Abstract
Emerging evidence indicates that microRNAs (miRNAs) may play an important role in the pathogenesis of Huntington’s disease (HD). To identify the individual miRNAs that are altered in HD and may therefore regulate a gene network underlying mutant huntingtin-induced neuronal dysfunction in HD, we performed miRNA array analysis combined with mRNA profiling in the cerebral cortex from N171-82Q HD mice. Expression profiles of miRNAs as well as mRNAs in HD mouse cerebral cortex were analyzed and confirmed at different stages of disease progression; the most significant changes of miRNAs in the cerebral cortex were also detected in the striatum of HD mice. Our results revealed a significant alteration of miR-200 family members, miR-200a and miR-200c in the cerebral cortex and the striatum, at the early stage of disease progression in N171-82Q HD mice. We used a coordinated approach to integrate miRNA and mRNA profiling, and applied bioinformatics to predict a target gene network potentially regulated by these significantly altered miRNAs that might be involved in HD disease progression. Interestingly, miR-200a and miR-200c are predicted to target genes regulating synaptic function, neurodevelopment and neuronal survival. Our results suggest that altered expression of miR-200a and miR-200c may interrupt the production of proteins involved in neuronal plasticity and survival, and further investigation of the involvement of perturbed miRNA expression in HD pathogenesis is warranted, and may lead to reveal novel approaches for HD therapy.
Keywords: miRNA array, Huntington’s disease, gene array, miR-200, Trim2
Introduction
Huntington’s disease (HD) is a fatal neurodegenerative disease caused by mutation of the huntingtin (HTT) gene. The causative mutation in the HTT gene was shown to be a CAG expansion within the first coding exon, leading to a polyglutamine expansion in the HTT protein. The polyglutamine expansion converts HTT from a neuroprotective to a neurotoxic protein (Cattaneo et al. 2005). HTT is expressed ubiquitously in humans and rodents, with the highest levels found in neurons and the testes. Complete knockout of the mouse HTT gene (Hdh) causes embryonic death before day 8.5 (E8.5, before gastrulation and the formation of the nervous system) (Trottier et al. 1995, Ferrante et al. 1997). After gastrulation, HTT becomes important for neurogenesis. In addition to its function in development, HTT may play a role in neurodevelopment, regulation of apoptosis, control of BDNF production, vesicular and mitochondrial transport, neuronal gene transcription, and synaptic transmission (Cattaneo et al. 2005, Zuccato et al.2010). The exact function of HTT in cells still remains largely unknown however. Despite ubiquitous HTT expression in neurons throughout the brain, striatal and cortical neurons are most vulnerable in HD (Fusco et al. 1999). Although a vast amount of literature indicate possible molecular mechanisms are involved in the neuronal dysfunction and neuronal degeneration observed in HD, such as loss of BDNF, excitotoxicity and corticalstriatal dysfunction, proteolysis, misfolding aggregation and clearance of mutant HTT, mitochondrial dysfunction, transcriptional dysregulation, and autophagy, the pathogenic mechanisms caused by the mutant HTT remain incompletely characterized (Zuccato et al. 2010). Various molecular changes precede behavioral symptoms in HD, including deregulation of gene expression (Augood et al. 1997b, Augood et al. 1997a), and histone modifications(Gray 2011). Such changes suggest that understanding regulation of gene expression may provide potential therapeutic strategies to forestall the disease process in HD.
The discovery of microRNAs (miRNAs) has broadened our overall understanding of the mechanisms regulating gene expression, by revealing an entirely novel level of gene regulatory control. MiRNAs are endogenous, single-stranded, noncoding RNA molecules, typically 22 nucleotides in length that negatively regulate gene expression. MiRNAs are expressed in all tissues, but the brain appears to have the greatest diversity of miRNAs (Thomson et al. 2004). There is emerging evidence that miRNAs may play an important role in neuronal development as well as in the pathogenesis of neurodegenerative disorders (Conrad et al. 2006, Trivedi & Ramakrishna 2009, Bilen et al. 2006, Kim et al. 2007). A study shows that ablating the miRNA processing protein dicer in dopaminoceptive neurons produces behavioral and neuroanatomical phenotypes before neurodegeneration (Cuellar et al. 2008), highlighting the contribution of deregulated miRNAs to neurological dysfunctional consequence before cell death. Indeed, several miRNAs regulated by repressor element-1 transcription factor (REST) have significantly altered expression levels in HD (Johnson et al. 2008), and another study reported that miR9/miR9* regulates REST and CoREST and is downregulated in HD (Packer et al. 2008, Johnson et al. 2008). These findings suggest that dysregulation of neuronal miRNAs might be involved in HD pathogenesis; however, how the network of miRNAs is altered in the early stages of the disease, and how these changes contribute to dysregulation of gene expression remain unknown.
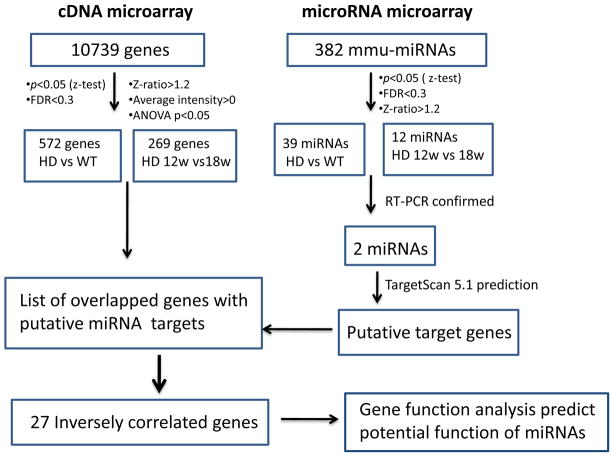
Our objective was to identify miRNA(s) that altered before neuronal death, and may contribute to mutant HTT-induced neuronal dysfunction and further degeneration in HD. This was accomplished by interrogation of miRNA signature and mRNA expression profiles in cerebral cortex of a mouse model of HD, and further investigating the significantly changed miRNAs in the striatum. By obtaining expression-profiling data from HD mice at different ages, we were able to analyze the dynamic changes in miRNA and mRNA expression that occur at the early and middle stages of disease progression in HD mice. Figure 1 shows a schematic diagram describing the experimental procedures and data analysis approaches used in this study.
Fig. 1.
Schematic flow diagram describing procedures used to identify inversely correlated putative target genes of the differentially expressed miRNAs in HD mice.
Here we present a comprehensive analysis of miRNA expression signatures integrated with gene expression profiles in the cerebral cortex of an HD mouse model. By interrogating the data from miRNA and mRNA profiling, we identified the significantly altered miRNAs in the brain of HD mice at early stage of disease and the targeted genes by these altered miRNAs. The results reveal a subset of miRNAs that are specifically deregulated at an early stage of disease in HD mice, and predict possible target genes that are negatively correlated with significantly altered miRNAs in these mice, suggesting that these miRNAs may be part of a gene-regulatory network that contributes to mutant HTT-induced neuronal dysfunction and degeneration.
Materials and Methods
Animals
N171-82Q HD mice express a human N-terminal-truncated HTT with 82 polyglutamine repeats driven by a mouse prion protein promoter; these exhibit the motor phenotypes around 12 weeks of age (Cheng et al. 2011) before much neurodegeneration occurs. Apoptotic neurons were detected in the cortex and striatum at 4.5 months of age, and TUNEL-positive neurons in N171-82Q mice appeared as the animals became older at 5 months of age (Yu et al. 2003).
Transgenic mice were identified by PCR analysis of genomic DNA extracted from mouse tail. Genotyping was conducted using a three-way PCR analysis: two primers were complementary to the prion protein genomic DNA sequence (PrP-sense 5′-CCTCTTGTGACTATGTGGACTGATGTCGG-3′ and PrP-antisense 5′-GTGGATACCCCCTCCCCCAGCCTAGACC-3′). The amplified product of this reaction is 700 bp in length. The antisense primer is also complementary to the 3′-untranslated portion of the PrP vector and, in combination with a third sense primer to the HD sequence (HD-591-5′: 5′-GAACTTTCAGCTACCAAGAAAGACCGTGT-3′), generated a transgene-specific product that is 350 bp in length. Transgenic mice were mated continuously to hybrid (C3H/HEJ×C57 BL/6J F1) mice, and the mice were maintained on the hybrid background. Because we previously reported that we found a sex-dependent difference between males and females as we reported previously (Duan et al. 2003, Duan et al. 2008), we used male mice in the present study. The mice at different stages were anesthetized by isofluorane and the brains were dissected quickly on ice and different brain regions were stored at −80°C in RNA later solution to prevent RNA degradation. All animal experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by Johns Hopkins University Animal Care Committees. Body weight was monitored weekly as well as survival.
RNA preparation
Total RNA including small RNA was extracted with TRIZOL (Invitrogen) according to the manufacturer’s instruction. Briefly, brain tissue samples were homogenized in Trizol reagent, chloroform was added to the tissue lysate and tubes were shaken vigorously for 15 sec to mix well and incubated at room temperature for 5 min. The homogenate was separated into aqueous and organic phases by centrifugation. The upper, aqueous phase containing RNA was then extracted. RNA was eluted with nuclease-free water. RNA quantification and quality were assessed by using the RNA 6000 Nano Kit and 2100 Bioanalyzer (Agilent Technologies UK Ltd, West Lothian, UK).
miRNA microarray hybridization
miRNA labeling and hybridization were performed as previously described (Cunningham et al. 2009). One microgram of total RNA containing miRNAs was first polyadenylated, the introduced polyA tail was used as a priming site for cDNA synthesis. The primer used for cDNA synthesis is biotinylated and contains a universal PCR primer sequence. cDNA was added to the multiplexed miRNA specific oligo pool (Illumina, San Diego, CA) heated to 70°C, and the oligos were allowed to anneal as the temperature of the cDNAs was slowly decreased to 40°C. During the annealing step the oligo/cDNA hybrids were bound to streptavidin-coated paramagnetic particles and, after washing, the bound annealed oligos were extended through the cDNA primer, forming an amplifiable product. The extended oligos were eluted from the beads and added to a PCR reaction, in which one of the universal primers was fluorescently labeled and the other biotinylated. These amplified PCR products were captured on streptavidin-coated beads, washed, and then denatured to yield single-stranded fluorescent molecules. These were hybridized to arrays containing the captured oligos for 30 min at 60°C, then overnight at 45°C (Universal 12 BeadChip, Illumina, San Diego, CA). Arrays were washed and scanned on the Illumina BeadArray Reader, and automatic image registration and intensity extraction software was used to derive intensity data. The array intensity data were imported into BeadStudio v3.2 (Illumina).
miRNA microarray data analysis
Average values of the replicate spots of each miRNA on the microarray were normalized by global normalization. The correction factor was calculated by dividing the sum of intensities of each sample by the average of all the samples. The normalized values were calculated by multiplying average intensities of each miRNA with the correction factor. Raw intensity data for each experiment were transformed to log10, and used for the calculation of Z-scores. Significant changes in miRNA expression were calculated in the form of Z- ratios and/or Z-test values, by using Z-score values in all calculations. Z-ratios constitute a measure of the change in miRNA expression of a given gene from control group value, expressed in units of standard deviation from the average change of all genes for that comparison. The Z-ratio is a measure of fold change between comparisons, and the p values test for reproducibility of the intensity of a gene among biological replicate arrays: Z-ratio (between condition A and B) = z(A) − z(B)/SD deviation). Remaining genes were analyzed by two-way ANOVA to establish the statistical significance of differential levels of expression between ages and genotypes (p < 0.05). Comparisons between Z-ratios, however, test for equivalence of significant changes between the HD group and control (WT group). All miRNA expression changes were assessed through comparison with control samples. A Z-ratio value of ± 1.50 and/or a Z-test value p < 0.01 were the significance thresholds used in this study. Hierarchical clustering was performed with the software package DIANE 6.0, a spreadsheet-based microarray analysis program based on the SAS JMP7.0 system.
mRNA microarray hybridization
Total RNA was used to generate biotin-labeled cRNA by using the Illumina TotalPrep RNA Amplification Kit (Ambion; Austin, TX, cat #IL1791). A total of 0.75 μg of biotin-labeled cRNA was hybridized at 58 °C for 16 h to Illumina’s Sentrix MouseRef-8 Expression Bead-Chips (Illumina, San Diego, CA). The arrays were washed and blocked, and the labeled cRNA was detected by staining with streptavidin-Cy3. The arrays were scanned with an Illumina BeadStation 500× Genetic Analysis Systems scanner and the image data were extracted using the Illumina BeadStudio software, Version 3.0. mRNA microarray data were analyzed by a method similar to miRNA analysis by using DIANE 6.0.
qRT-PCR
The miRCURY LNA microRNA PCR system (Exiqon) was used to detect levels of mature mmu-miR-200a and mmu-miR-200c in brain tissue. U6 was used as control for normalization. Briefly, 10 ng of total RNA were used to synthesize first-strand cDNA with the first-strand cDNA synthesis kit and the miR-specific or control RT primers. Each individual miRNA or U6 was amplified using the miRCURY LNA SYBRGreen master mix, miRNA specific LNA PCR primer, the universal primer or the control PCR primers. ABI 7900HT Fast Real-Time PCR System was used to perform PCR amplification. Data were analyzed by applying the comparative quantification method.
Construction of mouse pre-miR-200a expressing vector and Trim2-3′UTR reporter vector
A pPRIME-CMV-GFP-FF3 vector (kindly provided by Dr. Stephen Elledge, Harvard University) was used to generate a construct expressing mouse pre-miR-200a. The cDNA containing mouse pre-miR-200a was amplified from mouse genomic DNA by PCR. The EcoR I restriction enzyme site was added to the reverse primer. The sequence of mouse pre-miR-200a was confirmed by DNA sequencing. Full length 3′UTR of Trim2 with predicted miR-200a binding sites were amplified from genomic DNA and inserted into a pMIR-REPORT luciferase vector and the sequence was confirmed by DNA sequencing.
Striatal cell model of HD
Immortalized striatal precursor cells expressing normal HTT (STHdhQ7/Q7) or mutant HTT (STHdhQ111/Q111) were initially derived from huntingtin knock-in mouse model, the cells were provided by Dr. Marcy McDonald at MGH and were prepared as described previously (Trettel et al. 2000). The cells were maintained at 33°C in high glucose DMED (Invitrogen, Cat #11995065) medium with 10% of fetal bovine serum (FBS, GIBCO, Cat# 16140071), 1% of Penicillin-Streptomycin (Invitrogen, Cat# 15140122), 1% of L-glutamate (Invitrogen, Cat #25030081) and 400ug/ml of G418 (Mediatech, Cat# 30-234-CR), in a humidified atmosphere of 95% air and 5% of CO2.
MiR-200a expression and pMIR-REPORT luciferase assay
Full length 3′-UTR of Trim2 and miR-200a were transfected into immortalized striatal cells expressing mutant HTT (STHdhQ111/Q111) by using Lipofectamine 2000 (Invitrogen Inc) according to the manufacturer’s instruction. Twenty-four hours after transfection, the total RNA was extracted and mature miR-200a expression was determined. Cells co-transfected with Trim2 3′UTR reporter vector and vector expressing pre-miR-200a were used for luciferase assay by ONE-Glo™ System (Promega).
Results
MiRNAs and mRNAs are dynamically regulated along disease progression in HD mice
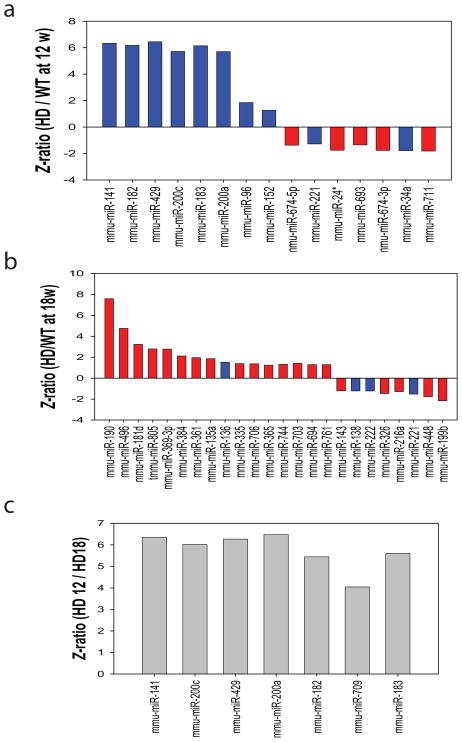
To determine how mutant HTT modulates miRNA expression along disease progression in HD mice, we analyzed miRNAs profiles in the cerebral cortex of N171-82Q HD mice and their littermate controls at 12 weeks and 18 weeks of age, 12 weeks of age represents the onset of motor phenotype, and 18 weeks of age represent the middle state of disease progresssion. Cerebral cortex samples were collected from male N171-82Q HD mice and age-matched littermate control mice. The male N171-82Q mice live to about 22–24 weeks of age on average, and have onset of motor impairment around 12 weeks. Our analysis revealed 15 miRNAs that were significantly deregulated at 12 weeks of age (Fig. 2a) and 24 miRNAs that were altered significantly at 18 weeks of age (Fig. 2b). Three criteria were used to establish significance of a difference in levels of a miRNA between wild type and HD mice: Z-ratio > 1.2; false discovery rate (FDR) <0.3; and p<0.05 (Fig. 2). Among these 39 miRNAs that were differentially expressed between HD mice and wild type control mice, 14 miRNAs were previously reported to be enriched in brain (blue bars in Fig. 2) (Bak et al. 2008) and 25 miRNAs are newly described miRNAs in brain (red bars in Fig. 2). Interestingly, seven dramatically upregulated miRNAs in 12-week-old HD mice belong to two families: miR-200a, miR-200c, miR-141 and miR-429 belong to one family, and miR-96, miR-182 and miR-183 belong to another family. To further determine the expression pattern of miRNAs with disease progression, we compared miRNA expression profiles of cortical tissue samples from 12- and 18-week-old HD mice. Interestingly, we found that seven miRNAs were specifically upregulated at the early stage of disease progression in HD mice (Fig 2c), suggesting that alteration of these miRNAs might contribute to the disease pathogenesis by post-transcriptional regulation of gene expression.
Fig. 2. Significantly altered miRNAs in the cerebral cortex of N171-82Q HD mice by miRNA microarray analysis.
From 382 distinct miRNAs included on our array, (a) 8 miRNAs were significantly upregulated and 7 miRNAs were downregulated in cerebral cortex of 12-week-old HD mice compared to those in their age-matched littermate nontransgenic controls. (b) 16 miRNAs were upregulated and 8 miRNAs were downregulated in cerebral cortex of 18-week-old HD mice compared to those in their age-matched littermate controls. Blue bars represent previously described brain-enriched miRNAs, and red bars represent newly identified miRNAs in brains in our study. (c) miRNAs were significantly upregulated in the cerebral cortex of 12-week-old HD mice compared to those in 18-week-old HD mice. The criteria for the selection of significantly changed miRNAs is fold change >1.2, FDR<0.3, and p<0.05 (Z-test). N=3 samples each group.
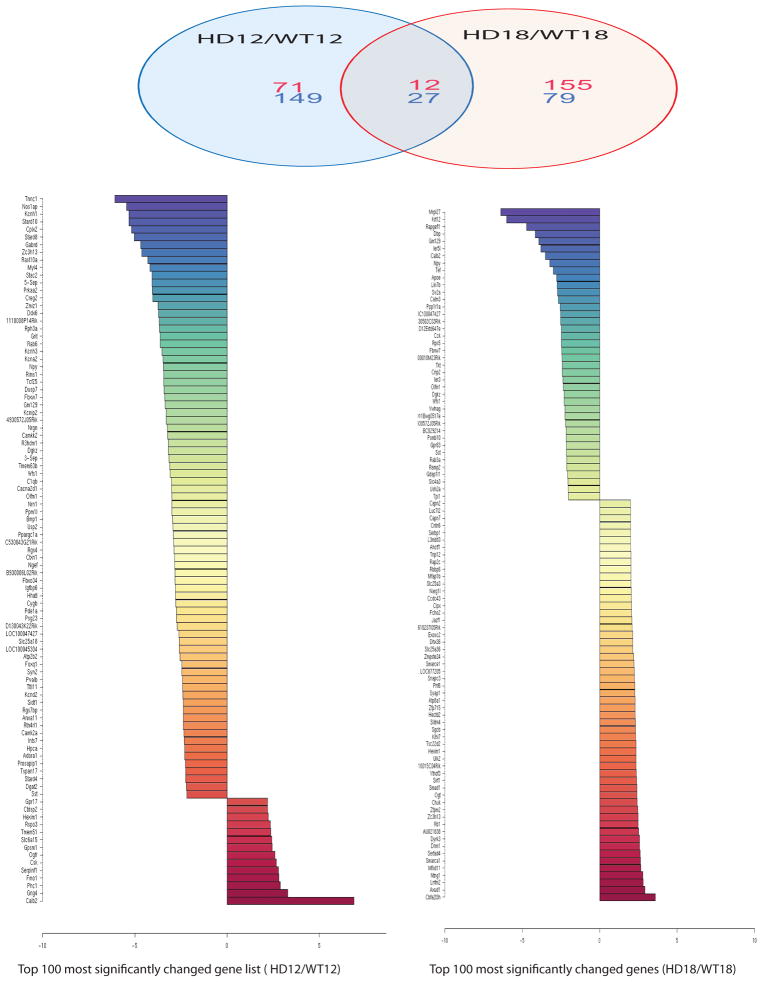
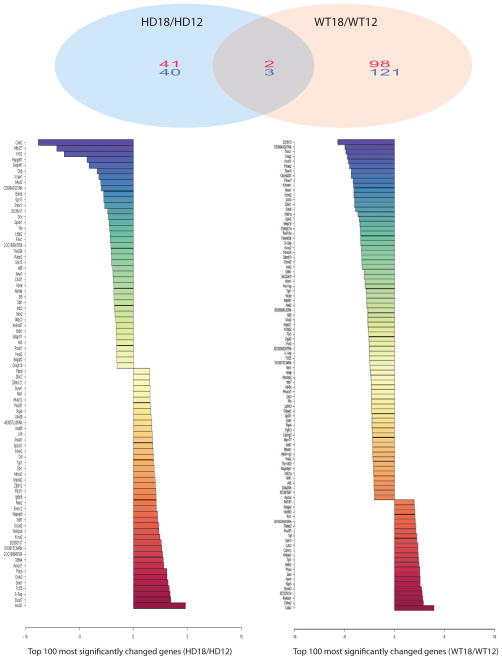
Gene microarray analysis from the same samples that we examined for miRNA profiles identified 83 upregulated mRNAs (numbers in red) and 176 downregulated mRNAs (numbers in blue) in 12-week-old HD mice compared to littermate control mice (Fig. 3 and Suppl. Table 1). There were 167 transcripts upregulated (numbers in red) and 106 transcripts downregulated (numbers in blue) in 18-week-old HD mice compared to their age-matched controls (Fig. 3 and Suppl Table 2). Levels of several gene transcripts were also altered with disease progression; 43 genes were upregulated (numbers in red) and 43 genes were downregulated (numbers in blue) in the cerebral cortex of 12-week-old HD mice compared to 18-week-old HD mice (Fig. 4 and Suppl. Table 3). The majority of mRNAs altered in HD mice did not overlap with the changes in control mice, suggesting that these changes are specific to HD (Fig. 4 and Suppl Table 4).
Fig 3. Significantly altered gene expression and Z-ratio values were identified between HD mice and wild type controls by microarray analysis.
Top panel: Venn diagram summarizes distinct and overlapping expression of mRNAs in cerebral cortex of HD mice compared to their controls at 12 and 18 weeks of age. Numbers of upregulated gene (Red) and downregulated gene (Blue). Note that there are 71 upregulated genes and 149 downregulated genes present only in 12-week-old HD mouse brains; and 155 genes are upregulated, and 79 genes are downregulated only in 18-week-old HD mice. Twelve upregulated genes and 27 downregulated genes are commonly present in both 12-week-old and 18-week-old HD mouse brains. Bottom colored bar graphs represent Z-ratios of top 100 significantly altered mRNAs. Criteria for the selection of significantly changed mRNAs are: fold change >1.2, FDR<0.3, and p<0.05 (Z-test). n=3
Fig. 4. Significantly altered gene expression and Z-ratio were identified in cerebral cortex of 12-week-old mice and 18-week-old mice by microarray analysis.
Top panel: Venn diagram summarizes distinct mRNA expression profiles and overlapping genes in HD mice along with disease progression or wild type (WT) control mice along with age. Numbers represent upregulated gene (Red), and downregulated gene (Blue). Note that there are 41 genes that are upregulated and 40 genes are downregulated in 18-week-old HD mouse brains compared to those in 12-week-old HD mouse brains, while 98 genes are upregulated and 121 genes are downregulated in 18-week-old wild type mouse brains compared to those in 12-week-old wild type mouse brains. Only 2 genes that are upregulated and 3 genes that are downregulated commonly exist in both HD and wild type mouse brains as age increases from 12 weeks to 18 weeks. Bottom Z-ratio graphs show top 100 significantly altered genes between 18-week-old HD mouse brains and 12-week-old HD brains (left) or between 18-week-old wild type mouse brains versus 12-week-old wild type mouse brains, according to the criteria for the selection of significantly changed mRNAs, i.e, fold change >1.2, FDR<0.3, and p<0.05 (Z-test). n=3.
Significantly altered miRNAs are detected by qRT-PCR in the cortex and striatum of N171-82Q HD mice
To further confirm the changes of miRNAs identified in the miRNA array analysis, we used quantitative RT-PCR (qRT-PCR) to determine their levels in tissue samples from the cerebral cortex. We confirmed the upregulation of miR-200a and miR-200c in the cortex of 12 weeks old HD mice (Fig. 5); the changes of miR-200a and miR-200c are also detected in the striatum of N171-82Q mice (Fig. 5). The expression levels of other miRNAs, including miR-182, miR-183, miR-96, miR-141, and miR-429, were too low to be detected in these brain regions by qRT-PCR (data not shown). Furthermore, we detected dynamic changes of miR-200a (Fig 6a) and miR-200c (Fig 6b) along disease progression in the cortex and striatum of HD mice by qRT-PCR; levels of both miR-200a and miR-200c were significantly elevated at the early stage of disease progression (8- and 12-week-old HD mice), but did not differ at the later stage of disease progression (18-week-old HD mice) compared to age-matched wild type mice (Fig. 6). These results further indicated that miRNA regulation is altered dynamically as disease progresses.
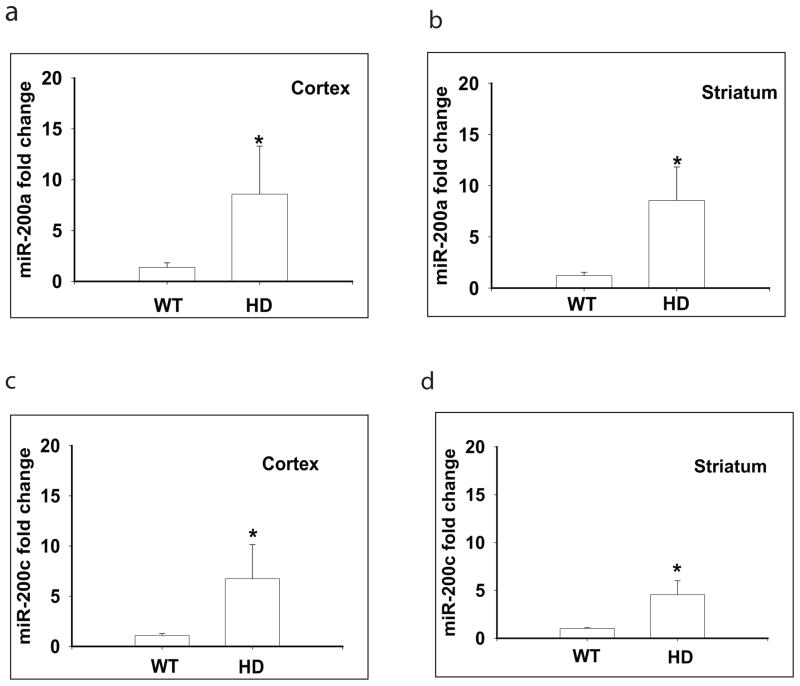
Fig. 5. Significantly altered miR-200a and miR-200c were detected in the cortex and striatum of N171-82Q HD mice by quantitative RT-PCR.
MiR-200a levels were significantly upregulated in the cortex (a) and striatum (b) of 12-week-old HD mice; miR-200c levels were significantly upregulated in the cortex (c) and striatum (d) of 12-week-old HD mice. Bars represent Mean and STD, n= 5–6 mice in each group. *p<0.05 vs the levels of age-matched littermate controls by Student’s t-tests.
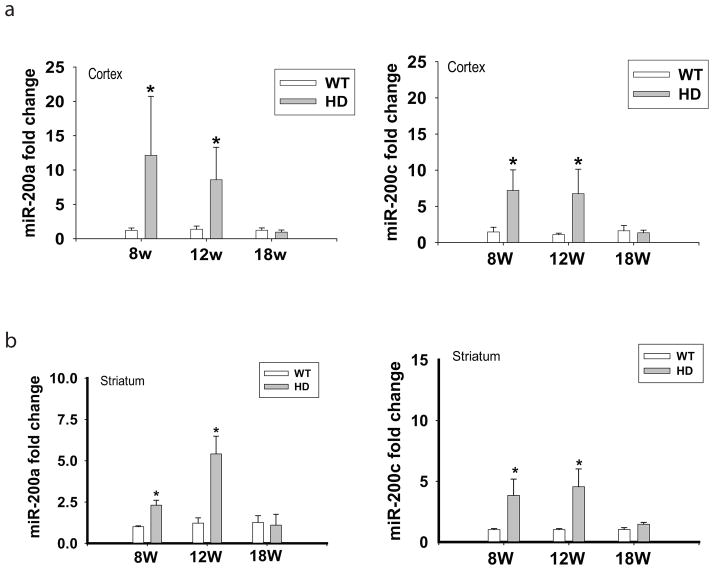
Fig. 6. Dynamic changes of miR-200a and miR-200c from presymptomatic stage to late disease progression in the cortex and striatum of N171-82Q.
(a) Levels of miR-200a and miR-200c in the cortex of N171-82Q at different ages. (b) Levels of miR-200a and miR-200c in the striatum of N171-82Q mice at different ages. Bars represent Mean and STD, n= 5–6 mice in each group. *p<0.05 vs the levels of age-matched littermate controls by Student’s t-tests.
Computational predictions of putative targets of miRNAs altered in HD mice suggest potential mechanisms underlying synaptic dysfunction and compromised neurogenesis in HD
We used the computational algorithm Target Scan 5.1 to identify putative targets for confirmed deregulated miR-200a and miR-200c on the basis of “seed sequence” homology with 3′-UTRs of mRNA transcripts (Krek et al. 2005, Grimson et al. 2007). The total number of target genes predicted is 680 for miR-200a and 462 for miR-200c. These two miRNAs have considerable sequence similarity; in particular, the “seed sequences” of these two miRNAs (UAACACUGU) are identical. This similarity predicts that these two miRNAs would have overlapping sets of target genes. We then filtered the list of predicted targets with the mRNA profile analyzed in the same set of RNA samples. We identified 10 predicted target genes of miR-200a and 12 predicted target genes of miR-200c that were indeed downregulated in the cerebral cortex of HD mice in our mRNA array data (Table 1). The mRNAs encoding ATP2A2, ATXN, and NRXN1 are shared targets for miR-200a and miR-200c. Interestingly, these downregulated target genes of miR-200a and miR-200c have been suggested to play important roles in synaptic function, axonal trafficking, neurotransmitter release, neurogenesis, and neuronal survival (Table 1). Upregulation of miR-200a and miR-200c might repress these genes involved in progressive neuronal dysfunction and neurodegeneration in HD.
Table 1.
Significantly inversely-regulated genes of predicted targets of miR-200a and miR-200c in the cortex of 12-week-old N171-82Q mice.
| Genes | Fold change (HD/WT) | z-ratio | P value | Target of | Function |
|---|---|---|---|---|---|
| Genes involved in synaptic function | |||||
| KIF3a | −1.42 | −1.97 | 0.0234 | 200a | Involved in anterograde transport |
| NRXN1 | −1.48 | −2.23 | 0.018 | 200a, 200c | Encodes neurexin 1α, function in the vertebrate nervous system as cell adhesion molecules and receptors, required for synapse formation, mediates the assembly of presynaptic terminals |
| PTPRD | −1.43 | −1.96 | 0.0005 | 200a | Promotes neurite growth, regulates axon guidance |
| RTN4RL1 | −1.53 | −2.37 | 0.00003 | 200a | Localizes to the surface of neurons including axons, may play a role in regulating axonal regeneration and plasticity in the adult CNS |
| DNAJC5 | −1.31 | −1.53 | 0.0119 | 200c | May have an important role in presynaptic function. May be involved in calcium-dependent neurotransmitter release at nerve endings |
| KCNA2 | −1.92 | −3.47 | 0.00002 | 200c | Potassium channel, regulates neurotransmitter release and neuronal excitability |
| Lin7b | −1.52 | −2.29 | 0.002 | 200c | Ensures proper localization of GRIN2B (subunit 2B of the NMDA receptor) to neuronal postsynaptic density and may function in localizing synaptic vesicles at synapses where it is recruited by beta-catenin and cadherin |
| PPP1R9B | −1.56 | −2.53 | 0.0275 | 200c | Acts as a scaffold protein in multiple signaling pathways. Modulates excitatory synaptic transmission and dendritic spine morphology. May establish a signaling complex for dopaminergic neurotransmission through D2 receptors by linking receptors to downstream signaling molecules and the actin cytoskeleton |
| DRD2 | −1.33 | −1.72 | 0.042 | 200a | Dopamine receptor D2, dopamine transmission |
| Genes involved in development and neurogenesis | |||||
| NFIC | −1.76 | −3.12 | 0.0207 | 200c | DNA binding transcription factor, regulates neural progenitor differentiation |
| NGEF | −1.71 | −2.84 | 0.00002 | 200c | Plays a role in axon guidance regulating ephrin-induced growth cone collapse |
| PCTK1 | −1.37 | −1.74 | 0.0006 | 200c | May play a role in signal transduction cascades in terminally differentiated cells |
| Genes highly expressed in brain, functions not well-known, or play roles in neuronal survival and function | |||||
| ATXN1 | −1.82 | −3.32 | 0.0003 | 200a, 200c | Involved in RNA metabolism, ATXN1 functions as a genetic risk modifier that contributes to Alzheimer’s disease pathogenesis through a loss-of-function mechanism by regulating β-secretase cleavage of APP and Aβ levels |
| PRKCB1 | −1.47 | −2.08 | 0.00002 | 200c | A calcium-activated, phospholipid-dependent, serine- and threonine-specific enzyme, may also regulate neuronal functions |
| TRIM2 | −1.4 | −1.77 | 0.0051 | 200a | Ubiquitin ligase, the protein localizes to cytoplasmic filaments, binds to neurofilament light subunit (NF-L) and regulates NF-L ubiquitination, dysregulation triggers neurodegeneration |
| OLFM1 | −1.76 | −3.00 | 0.0003 | 200a | Abundant expression in brain, function not known |
| ST3GAL5 | −1.37 | −1.77 | 0.0001 | 200a | The encoded protein is a member of glycosyltransferase family 29 and may be localized to the Golgi apparatus, Catalyzes formation of ganglioside GM3. Participates in the induction of cell differentiation, |
Trim2 is a confirmed target of miR-200a
As an additional approach to identifying the predicted target mRNAs of miRNAs, we constructed pre-miR-200a in a pPRIME-CMV-GFP-FF3 viral vector. Expression of miR-200a in striatal cells expressing mutant HTT (STHdhQ 111/Q111) (Trettel et al. 2000) was identified by GFP fluorescence (Fig. 7a). Striatal cells transfected with pre-miR-200a produced mature miR-200a (Fig. 7b–c). We selected the gene encoding Trim2, as Trim2 plays important roles in neurite outgrowth and is involved in neurodegeneration (Balastik et al. 2008, Khazaei et al.). Luciferase reporter vectors with full-length 3′-UTR regions of Trim2 and mutation of the miR-200a binding sites were constructed (Fig. 7d). Overexpression of miR-200a significantly inhibited luciferase activity in Trim2 3′-UTR-transfected striatal cells (Fig. 7e). In contrast, mutation of miR-200a binding sites in 3′UTR region of Trim2 abolished this inhibitory effect (Fig 7e), suggesting that Trim2 is indeed a target of miR-200a. Furthermore, striatal cells transfected with miR-200a exhibited decreased Trim2 protein levels (Suppl Fig. 2), whereas striatal cells transfected with miR-200a antisense displayed increased Trim2 levels (Fig 7f). Our results confirm that Trim2 is a target of miR-200a.
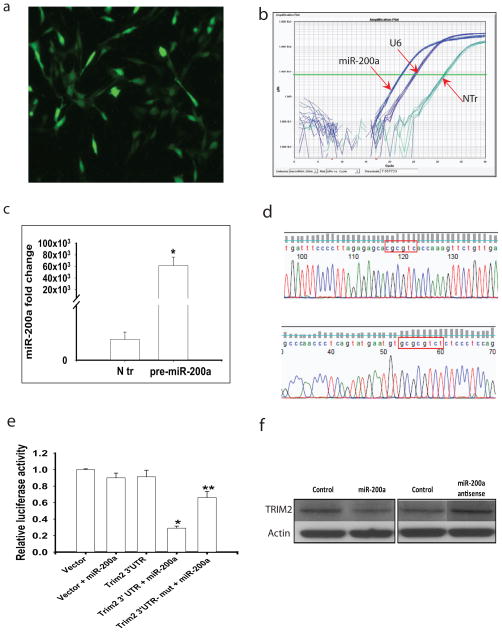
Fig. 7. Trim2 is a target of miR-200a.
(a) Immortalized striatal cells expressing mutant HTT (STHdhQ111/Q111 cells) were transfected with pPRIME-CMV-GFP-FF3-pre miR-200a. Cells with green fluorescence represent transfected cells. (b) STHdhQ111/Q111 cells transfected with pre-miR-200a produce mature miR-200a detected by real-time qPCR. (c) Quantification of miR-200a levels in STHdhQ111/Q111 cells transfected with pre-miR-200a. (d) DNA sequencing confirmed the mutation of two binding sites of mouse Trim 2 full length 3′UTR: from ‘agtgtt’ to ‘CgCgtC’ and from ‘cagtgttt’ to ‘GCgCgtCt’, respectively, by site mutagenesis. (e) Luciferase activity assayed in STHdhQ111/Q111 cells transfected with miR-200a and 3′UTR regions of Trim2 or 3′-UTR with mutation of miR-200a binding sites (Trim2 3′UTR-mut). Mean ± STD, n= 3. *p<0.05 vs Trim2 3′UTR transfected group; **p<0.05 vs Trim2 3′UTR+miR-200a transfected group by ANOVA followed by Scheffe’s post-hoc analysis. (f) Western blots of Trim2 levels in striatal cells transfected with miR-200a or antisense to miR-200a. Cells were collected 24 h after transfection.
Discussion
HD brain cells display profound abnormalities in gene transcription (Hodges et al. 2006). Microarray studies have demonstrated that large numbers of mRNAs are diminished in mouse models of HD (Luthi-Carter et al. 2000, Luthi-Carter et al. 2002), with the most profound changes occurring in the striatum and motor cortex (Hodges et al. 2006). Selective gene dysregulation in HD occurs before overt neurological symptoms (Augood et al. 1997b), strongly suggesting that the gene dysregulation is an important causative factor in HD. Therefore, understanding the cause of the gene dysregulation may accelerate the discovery of promising therapeutic targets for HD (Leone et al. 2008). MiRNAs represent an elegant gene-regulatory mechanism by which mRNAs are repressed or degraded in a sequence-specific manner; miRNAs are essential gene regulators in the central nervous system where they control the development and function of neuronal circuits (Kosik 2006). We have identified two miRNAs, miR-200a and miR-200c, that are significantly upregulated at the presymptomatic stage of the disease process in an HD mouse model. Comparative analysis of networks suggests that these two miRNAs may control genes involved in mutant HTT-induced neuronal dysfunction, including abnormal synaptic transmission and disturbed neurogenesis.
The functions of miR-200a and miR-200c in brain remains largely unknown; there is a report that miR-200a is involved in regulating olfactory neurogenesis (Choi et al. 2008), and recent studies indicated that miR-200a upregulated in a cell model of HD (Sinha et al.) and an HD mouse model (Lee et al.). Interestingly, the predicted target genes of miR-200a and miR-200c that are altered in HD mouse brains can be divided into following categories (Table 1). The first category includes genes involved in synaptic function, including KCNA2 (Dodson et al. 2003, Dodson et al. 2002), KIF3a (Chung et al. 2009), NRXN1(Etherton et al. 2009, Fairless et al. 2008), PTPRD (Pulido et al. 1995a, Pulido et al. 1995b, Wallace et al. 1998), RTN4RL1(Barton et al. 2003, Wang et al. 2002), DNAJC5(Miller et al. 2003, Chamberlain et al. 2001), Lin7b (Butz et al. 1998, Sudo et al. 2006), PPP1R9B (Meng et al. 2009); these molecules play important roles in regulating neurotransmitter release, axonal transport, synaptogenesis, and synaptic plasticity. The study by Hodges et al (Hodges et al. 2006) shows that the downregulation of multiple genes in HD may underlie a defect in the capacity to maintain axonal projections. In agreement with these findings, our analysis revealed that genes controlling axonal guidance are enriched in the list of predicted targets of the miR-200a and miR-200c. The second category includes genes involved in development and neurogenesis, including NFIC (Mason et al. 2009), NGEF ((Rodrigues et al. 2000), PCTK1 (Tang et al. 2006), and PDPK1(Pahnke et al. 2004). The third category includes genes highly expressed in the brain that play important roles in neuronal survival, including ATXN1(Lam et al. 2006, Goold et al. 2007), PRKCB1(Lintas et al. 2009), TRIM2 (Balastik et al. 2008), OLFM1(Nakaya et al. 2008), and ST3GAL5 (Guillerme-Bosselut et al. 2009). It is worth to mention that a couple of significantly downregulated target genes of miR-200a, DRD2 and NEGF, are highly expressed in the most vulnerable brain regions in HD (Table 1 & Suppl Table 1). It has been reported that DRD2 mRNA levels decreased in neurons expressing mutant HTT (Luthi-Carter et al. 2000), and dysregulation of DRD2 is a sensitive measure for HD pathology in mice (Crook & Housman 2012). DRD2 shows high expression levels in medium spiny neurons of the indirect pathway (striatopalladial) of the basal ganglion, which are among the earliest to die in HD. Our data indicated that DRD2 is a target of miR-200 and its expression decreased in the cortex of N171-82Q mice at 12 weeks of age, suggesting that tartgeting miR-200 might provide a protective approach to rescue DRD2 levels and preserve the normal function of the striatopalladial pathway. NGEF is a protein predominantly expressed in brain, with the strongest signal in the caudate nucleus (Rodrigues et al. 2000) where selective neurodegeneration occurs in HD, previous studies also revealed decline of NGEF in striatal neurons expressing HTT 171-82Q and human HD caudate (Runne et al. 2008). Further study of these target genes and its regulation by miRNA may provide new approaches to treat HD.
Previous studies have shown that transcription factor REST regulates miRNAs in HD (Johnson et al. 2008, Packer et al. 2008, Lau & de Strooper). It is noteworthy that levels of a set of miRNAs regulated by REST did not differ in the N171-82Q HD model in the present study (Suppl Fig. 1). The differences between our study and other studies might be due to the disease stages, since we examined the early changes of disease progression, whereas most previous studies used postmortem brain tissues which are at the end stage of disease progression, or at the late stage in HD mouse model (Packer et al. 2008, Johnson et al. 2008). As we found in the current study, miRNAs were altered dynamically during disease progression. Therefore, it is possible that regulation of miRNAs varies at different stages of disease. These differences also suggest considerable heterogeneity among different mouse models as well as between mouse and human, which warrants further detailed study of these possibilities. Nevertheless, we have identify miR-200a, which is consistently altered in other HD models (Lee et al., Sinha et al.), suggesting that it may also play an important role in the pathogenesis of this disease. Moreover, we confirmed that Trim2 is a target of miR-200a, and its levels are decreased in cells expressing mutant HTT in our present study. This is consistent with upregulation of miR-200a in HD models. Trim2 is an E3-ubiquitin ligase and plays a role in regulate neuronal polarization and axon outgrowth (Khazaei et al.), and Trim2 knock-out mice develop intention tremor, spontaneous seizure, and early-onset neurodegeneration (Balastik et al. 2008). Our data provided the first evidence in dysregulation of miR-200a and Trim2 in HD, further investigation of the role of miR-200a and Trim2 in mutant HTT-induced neuronal dysfunction and degeneration may provide insight on developing potential therapeutic targets for HD.
In summary, our findings suggest that dysregulation of miR-200a and miR-200c occurs early in the disease process and may result in reduced production of several different proteins critical for neuronal plasticity and survival in N171-82Q HD model. Further understanding how miRNAs are selectively affected and the possible specific role of these deregulated miRNAs in the pathological progression of HD, may reveal novel aspects of the molecular underpinnings of HD, and may also lead to the development of novel therapeutic approaches for preventing or delaying the neurodegenerative process.
Supplementary Material
Acknowledgments
We thank Dr. Stephen Elledge at Harvard University for providing us pPRIME-CMV-GFP-FF3 vector, and we acknowledge Dr. Pamela Talalay for dedicated editorial assistance. Dr. Marcy Macdonald provided us striatal cells. This work was supported by NINDS (WD), CHDI (W.D.), NIA Intramural Research Program (M.P.M, K.G.B).
Footnotes
All authors have no conflict of interest to declare.
References
- Augood SJ, Faull RL, Emson PC. Dopamine D1 and D2 receptor gene expression in the striatum in Huntington’s disease. Ann Neurol. 1997a;42:215–221. doi: 10.1002/ana.410420213. [DOI] [PubMed] [Google Scholar]
- Augood SJ, Westmore K, Emson PC. Phenotypic characterization of neurotensin messenger RNA-expressing cells in the neuroleptic-treated rat striatum: a detailed cellular co-expression study. Neuroscience. 1997b;76:763–774. doi: 10.1016/s0306-4522(96)00449-6. [DOI] [PubMed] [Google Scholar]
- Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Tommerup N, Kauppinen S. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14:432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balastik M, Ferraguti F, Pires-da Silva A, Lee TH, Alvarez-Bolado G, Lu KP, Gruss P. Deficiency in ubiquitin ligase TRIM2 causes accumulation of neurofilament light chain and neurodegeneration. Proc Natl Acad Sci U S A. 2008;105:12016–12021. doi: 10.1073/pnas.0802261105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton WA, Liu BP, Tzvetkova D, Jeffrey PD, Fournier AE, Sah D, Cate R, Strittmatter SM, Nikolov DB. Structure and axon outgrowth inhibitor binding of the Nogo-66 receptor and related proteins. EMBO J. 2003;22:3291–3302. doi: 10.1093/emboj/cdg325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilen J, Liu N, Bonini NM. A new role for microRNA pathways: modulation of degeneration induced by pathogenic human disease proteins. Cell Cycle. 2006;5:2835–2838. doi: 10.4161/cc.5.24.3579. [DOI] [PubMed] [Google Scholar]
- Butz S, Okamoto M, Sudhof TC. A tripartite protein complex with the potential to couple synaptic vesicle exocytosis to cell adhesion in brain. Cell. 1998;94:773–782. doi: 10.1016/s0092-8674(00)81736-5. [DOI] [PubMed] [Google Scholar]
- Cattaneo E, Zuccato C, Tartari M. Normal huntingtin function: an alternative approach to Huntington’s disease. Nat Rev Neurosci. 2005;6:919–930. doi: 10.1038/nrn1806. [DOI] [PubMed] [Google Scholar]
- Chamberlain LH, Graham ME, Kane S, Jackson JL, Maier VH, Burgoyne RD, Gould GW. The synaptic vesicle protein, cysteine-string protein, is associated with the plasma membrane in 3T3-L1 adipocytes and interacts with syntaxin 4. J Cell Sci. 2001;114:445–455. doi: 10.1242/jcs.114.2.445. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Peng Q, Hou Z, Aggarwal M, Zhang J, Mori S, Ross CA, Duan W. Structural MRI detects progressive regional brain atrophy and neuroprotective effects in N171-82Q Huntington’s disease mouse model. Neuroimage. 2011;56:1027–1034. doi: 10.1016/j.neuroimage.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi PS, Zakhary L, Choi WY, et al. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron. 2008;57:41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CY, Koprich JB, Siddiqi H, Isacson O. Dynamic changes in presynaptic and axonal transport proteins combined with striatal neuroinflammation precede dopaminergic neuronal loss in a rat model of AAV alpha-synucleinopathy. J Neurosci. 2009;29:3365–3373. doi: 10.1523/JNEUROSCI.5427-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R, Barrier M, Ford LP. Role of miRNA and miRNA processing factors in development and disease. Birth Defects Res C Embryo Today. 2006;78:107–117. doi: 10.1002/bdrc.20068. [DOI] [PubMed] [Google Scholar]
- Crook ZR, Housman DE. Dysregulation of dopamine receptor D2 as a sensitive measure for Huntington disease pathology in model mice. Proc Natl Acad Sci U S A. 2012;109:7487–7492. doi: 10.1073/pnas.1204542109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar TL, Davis TH, Nelson PT, Loeb GB, Harfe BD, Ullian E, McManus MT. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc Natl Acad Sci U S A. 2008;105:5614–5619. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JM, Oberg AL, Borralho PM, et al. Evaluation of a new high-dimensional miRNA profiling platform. BMC Med Genomics. 2009;2:57. doi: 10.1186/1755-8794-2-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson PD, Barker MC, Forsythe ID. Two heteromeric Kv1 potassium channels differentially regulate action potential firing. J Neurosci. 2002;22:6953–6961. doi: 10.1523/JNEUROSCI.22-16-06953.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson PD, Billups B, Rusznak Z, Szucs G, Barker MC, Forsythe ID. Presynaptic rat Kv1.2 channels suppress synaptic terminal hyperexcitability following action potential invasion. J Physiol. 2003;550:27–33. doi: 10.1113/jphysiol.2003.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan W, Guo Z, Jiang H, Ware M, Mattson MP. Reversal of behavioral and metabolic abnormalities, and insulin resistance syndrome, by dietary restriction in mice deficient in brain-derived neurotrophic factor. Endocrinology. 2003;144:2446–2453. doi: 10.1210/en.2002-0113. [DOI] [PubMed] [Google Scholar]
- Duan W, Peng Q, Masuda N, et al. Sertraline slows disease progression and increases neurogenesis in N171-82Q mouse model of Huntington’s disease. Neurobiol Dis. 2008;30:312–322. doi: 10.1016/j.nbd.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etherton MR, Blaiss CA, Powell CM, Sudhof TC. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc Natl Acad Sci U S A. 2009;106:17998–18003. doi: 10.1073/pnas.0910297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairless R, Masius H, Rohlmann A, Heupel K, Ahmad M, Reissner C, Dresbach T, Missler M. Polarized targeting of neurexins to synapses is regulated by their C-terminal sequences. J Neurosci. 2008;28:12969–12981. doi: 10.1523/JNEUROSCI.5294-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante RJ, Gutekunst CA, Persichetti F, McNeil SM, Kowall NW, Gusella JF, MacDonald ME, Beal MF, Hersch SM. Heterogeneous topographic and cellular distribution of huntingtin expression in the normal human neostriatum. J Neurosci. 1997;17:3052–3063. doi: 10.1523/JNEUROSCI.17-09-03052.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco FR, Chen Q, Lamoreaux WJ, et al. Cellular localization of huntingtin in striatal and cortical neurons in rats: lack of correlation with neuronal vulnerability in Huntington’s disease. J Neurosci. 1999;19:1189–1202. doi: 10.1523/JNEUROSCI.19-04-01189.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goold R, Hubank M, Hunt A, Holton J, Menon RP, Revesz T, Pandolfo M, Matilla-Duenas A. Down-regulation of the dopamine receptor D2 in mice lacking ataxin 1. Hum Mol Genet. 2007;16:2122–2134. doi: 10.1093/hmg/ddm162. [DOI] [PubMed] [Google Scholar]
- Gray SG. Targeting Huntington’s disease through histone deacetylases. Clin Epigenetics. 2011;2:257–277. doi: 10.1007/s13148-011-0025-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillerme-Bosselut F, Forestier L, Jayat-Vignoles C, et al. Glycosylation-related gene expression profiling in the brain and spleen of scrapie-affected mouse. Glycobiology. 2009;19:879–889. doi: 10.1093/glycob/cwp062. [DOI] [PubMed] [Google Scholar]
- Hodges A, Strand AD, Aragaki AK, et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum Mol Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- Johnson R, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ. A microRNA-based gene dysregulation pathway in Huntington’s disease. Neurobiol Dis. 2008;29:438–445. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Khazaei MR, Bunk EC, Hillje AL, Jahn HM, Riegler EM, Knoblich JA, Young P, Schwamborn JC. The E3-ubiquitin ligase TRIM2 regulates neuronal polarization. J Neurochem. doi: 10.1111/j.1471-4159.2010.06971.x. [DOI] [PubMed] [Google Scholar]
- Khazaei MR, Bunk EC, Hillje AL, Jahn HM, Riegler EM, Knoblich JA, Young P, Schwamborn JC. The E3-ubiquitin ligase TRIM2 regulates neuronal polarization. J Neurochem. 117:29–37. doi: 10.1111/j.1471-4159.2010.06971.x. [DOI] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lam YC, Bowman AB, Jafar-Nejad P, et al. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127:1335–1347. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- Lau P, de Strooper B. Dysregulated microRNAs in neurodegenerative disorders. Semin Cell Dev Biol. 21:768–773. doi: 10.1016/j.semcdb.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Lee ST, Chu K, Im WS, et al. Altered microRNA regulation in Huntington’s disease models. Exp Neurol. 227:172–179. doi: 10.1016/j.expneurol.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Leone S, Mutti C, Kazantsev A, Sturlese M, Moro S, Cattaneo E, Rigamonti D, Contini A. SAR and QSAR study on 2-aminothiazole derivatives, modulators of transcriptional repression in Huntington’s disease. Bioorg Med Chem. 2008;16:5695–5703. doi: 10.1016/j.bmc.2008.03.067. [DOI] [PubMed] [Google Scholar]
- Lintas C, Sacco R, Garbett K, et al. Involvement of the PRKCB1 gene in autistic disorder: significant genetic association and reduced neocortical gene expression. Mol Psychiatry. 2009;14:705–718. doi: 10.1038/mp.2008.21. [DOI] [PubMed] [Google Scholar]
- Luthi-Carter R, Strand A, Peters NL, et al. Decreased expression of striatal signaling genes in a mouse model of Huntington’s disease. Hum Mol Genet. 2000;9:1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- Luthi-Carter R, Strand AD, Hanson SA, et al. Polyglutamine and transcription: gene expression changes shared by DRPLA and Huntington’s disease mouse models reveal context-independent effects. Hum Mol Genet. 2002;11:1927–1937. doi: 10.1093/hmg/11.17.1927. [DOI] [PubMed] [Google Scholar]
- Mason S, Piper M, Gronostajski RM, Richards LJ. Nuclear factor one transcription factors in CNS development. Mol Neurobiol. 2009;39:10–23. doi: 10.1007/s12035-008-8048-6. [DOI] [PubMed] [Google Scholar]
- Meng X, Kanwar N, Du Q, Goping IS, Bleackley RC, Wilkins JA. PPP1R9B (Neurabin 2): involvement and dynamics in the NK immunological synapse. Eur J Immunol. 2009;39:552–560. doi: 10.1002/eji.200838474. [DOI] [PubMed] [Google Scholar]
- Miller LC, Swayne LA, Chen L, Feng ZP, Wacker JL, Muchowski PJ, Zamponi GW, Braun JE. Cysteine string protein (CSP) inhibition of N-type calcium channels is blocked by mutant huntingtin. J Biol Chem. 2003;278:53072–53081. doi: 10.1074/jbc.M306230200. [DOI] [PubMed] [Google Scholar]
- Nakaya N, Lee HS, Takada Y, Tzchori I, Tomarev SI. Zebrafish olfactomedin 1 regulates retinal axon elongation in vivo and is a modulator of Wnt signaling pathway. J Neurosci. 2008;28:7900–7910. doi: 10.1523/JNEUROSCI.0617-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahnke J, Mix E, Knoblich R, et al. Overexpression of glial cell line-derived neurotrophic factor induces genes regulating migration and differentiation of neuronal progenitor cells. Exp Cell Res. 2004;297:484–494. doi: 10.1016/j.yexcr.2004.03.037. [DOI] [PubMed] [Google Scholar]
- Pulido R, Krueger NX, Serra-Pages C, Saito H, Streuli M. Molecular characterization of the human transmembrane protein-tyrosine phosphatase delta. Evidence for tissue-specific expression of alternative human transmembrane protein-tyrosine phosphatase delta isoforms. J Biol Chem. 1995a;270:6722–6728. doi: 10.1074/jbc.270.12.6722. [DOI] [PubMed] [Google Scholar]
- Pulido R, Serra-Pages C, Tang M, Streuli M. The LAR/PTP delta/PTP sigma subfamily of transmembrane protein-tyrosine-phosphatases: multiple human LAR, PTP delta, and PTP sigma isoforms are expressed in a tissue-specific manner and associate with the LAR-interacting protein LIP.1. Proc Natl Acad Sci U S A. 1995b;92:11686–11690. doi: 10.1073/pnas.92.25.11686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues NR, Theodosiou AM, Nesbit MA, Campbell L, Tandle AT, Saranath D, Davies KE. Characterization of Ngef, a novel member of the Dbl family of genes expressed predominantly in the caudate nucleus. Genomics. 2000;65:53–61. doi: 10.1006/geno.2000.6138. [DOI] [PubMed] [Google Scholar]
- Runne H, Regulier E, Kuhn A, et al. Dysregulation of gene expression in primary neuron models of Huntington’s disease shows that polyglutamine-related effects on the striatal transcriptome may not be dependent on brain circuitry. J Neurosci. 2008;28:9723–9731. doi: 10.1523/JNEUROSCI.3044-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha M, Ghose J, Das E, Bhattarcharyya NP. Altered microRNAs in STHdh(Q111)/Hdh(Q111) cells: miR-146a targets TBP. Biochem Biophys Res Commun. 396:742–747. doi: 10.1016/j.bbrc.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Sudo K, Ito H, Iwamoto I, Morishita R, Asano T, Nagata K. Identification of a cell polarity-related protein, Lin-7B, as a binding partner for a Rho effector, Rhotekin, and their possible interaction in neurons. Neurosci Res. 2006;56:347–355. doi: 10.1016/j.neures.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Tang X, Guilherme A, Chakladar A, Powelka AM, Konda S, Virbasius JV, Nicoloro SM, Straubhaar J, Czech MP. An RNA interference-based screen identifies MAP4K4/NIK as a negative regulator of PPARgamma, adipogenesis, and insulin-responsive hexose transport. Proc Natl Acad Sci U S A. 2006;103:2087–2092. doi: 10.1073/pnas.0507660103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F, Cattaneo E, MacDonald ME. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet. 2000;9:2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- Trivedi S, Ramakrishna G. miRNA and neurons. Int J Neurosci. 2009;119:1995–2016. doi: 10.1080/00207450903139788. [DOI] [PubMed] [Google Scholar]
- Trottier Y, Lutz Y, Stevanin G, et al. Polyglutamine expansion as a pathological epitope in Huntington’s disease and four dominant cerebellar ataxias. Nature. 1995;378:403–406. doi: 10.1038/378403a0. [DOI] [PubMed] [Google Scholar]
- Wallace MJ, Fladd C, Batt J, Rotin D. The second catalytic domain of protein tyrosine phosphatase delta (PTP delta) binds to and inhibits the first catalytic domain of PTP sigma. Mol Cell Biol. 1998;18:2608–2616. doi: 10.1128/mcb.18.5.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Koprivica V, Kim JA, Sivasankaran R, Guo Y, Neve RL, He Z. Oligodendrocyte-myelin glycoprotein is a Nogo receptor ligand that inhibits neurite outgrowth. Nature. 2002;417:941–944. doi: 10.1038/nature00867. [DOI] [PubMed] [Google Scholar]
- Yu ZX, Li SH, Evans J, Pillarisetti A, Li H, Li XJ. Mutant huntingtin causes context-dependent neurodegeneration in mice with Huntington’s disease. J Neurosci. 2003;23:2193–2202. doi: 10.1523/JNEUROSCI.23-06-02193.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.