang Z, Papenfuss TJ (1997) Two novel gene orders and the role of light-strand replication in rearrangement of the vertebrate mitochondrial genome. Mol Biol Evol 14: 91–104. [DOI] [PubMed] [Google Scholar]
- 79. Murrell A, Barker SC (2003) Synonymy of Boophilus Curtice, 1891 with Rhipicephalus Koch, 1844 (Acari: Ixodidae). Syst Parasitol 56: 169–172. [DOI] [PubMed] [Google Scholar]
- 80. Meusemann K, von Reumont BM, Simon S, Roeding F, Strauss S, et al. (2010) A phylogenomic approach to resolve the arthropod tree of life. Mol Biol Evol 27: 2451–2464. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Putative secondary structure of the 22 tRNAs of I. ricinus .
(PDF)
Conserved motifs and secondary structures of the tick control region, mapped on the I. ricinus sequence.
(PDF)
Accession numbers of 98 mitochondrial sequences belonging to 68 species analysed in this study.
(XLS)
Amplification strategy and general features of tRNAs, control region, and small non-coding regions of the I. ricinus mtDNA.
(DOC)