SUMMARY
Myosin VI is the only known reverse-direction myosin motor. It has an unprecedented means of amplifying movements within the motor involving rearrangements of the converter subdomain at the C-terminus of the motor and an unusual lever arm projecting from the converter. While the average step size of a myosin VI dimer is 30–36nm, the step size is highly variable, presenting a challenge to the lever arm mechanism by which all myosins are thought to move. Herein we present new structures of myosin VI that reveal regions of compliance that allow an uncoupling of the lead head when movement is modeled on actin. The location of the compliance restricts the possible actin binding sites and predicts the observed stepping behavior. The model reveals that myosin VI, unlike plus-end directed myosins, does not use a pure lever arm mechanism, but instead steps with a mechanism analogous to the kinesin neck-linker uncoupling model.
INTRODUCTION
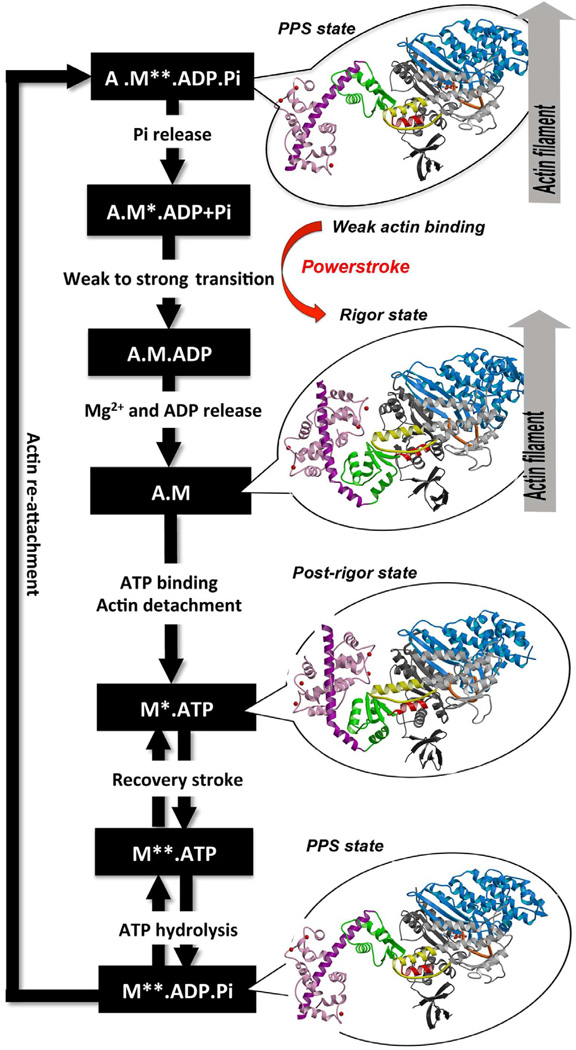
Myosin VI is unique in the superfamily of myosin motors in its ability to perform processive steps that are large on average, but highly variable in size, and in the reverse direction as compared to all other characterized myosin motors on actin (Rock et al., 2001; Nishikawa et al., 2002). Even though its directionality has been reversed, myosin VI undergoes the same kinetic cycle (Figure 1) for chemo-mechanical transduction as for all other characterized myosins (Sweeney and Houdusse, 2010). The cycle begins when myosin binds to actin in a state known as the pre-powerstroke state that has hydrolyzed ATP, but has trapped the hydrolysis products, MgADP and Pi. The interaction with actin causes a conformational change in the myosin that releases Pi and allows a strong binding interface to be created. This induces further conformational changes that increase the binding affinity for actin and trigger the release of MgADP to form a rigor conformation on actin. Concomitant with the changes in binding affinity and product release, these conformational changes cause a swing of the C-terminal subdomain of the motor, which is known as the converter. This drives the movement of an extended alpha-helix that binds multiple calmodulin (CaM) or calmodulin-like light chains. Both the converter and the CaM-/light chain-binding region form the myosin lever arm. The lever arm amplifies the conformational changes in the motor into a movement on the order of nanometers known as the powerstroke. The powerstroke ends with the formation of the rigor state in which myosin is strongly bound to actin and has released MgADP. ATP binding to the rigor state on actin induces conformational changes in the myosin motor that causes a large cleft at the actin interface to open, destroying high affinity actin binding and creating the post-rigor state, which rapidly dissociates from actin. Once the myosin has dissociated, there is a rapid and reversible isomerization between the post-rigor state, which cannot hydrolyze ATP, and the pre-powerstroke state that can rapidly hydrolyze ATP due to repositioning of a nucleotide-binding element known as switch II (Figure 1 or 2A). This movement of switch II is part of a conformational change in the motor that repositions the myosin lever arm, repriming the lever arm for movement on actin.
Figure 1. ATPase cycle of the myosin VI motor.
The different structural states that the myosin VI motor adopts along the ATP hydrolysis cycle are indicated with detached states on the left (Post-rigor, 2VAS and Pre-powerstroke (PPS), 4ANJ) and actin-binding states on the right (PPS-4ANJ and Rigor, 2BKH). The motor is composed of important connectors (SwII, orange – relay, yellow – SH1 helix, red) and four subdomains (Nter, grey – U50, blue – L50, white – converter, green). Insert-2 and its bound Ca2+-calmodulin are in purple and pink.
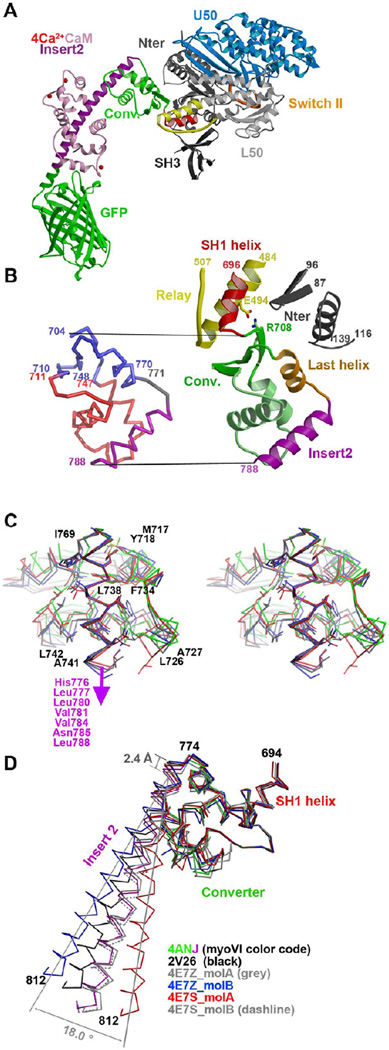
Figure 2. Variability in the converter of myosin VI in different pre-powerstroke state structures.
(A) The structure of the myosin VI MDins2-GFP prior to force generation (pre-powerstroke state) is shown with the motor sub-domains and important structural motifs indicated in colors (Switch II in orange, relay in yellow and SH1 helix in red). Insert-2 and calmodulin are shown in purple and pink, while the GFP fusion is in green. (B) (Left) The converter and its contacts with the Relay and the N-terminus subdomain are shown with the same color code as in A (except for the last helix shown in orange). (Right) A ribbon structure of the converter is displayed with the conserved regions indicated in blue and the variable regions in red. The hinge and the insert-2 are indicated in grey and purple, respectively. (C) Cross-eyed stereo view of the variable interactions between insert-2 and the rest of the converter in different PPS structures superimposed on insert-2 (4ANJ in green (converter) and purple (insert-2), 2V26 in black, 4E7Z_molB in blue and 4E7Z_molA in red). (D) The converter in the pre-powerstroke state of myosin VI. A superposition on the converter of the six different PPS structures is shown with the same color code as in C. Note that overall the variation in the P-fold conformation of the converters result in different orientations of the insert-2 proximal helix. Modeling of the full insert-2 helix is shown to highlight the degree of variability in the insert-2 orientation.
Although the motor domain of myosin VI undergoes similar rearrangements in its ATPase cycle as for plus-end motors, the movement of its lever arm is in the opposite direction. This is accomplished by a repositioning of the CaM-binding region on the converter. In order to reposition the myosin VI lever arm, myosin VI contains an insert (insert-2) found only in Class VI and XXX myosins (Odronitz and Kollmar, 2007) at the junction between the converter and the CaM-binding region. The first part of insert-2 interacts with the converter while the distal part of the insert-2, and the IQ motif that follows, each bind a calmodulin to create an unusual and repositioned CaM-binding region (Menetrey et al., 2005). The lever arm in turn is followed by a proximal tail region (PTD), which folds as a triple helix bundle in the monomeric head but opens in the dimeric molecule as an elongated lever arm extension (Mukherjea et al., 2009).
A further adaptation of myosin VI to enable a larger powerstroke to occur in the reverse direction is that during the transition from pre-powerstroke the converter subdomain undergoes rearrangement creating two distinct conformations of the converter (Menetrey et al., 2007), which we refer to herein as the R-fold and P-fold. The R-fold is found in the rigor state (R) at the end of the powerstroke, and is similar to the conformation that has been seen in plus-end-directed myosins. The P-fold found in the pre-powerstroke state (PPS) differs significantly. In the rigor state, the P-fold cannot be explored due to steric hindrance, but in the PPS state, modeling of a R-fold of the converter is possible without creating steric collision.
Myosin VI can dimerize upon interaction with its cargo proteins creating a dimer that moves processively along an actin filament with steps that are highly variable in length (Phichith et al., 2009). The lever arm extension appears to be less rigid than that of well characterized myosins such as those of class II and V, which initially was proposed to be the basis for variability of the myosin VI step size (Rock et al., 2005). However later work suggested that the variability of the step sizes appears to be derived from properties of the myosin converter itself, or to the nature of the converter interactions with the motor domain and/or the rest of the lever arm, since chimeric molecules with rigid lever arms after insert-2 also display the same degree of step size variability (Park et al., 2007; Bryant et al., 2007). We report herein structural investigations that provide insights into how myosin VI moves on actin. They demonstrate the location of compliance within the lever arm and show that it differs in different states of the motor. These structures explain the observed processive, but highly variable, movement of myosin VI in the reverse direction on actin.
RESULTS AND DISCUSSION
To attempt to explain the variability of the myosin VI step size on actin, we examined the possibility that there are multiple possible structures of the pre-powerstroke (PPS) state, the structural state that the lead head of a processive dimer is in when it rebinds to actin (Figure 1). We did this by solving six structures of the pre-powerstroke state of myosin VI in different crystal packing environments. Based on these structures, we modeled how variability in the converter conformation and/or in the lever arm position in the PPS state might influence where a lead head can dock on actin as well as the subsequent power stroke, thus contributing to the variability of the myosin VI dimer’s step size.
The myosin VI Pre-powerstroke state – Overall structure
As discussed above and depicted in Figure 1, the so-called pre-powerstroke (PPS) state of myosin is the state that re-attaches to actin with MgADP and Pi trapped in the active site, initiating the powerstroke and force generation. The first structure of the pre-powerstroke state of myosin VI was derived from a construct truncated prior to the CaM-binding site of insert-2 and thus was a motor domain (MD) construct without the CaM-binding region (Menetrey et al., 2007). Interestingly the converter, the C-terminal subdomain of the MD that controls the lever arm orientation, had adopted a new conformation (P-fold) compared to that previously described for the nucleotide-free state (R-fold). The R-fold conformation of the myosin VI converter is the same as seen for all other myosin structures to date. As described in the Supplemental Information, the transition between these two configurations of the converter is made possible due to conformational changes of loops, called hereafter hinge regions that connect together the converter secondary elements: the β-sheet to the helices and the helices to each other (Figure S1).
The six structures of the PPS state allowed identification of compliant elements in the motor in this state. In particular, the influence of the lever arm on the converter conformation was visualized at 2.6 Å resolution in a PPS crystal structure of a construct encompassing the first part of the lever arm (insert-2 bound to its CaM with four bound Ca2+) fused to a Green Fluorescent Protein (GFP) molecule (MDins2-GFP construct ; Figure 2A).
In order to generate new crystal packing forms of the myosin VI PPS, we created a new construct (MDPM) with modified surface residues (D23R, I24R and R569E), which would be predicted to prevent the packing seen in earlier MD crystals. We solved ~2.3 Å resolution structures of both the MD and MDPM constructs in two new crystal forms (see Table 1 and Methods) with two independent molecules in each asymmetric unit. In total, this allowed structural comparisons of six structures in the PPS state (PDB code 2V26 (Menetrey et al., 2007); and five new structures) crystallized in different packing environments. In all these structures, there are no significant differences to report for the regions of the motor domain prior to the converter (residues 5–705) with an average root mean square deviation (rmsd) lower than 0.68 Å on 662 Cα atoms. The MD structure is very similar to that found for plus-end motors in this state (Menetrey et al., 2007), except for the conformation of the converter. The motor is in a conformation that positions the converter appropriately for a “primed” lever arm position, characteristic of the PPS.
Table 1.
Data collection and refinement statistics
| MDins2-GFP | MDPM | MDcrystal form 2 | |
|---|---|---|---|
| Data Collection | |||
| Space group | C2 | P212121 | P21 |
| Cell dimensions | |||
| a, | 193.09, | 81.95 | 98.15 |
| b, | 62.66, | 135.14 | 93.32 |
| c (Å) | 156.04 | 196.54 | 101.87 |
| β (°) | 117.96 | 90.00 | 90.60 |
| Molecules per asym. unit | 1 | 2 | 2 |
| Resolution (Å) | 48.8-2.6 | 30-2.25 | 30-2.3 |
| (High. Res. Shell) | (2.74-2.60) | (2.30-2.25) | (2.35-2.3) |
| Rmeas | 11.4 (42.8) | 11.1 (46.2) | 7.1 (47.9) |
| I/σI | 7.2 (2.0) | 12.86 (3.44) | 13.61 (3.25) |
| Completeness (%) | 100 (100) | 98.8 (98.0) | 98.3 (98.3) |
| Redundancy | 4.8 (4.8) | 4.8 (3.68) | 4.0 (3.81) |
| Refinement | |||
| Resolution (Å) | 2.60 | 2.25 | 2.30 |
| No. reflections | 51,178 | 102,983 | 80,429 |
| Rwork/ Rfree | 23.8/28.7 | 17.9/21.6 | 17.6/23.0 |
| No. atoms | |||
| Protein | 9062 | 11964 | 11881 |
| Heterogen | 36 | 107 | 102 |
| Water | 91 | 469 | 456 |
| B-wilson (Å2) | 41.8 | 47.21 | 40.0 |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.007 | 0.010 | 0.009 |
| Bond angles (°) | 1.038 | 1.215 | 1.191 |
Data were collected from a single crystal. Values in parentheses are for the highest resolution shell.
The converter always adopts a P-fold conformation in the pre-powerstroke state
In all five of the new PPS structures of myosin VI, the converter adopts the P-fold conformation, as determined previously (Menetrey et al. 2007) (rmsd < 0.85 Å on 68 Cα atoms; residues 706–773) (Figure 2D). This conformation is markedly different from the one observed in the rigor state structure, which we refer to as the R-fold (rmsd > 3.5 Å on 68 Cα atoms; residues 706–773) (PDB code 2BKI, (Menetrey et al., 2005)) (Figure S1). It is important to note that one of the molecules of the MDPM structure adopts a well-defined P-fold even though the R-fold would be compatible with the packing environment of the crystal and there is sufficient space to allow the transition in the crystal (Figure S2A).
To remove any possible crystal packing constraints, we investigated the conformation of the MDins2-GFP construct in solution using Small-angle X-ray Scattering (SAXS). The molecule was trapped in the PPS state using MgADP.VO4. The SAXS studies clearly showed that, in solution, the converter adopts a P-fold in the PPS state and does not populate the R-fold in the PPS state (Figure S2).
While the position of the motor domain structural elements that control the overall position of the converter (known as the relay and the SH1 helix; shown in Figure 2A) and the beta sheet of the converter have interactions and orientations that are conserved in all of the PPS structures, variability in the relative position of the converter helices demonstrates that the P-fold conformation of the converter is not rigid and is in fact a compliant structure (Figure 2C). The second helix which is parallel to the surface of the beta sheet is found translated by more than 3.2 Å in these different PPS structures. Large variation also occurs for the loop that precedes and the proximal part of insert-2 which interacts with it. In contrast, the last helix of the converter adopts a conserved position in these structures allowing conserved interactions with the N-terminal subdomain of the motor to be maintained (Figure 2B and Supplemental Table 1). The variability in the P-fold of the converter indicates that the insert-2 orientation that defines the orientation of the lever arm of myosin VI differs by at least 18° in these structures (Figure 2D). This is likely an under estimate of the possible variability in lever arm orientation in a functioning myosin VI molecule, since strain would further perturb the lever arm position given the compliance of this region.
In contrast, comparison of the rigor and post-rigor state structures, which both have converters in the R-fold conformation (Menetrey et al., 2005, Menetrey et al., 2008), demonstrates that the proximal part of the insert-2 helix (residues 774–787) interacts with both the converter in this conformation as well as with the insert-2/CaM module. These multiple interactions should make the converter/insert2/CaM module much stiffer when the converter adopts the R-fold compared to when it adopts a P-fold, and thus greatly constrain the lever arm position (Figure S3). These structures further reveal that upon converter rearrangement from P-fold to R-fold, the proximal part of insert-2 undergoes a 90° rotation, but maintains interactions with the converter through hydrophobic interactions (Menetrey et al., 2007). However, these interactions do not restrict the insert 2 proximal helix to a precise position in the P-fold conformation (Figure 2D).
While steric hindrance with the N-terminal subdomain prevents the converter from adopting the P-fold when the motor domain is found in the rigor or in the post-rigor state (Figure S4D), the two converter conformations are theoretically compatible with the motor domain when it adopts the pre-powerstroke state (Menetrey et al., 2007). However, only the P-fold is populated in solution for myosin VI in the PPS state and all of the six PPS crystal structures we obtained adopt a P-fold. There are important interactions between the N-terminal subdomain of the motor, the end of the SH1 helix and the last helix of the converter that are conserved in all these PPS structures. These conserved interactions stabilize the P-fold of the converter (Figure S4 + Supplemental Table 1). A detailed description of these interactions is presented in the Supplemental Information.
Altogether, these observations reveal that the atypical conformation of the converter first described in the MD structure (Menetrey et al., 2007) is the sole conformation adopted in the PPS state and was not simply trapped in the original structure by the crystal packing or the lack of the myosin VI lever arm. Furthermore, these structures highlight that there is considerable compliance within the PPS conformation of the converter that will allow variability in lever arm positioning. Directed stress that would be generated between the distal part of the lever arm and the motor domain when an unbound lead head of a dimer is docking on actin during processive stepping can thus be partially accommodated by compliance associated with changes in the position of the converter helices.
Lever arm orientation and stroke size
The MDins2-GFP structure reveals that the lever arm orientation in the pre-powerstroke state is directed toward the barbed-end (plus-end) of the actin filament as previously modeled (Menetrey et al., 2007) (Figure 3A). The axis defined by the two positions observed for the pre- and post-powerstroke states of myosin VI is very similar to that of the actin filament axis. This PPS structure and the nucleotide-free (rigor like) structure of myosin VI (Menetrey et al., 2005) thus could allow a 135° swing of the lever arm, parallel to the actin filament, during the powerstroke in the absence of strain. As indicated in Figure 3B, this degree of lever arm rotation predicts the measured stroke sizes seen with different myosin VI length lever arms (Bryant et al., 2007; Rock et al., 2005; Sivaramakrishnan et al., 2009).
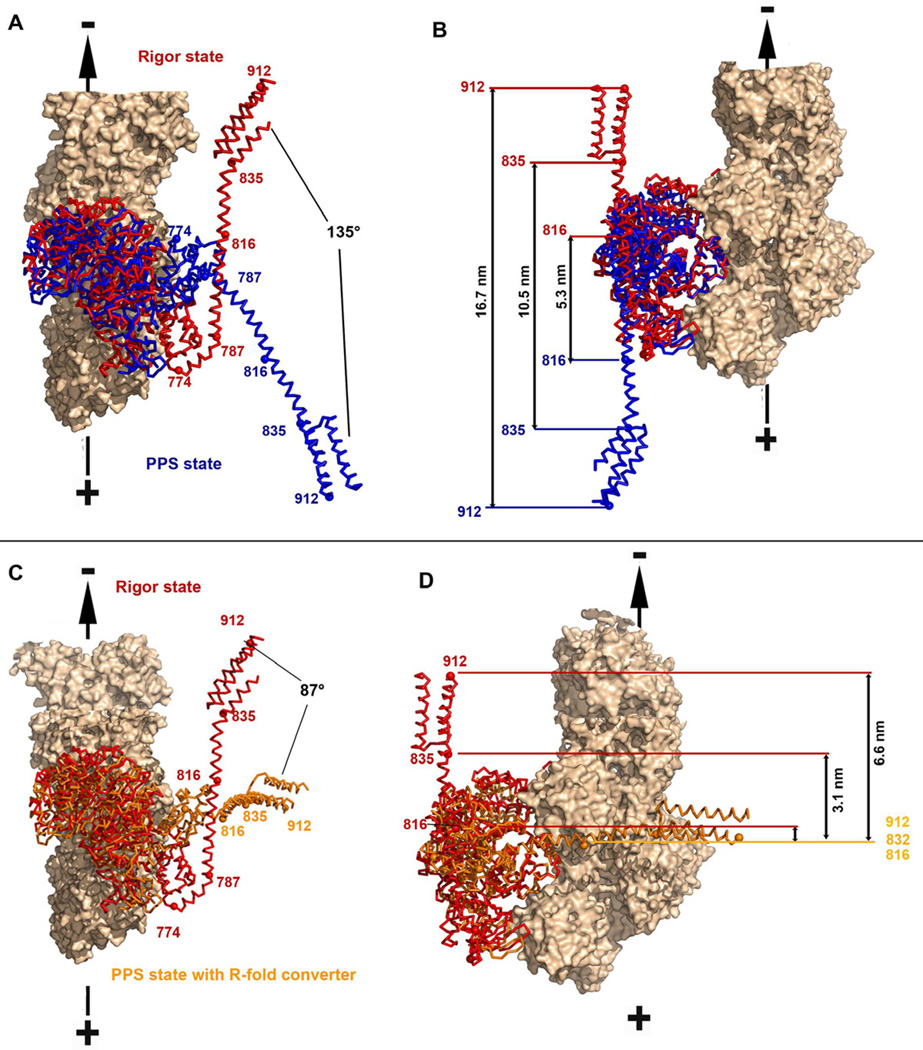
Figure 3. The lever arm orientation in the pre-powerstroke state.
A, B– Superposition on the lower 50kDa sub-domain of the MDins2-GFP pre-powerstroke (this study; blue) and the MDins2 rigor-like (2BKH, (Menetrey et al, 2005) ; red) structures is shown with two different orientations (A and B). The calmodulin molecules were omitted for clarity while the IQ and PTD region were modeled based on the structure of the lever arm (3GN4). (A) Relative rotations of the lever arms are indicated. (B) Distances at the extremity of the lever arm prior and after the powerstroke (measured at the end of insert-2 (816), at the end of the IQ motif (835) and at the end of the folded triple helix bundle (912) are indicated. (C, D) In the same view as in A and B, is a model of the pre-powerstroke state if the converter would adopt the rigor fold in its primed position (orange), as compared to the rigor state of myosin VI [2BKH, (Menetrey et al, 2005); red] to evaluate the stroke size that would be produced if there was no rearrangement of the converter.
A model of the pre-powerstroke state in which the converter would adopt a R-fold can easily be constructed by super-imposition of the beta sheet of the converter. In this case, the lever arm would be directed perpendicularly compared to the actin filament axis (Figures 3C, 3D). The predicted stroke would then be of ~3.1 nm for a lever arm consisting of both the insert-2 CaM and IQ CaM, and ~6.6 nm with the addition of the folded 3-helix bundle. No movement would be predicted for a lever arm of only the insert-2 CaM. The stroke size measured for these various constructs are much larger than these predictions, but similar to the predictions based on a P-fold, consistent with the converter being solely in a P-fold conformation in the PPS at the beginning of the powerstroke. It is unlikely that any isomerization of the converter conformation occurs in the PPS state since this would manifest as a much wider distribution and a smaller average of stroke sizes. This is an important insight, because it implies that variability in the myosin VI step size is not due to an isomerization of the converter conformation occurring in the PPS state prior to the lead head of a dimer binding to the actin filament.
A pliant region within insert-2 further increases the compliance of the myosin VI lever arm in the PPS state
The proximal part of the myosin VI lever arm, consisting of the converter and the beginning of the insert-2 helix, is followed by a part of insert-2 to which a Ca2+-bound calmodulin is bound. In the PPS conformation, the MDins2-GFP structure reveals that the converter makes no interactions with the calmodulin bound to the distal part of insert-2 (Figure 4A). This is in contrast to what has been described for the rigor-like and post-rigor states, in which strong interactions with the R-fold converter maintain a specific position for the insert-2/CaM (Menetrey et al., 2005) ; Figure 4B and S3). For these interactions to occur, insert-2 must adopt a kink at position Trp787, at the junction between the proximal helix that interacts with the converter and the distal helix that binds calmodulin. The converter and the insert-2/CaM form a rigid module in the rigor state that would minimize the compliance of this proximal region of the lever arm in the rigor state, which may be critical for stepping while bearing load.
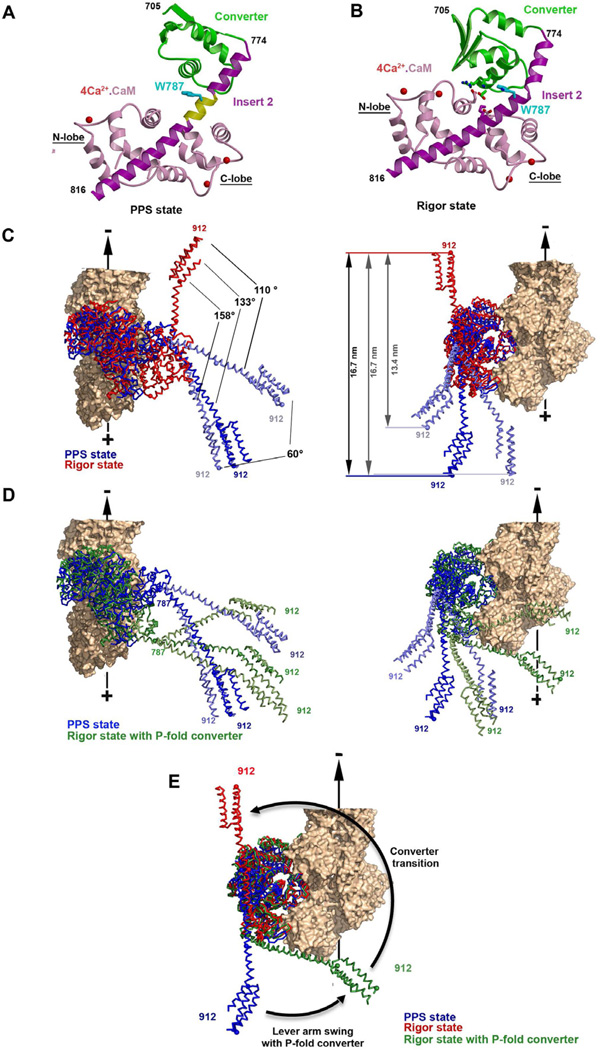
Figure 4. The converter-insert2-CaM structural module.
Structures of the converter-insert-2-CaM module for the pre-powerstroke (A) and the rigor-like (B) states are indicated with the same color code as in Figure 1. These show the different conformations of the converter (green), the different bends of the insert-2 helix and the different position of the N-lobe of the CaM relative to the converter. Note that the insert-2 helix is straight in the pre-powerstroke state (A) and no interactions between the converter and the N-terminal CaM takes place. Eight residues (N785 to R792, yellow) of the insert-2 helix are not involved in any interactions. (B) In contrast, the helix is kinked at residue Trp787 in the rigor state, allowing interactions between the converter and the N-lobe of CaM to take place. Some of the salt bridges between the converter and the calmodulin are also shown – see also Supplemental Figure 3 for comparison with the post-rigor state. (C) Incorporating the observed bending in this pliant region allows modeling of lever arm positions (two extremes in light blue) that differ by as much as 60° without creating any steric hindrance. The resulting variability in the stroke size is indicated for a MD-Ins2-IQ-PTD fragment. (D) Using the pre-powerstroke (blue) positions of the lever from C, the post-stroke state (green) is modeled using a converter that has rotated to adopt a position close to that found in MDins2 rigor-like (2BKH, Menetrey et al, 2005), but in the PPS fold was maintained. The results of bending of the pliant region are modeled in light green. Note that the converter rotation without a converter rearrangement allows the lever arm to stay close to its position in the prepowerstroke state (very small stroke size), while a larger azimuthal component would occur for the stroke. However this azimuthal component could be greatly diminished by changes in the pliant region since a ~60° amplitude of movement (light green) is possible. This model shows that the lead head can undergo this converter rotation without much change in the lever arm orientation and position when the rear head impose strain on the lead head.
In contrast, the absence of interactions between the converter and the insert-2 bound calmodulin in the pre-powerstroke state (Figure 4A) creates flexibility in the positioning of the lever arm by freeing a pliant region in insert-2. No interactions stabilize the insert-2 helix residues N785 to R792, allowing highly variable bending of the helix. This in turn allows multiple positions of the myosin VI lever arm (CaM-Ins2/CaM + IQ/CaM + 3-helix bundle) for a particular MD/converter pre-powerstroke state (Figure 4C). We also performed experiments to monitor the exchange of the calmodulin bound to insert-2, which would be greatly inhibited if interaction occur between this CaM and the converter. These experiments confirm that almost no exchange occurs in the rigor-like state of the motor while CaM exchange does occur in the pre-powerstroke state (see Supplemental Table 2). This is consistent with the absence of interactions involving this calmodulin and the P-fold of the converter and thus the presence of a pliant region within insert-2 in the PPS state. Thus the exact orientation of the lever arm of the head that rebinds to actin can vary greatly compared to the rest of the motor domain due to large variations in this pliant region. As modeled below, this contributes even more variability to the myosin VI step size than does the variable positioning of the converter helices orientations that gives rise to the compliance within the P-fold of the converter described above.
Compliance in the lever arm greatly differs in the lead head and the rear head
For a monomeric motor, the strokes measured are consistent with a large swing of the lever arm direction derived from a combination of a converter swing coupled to a change in the converter conformation (P-fold to R-fold). The converter swing itself is similar to that of plus-end motors, but insert-2 re-directs the lever arm so that the rotation axis of the lever arm differs from that of plus-end motors and results in a large azimuthal component for the lever arm swing. The lever arm would rotate by ~70° due solely to a converter swing in the P-fold conformation, to adopt an orientation that would be perpendicular from the actin axis (Figure 4E). A subsequent isomerization of the converter towards the R-fold would further rotate the lever arm by ~90° in the same plane. Thus a large azimuthal torque develops during the stroke of myosin VI.
For a dimeric molecule, compliance in the myosin VI converter and at the junction with the lever arm could allow the lead head to attach strongly to actin without developing strain on the lever arm. The new structures presented herein highlight that dynamics in the converter as well as at the pliant region within insert-2 controls the relative position of the converter and the rest of the lever arm. Lever arm compliance is much greater in the pre-powerstroke state (P-fold) than in the rigor or post-rigor states, since these adopt a R-fold converter, which has interactions that markedly constrain the lever arm position. The dynamics in the myosin VI lever arm are thus quite different in the lead head of a stepping dimer (that rebinds to actin with a converter in the P-fold), as compared to that of the rear head (that is strongly bound to actin in a rigor state). In the rear head, the CaM/converter interactions limit the compliance in the lever arm and would enable it to bear load as well as to efficiently direct the lever arm (and thus the other head) towards the minus end of the actin filament.
Modeling step sizes and converter conformations
To ascertain to what extent the compliance due to the pliant region of insert-2 and the variable positioning of the converter helices can explain the observed stepping behavior of a myosin VI dimer, we first modeled a myosin VI lead head docked on actin with the range of lever arm positions that would be specified by the variable lever arm positions found in our six PPS structures (Figure 5). For modeling of dimers with both heads docked on actin, we used the general model of a myosin VI dimer proposed by Mukherjea et al., (2009), which has a lever arm that is extended by an unfolded three-helix bundle and dimerizes using the sequence immediately following this region. (See Supplemental Data for details).
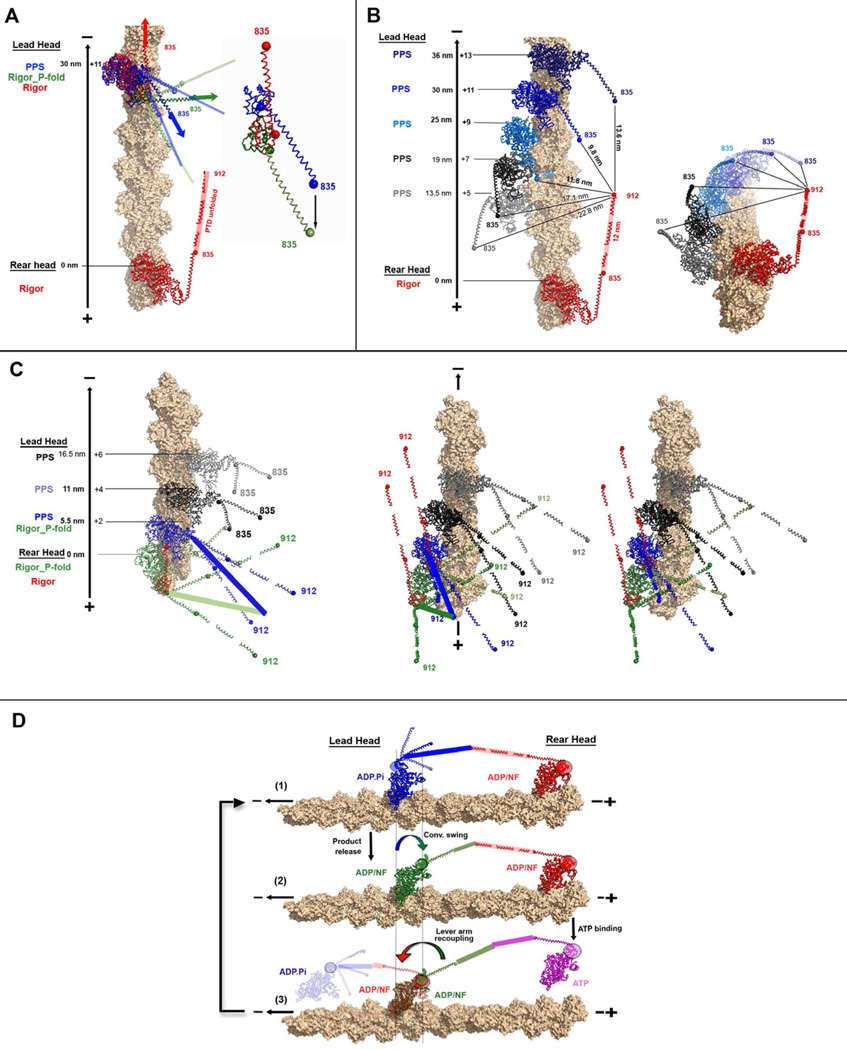
Figure 5. The Myosin VI walking mechanism.
(A) The converter of the lead head rotates while staying in the PPS fold (blue to green) to promote strong attachment. The transition towards the rigor fold of the converter (red) cannot occur since it would generate too much torque. (B and C), various possible dockings of the lead head on an actin filament are indicated. (B left) The rear head (red) is indicated using the rigor state of myosin VI (2BKH). This corresponds to the state that the rear head adopts after recoupling of the lever arm (R-fold converter). Assuming a rigid lever arm for the rear head, possible steps correspond to distances between the end of the rigid lever arm of the rear head (ending at residue 912) and the end of the of the lead head lever arm (which incorporates both the IQ motif ending at residue 835 and ~12 nm from the unfolded three-helix bundle lever arm extension). Possible dockings for the PPS lead head are indicated with a MDins2-IQ construct (with converter in the P-fold) – the possible dockings are in blue and those that are too far are in black or grey. (Right) Same model in a different orientation to highlight the azimuthal components of the various dockings. (C) The rear head (green) is indicated using the rigor state of myosin VI (2BKH) in which the converter is modeled with a P-fold and the lever arm after the IQ motif is indicated assuming a rigid lever arm. This corresponds to the state the rear head likely adopts after the swing of its converter but prior to the recoupling of its lever arm. Possible dockings for the PPS lead head are indicated with a MDins2-IQ-PTD construct (with converter in the P-fold). Only the site at 5.5 nm would allow the converter rotation of the lead and a transition of both heads together toward the rigor state (red). The docking sites of the lead head in black or grey cannot lead to a productive step. (Right) Same model in a different orientation to highlight the azimuthal components of the various dockings. (D) Different phases for the step of a dimeric myosin VI molecule: (1) Docking of the lead head, (2) Swing with a PPS converter of the lead head maintains the orientation of its lever arm without significant strain, allowing strong binding to actin and phosphate and MgADP release (MgADP can rebind, but MgATP cannot bind), (3) Detachment of the rear head by MgATP binding allows the converter fold transition and recoupling of the lever arm in the lead head to bias rebinding of the former rear head towards the minus end.
The compliance we observed in the lead head (PPS) of myosin VI allows converter rotation in the P-fold conformation to occur with minimal change in the orientation of the distal part of the lever arm (Figure 4D). Further modeling reveals that the isomerization between the P- and R-fold of the lead head converter cannot occur while the rear head is strongly attached, since this would result in considerable torque and strain within the lever arm (Figure 5A). Modeling of the lead head undergoing 19–36 nm steps (Figure 5) reveals that the lever arm compliance we detect is sufficient to allow the converter to swing to the post-stroke position if it remains in the P-fold conformation. This explains why the motor domain of the lead head is not slowed in releasing Pi and ADP by the attached rear head. Instead, the lead head undergoes the necessary conformational rearrangements promoted by strong binding to actin and release its products as if it is not influenced by the bound rear head (Sweeney et al., 2007). This P-fold converter swing is also consistent with the fact that reverse strain has been shown to facilitate strong binding of a myosin VI head (Iwaki et al., 2009) since the center of mass of the converter is moved towards the plus-end of the filament in this transition (Figure 5A).
Note that the rear head prevents the lead head from undergoing the further ~90° rotation that would occur upon transition to the R-fold of the converter after its swing (Figure 5A). Although the lead head is not prevented from releasing or rebinding MgADP with the rear head attached, it is prevented in binding ATP until the rear head detaches (Sweeney et al., 2007). A possible implication is that rapid binding of ATP can only occur if the converter assumes its R-fold conformation. How this converter isomerization is coupled to elements of the nucleotide-binding site is unclear. However these coordinated changes in the converter and nucleotide binding site must underlie the slow isomerization that was detected and required to allow ATP binding to the lead head following detachment of the rear head (Sweeney et al., 2007).
With modeling, we next investigated what structural states and properties of myosin VI account for the variable step sizes that have been observed for the processive dimer (see Figure 5B). We considered both the possibility that the unfolded 3-helix bundle is flexible and the possibility that it is somewhat rigid, since there is no detailed characterization of this region. For both possibilities we report the possible size steps that can be produced for a myosin VI dimer depending on the converter fold that the lead head adopts. Note that in comparing these values to published step sizes obtained for myosin VI, in many cases experiments have been done by placing a fluorescent probe on only one head of a myosin VI dimer in order to use the FIONA (Yildiz et al., 2004) technique to track its position with ultra-high resolution. This means that what is actually observed is movement achieved by two sequential steps, which is then reported as either the summation of these steps (i.e. two sequential steps of 30nm would be reported as 60nm) or as their average. We comment on this point when relevant below.
First we modeled the rear head in the rigor conformation with a ~18 nm long lever arm (2.5 nm for the converter/insert-2 module, 3.5 nm for the IQ motif/CaM and ~12 nm for the unfolded 3-helix bundle) assuming that they constitute a rather rigid lever arm. We consider this the most likely possibility, since replacement of the IQ-CaM and 3-helix bundle of myosin VI with the relatively stiff lever arm of myosin V does not alter the observed step size distribution of the dimer (Park et al., 2007). Our modeling revealed that if the converter of the lead head adopts a P-fold conformation, as in all of our PPS structures, then three actin-binding sites can be easily reached. This would allow the swing of the converter to take place without much strain due the pliant region of insert-2. 30 nm (11 actin subunits) away from the rear head seems to be the most easily reached and would position the lever of the lead head parallel to the actin axis, but 36 nm (13 subunits) and 25 nm (9 subunits) would be allowed and have only a small azimuthal component (see Figure 5B). The actin-binding site 19 nm (7 subunits) from the rear head might also be reached if a large bend is applied in the pliant region of insert-2. This would orient the lead head lever arm perpendicularly to the actin filament axis. The actin binding site 13.5 nm (5 subunits) from the rear head cannot be reached since it would be too far from the end of the rigid lever arm of the rear head. Note that the even numbered actin binding sites are not accessible since they are found on the opposite side of the actin filament helix. (In many cases experiments with myosin VI dimers have been performed with a probe on one head only, and thus the observed steps are the summation of two successive stepping events. Since modeling suggests that the favored spacing between heads will be 25, 30 or 36 nm (see Figure 5B), the modeling accounts for the 50–72 nm steps that have been observed experimentally with probes on one of the two heads).
We also modeled the situation in which the distal part of the myosin VI lever arm, created by unfolding of the three-helix bundle, is compliant rather than rigid. The main impact of this would be to decrease the average step size by favoring attachment of the lead head to actin-binding sites 19, 25 or 30nm from the rear head (see Supplemental discussion and Figure S5A). As shown in Supplemental discussion, note also that the formal possibility that the lead head would adopt a R-fold converter upon attachment must be excluded since the head could not release its products without creating a large torque on the lever arm (Figure S5B, S5C and S5D).
Generation of very small steps
Very small steps (~5.5nm) have also been measured for myosin VI and led to the suggestion that myosin VI has an inch worm-like component to its stepping behavior that alternates with more conventional hand-over-hand stepping (Nishikawa et al., 2010). (By inch worm mechanism, the authors suggest that the rear head sometimes moves to an actin binding site adjacent to the former lead head, much as an inch worm pulls the rear half of his body towards the front half.) Modeling indicates a simple explanation for this observed behavior. These small steps can occur if rebinding of the lead head takes place while the rear head converter is still in a P-fold conformation, and thus its lever arm is not directed towards the minus end. While this maintains compliance and variability in the position of the rear head lever arm, the lever arm is nonetheless mainly oriented perpendicular to the actin filament axis by the overall converter position in the poststroke position. Modeling shows that in this case, binding at 5.5 nm (a+2) from this attached rear head followed by a converter swing would be possible if the lead head would have its converter in the P-fold thus mainly oriented parallel to the actin filament axis (Figure 5C). Both heads would then likely swing together and end up with both converters in the R-fold conformation. This would explain why the next step following a 5.5 nm step has to be a large step, as has been experimentally observed (Nishikawa et al., 2010).
Note that if such small steps occur and one is tracking the movements of only one head of a dimer experimentally, the step size observed will depend on the distance between the heads prior to detachment to which 5.5 nm would be added. If we assume the prior distance to be 25–36 nm, which is the favored spacing (see Figure 5B), this modeling accounts for the 30–42 nm steps that have been observed. The fact that the frequency of small steps increases when the ADP concentration is raised (Nishikawa et al., 2010) indicates that the isomerization from P-fold to R-fold of the rear head may be slower if ADP is bound to the head, which is consistent with the converter rearrangement towards the R-fold being promoted by the rear head being free of nucleotide. [Note that, based on the kinetic measurements on myosin VI dimers, when the lead head is constrained by an attached rear head, ADP can freely dissociate and rebind to the lead head (Sweeney et al., 2007). Thus as ADP concentration increases, the lead head is increasingly likely to be an ADP-bound state when the rear head detaches. While this lowers the likelihood of a processive run on actin being terminated due to ATP binding causing detachment of the lead head prior to strong reattachment of the rear head (Sweeney et al., 2007), it also will increase the frequency of small steps.
Predicted lever arm angles during stepping
From the modeling presented it is clear that compared to myosin V, the lever arms of myosin VI are oriented so that they stay closer to the actin filament, which had been suggested to allow myosin VI to tend to stay on the same actin track (Sweeney & Houdusse 2010). When the lead head of myosin VI rebinds to actin with a P-fold with its preferred step size of 25–36 nm, the IQ/CaM module of the lever arm would tend to stay mostly parallel to the actin filament axis during the converter swing. This point has been addressed experimentally, with two different groups examining the distribution of the lever arm angles relative to the actin filament axis during processive stepping of myosin VI (Reifenberger et al., 2009, Sun et al., 2007). However, their conclusions differ in terms of whether the lever arm is always parallel to the actin filament axis or not (i.e. whether or not there is a significant azimuthal component to the lever arm swing). From our analysis, when the most favored steps of 25–36nm occur, they would not lead to a significant azimuthal component to the lever arm position, as reported by Reifenberger et al., (2009). [Note that in the experiments ofReifenberger et al. (2009), a probe on only one head of a dimer was observed, thus they were measuring the distance of two sequential steps. For two sequential steps of 25–36nm, they would have observed steps of 50–72nm, which is what they reported.] However, when the distance between the heads is 19nm, then significant azimuthal angles would be predicted to be measurable for the lead head lever arm, since these distances correspond to binding to actin sites on the side of the filament. One could thus speculate that the differences in the data reported by Reifenberger et al. (2009) and Sun et al. (2007) might be accounted for by differing in vitro geometries if the assay ofSun et al. (2007) allowed myosin to spiral around the actin filament while the assay system of Reifenberger et al. (2009) did not (Figure 5B). In this case, the “short” steps (30–42nm) observed by Reifenberger et al. (2009) are likely occurring when the attached rear head converter has not undergone the P-fold to R-fold transition, promoting re-docking of the head in an orientation parallel to the actin filament axis at the site 5.5 nm from this uncoupled head (as discussed above). In the case of Sun et al. (2007), the small steps could in part correspond to stepping on the side of the actin filament with 19 nm spacing between heads and this stepping would also explain the frequent observation of azimuthal angles for the lever arm in this assay (Figure 5B).
State-dependent compliance in the myosin VI lever arm is essential for processive movement in the reverse direction
Based on this modeling, the compliance in the converter and pliant region insert-2 found in our PPS structures can completely account for the variable step sizes of the myosin VI dimer by allowing different actin binding sites to be reached (Figure 5). However, this step size variability may simply be a consequence of the design that is necessary to allow the re-attached lead head to bind strongly to actin without developing significant intramolecular strain so that the release of Pi and ADP can occur even though the lead head is constrained by the rear-head (Sweeney et al., 2007). Progression through the strong-binding states of the motor (accompanying product release) requires MD rearrangements that must involve the rotation of the converter due to movements of the relay. If no compliance were found in the lead head, this would result in an azimuthal torque on the lever arm that would likely either inhibit strong binding of the lead head until the rear head detaches or lead to high degrees of intramolecular strain if both heads were attached strongly. While intramolecular strain can be used to gate plus-end directed myosin motors, such as myosin V (Rosenfeld and Sweeney 2004; Veigel et al. 2005) by greatly slowing ADP release from the lead head. We have argued that due to the repositioning of the lever arm in myosin VI, intramolecular strain would not slow ADP release from the lead head, and could in fact slow ADP release from the rear head. Thus, processive movement would necessitate a new type of gating via a design that allows stepping without the development of intramolecular strain as well as a new means of preventing release of the lead head prior to release of the rear head (Sweeney et al., 2007; Sweeney and Houdusse, 2010). We have presented evidence that gating is accomplished in myosin VI by preventing ATP binding to the lead head until the rear head has detached Sweeney et al., 2007).
Dimeric Myosin VI does not generate force using a swinging lever arm mechanism
Based on our new structures and the prior structural knowledge of myosin VI, a mechanism emerges that involves the motor domain of myosin VI undergoing similar conformational changes as those proposed for plus-end motors, while the compliant properties of the P-fold converter and insert-2 elements are essential to decouple the rotation of the lever arm from that of the converter at the beginning of the myosin VI powerstroke. This ensures committed attachment of the lead head upon processive movement without the development of intramolecular strain in the lever arm, which allows the ATP hydrolysis products to be released from the lead head at the unstrained rate. Since the lever arm re-orientation is not coupled to the swing of the converter, the converter swing does not impose a large azimuthal torque.
Thus these results lead us to propose that dimeric myosin VI does not strictly direct its lever arm upon force generation (as postulated by the swinging lever arm mechanism), but rather has an initial phase of force generation in which its lever arm is essentially uncoupled from motor domain rearrangements due to lever arm compliance, which is then followed by a second phase in which the lever arm is directed by further MD rearrangement that includes a recoupling of the lever arm upon strain release when the rear head detaches (see Figure 5D). Fluctuations in the position of the IQ/CaM module have been reported when myosin VI walks (Yildiz et al., 2004) and are consistent with this model since it would correspond to the variability in lever arm position of the lead head upon the converter swing while in the P-fold compensated by a pliant region. In this regard, the processive movement of myosin VI is similar in mechanism to that proposed for kinesin motors (Rice et al., 1999; Vale and Milligan, 2000). This kinesin mechanism involves uncoupling of the neck linker/lever arm to allow strong attachment of the lead and recoupling of the linker/lever arm within the lead head upon dissociation of the rear head.
Summary
This study highlights the importance of the converter isomerization in myosin VI and the variable, state-dependent compliance within the converter subdomain and within the adjacent pliant region of insert-2. The puzzling stepping behavior of the myosin VI dimer can be easily explained by the variability in actin attachment created by these converter/lever arm properties. Compliance in the converter of a plus end motor such as myosin II is also likely to play a role for force generation of the motor under load, while lack of compliance in the lever arm and lack of global conformational changes in the converter would allow for more precise stepping behavior. Interestingly, mutations in the converter of cardiac myosin II that are linked to hypertrophic cardio-myopathies have been reported to influence force generation (Seebohm et al., 2009). Sequence variations for the converter in different muscle isoforms of Drosophila myosin II are also likely to be a way of tuning the motor properties of these myosins (Miller et al., 2003). For myosin VI, it appears that a high degree of compliance is necessary to prevent the generation of intramolecular strain within the dimer in order to allow processive movement in the opposite direction. Analogous to conventional kinesin motors, the mechanism involves strong attachment of the lead head with a lever arm poorly coupled to its motor, which after rear head detachment is followed by a recoupling of the lever arm to bias the rebinding of the new lead head towards the minus end of the track.
EXPERIMENTAL PROCEDURES
Details of the experimental procedures are indicated in supplemental material. Briefly, A series of truncations of porcine myosin VI cDNA allowed to purify different constructs of myosin VI with C-terminal truncations corresponding to amino acids 816 fused to GFP (MDins2-GFP), 839 (MDins2-IQ) and 789 (MD and the packing mutant MDPM). Crystals were obtained using the hanging drop method and were cryo-cooled prior to data collection at either the European Synchrotron Radiation Facility beamlines or the SOLEIL Proxima-1 beamline. The data sets were integrated and scaled with XDS and XSCALE (Kabsch et al., 2010) or Mosflm and SCALA (CCP4, 1994). Statistics on the data collection for these four new pre-powerstroke state myosin VI structures are indicated in Table 1. The structures were solved by molecular replacement with the myosin VI MD prepowerstroke model (PDB code 2V26, (Menetrey et al., 2007)) by using the program AmoRe (Navaza, 1994) or Phaser (McCoy, 2007). Model building was done as mentioned in supplemental material. Refinement was performed with Coot (Emsley & Cowtan, 2004) and Refmac5 (for MDins2-GFP and MDins2-IQ) (CCP4, 1994) or Phenix (for MDcrystal form 2 and MDPM) (Adams et al. 2010). Crystallographic statistics of the final models are summarized in Table 1. All diagrams for the figures were computed using MOLSCRIPT/Raster3D (Kraulis, 1991; Merritt and Bacon, 1997) or pymol (DeLano, 2002).
Supplementary Material
Highlights.
Myosin VI uses a kinesin-like mechanism to step
Compliance in the lever arm lead head is essential to attach strongly to actin
Myosin VI does not move using a simple swinging lever arm mechanism
ACKNOWLEDGMENTS
We thank Pierre Legrand and Andy Thomson at the PROXIMA 1 beamline of the SOLEIL synchrotron for assistance with data collection. Dr. Olena Pylypenko is the recipient of an ARC post-doctoral fellowship. Dr. Monalisa Mukherjea is the recipient of an American Heart Association fellowship. Author contributions: A.H. and H.L.S. designed research; J.M., T.I., V.R., M.M., O.P. and X.L. performed research; V.R., J.P. and P.V. analyzed the SAXS data, A.H. performed the modeling, J.M. and A.H. made the figures and A.H. and H.L.S. wrote the paper. This work was supported by a grant from NIH (DC009100) (HLS), the CNRS (AH), the ARC subvention fixe 1067XA0830F, and the ANR BLAN 2010 1504 01 (AH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers. Atomic coordinates and structure factors for the pre-powerstroke structures of the myosin VI constructs have been deposited in the Protein Data Bank under the accession numbers 4ANJ for the MDins2-GFP, 4E7Z for the P21 crystal form of the MD and 4E7S for the MDPM (packing mutant) construct, respectively.
References
- Adams PD, Zwart PH, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant Z, Altman D, Spudich JA. The power stroke of myosin VI and the basis of reverse directionality. Proc Natl Acad Sci U S A. 2007 Jan 16;104(3):772–777. doi: 10.1073/pnas.0610144104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4. The CCP4 suite: program for protein crystallography. Acta Cristallographica D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- DeLano WL. PyMOL molecular graphics system. 2002 Available at: http://www.pymol.org. [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Cryst. 2010;D66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis PJ. MOLSCRIPT : a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- Liao JC, Elting MW, Delp SL, Spudich JA, Bryant Z. Engineered myosin VI motors reveal minimal structural determinants of directionality and processivity. J Mol Biol. J Mol Biol. 2009 Oct 2;392(4):862–867. doi: 10.1016/j.jmb.2009.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetrey J, Bahloul A, Wells AL, Yengo CM, Morris CA, Sweeney HL, Houdusse A. The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature. 2005;435:779–785. doi: 10.1038/nature03592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetrey J, Llinas P, Mukherjea M, Sweeney HL, Houdusse A. The structural basis for the large powerstroke of myosin VI. Cell. 2007;131:300–308. doi: 10.1016/j.cell.2007.08.027. [DOI] [PubMed] [Google Scholar]
- Merritt EA, Bacon DJ. Raster3D: Photorealistic Molecular Graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- Miller BM, Nyitrai M, Bernstein SI, Geeves MA. Kinetic analysis of Drosophila muscle myosin isoforms suggests a novel mode of mechanochemical coupling. J Biol Chem. 2003 Dec 12;278(50):50293–50300. doi: 10.1074/jbc.M308318200. [DOI] [PubMed] [Google Scholar]
- Mukherjea M, Llinas P, Kim H-J, Travaglia M, Safer D, Zong AB, Ménétrey J, Franzini-Armstrong C, Selvin PR, Houdusse A, Sweeney HL. Myosin VI dimerization triggers an unfolding of a 3-helix bundle in order to extend its reach. Molecular Cell. 2009;35:305–315. doi: 10.1016/j.molcel.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallographica A. 1994;50:157–163. [Google Scholar]
- Nishikawa S, et al. Class VI myosin moves processively along actin filaments backward with large steps. Biochem Biophys Res Commun. 1994;290:311–317. doi: 10.1006/bbrc.2001.6142. [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Arimoto I, Ikezaki K, Sugawa M, Ueno H, Komori T, Iwane AH, Yanagida T. Switch between large hand-over-hand and small inchworm-like steps in myosin VI. Cell. 2010;142:879–888. doi: 10.1016/j.cell.2010.08.033. [DOI] [PubMed] [Google Scholar]
- Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007;8:R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Li A, Chen LQ, Houdusse A, Selvin PR, Sweeney HL. The unique insert at the end of the myosin VI motor is the sole determinant of directionality. Proc Natl Acad Sci USA. 2007;104:778–783. doi: 10.1073/pnas.0610066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phichith D, Travaglia M, Yang Z, Liu X, Zong AB, Safer D, Sweeney HL. Cargo binding induces dimerization of myosin VI. Proc Natl Acad Sci U S A. 2009;106:17320–17334. doi: 10.1073/pnas.0909748106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifenberger JG, Toprak E, Kim H, Safer D, Sweeney HL, Selvin PR. Myosin VI undergoes a 180 degrees power stroke implying an uncoupling of the front lever arm. Proc Natl Acad Sci U S A. 2009;106:18255–18260. doi: 10.1073/pnas.0900005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, Pate E, Cooke R, Taylor EW, Milligan RA, Vale RD. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- Rock RS, Rice SE, Wells AL, Purcell TJ, Spudich JA, Sweeney HL. Myosin VI is a processive motor with a large step size. Proc Natl Acad Sci USA. 2001;98:13655–13669. doi: 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock RS, Ramamurthy B, Dunn AR, Beccafico S, Rami BR, Morris C, Spink BJ, Franzini-Armstrong C, Spudich JA, Sweeney HL. A flexible domain is essential for the large step size and processivity of myosin VI. Mol Cell. 2005;17:603–609. doi: 10.1016/j.molcel.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Rosenfeld SS, Sweeney HL. A model of myosin V processivity. J. Biol. Chem. 2004;279:40100–40111. doi: 10.1074/jbc.M402583200. [DOI] [PubMed] [Google Scholar]
- Seebohm B, Matinmehr F, Köhler J, Francino A, Navarro-Lopéz F, Perrot A, Ozcelik C, McKenna WJ, Brenner B, Kraft T. Cardiomyopathy mutations reveal variable region of myosin converter as major element of cross-bridge compliance. Biophys J. 2004;97:806–824. doi: 10.1016/j.bpj.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaramakrishnan S, Spink BJ, Sim AY, Doniach S, Spudich JA. Dynamic charge interactions create surprising rigidity in the ER/K {alpha}-helical protein motif. Proc Natl Acad Sci U S A. 2008;105:13356–13361. doi: 10.1073/pnas.0806256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Schroeder HW, 3rd, Beausang JF, Homma K, Ikebe M, Goldman YE. Myosin VI walks "wiggly" on actin with large and variable tilting. Mol Cell. 2007;28:954–964. doi: 10.1016/j.molcel.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney HL, Park H, Zong AB, Yang Z, Selvin PR, Rosenfeld SS. How myosin VI coordinates its heads during processive movement. EMBO J. 2007;26:2682–2692. doi: 10.1038/sj.emboj.7601720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney HL, Houdusse A. Myosin VI rewrites the rules for myosin motors. Cell. 2010;141:573–582. doi: 10.1016/j.cell.2010.04.028. [DOI] [PubMed] [Google Scholar]
- Vale RD, Milligan RA. The way things move: looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- Veigel C, Schmitz S, Wang F, Sellers JR. Load-dependent kinetics of myosin V can explain its high processivity. Nat. Cell Biol. 2005;7:861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- Yildiz A, Park H, Safer D, Yang Z, Chen LQ, Selvin PR, Sweeney HL. Myosin VI steps via a hand-over-hand mechanism with its lever arm undergoing fluctuations when attached to actin. J Biol Chem. 2004;279:37223–37226. doi: 10.1074/jbc.C400252200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.