Abstract
The mammalian intestine harbors a complex microbial community that provides numerous benefits to its host. However, the microbiota can also include potentially virulent species, termed pathobionts, which can cause disease when intestinal homeostasis is disrupted. The molecular mechanisms by which pathobionts cause disease remain poorly understood. Here we describe a sepsis-like disease that occurs upon gut injury in antibiotic-treated mice. Sepsis was associated with the systemic spread of a specific multidrug-resistant E. coli pathobiont that expanded dramatically in the microbiota of antibiotic-treated mice. Rapid sepsis-like death required a component of the innate immune system, the Naip5-Nlrc4 inflammasome. In accordance with Koch's postulates, we found the E. coli pathobiont was sufficient to activate Naip5-Nlrc4 and cause disease when injected intravenously into unmanipulated mice. These findings reveal how sepsis-like disease can result from recognition of pathobionts by the innate immune system.
The intestinal microbiota is comprised of diverse microbial species that provide numerous benefits to their hosts1. However the microbiota can also contain species, termed `pathobionts', that can cause disease if not properly constrained1,2. A major mechanism constraining pathobionts is believed to be competition or suppression by the healthy microbiota1. Indeed, disruption of the healthy microbiota (for example, by antibiotics) can elicit pathobiont virulence and disease1, 3–6. Although pathobionts are believed to contribute to numerous disease states7–11, it has been challenging to identify the causal molecular mechanisms that are responsible for disease in vivo12–15.
A key orchestrator of innate immune responses are inflammasomes, multi-protein complexes that detect infection in the cytosol and are required for activation of the Caspase-1 protease, and downstream secretion of its substrates, the pro-inflammatory cytokines interleukin (IL)-1β and −1816. Several distinct inflammasomes have been described. For example, the Naip5-Nlrc4 inflammasome detects flagellin proteins from diverse bacteria17,18. Recently, several reports have established a protective role for inflammasomes in regulating microbiota composition and tissue reparative and regenerative responses to intestinal injury13,19–22. Although inflammasome responses in these models are protective against disease, it is also well-recognized that excessive inflammation has the potential to be pathological. However, a potential pathological role of inflammasome activation in response to a disrupted intestinal flora has not yet been investigated.
Here, we established a sepsis model triggered by a dysbiotic microbiota. In response to intestinal injury, antibiotic-treated mice succumbed rapidly to a sepsis-like death. Death was associated with the systemic presence of a multi-drug resistant E. coli pathobiont that we isolated from the organs of septic mice. Interestingly, we found that sepsis pathogenesis required activation of the Naip5-Nlrc4 inflammasome. Importantly, and in accordance with Koch's postulates, intravenous injection of the E. coli pathobiont into normal mice recapitulated the rapidly fatal Naip5-Nlrc4-dependent sepsis that was observed in antibiotic-treated mice in response to intestinal injury. Our results reveal a molecular mechanism by which disruptions of intestinal homeostasis can result in aberrant pathobiont-induced innate immune signaling and rapid sepsis-like death.
Results
Antibiotics and intestinal injury results in sepsis
To study the innate immune response to a disrupted intestinal microbiota, we established a disease model that couples antibiotic treatment with the dextran sulfate sodium (DSS)-induced intestinal injury. DSS is toxic to colonic epithelial cells, and typically results in a colitis-like disease23 characterized by severe weight loss, colonic bleeding and colonic shortening (Supplementary Fig 1a, b) . We wanted to understand how antibiotic-induced disruption of the microbiota (dysbiosis) would influence DSS-induced disease. Oral administration of a broad-spectrum antibiotic cocktail composed of ampicillin, vancomycin, neomycin and metronidazole (`AVNM') to colony-born C57BL/6 wild-type mice resulted in a change in the microbiota composition and reduction in the amount of 16S rDNA gene copy number along the intestinal-tract (Supplementary Fig. 1c – e). After this initial treatment, mice were given AVNM+5% DSS and we monitored survival. Consistent with previous reports24, AVNM-treatment rendered wild-type mice more susceptible to DSS (Fig. 1a and Supplementary Fig. 1f). Interestingly, ampicillin alone was sufficient to increase susceptibility to DSS treatment (Supplementary Fig. 1g, h). By contrast, mice treated with the broad-spectrum antibiotic streptomycin were just as susceptible to DSS as non-antibiotic treated mice (Fig. 1b).
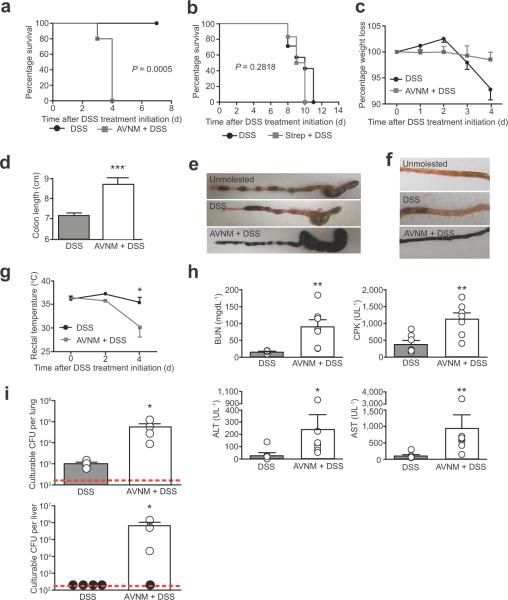
Figure 1. Antibiotic-treatment plus intestinal injury triggers a sepsis-like syndrome in wild-type mice.
(a) Survival of wild-type male and female mice that received AVNM+5% DSS (n = 5) and 5% DSS-only (n = 8). P = 0.0005 by Log rank analysis.
(b) Survival of wild-type male and female mice that received streptomycin+5% DSS (n = 6) and 5% DSS-only (n = 7). P = 0.2818 by Log rank analysis.
(c) Weight loss of wild-type male and female mice treated with 5% DSS compared to littermates that received drinking water supplemented with AVNM+5% DSS. Error bars indicate standard deviation; DSS-only (n = 9); AVNM+DSS (n = 9).
(d) Length of colons from wild-type male and female mice treated with 5% DSS compared to littermates that received drinking water supplemented with AVNM+5% DSS. Error bars indicate standard deviation. ***P = 0.0003 by Student's t-test. DSS-only (n = 4); AVNM+DSS (n = 4).
(e) Representative cecums and colons at 4 d post DSS treatment initiation of wild-type male mice treated with 5% DSS, AVNM+5% DSS or water-treated (unmolested) control littermates.
(f) Representative images of small intestine at 4 d post DSS treatment initiation of wild-type male mice treated with 5% DSS, AVNM+5% DSS or water-treated (unmolested) control littermates.
(g) Rectal temperature of wild-type male and female mice treated with AVNM+DSS compared to littermates that received 5% DSS only. Error bars represent standard deviation. *P < 0.05 by Student's t-test. DSS-only (n = 8); AVNM/DSS (n = 5).
(h) Serum levels of blood urea nitrogen (BUN), creatine phosphokinase (CPK), alanine transaminase and aspartate transaminase (AST) at 3 d post DSS treatment initiation of wild-type male and female mice. Data are representative of two combined experiments. **P < 0.01 and *P < 0.05 by Mann Whitney test. DSS-only (n = 7); AVNM/DSS (n = 7).
(i) Levels of culturable bacteria in the lung and liver of wild-type male and female mice 3 d post DSS treatment initiation. Data are representative of three independent experiments. *P < 0.05 by Student's t-test and (n = 4) for both conditions. Red dashed lines indicates limit of detection and black circles indicate individual mice below the limit of detection.
Although DSS typically induces colitis, we found that AVNM+DSS-treated mice did not exhibit the hallmark symptoms of colitis (e.g., weight loss, colonic inflammation/shortening, poor stool pellet formation; Fig. 1c – e and data not shown). Rather, AVNM+DSS-treated mice exhibited small-intestinal bleeding and hypothermia (Fig. 1f, g and Supplementary Fig. 2a), as well as multi-organ damage (MOD) and substantial serum levels of pro-inflammatory cytokines including TNF-α and IL-6 (Fig. 1h and data not shown). These results suggest that AVNM-induced changes to the microbiota trigger a systemic disease, distinct from colitis, in response to intestinal injury. Hypothermia and MOD are associated with sepsis in mice25,26. Consistent with this diagnosis, we found that antibiotic-treated mice exhibited significantly higher levels of culturable bacteria in the lung (P < 0.05) and the liver (P < 0.05) compared to nonantibiotic-treated mice (Fig. 1i, and Supplementary Fig. 2b, c).
To determine if the sepsis-like disease was specific to mice from our facility, we analyzed disease progression in AVNM+DSS-treated C57BL/6 specific pathogen free (SPF) mice from Jackson Laboratory (Jax mice) and Taconic farms (Taconic mice). Taconic mice developed hypothermia and were more susceptible to AVNM+DSS compared to DSS-only treated Taconic mice. Furthermore, AVNM attenuated DSS-induced colitis symptoms in Taconic mice (Supplementary Fig. 3a – b and data not shown). By contrast, disease in AVNM+DSS-treated Jax mice appeared to develop similarly to DSS-only treated Jax mice (Supplementary Fig. 3c – e and data not shown). Thus it appears that SPF mice from different facilities exhibit distinct disease symptoms in response to AVNM+DSS treatment.
Antibiotic-induced expansion of drug-resistant E. coli
Several non-exclusive models could explain how alterations in the microbiota can trigger disease. For example, antibiotic treatment could eliminate members of the microbiota that elicit protective host responses24. In addition, antibiotics could result in the overgrowth of AVNM-resistant pathobionts. Consistent with the latter possibility, after 7 d of AVNM-treatment, colony-born wild-type mice harbored an expanded population of AVNM-resistant bacteria along the intestinal-tract (Fig. 2a). We observed a similar expansion of an AVNM-resistant population in Taconic mice, but no detectable levels of culturable AVNM-resistant bacteria in Jax mice (Supplementary Fig. 3). Furthermore, we found that the AVNM-resistant bacteria colonized extraintestinal tissues of AVNM+DSS colony-born wild-type mice (Fig. 2b, c and Supplementary Fig. 2). Thus, sepsis-like disease correlated with the intestinal expansion and extraintestinal colonization of AVNM-resistant bacteria.
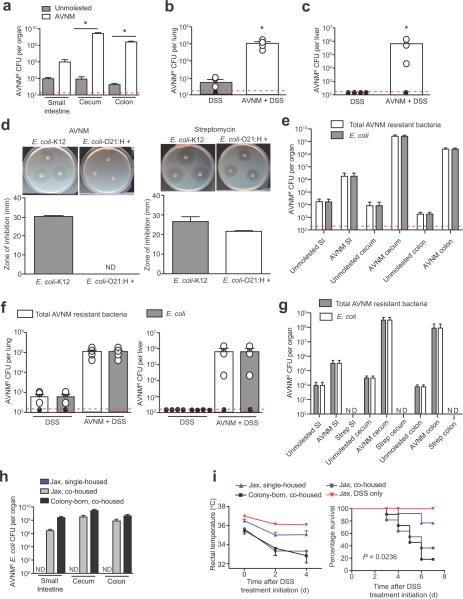
Figure 2. Expansion and extraintestinal colonization of a multi-drug resistant E. coli in response to intestinal injury in dysbiotic mice.
(a) Levels of AVNM-resistant bacteria in the intestinal-tract of AVNM-treated and water-treated wild-type male and female littermates 7 d post treatment-initiation. *P < 0.05 by Student's t-test and (n = 4) for both conditions
(b) Levels of AVNM-resistant bacteria in the lung of AVNM- plus DSS-treated and DSS-only treated male and female littermates 7 d post DSS treatment initiation. *P < 0.05 by Student's t-test and (n = 4) for both conditions. Red dashed line represents the limit of detection and black circles represent individual mice below the limit of detection.
(c) Levels of AVNM-resistant bacteria in the liver of AVNM- plus DSS-treated and DSS-only treated male and female littermates 7 d post DSS treatment initiation. *P < 0.05 by Student's t-test and (n = 4) for both conditions
(d) Representative images of disc diffusion assays and quantitation of the zone of inhibition to determine the susceptibility of an E. coli-O21:H+ isolate to AVNM or streptomycin. E. coli-K12 was used as a control. Error bars indicate standard deviation.
(e) Intestinal levels of total AVNM-resistant bacteria and AVNM-resistant E. coli-O21:H+ in AVNM-treated wild-type male and female mice compared to littermates that received a water control. DSS-treated mice (n = 4) and AVNM+DSS-treated mice (n = 4). Red dashed line represents the limit of detection and black circles represent individual mice below the limit of detection.
(f) Lung and liver levels of total AVNM-resistant bacteria and AVNM-resistant E. coli-O21:H+ in AVNM+DSS-treated wild-type male and female mice compared to littermates that received a DSS-only control. DSS-treated mice (n = 4) and AVNM+DSS-treated mice (n = 4).
(g) Intestinal levels of AVNM-resistant E. coli and streptomycin-resistant E. coli in AVNM and streptomycin-treated wild-type male and female mice were determined and compared to wild-type littermates that received a water control. Unmolested mice (n = 4); AVNM treated mice (n = 4) and streptomycin treated mice (n = 4).
(h) Intestinal levels of AVNM-resistant E. coli in single-housed female Jax mice and Jax mice co-housed with female colony-born (CB) mice after 7 d of AVNM treatment. Jax single-housed (n = 3); Jax co-housed (n = 3) and CB co-housed (n = 3).
(i) Temperature and survival of AVNM+DSS-treated female single-housed Jax mice and Jax mice co-housed with female colony-born mice. For survival, P = 0.0236 by Log rank analysis. Data represent two combined experiments; Jax single-housed (n = 11), Jax co-housed (n = 11); CB co-housed (n = 11) and DSS only (n = 5). Error bars indicate standard deviation.
We obtained an AVNM-resistant isolate from our mice that we identified as an O21:H+ Escherichia coli by16S rDNA sequencing, biochemical characterization, and serotyping (Fig. 2d, e and data not shown). An expansion of intestinal Enterobacteriaceae species in response to antibiotic treatment is consistent with previous reports27. AVNM-resistant E. coli was also recovered from the lung, liver, spleen and kidney of AVNM+DSS-treated colony-born mice (Fig. 2f and Supplementary Fig. 2e, f). To determine if E. coli was the predominant systemic AVNM-resistant species in our mice, we performed 16S rDNA cloning and sequencing analyses of livers and spleens from AVNM+DSS-treated colony-born mice. Although this approach identified clones representing multiple taxa, Escherichia was the most abundant taxon identified, consistent with our culturing experiments. In addition, E. coli was the only taxon represented by all tissues from AVNM+DSS-treated mice analyzed (data not shown). We also identified AVNM-resistant E. coli in the intestines of Taconic mice (Supplementary Fig. 3). Taken together, our data demonstrate that antibiotic treatment facilitates the intestinal overgrowth of AVNM-resistant O21:H+ E. coli that can translocate to extraintestinal sites upon intestinal injury.
The E. coli-O21:H+ was present in the microbiota of unmanipulated mice (Fig. 2e), but was not abundant, suggesting it was not able to compete efficiently for intestinal colonization with other members of the flora in the absence of antibiotics. Consistent with this hypothesis, we found that 1 week after cessation of AVNM treatment, the aberrant expansion of E. coli-O21:H+ was suppressed (Supplementary Fig. 4a), and importantly such mice no longer developed sepsis in response to DSS (Supplementary Fig. 4b, c).
As streptomycin-treated mice did not exhibit rapid sepsis-like death in response to DSS (Fig. 1b), we hypothesized that our AVNM-resistant E. coli isolate is sensitive to streptomycin. Indeed we found that our E. coli-O21:H+ isolate is sensitive to streptomycin and is eliminated from mice by streptomycin treatment (Fig. 2d, g). Taken together, these data clearly demonstrate that the increased sensitivity to AVNM+DSS and development of sepsis is associated with intestinal overgrowth and extraintestinal colonization of AVNM-resistant E. coli-O21:H+.
Because Jax mice do not appear to be colonized with AVNM-resistant E. coli-O21:H+, we could determine if this bacterium and its associated disease phenotype are transmissible. We co-housed Jax mice with colony-born mice and treated with AVNM for 7 d, after which co-housed Jax mice exhibited AVNM-resistant E. coli colonization levels comparable to colony-born co-housed mice. By contrast, single-housed Jax mice had no detectable levels of AVNM-resistant E. coli (Fig. 2h). Importantly, upon induction of intestinal injury, co-housed Jax mice exhibited increased susceptibility to DSS, similar to colony-born mice, and unlike single-housed Jax mice (Fig. 2i). These results demonstrate that E. coli-O21:H+ and the associated sepsis phenotype are transmissible.
Systemic E. coli infection is sufficient to induce sepsis
To determine if our E. coli-O21:H+ isolate is sufficient to cause disease in wild-type mice, we orally infected AVNM-treated Jax mice with E. coli-O21:H+. Oral administration of E. coli-O21:H+ to antibiotic treated Jax mice resulted in high levels of colonization (Supplementary Fig. 4d). Furthermore, colonized Jax mice developed hypothermia that was associated with reduced colitis symptoms compared to AVNM+DSS-treated mice that received a PBS oral challenge (Supplementary Fig. 4e).
To determine if the E. coli-O21:H+ isolate is sufficient to cause disease in wild-type mice with a normal (non-antibiotic treated) microbiota, we intravenously injected normal B6 mice with live or dead E. coli-O21:H+ and monitored disease. Mice were highly susceptible to a systemic challenge of live but not heat-killed bacteria (Fig. 3a). In addition, systemic live E. coli-O21:H+ infection was sufficient to recapitulate the specific pathology associated with AVNM+DSS treatment (Fig. 3b, c). Furthermore, live E. coli-infected mice exhibited tissue colonization patterns and levels similar to what was observed in AVNM+DSS-treated wild-type mice (Fig. 1i, 3d and Supplementary Fig. 2b, c). Thus, in fulfillment of Koch's postulates, a live systemic infection with E. coli-O21:H+ is sufficient to recapitulate the mortality and morbidity associated with AVNM+DSS-induced sepsis.
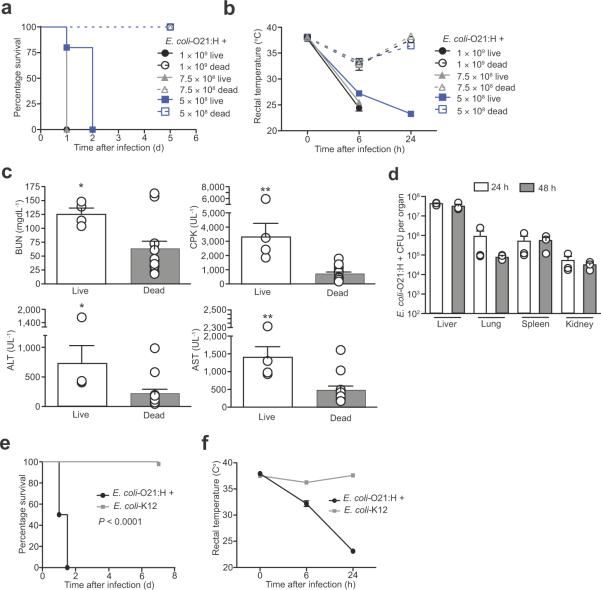
Figure 3. Systemic E. coli-O21:H+ infection is pathogenic in wild-type mice.
(a) Survival of wild-type female mice injected with live or dead 5×108, 7.5×108 or 1×109 live E. coli-O21:H+. P < 0.0001 for live vs. dead comparisons by Log rank analysis, (n = 5) females for all conditions.
(b) Rectal temperature of wild-type female mice injected with 5×108, 7.5×108 or 1×109 live or dead E. coli-O21:H+. At 6 h PI 1×109 live vs. 1×109 dead P = 0.0079, 7.5×108 live vs. 7.5×108 dead P = 0.0079, at 24 h PI 5×108 live vs. 5×108 dead P = 0.0195. (n = 5) for all conditions.
(c) Wild-type female mice were inoculated with 5×108 live or dead E. coli-O21:H+ and serum levels of BUN, CPK, ALT and AST were analyzed 24 h post infection. (n = 4) for live infection and (n = 12) females for dead infection. *P < 0.05, **P < 0.01 and *** P < 0.0005 by Student t-test and error bars indicate standard deviation.
(d) E. coli-O21:H+ CFUs in liver, lung, spleen and kidneys of wild-type female mice infected with 5×108 live bacteria at 24 and 48 h post infection. (n = 3) at both 24 h and 48 h PI and error bars indicate standard deviation.
(e) Survival of wild-type female mice infected with 5×108 live E. coli-O21:H+ or E. coli-K12. P < 0.0001 by Log rank analysis. (n = 8) for both E. coli-O21:H+ and E. coli-K12 injections.
(f) Rectal temperature of wild-type female mice infected with 5×108 live E. coli-O21:H+ or E. coli-K12. (n = 8) for both E. coli-O21:H+ and E. coli-K12 injections.
E. coli-O21:H+ harbors virulence factors
E. coli O21 isolates have been obtained from patients with extraintestinal infections including bacteraemia/sepsis28–32. We therefore hypothesized that the E. coli pathobiont encodes virulence factors mediating its pathogenicity in wild-type mice. Consistent with this hypothesis, disease was attenuated in mice infected with a non-pathogenic E. coli-K12 strain (MG1655), as compared to our E. coli-O21:H+ isolate, (Fig. 3 e, f), suggesting that detection of viable, nonpathogenic strains of E. coli by the immune system33 is not sufficient to trigger the sepsis-like disease in wild-type mice.
To identify further virulence-associated determinants, we sequenced the genome of our E. coli-O21:H+ isolate and aligned the reads to reference E. coli genomes (Supplementary Table 1). The fliC gene, encoding flagellin (H-antigen), was most similar to that of H21 isolates (Supplementary Fig. 5a). In addition, alignment to the E. coli O157:H7 strain Sakai reference genome revealed that our O21:H+ isolate harbors a cluster of genes exhibiting similarity to the ETT2 type III secretion system (T3SS)34 (Supplementary Fig. 5b). Many septicemic E. coli isolates encode a unique version of the ETT2 gene cluster called ETT2sepsis35 that contains a large deletion in the eiv gene cluster (cluster E in Supplementary Fig. 5b), creating deletions and truncations for the majority of eiv genes35. Despite this, ETT2sepsis is essential for sepsis pathogenesis, as mutations in ETT2sepsis render septicemic E. coli avirulent35. Interestingly, we were unable to align genomic content of the E. coli-O21:H+ genome to an 8.7 kb region of the ETT2 gene cluster that contains the eiv genes, suggesting that E. coli-O21:H+ harbors a form of the ETT2sepsis gene cluster (Supplementary Fig. 5b).
We also found that E. coli-O21:H+ encodes additional factors associated with bacteremic/septic E. coli clinical isolates including those important for adhesion (papC, fim gene cluster and pil gene cluster), internalization (csg genes) and iron acquisition (enterobactin and ferrichrome gene clusters)35–43. The genome also encodes the malX (marker for pathogenicity associated island from strain CFT073)38. Thus, the genome of E. coli-O21:H+ encodes a number of factors associated with bacteremic/septicemic clinical E. coli isolates.
Naip5-Nlrc4 inflammasome mediates AVNM/DSS–induced sepsis
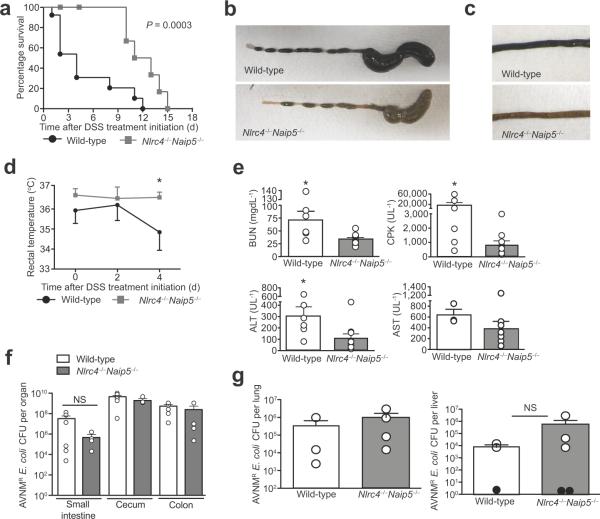
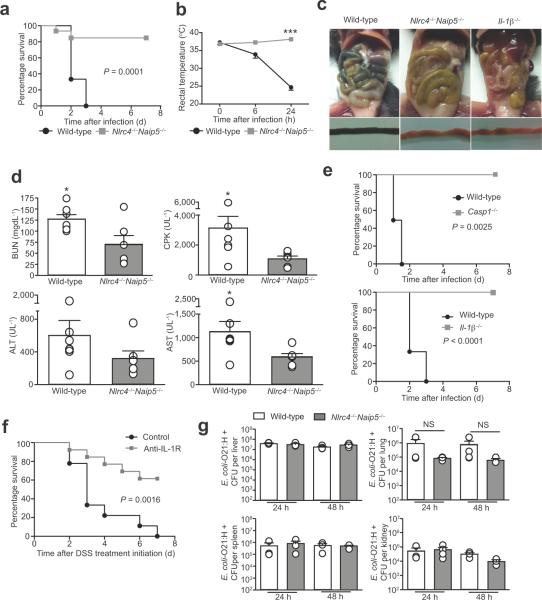
Since pathogenic E. coli were previously reported to activate inflammasomes containing Naip and Nlrc4 proteins44,45, we hypothesized that the Naip5-Nlrc4 inflammasome might mediate disease in our sepsis model. Indeed, we found AVNM+DSS-induced disease progression was highly attenuated in Nlrc4−/−Naip5−/− mice (Fig. 4a – e). Importantly, AVNM-treated Nlrc4−/− Naip5−/− mutant mice exhibited similar levels of AVNM-resistant E. coli overgrowth in the intestinal tract as wild-type fostermates prior to DSS treatment, implying that inflammasome-deficiency did not exert its protective effects by modulating bacterial growth (Figure 4f and Supplementary Fig. 6a, b). Moreover, the organs of AVNM+DSS-treated Nlrc4−/−Naip5−/− were colonized with similar levels of AVNM-resistant E. coli as wild-type mice (Figure 4g and Supplementary Fig. 6c, d). These data indicate that the absence of Naip5-Nlrc4 function protects AVNM+DSS-treated mice from developing sepsis at a step after extraintestinal dissemination of E. coli-O21:H+. Since Nlrc4−/−Naip5−/− mice eventually succumb to AVNM+DSS-induced sepsis, it is likely that additional host factors also contribute to disease.
Figure 4. Naip5-Nlrc4 mediates the pathogenesis of a sepsis-like syndrome in response to intestinal injury in dysbiotic mice.
(a) Survival of wild-type and Nlrc4−/−Naip5−/− male and female fostermates treated with AVNM+5% DSS. Kaplan-Meier plot is representative of two experiments and P = 0.0003 by log-rank analysis. Wild-type (n = 13) and Nlrc4−/−Naip5−/− (n = 14).
(b) Representative images of cecums and colons from wild-type and mutant female fostermates treated with AVNM+5% DSS at 3 d post DSS treatment initiation.
(c) Representative images of small intestines from AVNM+5% DSS treated wild-type and mutant female fostermates at 3 d post DSS treatment initiation.
(d) Rectal temperatures of wild-type and Nlrc4−/−Naip5−/− male and female fostermates treated with AVNM+5% DSS. Data are representative of three experiments. P < 0.05 by Student t-test. Wild-type (n = 10) and Nlrc4−/−Naip5−/− (n = 7).
(e) Serum BUN, CPK, AST and ALT levels at 2 d post DSS treatment initiation of wild-type and Nlrc4−/−Naip5−/− male and female fostermates treated with AVNM+5% DSS. Data are from three experiments and analyzed by Mann Whitney t-test. * P < 0.05. Wild-type (n = 3) and Nlrc4−/− Naip5−/− (n = 3).
(f) Levels of AVNM-resistant E. coli in the intestinal tracts of wild-type and Nlrc4−/−Naip5−/− male and female fostermates after 7 d of AVNM treatment. Wild-type (n = 6) and Nlrc4−/−Naip5−/− (n = 4).
(g) Lung and liver levels of total AVNM-resistant E. coli-O21:H+ in AVNM+DSS-treated wild-type mice and AVNM+DSS-treated Nlrc4−/− Naip5−/− male and female fostermates 3 d post DSS treatment initiation. Black dots represent tissues isolated from mice that had no bacteria above the limit detection.
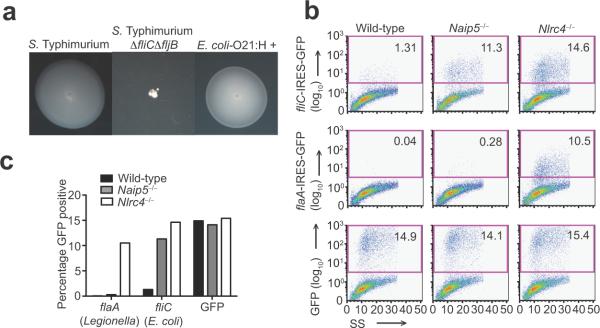
The Naip5-Nlrc4 inflammasome generally detects flagellin translocated to the host cell cytosol via type III or IV secretion systems44. Because E. coli-O21:H+ encodes a T3SS and a functional flagellin (FliC) (Fig. 5a and Supplementary Fig. 5) similar to pathogenic E. coli species, we hypothesized that the cytosolic presence of flagellin from this pathobiont can activate the Naip5-Nlrc4 inflammasome. We tested this hypothesis using a previously described retroviral `lethality' assay44 in which flagellin is expressed from a retroviral promoter directly in host cells, allowing for analysis of the effects of flagellin in the absence of other bacterial factors. Wild-type, Nlrc4−/− and Naip5−/− bone marrow derived macrophages were retrovirally transduced with fliC-IRES-GFP, control GFP, or control L. pneumophila flagellin (flaA-IRES-GFP). Activation of Naip5-Nlrc4 by flagellin results in cell death, as indicated by the loss of GFP positive cells. As previously reported, cells expressing L. pneumophila flaA-IRES-GFP could only be recovered from Nlrc4−/− macrophages46. Detection of FlaA by Naip6 likely accounts for the residual responsiveness of Naip5−/− cells. However cells expressing E. coli-O21:H+ fliC-IRES-GFP could be recovered from both Nlrc4−/− and Naip5−/− macrophages, but not wild-type macrophages (Fig. 5b, c). These data demonstrate that cytosolic flagellin derived from E. coli-O21:H+ activates the Naip5-Nlrc4 inflammasome and triggers host cell death.
Figure 5. E. coli-O21:H+ activates the Naip5-Nlrc4 inflammasome.
(a) Motility agar was inoculated with wild-type, motile S. Typhimurium LT2, flagellin-deficient S. Typhimurium ΔfliCΔfljB or E. coli-O21:H+ and incubated at 37 °C overnight. Representative images of the diffusion of motile bacteria are shown.
(b) Retroviral constructs expressing fliC from E. coli-O21:H+ or flaA from L. pneumophila followed by an IRES-GFP element or GFP alone were transduced into wild-type, Naip5−/−, or Nlrc4−/− macrophages, and the percentage of GFP-positive cells was enumerated by flow cytometry 4 d post-transduction.
(c) The percentage of cells expressing GFP from the retroviral lethality assay in (b) is indicated and charted.
Data are representative of three independent experiments.
Naip5-Nlrc4 inflammasome reduces host tolerance of E. coli
To address Koch's Postulates we infected wild-type and Nlrc4−/−Naip5−/− mice intravenously with E. coli-O21:H+. Systemic E. coli-O21:H+ infection was significantly less lethal in the absence of Naip5-Nlrc4 function (P = 0.0001) (Fig. 6a and Supplemental Fig. 7a, b). The absence of Naip5-Nlrc4 function also alleviated the pathologies associated with E. coli-O21:H+ infection (Fig. 6b – d). Furthermore, E. coli-O21:H+ infection was attenuated in Caspase1−/− and Il-1β−/− mice (Fig. 6e and Supplementary Fig. 7c, d). Additionally, systemic administration of anti-IL-1 receptor antibody protected mice from disease AVNM+DSS-induced disease (Fig. 6f). Thus, the absence of IL-1β is sufficient to protect mice from a systemic inflammatory response induced by a systemic E. coli-O21:H+ infection (Fig. 6e and Supplementary Fig. 7d). Taken together, our data clearly demonstrate that Naip5-Nlrc4 inflammasome signaling through IL-1β in response to a systemic E. coli-O21:H+ infection leads to sepsis.
Figure 6. The Naip5-Nlrc4 inflammasome reduces host tolerance of a systemic E. coli infection.
(a) Survival of female wild-type and Naip5−/−Nlrc4−/− mutant mice infected with 5×108 live E. coli. P = 0.0001 by Log rank analysis. Wild-type (n = 9) and Naip5−/−Nlrc4−/− (n = 11).
(b) Rectal temperature of female wild-type and Naip5−/−Nlrc4−/− mutant mice infected with 5×108 live E. coli. Wild-type (n = 9) and Naip5−/−Nlrc4−/− (n = 11). *** P = 0.0003 by Mann-Whitney test.
(c) Intestinal pathology at 48 h post-infection of wild-type and Naip5−/−Nlrc4−/− mutant mice infected with 5×108 live E. coli.
(d) Serum levels of BUN, CPK, ALT and AST female wild-type and Naip5−/−Nlrc4−/− mutant mice infected with 5×108 live E. coli. at 24 h post-infection. *P < 0.05 by Mann-Whitney test. Wild-type (n = 7) and Naip5−/−Nlrc4−/− (n = 6).
(e) in vivo infection of Caspase-1−/− and Il-1β−/− female mutant mice. Survival was monitored and compared to infected wild-type female mice. Infectious dose was 5×108 live E. coli O21:H+. For Caspase-1−/− P = 0.0025 and for Il-1β−/− P < 0.0001 by Log rank analysis. Wild-type (n = 9), Caspase-1−/− (n = 6) and Il-1β−/− (n = 12).
(f) Wild-type male and female mice were treated with AVNM for 7 d and then given AVNM+5% DSS. At 1, 24 and 48 h post DSS treatment initiation, mice were injected i.p. with 100 μg of α-IL-1R (n = 13) or control IgG antibody (n = 8) and survival was monitored. P = 0.0016 by Log rank analysis
(g) Wild-type and Naip5−/−Nlrc4−/− mutant female mice were infected with 5×108 live E. coli. At 24 and 48 h post infection, tissues were harvested, homogenized and analyzed for levels of E. coli colonization. Data were analyzed by Student t-test.
The greater susceptibility of wild-type (as compared to Naip5−/−Nlrc4−/−) mice to E. coli-O21:H+ infection was not associated with higher levels of extraintestinal bacterial colonization (Fig. 6g). In fact, colonization levels were similar in both genotypes. Thus we conclude that mice lacking Naip5-Nlrc4 fare better in response to a systemic E. coli-O21:H+ infection because they exhibit increased tolerance of the infection47, rather than because of negative effects on pathogen fitness.
Discussion
We describe a mouse model of sepsis triggered by dysbiosis and intestinal injury. This model resembles the clinically significant sepsis that is a major problem in human patients undergoing combination therapies involving antibiotics and cytotoxic treatments that damage the gut epithelium48. Consistent with the defining features of sepsis, we found that disease in AVNM+DSS-treated mice is characterized by systemic dissemination of bacteria, multi-organ damage, and systemic responses, such as hypothermia. A key conclusion of our study is that sepsis in our AVNM+DSS-treated mice is due to inflammasome activation by a specific multi-drug resistant E. coli-O21:H+ pathobiont. This pathobiont exists in the normal unmanipulated flora, but expands dramatically in the gut upon antibiotic treatment, and reaches systemic sites following DSS-induced intestinal injury (Supplementary Fig. 7e). In accordance with Koch's postulates, we show definitively that the systemic presence of this pathobiont is sufficient to induce sepsis-like disease.
Similar to our findings, a previous report demonstrated that antibiotic pre-treatment rendered mice more susceptible to DSS treatment24. However, this report proposed that disease in AVNM+DSS-treated mice is not associated with systemic spread of bacteria, but is instead due to a failure to receive appropriate TLR-dependent signals needed to repair gut epithelial injury. Consistent with this interpretation, it was found that disease could be alleviated by the oral administration of LPS, suggesting that TLR signaling plays an important protective role24. By contrast, in our model, mice given AVNM+DSS exhibited symptoms of sepsis, not colitis, and oral administration of LPS was not sufficient to prevent sepsis (Supplementary Fig. 2d). Indeed, dysbiosis in the AVNM+DSS model may both reduce the levels of beneficial microbes, as well as result in an expansion of pathobionts. The apparent discrepancy between our work and the prior study24 may be explained by differences in microbial flora in the mice, as well by methodological differences. For example, the previous study utilized a 7 d treatment of 2% DSS, which produces milder damage that mice are able to repair. In our model, we used a continuous dose of 5% DSS. Nevertheless, since systemic infection of normal mice with E. coli-O21:H+ is sufficient to recapitulate the AVNM+DSS disease model, we favor the hypothesis that pathology in our mice is driven primarily by an E. coli pathobiont.
Prior studies have demonstrated a protective role for the inflammasome in mediating pathogen resistance45,49 and in inducing tissue repair20,21. By contrast, our study demonstrates that excessive or systemic inflammasome-mediated recognition of a bacterium can lead to pathology and death. In our model, inflammasome activation results in IL-1β-driven immunopathology that the host cannot tolerate (Supplementary Fig. 7). The dependence on IL-1β distinguishes our sepsis model from others including LPS endotoxemia (Supplementary Fig. 7)50–52. Blockade of IL-1 signaling has long been considered a potential therapeutic treatment for human sepsis, but has met with little success in clinical trials53,54. This is likely because sepsis is a complex disease involving diverse microbes and host immune pathways. Our results highlight the importance of experimentally defining the relevant pathogenic pathways for different microbial causes of sepsis, and identifying the causative agents of sepsis in human patients.
Antibiotic-resistant E. coli present a challenge to antimicrobial medical interventions4. Our finding that mice lacking inflammasome components exhibit increased tolerance of systemic E. coli infection may have important therapeutic implications. Tolerance is a defense strategy that allows a host to endure an infection without influencing microbial load47,55. Tolerance contrasts with resistance, which protects a host by decreasing pathogen burden47,55. We propose that the inflammasome may be an alternative therapeutic target for patients exhibiting pathology due to antibiotic-resistant pathobionts. An important benefit of therapeutic modulation of host tolerance is that microbes are not predicted to evolve antagonistic traits to such therapeutics, in contrast to standard antimicrobials47,56,57. Taken together, our results provide insight into the mechanisms by which a specific constituent of a dysregulated flora can cause disease by inappropriate inflammasome activation.
Online Methods
Mice
Naip5−/− mice were generated as described previously44. Nlrc4−/− mice were from S. Mariathasan and V. Dixit58. We crossed Naip5−/− and Nlrc4−/− mice to each other to generate double knockout mice. Caspase-1-deficient (Casp1−/−) mice were a gift from A. Van der Velden and M. Starnbach59. Il-1β−/− mice were from the Zychlinsky Lab at Max Planck Institute. Gene targeted mice were on a C57BL/6 (B6) background. Wild-type B6 mice were originally obtained from the Jackson Laboratories, but all colony-born mice used were bred in our mouse facility for at least 10 generations. All mice were specific pathogen free, maintained under a 12 h light-dark cycle (7 a.m. to 7 p.m.) and given a standard chow diet (Harlan irradiated laboratory animal diet) ad libitum. For experiments involving littermate mice, we crossed Naip5+/−Nlrc4+/− mice to generate Naip5−/− Nlrc4−/− and Naip5+/+Nlrc4+/+ littermates. For fostermate mice, we placed wild-type pups with the mutant litter within 48 h of birth to nurse on the Naip5−/− Nlrc4−/− mother. Pups were weaned at 21 d of age. Animal experiments were approved by the University of California, Berkeley, Animal Care and Use Committee.
Survival assays
We used time to moribund for all survival assays. We identified moribund mice as those mice exhibiting ataxia severe enough such that the animals could not recover (unable to move when gently touched, or experience trouble self-correcting when placed on their side). Moribund mice were euthanized in compliance with the Animal Care and Use Committee at UC Berkeley.
DSS colitis model
We transferred 6 w old littermates to new cages and gave them drinking water supplemented with 5% DSS (w/v) (M.W. = 36,000–50,000 Da; MP Biomedicals) continuously for the duration of the experiment. We measured disease severity by monitoring weight daily, and colon length was measured at 6 d post DSS treatment initiation and survival (time to moribund).
Antibiotic/DSS sepsis model
At 5 w of age, we transferred littermates or fostermates to new cages and gave them drinking water supplemented with a combination of ampicillin (1 g L−1), neomycin (1 g L−1), metronidazole (1 g L−1) and vancomycin (0.5 g L−1), ampicillin only (1 g L−1) or streptomycin only (2 g L−1) for 7–10 d (Sigma Aldrich). After this initial antibiotic treatment, we gave mice drinking water supplemented with the appropriate antibiotic at the abovementioned concentration plus 5% DSS (w/v) continuously over the course of the experiment. We monitored mice for signs of disease including weight loss and colonic shortening as described above, rectal temperature, and serum analysis of organ damage as well as bacterial extraintestinal tissue colonization. For LPS+AVNM+DSS administration, we gave mice drinking water supplemented with AVNM/5% DSS and 10 μg ml−1 LPS O55:B5 (Sigma) after an initial 10 d AVNM treatment.
E. coli in vivo infections
We grew E. coli overnight in LB media shaking at 37 °C. We transferred age-matched female mice to new cages and injected them intravenously with 5×108, 7.5×108 or 1×109 live bacteria in PBS and gave them food and water ad libitum. We monitored body temperature and survival (time to moribund). For injections with dead bacteria, we prepared inoculums at the appropriate concentration in PBS and incubated at 65 °C for 30 min to heat kill the bacteria. For E. coli K12 infections, we grew E. coli overnight in LB media shaking at 37 °C. We transferred age-matched female mice to new cages and injected them intravenously with 5×108 bacteria in PBS and gave them food and water ad libitum.
Body Temperature
We monitored body temperature using a rectal probe and microtherma thermometer (Braintree Scientific). We lubricated the probe with a water-based lubricant (Astroglide) prior to use.
Serum Analysis
We euthanized mice and obtained blood by cardiac puncture at the indicated time points. We then aliquoted blood into BD serum separator tubes, incubated at room temperature for 20 min and centrifuged at 2.6 rpm for 20 min. We stored serum at −80 °C until analysis. BUN, CPK, AST and ALT measurements were done by IDEXX Laboratories.
Bacterial culturing
For compositional analysis of the intestinal tract and extraintestinal tissues, we harvested whole tissues and homogenized them using a Polytron PT2100 homogenizer at 17000 rpm (Kinematica) in sterile thioglycolate media. We then serially diluted the homogenate and plated on LB agar and schaedler agar supplemented with 5% defibrinated sheep blood plates and grew at 37 °C aerobically and anaerobically for 24 h. We counted bacterial colonies and then separated based on colony appearance and performed colony 16S rDNA PCR and sequencing (see below).
For cultivation of Enterobacteriales species, we harvested and homogenized intestinal and extraintestinal tissues from AVNM+DSS mice as described above. We then serially diluted the homogenates and plated them on bacterial medias that support the growth of Enterobacteriales including LB, Maconkey, and EMB. We then incubated the plates aerobically at 37 °C for 24 h, after which we counted colonies, classified based on colony appearance and subjected them to 16S rDNA colony PCR and sequencing (see below). We then tested isolates for antibiotic susceptibility (see below).
For CFU analysis of E. coli in AVNM+DSS-treated and E. coli-O21:H+ infected mice, we harvested and homogenized intestinal and extraintestinal organs in sterile PBS, serially diluted and plated on EMB or LB plates containing AVNM and incubated them at 37 °C for 24 h.
Colony PCR
We resuspended colonies in sterile PBS, boiled for 10 min at 100 °C. We used this directly for PCR analysis with the universal bacterial primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′)60. We performed PCR on a Bio-Rad iCycler using an annealing temperature of 51 °C and the following conditions: 95 °C (5 min), followed by 30 cycles of 95 °C (30 s), annealing (1 min), 72 °C (2 min), and a final extension at 72 °C (10 min). Reactions were then subjected to a PCR cleanup using the QIAquick PCR Purification Kit (Qiagen) and sequenced using the 1492R primer (Elim Biopharm). We classified the sequences using the Michigan State University Ribosomal Database Project classifier function (http://rdp.cme.msu.edu/).
Antibiotic susceptibility assays
We determined E. coli-O21:H+ susceptibility to AVNM and streptomycin using a disc diffusion assay. We inoculated liquid cultures with E. coli and grown at 37 °C with shaking overnight. We placed aliquots of cultures onto LB agar plates and 6 mm Whatman discs loaded with the appropriate antibiotic onto the agar. We incubated plates overnight at 37 °C and measured the zone of bacterial growth inhibition. Antibiotic susceptibility profiling of the isolate was also performed by Jeffrey Schapiro and James LaPan of the Clinical Microbiology Laboratory at Kaiser Permanente (Berkeley, CA). E. coli DH5α was used as a control.
16S rDNA quantitative PCR analyses
We treated mice with regular drinking water or drinking water supplemented with AVNM for 7–10 d. We then harvested intestinal tissues, snap froze them in liquid nitrogen and stored them at −80 °C prior to analysis. We extracted DNA using the Qiagen Stool Kit per manufacturer's instructions with a bead-beating step. Specifically, we put whole tissues into Lysing Matrix E tubes (MP Biomedicals) with 1.4 ml of buffer ASL and subjected to three cycles of shaking for 3 min using a Vortex Genie 2. We placed samples on ice for 3 min in between cycles. We extracted DNA from the resulting supernatants per manufacturer's protocol. We quantified purified DNA on a Nanodrop 1000 (Thermo Scientific) and then performed quantitative PCR assays on the Step One Plus RT PCR system (Applied Biosystems) with the iTaq SYBR Green Supermix with Rox reagent (Bio–Rad). We normalized 16S gene copy numbers to Rps17 (mouse). We used the following primer sequences: mouse Rps17F (5′- CGCCATTATCCCCAGCAAG-3′) and Rps17R (5′-TGTCGGGATCCACCTCAATG-3′); bacteria universal 16S rDNA uniF340 (5′-ACTCCTACGGGAGGCAGCAGT-3′) and uniR514 (5′-ATTACCGCGGCTGCTGGC-3′)61; Enterobacteriaceace 16S rDNA specific 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 826R (5′-GCCTCAAGGGCACAACCTCCAAG-3′)61; Bacteroides 16S rDNA specific BactF285 (5′-GGTTCTGAGAGGAGGTCCC-3′) and UniR338 (5′-GCTGCCTCCCGTAGGAGT-3′)61; Actinobacteria 16S rDNA specific Actino235F (5′-CGCGGCCTATCAGCTTGTTG-3′) and Eub518R (5′-ATTACCGCGGCTGCTGG-3′)62.
Motility Assay
We inoculated single colonies of bacteria at a single point on soft motility LB agar (0.35%) and incubated overnight at 37 °C. As controls, we used S. Typhimurium strain LT2 and the flagellin mutant S. Typhimurium LT2 ΔfliCΔfljB (A. Van Der Velden and M. Starnbach).
Retroviral constructs and transductions
We produced retroviral constructs and executed transductions as previously described44. Briefly, we generated retroviral particles by the transient transfection of Phoenix–Eco packaging cells with MSCV2.2-based retroviral vectors. We cultured bone marrow–derived cells (1×106) for 48 h in a 6-well Falcon brand non-tissue culture treated plate in medium containing macrophage colony-stimulating factor (M-CSF) and then we transduced cells with 2 ml of retrovirus–containing packaging cell supernatant. 24 h after the initial infection, we infected bone marrow-derived cells once again with 2 ml of retrovirus–containing packaging cell supernatant. We analyzed macrophages 4 d post-transduction and analyzed for GFP expression on a Beckman Coulter FC-500 flow cytometer. We analyzed more than 25,000 cells per sample. We cloned the E. coli-O21:H+ fliC gene using the primers fliC XhoI (5′-AAAACTCGAGGCCACCATGGCACAAGTCATTAATAC-3′) and fliC NotI (5′- TTTTGCGGCCGCTTAACCCTGCAGCAGAAG-3′)
LPS injections
We injected age-matched female wild-type and Il-1β−/− mice intravenously with 5 mgkg−1 LPS O55:B5 (Sigma) and given food and water ad libitum. We monitored Survival (time to moribund).
E. coli-O21:H+ Serotyping
Serotyping was done at the E. coli reference center, Pennsylvania State University.
Experiments with commercial vendor mice
For disease analyses, we kept mice in autoclaved isolator cages and gave them sterile–filtered AVNM or water for 7 d followed by treatment with sterile-filtered AVNM+5% DSS or 5% DSS-only. We monitored weight, temperature and survival (time to moribund). For CFU analyses, upon receipt, we immediately dissected mice or put them in autoclaved isolator cages and gave them sterile-filtered drinking water with AVNM for 7 d, after which intestines were dissected. We plated homogenized tissues on AVNM+EMB plates, enumerated colony types and subjected them to 16S rDNA PCR and sequencing and genotyped for eprJ. Separate shipments of mice were received for each experimental replicate.
Horizontal Transfer Experiments
We obtained 5 w old C57BL/6 male mice were obtained from Jackson Laboratories (Sacramento, CA) and kept them single-housed in isolator cages or co-housed in isolator cages with age and sex-matched C57BL/6 mice bred in our colony. We then treated mice with AVNM ad libitum for 7 d, after which we harvested and homogenized intestinal tissues and plated them on EMB agar plates supplemented with AVNM to determine the levels of AVNM-resistant bacteria colonization. To analyze disease severity (temperature, body weight and time to moribund), we gave mice given drinking water supplemented with AVNM+5% DSS after the initial 7 d course of AVNM treatment. We compared disease severity to single–housed controls.
Oral infection of commercially purchased mice
We obtained 5 w old C57BL/6 female mice from Jackson Laboratory (Sacramento, CA), kept them in isolator cages and treated them with AVNM ad libitum for 7 d. On 4, 5 and 6 d, we gavaged mice with 200 μl of an overnight culture of E. coli-O21:H+, washed and resuspended to 2.5×109 in sterile PBS. We gavaged control mice with 200 μl of sterile PBS. On 7 d, we either dissected mice for CFU analysis or gave them drinking water supplemented with AVNM+5% DSS and monitored temperature and weight.
Whole genome sequencing of E. coli O21:H+
Library preparation, Illumina HiSeq 2000 SE50 sequencing and mapping were done by the Tufts Core Facility (TUCF) at Tufts Univeristy. For library preparation, we grew E. coli overnight in LB media shaking at 37 °C and isolated genomic DNA using the Qiagen DNeasy kit and submitted DNA to TUCF for library preparation and sequencing. Reads were aligned to eleven publicly available E. coli whole–genome sequences (E. coli HS CP000802, E. coli E24377A ETEC NC_009801, E. coli 53638 EIEC AAKB00000000, E. coli 042 EAEC FN554766, E. coli O127:H6 EPEC NC_011601, E. coli IAI39 ExPEC NC_011750, E. coli S88 ExPEC NC_011742, E. coli O157:H7 EHEC NC_002695, E. coli APEC O1 CP000468, E. coli UTI89 ExPEC CP000243, E. coli CFTO73 ExPEC AE014075) using the CLC Genomics workbench software. Annotation of the resulting consensus sequences was done using RAST rapid annotations using subsystem technology (http://rast.nmpdr.org).
Phylogenetic analysis of fliC
We performed analysis using Phylogeny.fr (http://www.phylogeny.fr) one–click mode63 and the publicly available E. coli fliC gene sequences (E. coli strain B2F1 DQ459008.1, E. coli strain U11a–44 AY250004.1, E. coli O111:H– str.11128 AP010960.1, E. coli O113:H21 DQ862122.1, E. coli ABU 83972 CP001671.1, E. coli VTH–15 GQ423574.1, E. coli CFT073 AE014075.1, E. coli 042 FN554766, E. coli O55:H7 str. CB961 CP001846.1, E. coli IHE3034 CP001969.1, E. coli O157:H7 str. Sakai NC_002695, E. coli HS CP000802, E. coli IAI39 NC_011750, E. coli UTI89 CP000243, E. coli S88 NC_011742, E. coli UMN026 CU928163.2, E. coli SMS–3–5 NC_010498, E. coli O127:H6 str. E2348 NC_011601, E. coli APEC O1 NC_008563) and Legionella pneumophila flaA gene sequence as the outgroup NC_006368.1.
IL-1R antibody experiments
We gave colony-born wild-type mice drinking water supplemented with AVNM for 7 d. We then gave mice given drinking water supplemented with AVNM and 5% DSS to induce gut injury. At 1, 24 and 48 h post DSS administration, we administered 100 μg of anti-IL-1R antibody (Amgen) i.p. or control IgG antibody to mice and and monitored survival (time to moribund) over the course of the disease.
Statistics
We analyzed Kaplan Meier plots using log rank analysis. All other data are expressed as mean +/− standard error of the mean and were analyzed using a Student's t-test or Mann Whitney test. We considered a P value less than 0.05 to be significant.
Supplementary Material
Acknowledgements
We thank J. Schapiro and J. LaPan for biochemical characterization of E. coli-O21:H+ and S. Brandt for helpful discussions about the isolate; K. Bodi for helpful advice about whole-genome sequencing analysis; G. Barton, D. Portnoy and K. Barry for helpful discussions and critical reading of the manuscript; E. Kofoed for the flaA-IRES-GFP construct; K. Sotelo-Troha for technical help and L. Lopez for support in our mouse facility. This work was supported in part by NIH grants AI075039 and AI080749 awarded to R.E.V. and NIH Ruth L. Kirschstein NRSA fellowship AI091068 awarded to J.S.A. R.E.V. is an Investigator of the Cancer Research Institute and a Burroughs Welcome Fund Investigator in the Pathogenesis of Infectious Disease.
Footnotes
Author Contributions J.S.A. and R.E.V. conceived of the study, designed experiments and wrote the manuscript. J.S.A. directed the study and performed all experiments. N.J.T. did all mouse intravenous injections and performed the retroviral lethality assay.
References
- 1.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chow J, Mazmanian SK. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe. 2010;7:265–276. doi: 10.1016/j.chom.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voth DE, Ballard JD. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev. 2005;18:247–263. doi: 10.1128/CMR.18.2.247-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oteo J, Perez-Vazquez M, Campos J. Extended-spectrum [beta]-lactamase producing Escherichia coli: changing epidemiology and clinical impact. Curr Opin Infect Dis. 2010;23:320–326. doi: 10.1097/qco.0b013e3283398dc1. [DOI] [PubMed] [Google Scholar]
- 5.Russell DA, Thompson RC. Targets for sepsis therapies: tumor necrosis factor versus interleukin-1. Curr Opin Biotechnol. 1993;4:714–721. doi: 10.1016/0958-1669(93)90055-2. [DOI] [PubMed] [Google Scholar]
- 6.Salyers AA, Gupta A, Wang Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004;12:412–416. doi: 10.1016/j.tim.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Penders J, Stobberingh EE, van den Brandt PA, Thijs C. The role of the intestinal microbiota in the development of atopic disorders. Allergy. 2007;62:1223–1236. doi: 10.1111/j.1398-9995.2007.01462.x. [DOI] [PubMed] [Google Scholar]
- 8.Lakhan SE, Kirchgessner A. Gut inflammation in chronic fatigue syndrome. Nutr Metab (Lond) 2010;7:79. doi: 10.1186/1743-7075-7-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vijay-Kumar M, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 12.Cadwell K, et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elinav E, et al. NLRP6 Inflammasome Regulates Colonic Microbial Ecology and Risk for Colitis. Cell. 2011 doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bloom SM, et al. Commensal bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schroder K, Tschopp J. The inflammasomes. Cell. 140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 17.Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 19.Allen IC, et al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med. 2010;207:1045–1056. doi: 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupaul-Chicoine J, et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity. 2010;32:367–378. doi: 10.1016/j.immuni.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 21.Zaki MH, et al. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–391. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu B, et al. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1016814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okayasu I, et al. A Novel Method in the Induction of Reliable Experimental Acute and Chronic Ulcerative-Colitis in Mice. Gastroenterology. 1990;98:694–702. doi: 10.1016/0016-5085(90)90290-h. [DOI] [PubMed] [Google Scholar]
- 24.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Sherwood ER, Enoh VT, Murphey ED, Lin CY. Mice depleted of CD8+ T and NK cells are resistant to injury caused by cecal ligation and puncture. Lab Invest. 2004;84:1655–1665. doi: 10.1038/labinvest.3700184. [DOI] [PubMed] [Google Scholar]
- 26.Larsen R, et al. A central role for free heme in the pathogenesis of severe sepsis. Sci Transl Med. 2010;2:51ra71. doi: 10.1126/scitranslmed.3001118. [DOI] [PubMed] [Google Scholar]
- 27.Ubeda C, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest. 2010;120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren Y, et al. Characterization of Escherichia coli O3 and O21 O antigen gene clusters and development of serogroup-specific PCR assays. J Microbiol Methods. 2008;75:329–334. doi: 10.1016/j.mimet.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 29.Evans DJ, Jr., Evans DG, Young LS, Pitt J. Hemagglutination typing of Escherichia coli: definition of seven hemagglutination types. J Clin Microbiol. 1980;12:235–242. doi: 10.1128/jcm.12.2.235-242.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevens P, Young LS, Adamu S. Opsonization of various capsular (K) E. coli by the alternative complement pathway. Immunology. 1983;50:497–502. [PMC free article] [PubMed] [Google Scholar]
- 31.Evans DJ, Jr., et al. Hemolysin and K antigens in relation to serotype and hemagglutination type of Escherichia coli isolated from extraintestinal infections. J Clin Microbiol. 1981;13:171–178. doi: 10.1128/jcm.13.1.171-178.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orskov I, Orskov F. Escherichia coli in extra-intestinal infections. J Hyg (Lond) 1985;95:551–575. doi: 10.1017/s0022172400060678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sander LE, et al. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature. 2011;474:385–389. doi: 10.1038/nature10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren CP, et al. The ETT2 gene cluster, encoding a second type III secretion system from Escherichia coli, is present in the majority of strains but has undergone widespread mutational attrition. J Bacteriol. 2004;186:3547–3560. doi: 10.1128/JB.186.11.3547-3560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ideses D, et al. A degenerate type III secretion system from septicemic Escherichia coli contributes to pathogenesis. J Bacteriol. 2005;187:8164–8171. doi: 10.1128/JB.187.23.8164-8171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong WT, Bettelheim KA, Cheng FC, Ong GB. Serotypes of Escherichia coli isolated from patients with recurrent pyogenic cholangitis. J Hyg (Lond) 1982;88:513–517. doi: 10.1017/s0022172400070364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sannes MR, Kuskowski MA, Owens K, Gajewski A, Johnson JR. Virulence factor profiles and phylogenetic background of Escherichia coli isolates from veterans with bacteremia and uninfected control subjects. J Infect Dis. 2004;190:2121–2128. doi: 10.1086/425984. [DOI] [PubMed] [Google Scholar]
- 39.Mokady D, Gophna U, Ron EZ. Virulence factors of septicemic Escherichia coli strains. Int J Med Microbiol. 2005;295:455–462. doi: 10.1016/j.ijmm.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Ramos NL, et al. Genetic relatedness and virulence gene profiles of Escherichia coli strains isolated from septicaemic and uroseptic patients. Eur J Clin Microbiol Infect Dis. 29:15–23. doi: 10.1007/s10096-009-0809-2. [DOI] [PubMed] [Google Scholar]
- 41.Korczak B, et al. Use of diagnostic microarrays for determination of virulence gene patterns of Escherichia coli K1, a major cause of neonatal meningitis. J Clin Microbiol. 2005;43:1024–1031. doi: 10.1128/JCM.43.3.1024-1031.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mapes S, Rhodes DM, Wilson WD, Leutenegger CM, Pusterla N. Comparison of five real-time PCR assays for detecting virulence genes in isolates of Escherichia coli from septicaemic neonatal foals. Vet Rec. 2007;161:716–718. doi: 10.1136/vr.161.21.716. [DOI] [PubMed] [Google Scholar]
- 43.Henderson JP, et al. Quantitative metabolomics reveals an epigenetic blueprint for iron acquisition in uropathogenic Escherichia coli. PLoS Pathog. 2009;5:e1000305. doi: 10.1371/journal.ppat.1000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lightfield KL, et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat Immunol. 2008;9:1171–1178. doi: 10.1038/ni.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miao EA, et al. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci U S A. 2010;107:3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lightfield KL, et al. Differential requirements for NAIP5 in activation of the NLRC4 inflammasome. Infect Immun. 2011;79:1606–1614. doi: 10.1128/IAI.01187-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider DS, Ayres JS. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat Rev Immunol. 2008;8:889–895. doi: 10.1038/nri2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russell JA. Drug therapy: Management of sepsis. New England Journal of Medicine. 2006;355:1699–1713. doi: 10.1056/NEJMra043632. [DOI] [PubMed] [Google Scholar]
- 49.Lamkanfi M, Dixit VM. The inflammasomes. PLoS Pathog. 2009;5:e1000510. doi: 10.1371/journal.ppat.1000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarkar A, et al. Caspase-1 regulates Escherichia coli sepsis and splenic B cell apoptosis independently of interleukin-1beta and interleukin-18. Am J Respir Crit Care Med. 2006;174:1003–1010. doi: 10.1164/rccm.200604-546OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fantuzzi G, et al. Effect of endotoxin in IL-1 beta-deficient mice. J Immunol. 1996;157:291–296. [PubMed] [Google Scholar]
- 52.O'Reilly M, Silver GM, Davis JH, Gamelli RL, Hebert JC. Interleukin 1 beta improves survival following cecal ligation and puncture. J Surg Res. 1992;52:518–522. doi: 10.1016/0022-4804(92)90321-p. [DOI] [PubMed] [Google Scholar]
- 53.Opal SM, et al. Confirmatory interleukin-1 receptor antagonist trial in severe sepsis: A phase III, randomized, double-blind, placebo-controlled, multicenter trial. Critical Care Medicine. 1997;25:1115–1124. doi: 10.1097/00003246-199707000-00010. [DOI] [PubMed] [Google Scholar]
- 54.Fisher CJ, et al. Initial Evaluation of Human Recombinant Interleukin-1 Receptor Antagonist in the Treatment of Sepsis Syndrome - a Randomized, Open-Label, Placebo-Controlled Multicenter Trial. Critical Care Medicine. 1994;22:12–21. doi: 10.1097/00003246-199401000-00008. [DOI] [PubMed] [Google Scholar]
- 55.Raberg L, Graham AL, Read AF. Decomposing health: tolerance and resistance to parasites in animals. Philos Trans R Soc Lond B Biol Sci. 2009;364:37–49. doi: 10.1098/rstb.2008.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roy BA, Kirchner JW. Evolutionary dynamics of pathogen resistance and tolerance. Evolution. 2000;54:51–63. doi: 10.1111/j.0014-3820.2000.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 57.Boots M. Fight or learn to live with the consequences? Trends Ecol Evol. 2008;23:248–250. doi: 10.1016/j.tree.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 58.Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 59.Li P, et al. Mice deficient in IL-1 beta-converting enzyme are defective in production of mature IL-1 beta and resistant to endotoxic shock. Cell. 1995;80:401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 60.DeSantis TZ, et al. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb Ecol. 2007;53:371–383. doi: 10.1007/s00248-006-9134-9. [DOI] [PubMed] [Google Scholar]
- 61.Croswell A, Amir E, Teggatz P, Barman M, Salzman NH. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect Immun. 2009;77:2741–2753. doi: 10.1128/IAI.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fierer N, Jackson JA, Vilgalys R, Jackson RB. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl Environ Microbiol. 2005;71:4117–4120. doi: 10.1128/AEM.71.7.4117-4120.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dereeper A, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.