Abstract
The promoter from rice tungro bacilliform badnavirus (RTBV) is expressed only in phloem tissues in transgenic rice plants. RF2a, a b-Zip protein from rice, is known to bind to the Box II cis element near the TATA box of the promoter. Here, we report that the full-length RTBV promoter and a truncated fragment E of the promoter, comprising nucleotides −164 to +45, result in phloem-specific expression of β-glucuronidase (GUS) reporter genes in transgenic tobacco plants. When a fusion gene comprising the cauliflower mosaic virus 35S promoter and RF2a cDNA was coexpressed with the GUS reporter genes, GUS activity was increased by 2–20-fold. The increase in GUS activity was positively correlated with the amount of RF2a, and the expression pattern of the RTBV promoter was altered from phloem-specific to constitutive. Constitutive expression of RF2a did not induce morphological changes in the transgenic plants. In contrast, constitutive overexpression of the b-ZIP domain of RF2a had a strong effect on the development of transgenic plants. These studies suggest that expression of the b-Zip domain can interfere with the function of homologues of RF2a that regulate development of tobacco plants.
Regulation of transcription is achieved by the activity of multiple proteins that bind to regulatory elements, many of which are upstream of the promoters and alter basal rates of transcription initiation and/or elongation (1, 2). To understand the mechanisms of tissue-specific and constitutive gene expression in plants, a number of promoters and transcription factors have been studied in recent years (3–16). It was shown that constitutive promoters, such as the cauliflower mosaic virus 35S promoter (17) and the promoter from cassava vein mosaic virus (14) are modular in organization and multiple cis elements. These elements, with specific transcription factors, apparently interact in an additive and/or synergistic manner to confer gene expression in all plant tissues. Similarly, tissue-specific promoters contain multiple elements that contribute to promoter activity in both positive and negative ways (4–6, 8, 12, 18).
The rice tungro bacilliform badnavirus (RTBV) promoter (19, 20) and the transcription factors that interact it may serve as a model system to study plant tissue-specific gene expression. The virus, which replicates solely in phloem tissues, has a single promoter that is active in transfected protoplasts and is phloem-specific in transgenic rice plants (7, 10, 21, 22).
Within the fragment E of the promoter (nucleotides −164 to +45), multiple cis elements were identified as being required for phloem-specific gene expression (7, 10, 15, 23). A b-ZIP type transcription factor, RF2a, was isolated from rice and bound to Box II, a crucial cis element of the promoter. RF2a activates transcription from the RTBV promoter in an in vitro transcription system derived from rice cell cultures (11). Moreover, studies in transgenic rice plants suggested that RF2a is involved in the development of vascular tissues (11).
We report here the function of the RTBV promoter in transgenic tobacco plants and functional interactions between RF2a and the promoter. When RF2a was constitutively expressed in transgenic plants that contain the RTBV promoter, expression of the promoter was altered from phloem-specific and became constitutive. Although overexpression of RF2a did not cause morphological changes in transgenic plants, constitutive expression of the b-Zip domain of RF2a had a negative effect on plant development.
Materials and Methods
Plasmid Constructions.
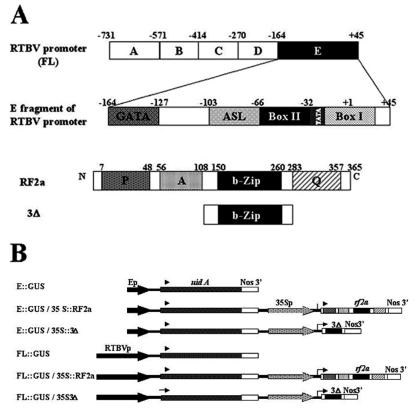
The fusion gene comprising the full-length RTBV promoter (FL; nucleotides −731 to +45) with the uidA coding sequence was released from plasmid pMB9089 (7) with XbaI and KpnI. The chimeric gene comprising the E fragment (nucleotides −164 to +45) and uidA was released from plasmid pRTBV-E (7) with HindIII and KpnI. Inserts were ligated into the binary vector pGA482 through XbaI and KpnI or HindIII and KpnI sites. The resultant plasmids are named pGA-FL∷GUS and pGA-E∷GUS, respectively (Fig. 1B).
Figure 1.
(A) Schematic representation of the RTBV full-length promoter and of the E fragment of the promoter. The cis elements GATA, ASL Box, Box II, and Box I of E, as described by Yin et al. (10), are also indicated. Box II, the cis element recognized by RF2a, is indicated in black. Schematic representations are shown of the RF2a transcription factor and of the 3Δ mutant. The potential activation domains are indicated as follows: P, proline-rich region; A, acidic region; Q, glutamine-rich region. The dimerization and DNA-binding domain is indicated as bZIP. (B) Diagram of the constructs used for Agrobacterium-mediated transformation of tobacco (N. tabacum). The uidA gene (GUS-encoding gene) is driven either by the full-length RTBV promoter or E fragment (−164 to +45). The genes encoding the rice transcription factor RF2a and 3Δ mutant are driven by the 35S promoter of cauliflower mosaic virus.
To construct plasmids for expression in Escherichia coli of RF2a and the b-Zip DNA binding domain of RF2a (referred to as 3Δ) (Fig. 1A), the coding sequences were amplified by PCR from plasmid pET-12–2a, [which contains the cDNA encoding for RF2a (11)]. To amplify RF2a, two primers, 5′RF2a (GCCGCCCATATGGAGAAGATGAACAGGGAGAAATCC) and 3′RF2a (CGCGGATCCTCAGTTGCCGCTGCTTCCTGA), were used. Primers 5′ΔPΔA-RF2a (GCCGCCCATATGGAGAAGATGTCCGCCGCCGCCCA) and 3′ΔQ-RF2a (CGCGGATCCTCAGTGTGGCATGCCACCGAA) were used to amplify 3Δ (amino acids 108–283 of RF2a) (11). NdeI and BamHI sites were introduced into the 5′ and 3′ primers, respectively (underlined). The PCR products were digested with NdeI and BamHI and inserted into the expression vector pET28a (Novagen) restricted by the same enzymes. The new constructs were named pET-RF2a and pET-3Δ. The sequence of inserts in each plasmid was confirmed.
Plant transformation plasmids were constructed for coexpression of the reporter genes and the RF2a effector proteins. The coding sequences of RF2a and 3Δ were released from pET-RF2a and pET-3Δ with NdeI (blunted) and BamHI and were cloned into pMON999 (a gift from Monsanto), a cassette with the cauliflower mosaic virus 35S promoter (P-35S) and Nos terminator of nopline synthase, after restriction by XbaI (blunted) and BamHI. The fusion genes, named P-35S∷RF2a and P-35S∷3Δ, were then released by NotI (blunted) and cloned into binary vectors pGA-E∷GUS or pGA-FL∷GUS through the blunted ClaI site. In the coexpression vectors, the reporter gene and effector genes are in head-to-tail orientation. The final plasmids were named pGA-E∷GUS/P-35S∷RF2a, pGA-FL∷GUS/P-35S∷RF2a, pGA-E∷GUS/P-35S∷3Δ, and pGA-FL∷GUS/P-35S∷3Δ (Fig. 1B).
Tobacco Transformation.
Gene constructs containing pGA482-derived plasmids (Fig. 1B) were introduced into Agrobacterium tumefaciens strain LBA4404 and used for Agrobacterium-mediated transformation. Leaf discs from Nicotiana tabacum cv. Xanthi NN were used for transformation following the protocol described by Horsch et al. (24). At least 14 independent transgenic lines for each construct were produced and grown in a greenhouse. Self-fertilized seeds were collected from T0 plants and were germinated on Murashige and Skoog (MS) medium (25) with or without kanamycin (100 mg/liter) selection, and the seedlings were grown in a greenhouse.
Analysis of β-Glucuronidase (GUS) Activity.
Histochemical analysis of GUS activity was performed essentially as described by Jefferson et al. (26). Hand-cut fresh tissue sections of leaves and stems of primary transformants (T0) or T1 progeny were incubated at 37°C for 4–12 h in reaction buffer containing 1 mM X-gluc (Research Organic), 100 mM sodium phosphate buffer, pH 7.0, 2 mM K3Fe(CN)6, and 2 mM K4Fe(CN)6/0.1% (vol/vol)/Triton X-100/20% (vol/vol) methanol. For analysis of young T1 seedlings, whole plantlets were collected about 1 week after germination and immersed in the buffer containing X-gluc followed by vacuum infiltration and incubation overnight at 37°C. Samples were cleared by several washes with 70% ethanol and visualized with a Nikon or Olympus microscope.
Quantitative GUS assays by using the substrate 4-methylum-belliferyl-β-d-glucuronide (MUG) were performed as described by Jefferson et al. (26).
ELISA.
To quantify RF2a in tissue extracts, sandwich ELISAs were performed. Microtiter plates (96-well, Nunc, MaxiSorp) were coated overnight at 4°C with 100 μl/well of 1 μg/ml protein A in PBS, and 1 h at room temperature in blocking buffer [10% (vol/vol) of FBS in PBS]. The plates were then incubated with 1 μg/ml purified anti-RF2a antibody in PBS plus 1% FBS for 1 h at room temperature. Plant extracts in 50 mM Tris-HCl, pH 7.5/0.1% (vol/vol) Triton X-100 were added, and the plates were incubated overnight at 4°C and then blocked with 1 μg/ml protein A in PBS plus FBS. Plates were incubated again with 1 μg/ml anti-RF2a antibody in 1 × PBS plus FBS for 1 h at room temperature and then with 1 μg/ml protein-A–horseradish peroxidase (Pierce) in PBS plus FBS and developed with TMB (Kirkegaard & Perry Laboratories). Plates were read at 650 nm with an absorbency plate reader. Purified RF2a was used to develop a standard curve of reactivity.
Production of RF2a and 3Δ in E. coli.
pET-RF2a and pET-3Δ were transformed into E. coli BL21 (DE3)/pLysE. Cells were grown at 37°C to an OD600 of 0.5–0.6 and induced by 0.5 mM isopropyl β-D-thiogalactopyranoside for 3 h at 25°C. The cell pellets were resuspended in 20 mM Tris-HCl, pH 8/500 mM NaCl/0.1% (vol/vol) Nonidet P-40/1 mM PMSF/1 mg/ml lysozyme/10 μg/ml DNase/10 μg/ml RNase. Lysis was completed by using a French press. The proteins were dialyzed against 20 mM Tris-HCl, pH 8, and stored at −80°C.
Electrophoretic Mobility-Shift Assays (EMSAs).
EMSAs were carried out essentially as described by Yin and Beachy (7). E. coli protein extracts (500 ng) were incubated with 32P-labeled DNA probes comprising Box IIml that was constructed by using annealed oligonucleotides (10). For competition EMSA, unlabeled oligonucleotides were added to the binding reactions at 80-fold molar excess relative to the labeled probe.
Results
Expression Pattern of the RTBV Promoter in Transgenic Tobacco Plants.
The RTBV full-length promoter as well as the E fragment are expressed exclusively in phloem tissues in transgenic rice plants (7, 10, 22). Within the E fragment, four cis sequence elements that contribute to phloem-specific gene expression have been described, including the GATA motif, ASL (AS-1 like) element, Box II, and Box I (Fig. 1A) (10). We wanted to determine whether the RTBV promoter is functional and maintains tissue specificity in transgenic tobacco plants. Plasmids pGA-FL∷GUS and pGA-E∷GUS (Fig. 1A) were introduced into tobacco through Agrobacterium-mediated transformation. Seventeen and 23 independent transgenic tobacco plants were developed for the P-FL∷GUS and P-E∷GUS genes, respectively.
A detailed histochemical analysis of GUS expression patterns in transgenic tobacco plants showed that expression of P-E∷GUS and P-FL∷GUS was essentially the same. In leaves of transgenic plants with either construct, strong GUS activity was observed in vascular tissues (Fig. 2 A and C), although in very young leaves there was a low amount of GUS activity in mesophyll cells (Fig. 2A). Cross sections through the midrib of more mature leaves showed GUS activity only in phloem cells (Fig. 2G). These results are similar to those reported from studies of transgenic rice plants (10).
Figure 2.
Histochemical localization of GUS in tissues from transgenic tobacco plants. The results shown are representative of those observed in the 15 independent transgenic lines developed for each gene construct. Results with P-E∷GUS and P-FL∷GUS constructs were identical, and the figure is compiled from both sets of transgenic plants. (A, C, E, G, I, and K) Plants containing either P-E∷GUS or P-FL∷GUS gene. (B, D, F, H, J, and L) Plants containing either P-E∷GUS/P-35S∷RF2a or P-FL∷GUS/P-35S∷RF2a. GUS activity is indicated in transgenic tissue by an indigo dye precipitate after staining with X-Gluc. (A and B) Seedlings. (C and D) Juvenile leaves. (E and F) Roots. (G and H) Leaf sections showing vascular tissues of the midrib and leaf lamina. (I and J) Vascular tissue of the leaf midrib. (K and L) Cross section of lamina. c, cotyledon; cr, cortex; e, epidermis; ep, external phloem; g, guard cell; ip, internal phloem; l, leaf; m, mesophyll; p, phloem; pm, palisade mesophyll; py, parenchyma; rt, root tip; rv, root vein; sm, spongy mesophyll; t, trichome; vb, vascular bundle; x, xylem.
Modulation of Phloem-Specific Expression of the RTBV Promoter by RF2a.
RF2a was isolated by virtue of its interaction with Box II DNA (10). RF2a is a b-Zip protein with three potential functional domains: a proline-rich domain, an acidic domain, and a glutamine-rich domain (Fig. 1A) (11). A mutant of RF2a that lacks these three domains was constructed and is referred to as “3Δ” (Fig. 1 A and B). The 3Δ protein contains the DNA-binding and leucine-zipper domains (b-Zip) of RF2a, including the putative nuclear localization signal. To determine the effect of RF2a and 3Δ on expression of the FL and E promoters, at least 15 independent transgenic tobacco plants were developed for the following constructs: pGA-E∷GUS/P-35S∷RF2a, pGA-E∷GUS/P-35S∷3Δ, pGA-FL∷GUS/P-35S∷RF2A, and pGA-FL∷GUS/P-35S∷3Δ (Fig. 1B). Integration of the full-length T-DNA in transgenic plants was confirmed by Southern blot hybridization (data not shown).
Histochemical analysis of GUS activity in the T1 progeny showed that expression of the reporter genes was substantially altered by coexpression with RF2a. In the plants that contained either P-E∷GUS/P-35S∷RF2a or P-FL∷GUS/P-35S∷RF2a, GUS activity was detected throughout the cotyledonary leaves and true leaves (Fig. 2 B and D). Cross sections through the leaf midrib of these plants showed very strong GUS activity in phloem cells, the epidermis, and trichomes (compare Fig. 2 G and H). The palisade and spongy mesophyll cells of the leaf lamina also exhibited intense staining (compare Fig. 2 K and L) as did parenchyma cells of the midrib (compare Fig. 2 I and J). Guard cells also showed strong GUS activity when the reporter and effector genes were coexpressed (not shown). In the root tissues of transgenic plants with P-E∷GUS/P-35S∷RF2a or P-FL∷GUS/P-35S∷RF2a, there was a high level of GUS activity throughout the cortex (Fig. 2F) and very strong levels of GUS activity in root tips. In contrast, root tissues of transgenic plants with the reporter genes only revealed GUS activity solely in the vascular cylinder (Fig. 2E).
GUS activity in the leaves of T0 transgenic tobacco plants was quantified. As anticipated, there were variations in the amount of GUS activity among transgenic lines for each construct. The GUS activity of transgenic plants with P-FL∷GUS was higher than the activity in plants with P-E∷GUS genes (Fig. 3). When RF2a was coexpressed with either P-E∷GUS or P-FL∷GUS, the GUS activity was increased by 2–20-fold compared with plants that lacked RF2a (Fig. 3). The increase of GUS activity is in agreement with the observation that the pattern of GUS activity was constitutive when RF2a was coexpressed with the reporter gene (see Fig. 2).
Figure 3.
GUS activity in extracts of leaves from transgenic tobacco plants carrying the P-E∷GUS gene, P-E∷GUS/P-35S∷RF2a, P-FL∷GUS, or P-FL∷GUS/P35S∷RF2a. The amount of GUS (pmol MUG min−1 mg−1 protein) is indicated. The mean level of GUS activity for each construct is indicated by the thick, horizontal bar.
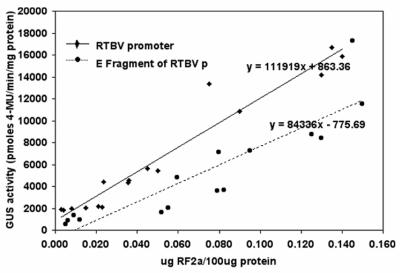
To establish a correlation between GUS activity and the amount of RF2a, ELISA reactions were carried out to quantify RF2a in individual transgenic plants. Soluble protein extracts of T0 plants were tested in quantitative ELISA reactions by using anti-RF2a antibodies. Aliquots of the same extracts were also used in quantitative analyses of GUS activity. As shown in Fig. 4, the results of these assays showed a positive correlation between the amount of RF2a in the samples and GUS activity.
Figure 4.
Correlation between GUS activity and the amount of RF2a determined by ELISA for transgenic T0 plant lines that carry P-E∷GUS/P-35S∷RF2a (●) or P-FL∷GUS/P-35S∷RF2a (⧫).
DNA Binding and Heterodimerization of RF2a and 3Δ.
The gene encoding the mutant protein 3Δ, which lacks the three putative regulatory domains of RF2a, was constructed to determine whether it could restrict expression of the RTBV promoter. It has been reported that sequences outside of the leucine-zipper region of such proteins can contribute to stability of dimerization (27). In these cases, deletions of other (putative) domains can affect the ability of the protein to form homodimers or heterodimers and to bind DNA. To test the DNA binding ability of protein 3Δ, EMSAs were carried out (Fig. 5). Both RF2a and 3Δ bind 32P-labeled Box IIml by forming homodimers (lanes 2 and 5). When both proteins are added to a reaction, a band with intermediate mobility to the bands of RF2a and 3Δ homodimers was observed. This new band presumably corresponds to the binding of the heterodimer of RF2a/3Δ with the probe (lane 4). All of these complexes can be competed by 80× molar excess of unlabeled cold probe (lane 3).
Figure 5.
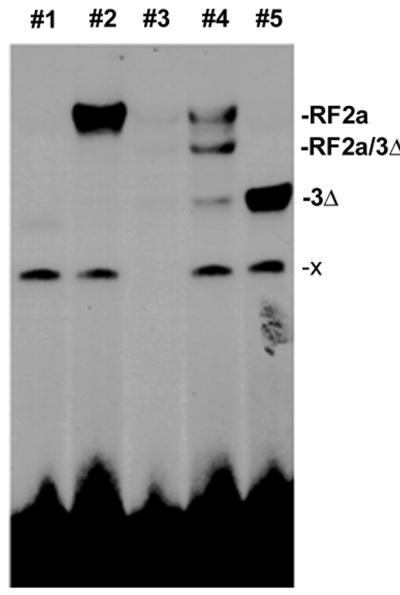
EMSA of RF2a and the 3Δ mutant of RF2a. Oligonucleotides containing the Box IIml sequences (10) were used as 32P-labeled probe or an unlabeled competitor in an EMSA with 500 ng of E. coli protein containing 3Δ or RF2a. Unbound probe is located near the bottom of the gel. The x band in lanes 1, 2, 4, and 5 is presumed to result from binding with an uncharacterized protein from E. coli. Lane 1, protein prepared from E. coli. Lane 2, reaction with extracts of E. coli that produce RF2a. Lane 3, reaction with equimolar amounts of RF2a and 3Δ plus 80× molar excess of unlabeled competitor probe relative to the labeled probe. Lane 4, reaction with equimolar amounts of RF2a and 3Δ. Lane 5, reaction with 3Δ.
Morphological Changes of Transgenic Plants with 3Δ.
In the 15 independent lines developed with P-E∷GUS/P-35S∷3Δ and P-FL∷GUS/P-35S∷3Δ genes, 7 lines with P-E∷GUS/P-35S∷3Δ genes and 6 lines with P-FL∷GUS/P-35S∷3Δ genes exhibited abnormal phenotypes. In contrast, no abnormal phenotypes were observed in transgenic lines with only the reporter gene alone or reporters plus the RF2a gene. The abnormal T0 transgenic lines carrying P-E∷GUS/P-35S∷3Δ or P-FL∷GUS/P-35S∷3Δ genes were characterized by downward curving of leaf mid-veins (Fig. 6 D and E), and the most severely affected plants were stunted (Fig. 6A).
Figure 6.
Phenotypes of plants that contain the gene encoding the 3Δ mutant of RF2a. (A) The plants on the left contain the transgene and grew more slowly than the plant that lacked the transgene due to segregation in the T1 generation (Right). (B) intermediate phenotype, showing shoot elongation, curvature of leaves, and a decrease in apical dominance. (C) Size of roots in plants with the 3Δ gene (Right) compared with nontransgenic T1 progeny (Left). (D and E) Close-up of leaves showing downward curvature. (F) Abnormal plant showing the phenotype of severe stunting with thick leaf lamina.
To confirm that the abnormal phenotypes were due to the transgene, we analyzed the T1 generation of the abnormal T0 plant lines that gave seeds. Two of the affected plant lines produced a few flower buds that never opened and did not produce seeds. Among each of the 11 lines tested, the abnormal phenotype was inherited to the second generation. Line l with P-FL∷GUS/P-35S∷3Δ showed an abnormal segregation pattern, whereas the segregation of the phenotype in other lines, for the most part, followed classical, single-locus segregation patterns (Table 1). Abnormal plants grew much more slowly than the normal plants in T1 progeny and were characterized by stunting of shoots and roots (Fig. 6 A and C). The abnormal phenotype of T1 plants with P-E∷GUS/P-35S∷3Δ or P-FL∷GUS/P-35S∷3Δ genes can be characterized as either mild (Fig. 6B) or severe (Fig. 6F). Plants with the mild phenotype grew much more slowly than nontransgenic control plants, but they ultimately achieved normal height. These plants had wrinkled or distorted leaves (Fig. 6 D and E) with yellow and green mosaic leaf color and exhibited reduced apical dominance (Fig. 6B, arrows). Plants that exhibited a severe phenotype were stunted with very short internodes, thick leaf lamina, and increased number of side shoots (Fig. 6F). Some of the plants with severe developmental problems did not produce seeds.
Table 1.
Segregation of abnormal phenotype in T1 generation transgenic tobacco plants with b-Zip domain of RF2a gene
| Construct | T0 line | Phenotype in T1 progeny
|
|
|---|---|---|---|
| Abnormal | Normal | ||
| E∷GUS/35S∷3Δ | i | 7 | 3 |
| k | 16 | 4 | |
| l | 5 | 5 | |
| m | 5 | 0 | |
| n | 7 | 3 | |
| o | 10 | 12 | |
| FL∷GUS/35S∷3Δ | e | 7 | 3 |
| h | 7 | 3 | |
| k | 14 | 7 | |
| l | 2 | 8 | |
| n | 8 | 1 | |
Expression of the 3Δ gene in the T1 progeny was confirmed by Northern blot analysis (data not shown). The phenotype of normal and abnormal plants was correlated with low and high 3Δ mRNA levels, respectively. However, there was not a significant difference in the amount of 3Δ mRNA between plants with mild and severe abnormal phenotype.
GUS activity of transgenic lines with the P-E∷GUS/P-35S∷3Δ or P-FL∷GUS/P-35S∷3Δ genes was analyzed. The transgenic lines that did not show an abnormal phenotype had the same levels of GUS activity as the lines only with the reporter genes (i.e., P-E∷GUS or P-FL∷GUS), and there was no clear indication of GUS gene activation or repression. Some of the lines that exhibited abnormal morphology (i.e., lines i, k, l, and m of P-E∷GUS/P-35S∷3Δ and lines l and n of P-FL∷GUS/P-35S∷3Δ) had a similar level of GUS activity as well. No correlation was established between severity of plant phenotype and GUS activity. In some lines with severely abnormal phenotypes, GUS activity increased with the age of the plants. These plants were apparently physiologically different from normal plants, and we suggest that factors other than those related to the 3Δ mutant protein may affect expression of the RTBV promoter in these plant lines.
Discussion
Phloem-Specific Expression of the RTBV Promoter.
RTBV is known to accumulate in phloem tissues in infected rice plants (19). Similarly, the full-length RTBV promoter and the E fragment of the promoter are expressed only in phloem tissues in transgenic rice plants (7, 10, 22). Within the E fragment, there are several cis DNA sequence elements that are conserved among vascular tissue-specific promoters, including promoters from monocots and dicots (10). The present work shows that FL and E promoters retain phloem-specific expression in transgenic tobacco plants. This result indicates that there may be similar transcription factors in tobacco and rice that regulate expression of the promoter. It is known that some plant promoters retain specific expression patterns in different plant species (21–24, 26–31) while others do not (32, 33).
RF2a Can Activate the Promoter in Cell Types Other Than Phloem Cells.
As we reported previously, the RTBV promoter contains several important cis elements that contribute to expression of the promoter in vascular tissues. Box II is a cis element that is responsible for the basal behavior of the promoter and shares significant homology with cis elements from other phloem- and xylem-specific promoters (10). Data from 5′ deletion analysis of E promoter and the deletion or mutation of Box II in the context of the E fragment showed that the Box II cis element is critical to the promoter activity and phloem-tissue specificity (10, 15).
When P-FL∷GUS or P-E∷GUS reporter genes were cotransformed with P-35S∷RF2a, the pattern of GUS activity in transgenic plants was constitutive rather than phloem-specific. This is consistent with the known pattern of expression of the 35S promoter (17) and indicates that RF2a is involved in regulating expression of the RTBV promoter.
Many conditions can influence the activity of regulatory proteins in expression of a promoter, including affinity of binding of transcription factors with the DNA sequence element (34), the subcellular distribution of factors, posttranscriptional modifications of factors (35), and synergistic interactions with other proteins (36–40). Furthermore, it is common to use multiple activation sequences or multimerize activation domains of DNA binding protein to achieve strong activation. In our system, a single copy of Box II together with RF2a was sufficient to cause strong expression of the RTBV promoter in nonvascular cells. In addition, the level of expression was positively correlated with the amount of RF2a.
Previous reports indicate that expression of chimeric genes similar to P-35S∷GUS in transgenic plants varies from 32 units to 113,000 units of MUG (1 unit = 1 pmol of MUG min−1 mg−1) (17, 26, 41, 42). In our study, transgenic plants had GUS activities between 4,000 and 18,000 units when P-FL∷GUS or P-E∷GUS was activated by expression of the P-35S∷RF2a gene, representing a 2–20-fold increase in activity compared with activity of the reporter genes in absence of P-35S∷RF2a.
Although we do not yet know the basis for the strong activation of the FL and E promoters by RF2a, there are several notable characteristics of this system. First, Box II is located in close proximity to the TATA box (≈7 nt). It is not yet known whether the proximity of the cis element to the TATA sequence is important for the activation of this promoter. Second, the results of in vitro studies indicate that there is a direct physical interaction between TATA binding protein and RF2a (Q. Zhu, M. I. Ordiz, T. Dabi, R.N.B., and C. Lamb, unpublished results). Direct interaction with TATA binding protein may indicate that RF2a has a direct role in regulating transcription. Third, RF2a has three putative activation domains: acidic, glutamine-rich, and proline-rich (11), any or all of which may be important for the activity of RF2a. We propose that proximity of the cis element and the unique features of RF2a contribute to the strong activation of the promoter by RF2a.
In transgenic plants with reporter genes and RF2a, it is most likely that promoter activation is due to formation of homodimers of RF2a. However, the possible activation by heterodimerization of RF2a with homologous factors of tobacco cannot be excluded.
A Dominant Negative Mutant of RF2a Affects Plant Development.
In rice plants, RF2a acts as a transcription activator, and its biological function is linked to the development of the vascular system. This conclusion was based on the results of experiments in which the levels of RF2a were reduced by expression of an antisense gene (11). An alternative approach to determine gene function is to introduce dominant negative mutants. Mutants of b-Zip transcription factors have been used in such studies (43–45). Here, the mutant 3Δ, which contains only the DNA-binding domain and the leucine-zipper region, was created to test the biological function of RF2a-like transcription factors in tobacco and the effect of 3Δ on expression of the RTBV promoter. The 3Δ mutant formed homodimers and was heterodimerized with RF2a, and both dimers bind to Box II in vitro. In other systems, it has been shown that dominant negative mutant proteins can reduce the ability of endogenous factors to bind their target genes by heterodimerizing with endogenous factors or by competing for their binding sites (45, 49–51).
When the 3Δ mutant was constitutively expressed in transgenic tobacco plants, about 50% of the plants exhibited strong abnormal morphological phenotypes (i.e., loss of apical dominance, downward curling of leaf veins). In contrast, GUS activity was not repressed in plants that were cotransformed with P-35S∷3Δ and either P-E∷GUS or P-FL∷GUS. Contrary to expectations, there were increased levels of GUS activity in some plants. The increased GUS activity in these plants may be reflective of an inability of 3Δ to block the activity by an endogenous factor. Alternatively, an increase in GUS activity may be a consequence of the abnormal physiological state of these transgenic plants. We suggest that the 3Δ protein interferes with the activity of RF2a-like homologues in tobacco plants, which results in abnormal growth and development of the plants. In this model, the phenotypes caused by 3Δ may reflect primary or secondary effects of interactions of the 3Δ mutant with endogenous b-Zip and other transcription factors (44, 46–48).
An alternative explanation for the downward curling of the mid-vein and the decrease in apical dominance is that the curling is a secondary effect caused by alteration in auxin and other hormone levels and their transport. Recently, a b-Zip protein, RSG, which has a high sequence similarity (78%) with RF2a within the b-Zip domain, was isolated from tobacco (44). Furthermore, the putative functional domains of RF2a and RSG are similar to each other. Notwithstanding the high degree of similarity between RF2a and RSG, the dominant negative effects of the b-ZIP domains of RF2a and RSG are different from each other. Expression of the RSG b-ZIP domain in transgenic plants produced dwarfed plants with dark green leaves and altered levels of gibberellins in contrast to those induced by 3Δ (described above). The phenotype induced by overexpression of the b-Zip domain (3Δ) of RF2a may therefore indicate an effect on auxin-regulated or other pathways in plant development that are different from those affected by RSG.
Acknowledgments
S.P. was supported by a fellowship from Consejo Nacional de Investigaciones Científicas y Técnicas de Argentina. This work was also supported by Department of Energy Grants DEFG03-91ER20039 and DEFG02-99ER20355 (to R.N.B.).
Abbreviations
- RTBV
promoter from rice tungro bacilliform badnavirus
- GUS
β-glucuronidase
- MUG
4-methylumebelliferone-β-S glucuronide
- EMSA
electrophoretic mobility-shift assay
References
- 1.Roeder R G. Trends Biochem Sci. 1991;16:402–408. doi: 10.1016/0968-0004(91)90164-q. [DOI] [PubMed] [Google Scholar]
- 2.Yankulov K, Blau J, Purton T, Roberts S, Bentley D L. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 3.Benfey P N, Chua N-H. Science. 1989;244:174–181. doi: 10.1126/science.244.4901.174. [DOI] [PubMed] [Google Scholar]
- 4.Leyva A, Liang X, Pintor-Toro J A, Dixon R A, Lamb C J. Plant Cell. 1992;4:263–271. doi: 10.1105/tpc.4.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujiwara T, Beachy R N. Plant Mol Biol. 1994;24:261–272. doi: 10.1007/BF00020166. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki M, Koide Y, Hattori T, Nakamura K, Asahi T. Plant Cell Physiol. 1995;36:1067–1074. doi: 10.1093/oxfordjournals.pcp.a078849. [DOI] [PubMed] [Google Scholar]
- 7.Yin Y, Beachy R N. Plant J. 1995;7:969–980. doi: 10.1046/j.1365-313x.1995.07060969.x. [DOI] [PubMed] [Google Scholar]
- 8.Ellerstrom M, Stalberg K, Ezcurra I, Rask L. Plant Mol Biol. 1996;32:1019–1027. doi: 10.1007/BF00041385. [DOI] [PubMed] [Google Scholar]
- 9.Faktor O, Kooter J M, Dixon R A, Lamb C J. Plant Mol Biol. 1996;32:849–859. doi: 10.1007/BF00020482. [DOI] [PubMed] [Google Scholar]
- 10.Yin Y, Chen L, Beachy R. Plant J. 1997;12:1179–1188. doi: 10.1046/j.1365-313x.1997.12051179.x. [DOI] [PubMed] [Google Scholar]
- 11.Yin Y, Zhu Q, Dai S, Lamb C, Beachy R N. EMBO J. 1997;16:5247–5259. doi: 10.1093/emboj/16.17.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ringli C, Keller B. Plant Mol Biol. 1998;37:977–988. doi: 10.1023/a:1006030007333. [DOI] [PubMed] [Google Scholar]
- 13.Verdaguer B, de Kochko A, Beachy R N, Fauquet C. Plant Mol Biol. 1996;31:1129–1139. doi: 10.1007/BF00040830. [DOI] [PubMed] [Google Scholar]
- 14.Verdaguer B, de Kochko A, Fux C I, Beachy R N, Fauquet C. Plant Mol Biol. 1998;37:1055–1067. doi: 10.1023/a:1006004819398. [DOI] [PubMed] [Google Scholar]
- 15.He X, Hohn T, Futterer J. J Biol Chem. 2000;275:11799–11808. doi: 10.1074/jbc.275.16.11799. [DOI] [PubMed] [Google Scholar]
- 16.Niggeweg R, Thurow C, Weigel R, Pfitzner U, Gatz C. Plant Mol Biol. 2000;42:775–788. doi: 10.1023/a:1006319113205. [DOI] [PubMed] [Google Scholar]
- 17.Benfey P N, Ren L, Chua N-H. EMBO J. 1989;8:2195–2202. doi: 10.1002/j.1460-2075.1989.tb08342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauffe K D, Lee S P, Subramaniam R, Douglas C J. Plant J. 1993;4:235–253. doi: 10.1046/j.1365-313x.1993.04020235.x. [DOI] [PubMed] [Google Scholar]
- 19.Qu R D, Bhattacharyya M, Laco G S, De Kochko A, Rao B L, Kaniewska M B, Elmer J S, Rochester D E, Smith C E, Beachy R N. Virology. 1991;185:354–364. doi: 10.1016/0042-6822(91)90783-8. [DOI] [PubMed] [Google Scholar]
- 20.Hay J M, Jones M C, Blakebrough M L, Dasgupta I, Davies J W, Hull R. Nucleic Acids Res. 1991;19:2615–2621. doi: 10.1093/nar/19.10.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen G, Muller M, Potrykus I, Hohn T, Futterer J. Virology. 1994;204:91–100. doi: 10.1006/viro.1994.1513. [DOI] [PubMed] [Google Scholar]
- 22.Kloti A, Henrich C, Bieri S, He X, Chen G, Burkhardt P K, Wunn J, Lucca P, Hohn T, Potrykus I, Futterer J. Plant Mol Biol. 1999;40:249–266. doi: 10.1023/a:1006119517262. [DOI] [PubMed] [Google Scholar]
- 23.Meisel L, Lam E. Genet Eng. 1997;19:183–199. doi: 10.1007/978-1-4615-5925-2_10. [DOI] [PubMed] [Google Scholar]
- 24.Horsch R B, Fry J, Hoffmann N, Neidermeyer J, Rogers S G, Fraley R T. In: Plant Molecular Biology Manual. Gelvin S B, Schilperoort R A, editors. Dordrecht, The Netherlands: Kluwer; 1988. p. A5. , 1–9. [Google Scholar]
- 25.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 26.Jefferson R A, Kavanagh T A, Bevan M W. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katagiri F, Seipel K, Chua N H. Mol Cell Biol. 1992;12:4809–4816. doi: 10.1128/mcb.12.11.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medberry S L, Olszewski N E. Plant J. 1993;3:619–626. doi: 10.1046/j.1365-313x.1993.03040619.x. [DOI] [PubMed] [Google Scholar]
- 29.Torbert K A, Gopalraj M, Medberry S L, Olszewski N E, Somers D A. Plant Cell Rep. 1998;17:284–287. doi: 10.1007/s002990050393. [DOI] [PubMed] [Google Scholar]
- 30.John M, Petersen M. Plant Mol Biol. 1994;26:1989–1993. doi: 10.1007/BF00019509. [DOI] [PubMed] [Google Scholar]
- 31.Diaz I, Royo J, O'Connor A, Carbonero P. Mol Gen Genet. 1995;248:592–598. doi: 10.1007/BF02423455. [DOI] [PubMed] [Google Scholar]
- 32.Gallusci P, Salamini F, Thompson R D. Mol Gen Genet. 1994;244:391–400. doi: 10.1007/BF00286691. [DOI] [PubMed] [Google Scholar]
- 33.Medberry S L, Lockhart B E, Olszewski N E. Plant Cell. 1992;4:185–192. doi: 10.1105/tpc.4.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giniger E, Varnum S M, Ptashne M. Cell. 1985;40:767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- 35.Roberts S G, Green M R. Nature (London) 1995;375:105–106. doi: 10.1038/375105a0. [DOI] [PubMed] [Google Scholar]
- 36.Lin Y S, Carey M, Ptashne M, Green M R. Nature (London) 1990;345:359–361. doi: 10.1038/345359a0. [DOI] [PubMed] [Google Scholar]
- 37.Carey M, Lin Y S, Green M R, Ptashne M. Nature (London) 1990;345:361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- 38.Emami K H, Carey M. EMBO J. 1992;11:5005–5012. doi: 10.1002/j.1460-2075.1992.tb05607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohashi Y, Brickman J M, Furman E, Middleton B, Carey M. Mol Cell Biol. 1994;14:2731–2739. doi: 10.1128/mcb.14.4.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwechheimer C, Smith C, Bevan M W. Plant Mol Biol. 1998;36:195–204. doi: 10.1023/a:1005990321918. [DOI] [PubMed] [Google Scholar]
- 41.Sanger M, Daubert S, Goodman R M. Plant Mol Biol. 1990;14:433–443. doi: 10.1007/BF00028779. [DOI] [PubMed] [Google Scholar]
- 42.Weinmann P, Gossen M, Hillen W, Bujard H, Gatz C. Plant J. 1994;5:559–569. doi: 10.1046/j.1365-313x.1994.5040559.x. [DOI] [PubMed] [Google Scholar]
- 43.Eckardt N A, McHenry L, Guiltinan M J. Plant Mol Biol. 1998;38:539–549. doi: 10.1023/a:1006081009173. [DOI] [PubMed] [Google Scholar]
- 44.Fukazawa J, Sakai T, Ishida S, Yamaguchi I, Kamiya Y, Takahashi Y. Plant Cell. 2000;12:901–915. doi: 10.1105/tpc.12.6.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miao Z H, Lam E. Plant J. 1995;7:887–896. doi: 10.1046/j.1365-313x.1995.07060887.x. [DOI] [PubMed] [Google Scholar]
- 46.Qin X F, Holuigue L, Horvath D M, Chua N H. Plant Cell. 1994;6:863–874. doi: 10.1105/tpc.6.6.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ulmasov T, Hagen G, Guilfoyle T. Plant Mol Biol. 1994;26:1055–1064. doi: 10.1007/BF00040688. [DOI] [PubMed] [Google Scholar]
- 48.Pascuzzi P, Hamilton D, Bodily K, Arias J. J Biol Chem. 1998;273:26631–26637. doi: 10.1074/jbc.273.41.26631. [DOI] [PubMed] [Google Scholar]
- 49.Amaya E, Musci T J, Kirschner M W. Cell. 1991;66:257–270. doi: 10.1016/0092-8674(91)90616-7. [DOI] [PubMed] [Google Scholar]
- 50.Ransone L J, Visvader J, Wamsley P, Verma I M. Proc Natl Acad Sci USA. 1990;87:3806–3810. doi: 10.1073/pnas.87.10.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Inukai T, Inaba T, Yoshihara T, Look A T. Mol Cell Biol. 1997;17:1417–1424. doi: 10.1128/mcb.17.3.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]