Abstract
Cryptococcus neoformans, an opportunistic fungal pathogen, manifests an intrinsic adaptive mechanism of resistance toward fluconazole (FLC) termed heteroresistance. Heteroresistance is characterized by the emergence of minor resistant subpopulations at levels of FLC that are higher than the strain’s minimum inhibitory concentration. The heteroresistant clones that tolerate high concentrations of FLC often contain disomic chromosome 4 (Chr4). SEY1, GLO3 and GCS2 on Chr4 are responsible for ER integrity and important for Chr4 disomy formation under FLC stress. We sought an evidence of a direct relationship between ER morphology and Chr4 disomy formation. Deletion of the YOP1 gene on Chr7, which encodes an ER curvature-stabilizing protein that interacts with Sey1, perturbed ER morphology without affecting FLC susceptibility or the frequency of FLC-induced disomies. However, deletion of both YOP1 and SEY1, not only perturbed ER morphology more severely than in sey1Δ or yop1Δ strains but also abrogated the FLC-induced disomy. Although the heteroresistance phenotype was retained in the sey1Δyop1Δ strains, tolerance to FLC appeared to have resulted not from chromosome duplication but from gene amplification restricted to the region surrounding ERG11 on Chr1. These data support the importance of ER integrity in C. neoformans for the formation of disomy under FLC stress.
Keywords: Cryptococcus neoformans, disomy formation, fluconazole, anitifungal resistance, heteroresistance, endoplasmic reticulum
Introduction
Cryptococcosis has been effectively treated with amphotericin B induction therapy followed by maintenance regimens with azole drugs which target the biosynthetic pathway of ergosterol, an essential component of the fungal membrane (Kwon-Chung, et al., 2000, Akins, 2005, Sullivan, et al., 2006). Ergosterol is synthesized in the endoplasmic reticulum (ER) starting from acetyl-CoA through a series of enzymes encoded by various ERG genes (Akins, 2005). The sterol is then delivered to the cell membrane via both vesicular and non-vesicular routes (Sullivan, et al., 2006, Schulz & Prinz, 2007).
Fluconazole (FLC), a triazole drug, has been the most widely used azole antifungal in the maintenance therapy of cryptococcosis (Washton, 1989, Perfect, et al., 2010). Since the report on drug resistant cases of cryptococcosis has been infrequent, studies on the molecular mechanism of drug resistance in C. neoformans has been considerably limited compared to pathogenic species of other yeasts. Only few cases of missense mutations in ERG11 were reported from azole resistant clinical strains (Rodero, et al., 2003, Sionov, et al., 2011). A laboratory constructed C. neoformans strain that over expressed an ABC transporter, AFR1, was reported to be resistance to FLC (Posteraro, et al., 2003). In 1999, a novel azole resistance termed ‘heteroresistance’ was first reported in C. neoformans based on the behavior of two strains, one isolated from a patient in Italy and the other from a patient in Israel (Mondon, et al., 1999). Recently, heteroresistance was described as an intrinsic, adaptive resistance to FLC and it was found to be universal in the strains of C. neoformans and C. gattii thus far tested (Mondon, et al., 1999, Sionov, et al., 2009, Varma & Kwon-Chung, 2010). The phenomenon of heteroresistance was characterized as the emergence of minor subpopulations within a single colony of the susceptible strain that could adapt to FLC concentrations higher than the strain’s minimum inhibitory concentration (MIC). The acquired resistance in these subpopulations is transient since it is lost upon release from drug stress (Sionov, et al., 2009). The level of heteroresistance to fluconazole (LHF) of each strain was defined as the lowest concentration of FLC at which resistant subpopulations emerge. For the genome sequenced strain H99, the LHF is 32μg/ml FLC and the heteroresistant subpopulations (0.3–0.6%) that emerged at 32μg/ml FLC contained disomic Chr1 (Sionov, et al., 2010). When the FLC concentration was increased to 64 μg/ml or higher, Chr4 also became disomic (Sionov, et al., 2010, Ngamskulrungroj, et al., 2012). These extra copies of duplicated chromosomes were lost in a step-wise manner upon daily transfer in drug free media (Sionov, et al., 2010). Chr1 contains ERG11, the target of fluconazole, and the ABC drug transporter AFR1 which play a major role in C. neoformans azole resistance (Sionov, et al., 2010). It has been proposed that the increases in dosage of these genes enable the strains to overcome the drug stress. Moreover, a recent study showed that SEY1, GCS2 and GCS2 genes on Chr4 which are essential for endoplasmic reticulum integrity and/or function, are important for the FLC-induced Chr4 disomy (Ngamskulrungroj, et al., 2012).
In eukaryotic cells, ER plays an important role in several core processes of cell biology such as biosynthesis of secretory and membrane proteins, lipid synthesis, and Ca2+ storage. Unlike the extensive reticular structure of mammalian ER, the yeast tubular ER is closely located to the plasma membrane, thus, named as cortical ER (reviewed in (Hu, et al., 2011)). In addition to the key cellular processes, cryptococcal ER is believed to play an important role in azole resistance by influencing the process of disomy formation in response to fluconazole stress (Ngamskulrungroj, et al., 2012). However, the mechanism of such influence on disomy formation remains unknown. In the present study, we show that YOP1 which encodes a protein that interacts with Sey1 (Brands & Ho, 2002, Hu, et al., 2009), plays an important role in ER integrity. While deletion of YOP1 alone did not affect FLC susceptibility or the formation of chromosome disomy, the double deletion of sey1Δ/yop1Δ resulted in severe aberration of ER morphology and also abolished FLC-induced disomy formation.
Materials and Methods
Strains Media and level of FLC resistance
All deletant strains used in the study were constructed in the C. neoformans H99 background (Perfect, et al., 1993) and are listed in Table 1. Strains were maintained on YPD agar (1% yeast extract, 2% peptone, 2% glucose, 2% agar) or YPD supplemented with 8, 16, 32, 64 or 128 μg/ml FLC. For spot assays, a 2μl cell suspension with an optical density of 2 at 600 nm (OD600) and its 10-fold serial dilutions were spotted on YPD agar with or without drug supplements. The plates were incubated at 30°C for 3 – 5 days and photographed. As in the previous study (Ngamskulrungroj, et al., 2012), we defined the first level of heteroresistance to FLC (1LHF) as the lowest concentration of FLC at which minor resistant subpopulations emerge. We used arbitrary 2-fold increments of FLC concentration to define the subsequent LHF’s. For instance, 1LHF of the wild type strain H99 is 32 μg/ml, 2LHF is 64 μg/ml and 3LHF is 128 μg/ml (Table 1).
Table 1.
List of strains and their levels of heteroresistance.
| Strain | Description | Concentrations of FLC at 1, 2 and 3LHF (μg/ml) |
|---|---|---|
| H99 | wild type | 32, 64, 128 |
| yop1Δ | ER curvature-stabilizing protein | 32, 64, 128 |
| sey1Δ | GTPase interacts with ER-shaping protein | 16, 32, 64 |
| sey1Δyop1Δ | double deletion of SEY1 and GLO3 | 16, 32, 64 |
| yop1Δ::YOP1 | homologous complementation of yop1Δ | 32, 64, 128 |
| sey1Δ::SEY1 | homologous complementation of sey1Δ | 32, 64, 128 |
| sey1Δyop1Δ::YOP1 | Homologous complementation of yop1Δ in sey1Δyop1Δ | 16, 32, 64 |
| sey1Δyop1Δ::SEY1 | homologous complementation of sey1Δ in sey1Δyop1Δ | 32, 64, 128 |
Gene manipulations
The YOP1 homolog of S. cerevisiae in the C. neoformans strain H99 was identified by a BlastP search of the H99 genome database (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/MultiHome.html) and was disrupted by biolistic transformation (Toffaletti, et al., 1993). Briefly, disruption constructs were created by overlapping PCR technique (Davidson, et al., 2000) with Nourseothricin (NAT1), G418 (NEO) or Hygromycin (HGR) resistance genes as dominant selectable markers. These constructs were transformed into H99 cells by a BioRad model PDS-1000/He biolistic particle delivery system. Homologous integrations were confirmed by PCR and Southern hybridization. Complementation of each deletant was accomplished by homologous integration at the disrupted locus.
Quantitation of gene dosage
Quantitative real time PCR (qPCR) assays were performed to quantitate the copy number of genes on specific chromosomes as previously described (Sionov, et al., 2010). 6 – 8 independent clones of each strain were tested at 3LHF (128μg/ml FLC) (Ngamskulrungroj, et al., 2012).
Comparative Genome Hybridization (CGH)
Genomic DNA was extracted from 3–5 day old cultures grown on YPD plates with or without the drug as described previously (Sionov, et al., 2010). DNA was labeled by using the BioPrime®Array CGH Genomic Labeling System Kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s instruction. Briefly, genomic DNA was digested with DpnII (New England Biolabs, Ipswich, USA). Experimental and control samples were labeled with Alexa647 and Alexa555 dyes, respectively. The labeled samples were hybridized to JEC21-based 70mer slides (http://genome.wustl.edu/services/microarray/cryptococcus_neoformans) and analyzed as previously described (Sionov, et al., 2010).
Fluorescence microscopy
To visualize the ER within yeast cells, the ER-protein Sec61β/Sbh1 homolog, CNAG_06351, was tagged with GFP (Hu, et al., 2009) as described in the previous study(Ngamskulrungroj, et al., 2012). The GFP signals enabled visualization of the ER morphology by using the fluorescence Zeiss Axiovert microscope and Axiovision (version 4.0) software.
Transmission electron microscopy (TEM)
Cells were prepared and stained as previously described (Yoneda & Doering, 2006). Briefly, cells were grown overnight in YPD broth. Mid-log phase cells were harvested and washed once in 1 ml of the primary fixative (0.1 M sorbitol, 1 mM MgCl2, 1 mM CaCl2, 2% glutaraldehyde in 0.1M PIPES buffer, pH 6.8). Then, the cells were fixed with 2% KMnO4, dehydrated using a series of graded ethanol and propylene oxide prior to infiltrating and embedding in Epon. Cells were prepared for visualization of the membrane structures under TEM as described (Yoneda & Doering, 2006, Ngamskulrungroj, et al., 2012).
Focused Ion Beam-Scanning Electron Microscopy (FIB-SEM)
Cells were prepared as they were for the TEM mentioned above. Specimens were mounted on the FIB-SEM. All data sets were collected by a FEI Helios NanoLab 650 DualBeam™ equipped with Ga+ LMIS FIB used for milling (FEI, Hillsboro, OR) according to the method described previously(Ngamskulrungroj, et al., 2012).
Results
Deletion of YOP1 causes severe alterations in ER morphology
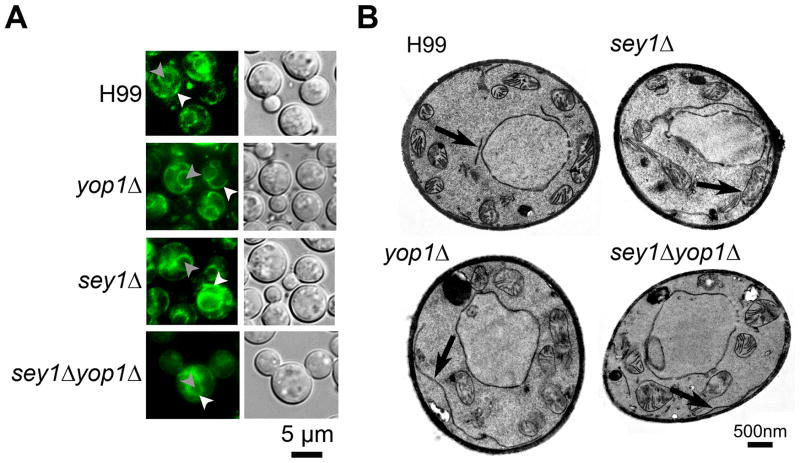
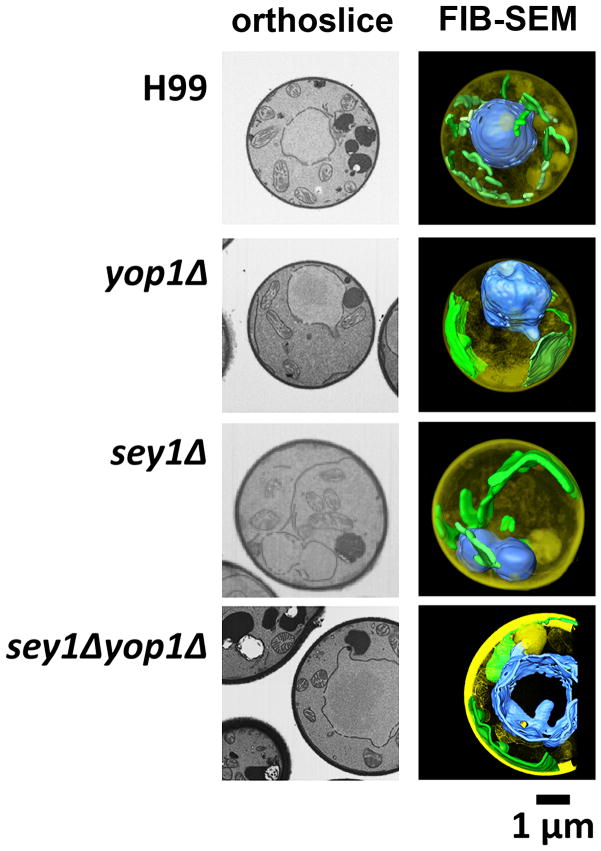
A previous study showed that deletions of the genes responsible for ER integrity reduced the frequency of Chr4 disomy. However, the correlation between normal ER morphology and chr4 disomy is controversial. To study the impact of ER morphology on FLC-induced disomy, we identified and deleted the homolog of Yop1, an ER curvature-stabilizing and Sey1 interacting protein (YOP1, CAN_06646) (Brands & Ho, 2002, Hu, et al., 2009). The YOP1 gene resides on Chr7 which has not been observed to undergo changes in copy number in response to elevated FLC concentrations irrespective of the tested strain’s genetic background [(Ngamskulrungroj, et al., 2012), data not shown]. The yop1 disruptants were characterized and compared with our previous data obtained with the wild type, sey1Δ and sey1Δ::SEY1 strains (Ngamskulrungroj, et al., 2012). When the GFP-Sec61β ER protein was used as a reporter, the wild type strain showed the GFP signal to be located near the nuclear envelope and an element near the plasma membrane connected with some strands in the cytoplasm; a typical ER pattern seen in Saccharomyces cerevisiae and C. neoformans (Figure 1A) (Hu, et al., 2009, Ngamskulrungroj, et al., 2012). In contrast, yop1Δ displayed an aberrant ER morphology similar to the sey1Δ strain under a fluorescent microscope. The ER strands of the sey1Δ and yop1Δ strains were portrayed as smooth threads in contrast to bead-like strands of the wild type. This ER aberrance was even more evident in the case of double deletant where the nuclear envelope was also distorted (Figure 1A). Like in sey1Δ, the ER in yop1Δ was clearly elongated as observed under TEM, which was almost never the case in the wild type strain (Figure 1B). These losses of the bead-like chartacteristics and elongation of the ER suggested the possible loss of ER reticulation. Finally, since ER is known to be a reticular network throughout the entire cell (English, et al., 2009), a FIB-SEM which provides a view of three dimensional structure was used to examine the ER network. The wild type cells showed the typical tubular ER extending throughout the cytoplasm. The ER (artificially labeled in green color) morphology in both yop1Δ and sey1Δ was aberrant and was observed mostly as an expanded sheet (Figure 2) in contrast to the reticulate ER of the wild type cell. Moreover, a double deletion of both YOP1 and SEY1 caused a more severe morphological abnormality of the ER as well as nuclear membrane (artificially labeled in blue color) than either of yop1Δ or sey1Δ (Figure 1 and 2).
Figure 1. Disruption of YOP1 causes ER abnormality as was seen in sey1Δ.
A) Localization of the ER protein GFP-Sec61β in each indicated strain. Cells were grown on YPD agar at 30°C and subsequently visualized by fluorescence microscopy. White arrow head = ER; grey arrow head = nucleus (determined by co-localization with Hoechst 3342 dye as published previously (Ngamskulrungroj, et al., 2012)). B) Transmission Electron Microscopy (TEM). Overnight cell cultures of indicated strains grown to mid log phase were prepared and examined by TEM. Arrow = ER.
Figure 2. The FIB-SEM shows aberration in ER morphology.
Orthoslices representing central sections are shown in the left column. Data collected by serial 3–10 nm cuts of each sample through the cell was used to generate 3D reconstructions. ER and nuclei were psuedo-colored with green and blue, respectively.
YOP1 is not important for FLC tolerance
Since YOP1 was shown to be required for the maintenance of ER structural integrity like SEY1, we investigated the effect of YOP1 deletion on FLC sensitivity. Figure 3 shows that sensitivity of yop1Δ to FLC is similar to that of the wild type H99. Disruption of YOP1 in combination with SEY1 exhibited FLC sensitivity similar to the levels in sey1Δ. Complementation of SEY1 alone restored FLC sensitivity to the wild type level. These results suggested that YOP1 does not play a role in FLC tolerance despite its similar function with SEY1 in the maintenance of ER morphology.
Figure 3. Spot assay for FLC tolerance.
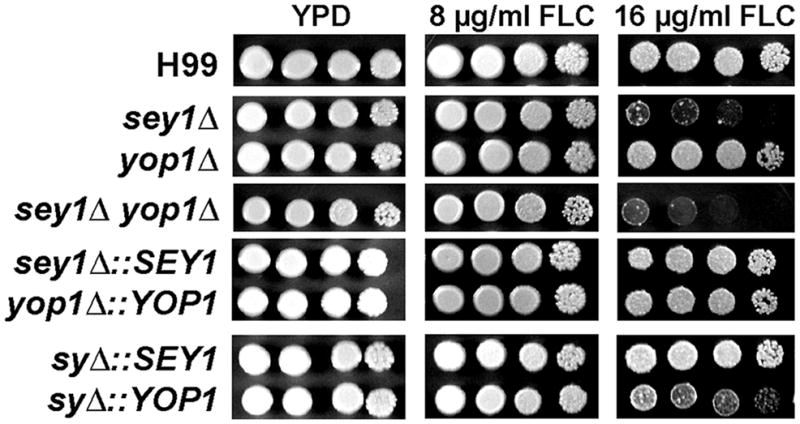
Cells of the indicated strains were spotted on YPD alone or YPD supplemented with either 8μg/ml or 16μg/ml FLC and incubated for 5 days at 30°C. sy = sey1Δyop1Δ.
Deletion of YOP1 alone does not affect l disomic chromosome formation in response to FLC stress
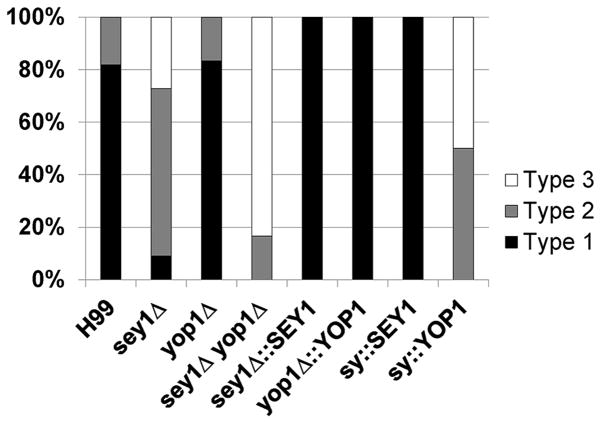
In this study, we used 3LHF (arbitrary 4-fold increments of FLC concentration from the 1LHF) to determine the frequency of disomic chromosomes since this FLC concentration was consistently shown to induce disomies of Chr1 and Chr4 in wild type strains (Ngamskulrungroj, et al., 2012). It has been shown that among the clones resistant to 3LHF derived from sey1Δ, only one of 11 clones (9%) showed the presumed Chr4 disomy (Figure 3,(Ngamskulrungroj, et al., 2012)). Surprisingly, although yop1Δ had a clearly altered ER morphology, the deletion of YOP1 alone did not have any impact on Chr1 or Chr4 disomy (Figure 4). However, if YOP1 was deleted in the sey1Δ background, Chr4 disomy was absent in the FLC resistant clones (Figure 4). Furthermore, the type-2 clones (see Figure 4 legend) of yop1Δ/sey1Δ resistant to 3LHF had a partial duplication of Chr1 exclusively in the region that contained ERG11 and no alteration was detected in the copy number of other chromosomes (Figure 5 and data not shown). These results suggested that, though not important for FLC tolerance, YOP1 plays a critical role in combination with SEY1 to form disomic chromosome in C. neoformans under FLC stress.
Figure 4. Frequency of Chr4 disomy formation in the deletants of the genes associated with ER integrity.
Clones resistant to 3LHF derived from each strain were isolated and the copy number of Chr1 and Chr4 in the genomes was inferred by qPCR of resident genes specific to each chromosome. The figure displays the percentage of each type of disomy in the FLC resistant strains. Type1 denotes strains that contain disomy of both Chr1 and Chr4, Type2 denotes strains that contain disomy of Chr1 only, and Type3 denotes strains that contain no disomies of either Chr1 or Chr4. 6 – 8 independent clones of each strain were tested. sy = sey1Δyop1Δ.
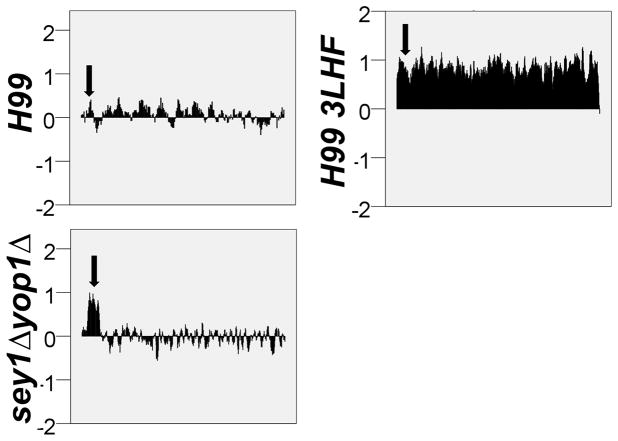
Figure 5. CGH analysis of the sey1Δyop1Δ double deletants isolated from 3LHF.
Genomic DNA extracted from a sey1Δyop1Δ double deletant with Type 2 duplication pattern at 3LHF shown in Figure 4 was used for CGH analysis. H99 and the resistant clones of H99 grown at 3LHF were included as controls. Only the Chr1 status is shown. Except for the 3LHF clone from H99, no disomy was found in any of the double mutant clones grown at 3LHF (data not shown). Arrows indicate the location of ERG11. Each bar represents the copy number of each gene residing on Chr1 in a log2 scale.
Discussion
In C. neoformans, the association between occurrence of aneuploidy and azole resistance by either partial or whole chromosome duplication has been documented (Sionov, et al., 2010, Ngamskulrungroj, et al., 2012). A number of genes on Chr1 and Chr4 that play critical roles in FLC-induced disomy formation have been characterized (Sionov, et al., 2010, Ngamskulrungroj, et al., 2012). Our previous study showed the functional importance of a SEY1, a GTPase, in FLC-induced disomy. Deletion of SEY1 decreased the frequency of Chr4 disomy and this disomy was abrogated when the gene was deleted in combination with GLO3, encoding an ADP-ribosylation factor GTPase activating protein (Ngamskulrungroj, et al., 2012). Interestingly, the inability to form FLC-induced disomy appears to be correlated with morphologically aberrant ER. Here, we show that the alteration of ER morphology resulting from the deletion of YOP1, an ER shaping protein, did not affect the frequency of FLC-induced disomy. This result indicates that intact ER morphology is not always required for disomy formation under FLC stress. However, severe alterations in ER morphology caused by deletion of both SEY1 and YOP1 resulted in a total loss of FLC-induced disomy.
Maintenance of ER integrity is essential for cellular homeostasis in eukaryotes. ER is often tightly associated with almost every other membrane-bound compartment in the cell including the plasma membrane, nuclear envelope, mitochondria, golgi, vacuoles and peroxisomes. Interactions between these organelles have shown to be functionally important (reviewed in (English, et al., 2009)). For example, proper alignment of the ER and mitochondria is indispensable for the function and survival of cells (Csordas, et al., 2006). Typically, the ER is comprised of ER tubules, ER sheets and the nuclear envelope (Shibata, et al., 2006). During mitosis, the ER and nuclear envelope undergo large structural and functional modifications in order to redistribute the organelles and the associated proteins to the daughter cells (Puhka, et al., 2007, English, et al., 2009). In yeast, the nuclear envelope has a direct role in chromosome segregation (Schmitt, 2010). In fact, a possible association between ER and disomy formation of C. neoformans has been shown in the present as well as in a previous study (Ngamskulrungroj, et al., 2012). Our present work underscores the importance of ER in chromosome disomy by showing that, in the sey1Δ/yop1Δ strain where the ER malformation is even more severe than in sey1Δ alone, FLC-induced disomy is further decreased. However, the deletion of the YOP1 alone did not show any defect in FLC-induced disomy despite the severe ER morphological defect. Since Sey1 is a GTPase and mediates homotypic ER fusion (Brands & Ho, 2002, Hu, et al., 2009, Hu, et al., 2011), it is possible that function of ER is more important than morphology alone in the formation of FLC- induced disomy in C. neoformans.
Yop1 is a member of curvature-stabilizing proteins along with Dp1 and reticulons (Rtn) (Shibata, et al., 2006). These proteins are required to form tubular structure of the ER. Depletion of these proteins in mammalian cells causes transformation of tubular ER mostly into the sheets (Voeltz, et al., 2006). In S. cerevisiae, a deletion of either of these proteins did not alter the ER morphology when grown in regular yeast media (Voeltz, et al., 2006). Similarly, a deletion of only the interacting protein, Sey1, had no effect on S. cerevisiae ER morphology, suggesting the existence of additional factor(s) required for the maintenance of ER morphology (Hu, et al., 2009). In contrast, deletion of either YOP1 or SEY1 resulted in a significant loss of tubular ER in C. neoformans. Since no homolog of Rtn was found in the C. neoformans genome by Blast search, our results suggest that the role of these proteins in ER shaping may be more restrictive in C. neoformans.
It has been shown that all yeast strains carrying extra copies of chromosome exhibit genomic instability (Niwa, et al., 2006). In 2007, Torres et al. reported that aneuploid strains of S. cerevisiae shared a number of defective traits including cell cycle progression (Torres, et al., 2007). Likewise, disomy formation in C. neoformans may result in some degree of genomic instability. Moreover, maintenance of extra chromosome copies would be significant burden to cells. Perhaps, the sey1Δyop1Δ strain with the severe defects in ER integrity is not able to overcome the burden of extra chromosome maintenance. Thus, only cells that amplified the region of ERG11 gene on Chr1 managed to survive the FLC stress without disomy formation.
Both Chr1 and Chr4, the two chromosomes most frequently found to be duplicated under FLC stress, contain genes which play critical roles in FLC-induced disomy (Sionov, et al., 2010). For example, relocation of ERG11 to Chr3 increases the frequency of Chr3 disomy while decreasing the frequency of Chr1 disomy under FLC stress. Similarly, relocation of SEY1 and/or GLO3 to Chr3 from Chr4 increased FLC-induced Chr3 disomy and reduced the frequency of Chr4 disomy ((Ngamskulrungroj, et al., 2012), unpublished data). However, the relationship between the importance of certain genes in FLC-induced disomy formation and the gene’s impact on FLC sensitivity is incongruent. For example, ERG11, AFR1, GLO3 and SEY1 all contribute to resistance of C. neoformans to fluconazole and are important for FLC-induced disomy (Sionov, et al., 2010) while GCS2 is involved in FLC-induced Chr4 disomy without any impact on FLC resistance (Ngamskulrungroj, et al., 2012). The mechanism by which azoles cause disomy remains elusive. Our results provide further evidence of the relationship between ER integrity and chromosomal disomy under FLC stress. How ER integrity dictates disomy formation remains unclear and requires further studies to resolve this important question of cell biology.
Acknowledgments
This study was supported by funds from the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. We thank A. Varma for critical discussions and reading of the manuscript.
References
- Akins RA. An update on antifungal targets and mechanisms of resistance in Candida albicans. Med Mycol. 2005;43:285–318. doi: 10.1080/13693780500138971. [DOI] [PubMed] [Google Scholar]
- Brands A, Ho TH. Function of a plant stress-induced gene, HVA22. Synthetic enhancement screen with its yeast homolog reveals its role in vesicular traffic. Plant Physiol. 2002;130:1121–1131. doi: 10.1104/pp.007716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordas G, Renken C, Varnai P, et al. Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol. 2006;174:915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, Cruz MC, Sia RA, Allen B, Alspaugh JA, Heitman J. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet Biol. 2000;29:38–48. doi: 10.1006/fgbi.1999.1180. [DOI] [PubMed] [Google Scholar]
- English AR, Zurek N, Voeltz GK. Peripheral ER structure and function. Curr Opin Cell Biol. 2009;21:596–602. doi: 10.1016/j.ceb.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Prinz WA, Rapoport TA. Weaving the web of ER tubules. Cell. 2011;147:1226–1231. doi: 10.1016/j.cell.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Zhu PP, et al. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138:549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Sorrell TC, Dromer F, Fung E, Levitz SM. Cryptococcosis: clinical and biological aspects. Med Mycol. 2000;38(Suppl 1):205–213. [PubMed] [Google Scholar]
- Mondon P, Petter R, Amalfitano G, Luzzati R, Concia E, Polacheck I, Kwon-Chung KJ. Heteroresistance to fluconazole and voriconazole in Cryptococcus neoformans. Antimicrob Agents Chemother. 1999;43:1856–1861. doi: 10.1128/aac.43.8.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngamskulrungroj P, Chang Y, Hansen B, Bugge C, Fischer E, Kwon-Chung KJ. Characterization of the chromosome 4 genes that affect fluconazole-induced disomy formation in Cryptococcus neoformans. PLoS One. 2012;7:e33022. doi: 10.1371/journal.pone.0033022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O, Tange Y, Kurabayashi A. Growth arrest and chromosome instability in aneuploid yeast. Yeast. 2006;23:937–950. doi: 10.1002/yea.1411. [DOI] [PubMed] [Google Scholar]
- Perfect JR, Ketabchi N, Cox GM, Ingram CW, Beiser CL. Karyotyping of Cryptococcus neoformans as an epidemiological tool. J Clin Microbiol. 1993;31:3305–3309. doi: 10.1128/jcm.31.12.3305-3309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect JR, Dismukes WE, Dromer F, et al. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the infectious diseases society of america. Clin Infect Dis. 2010;50:291–322. doi: 10.1086/649858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posteraro B, Sanguinetti M, Sanglard D, et al. Identification and characterization of a Cryptococcus neoformans ATP binding cassette (ABC) transporter-encoding gene, CnAFR1, involved in the resistance to fluconazole. Mol Microbiol. 2003;47:357–371. doi: 10.1046/j.1365-2958.2003.03281.x. [DOI] [PubMed] [Google Scholar]
- Puhka M, Vihinen H, Joensuu M, Jokitalo E. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J Cell Biol. 2007;179:895–909. doi: 10.1083/jcb.200705112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodero L, Mellado E, Rodriguez AC, et al. G484S amino acid substitution in lanosterol 14-alpha demethylase (ERG11) is related to fluconazole resistance in a recurrent Cryptococcus neoformans clinical isolate. Antimicrob Agents Chemother. 2003;47:3653–3656. doi: 10.1128/AAC.47.11.3653-3656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt HD. Dsl1p/Zw10: common mechanisms behind tethering vesicles and microtubules. Trends Cell Biol. 2010;20:257–268. doi: 10.1016/j.tcb.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Schulz TA, Prinz WA. Sterol transport in yeast and the oxysterol binding protein homologue (OSH) family. Biochim Biophys Acta. 2007;1771:769–780. doi: 10.1016/j.bbalip.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Voeltz GK, Rapoport TA. Rough sheets and smooth tubules. Cell. 2006;126:435–439. doi: 10.1016/j.cell.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Sionov E, Chang YC, Garraffo HM, Kwon-Chung KJ. Heteroresistance to fluconazole in Cryptococcus neoformans is intrinsic and associated with virulence. Antimicrob Agents Chemother. 2009;53:2804–2815. doi: 10.1128/AAC.00295-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sionov E, Lee H, Chang YC, Kwon-Chung KJ. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog. 2010;6:e1000848. doi: 10.1371/journal.ppat.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sionov E, Chang YC, Garraffo HM, Dolan MA, Ghannoum MA, Kwon-Chung KJ. Identification of a Cryptococcus neoformans Cytochrome P450 Lanosterol 14alpha-Demethylase (Erg11) Residue Critical for Differential Susceptibility between Fluconazole/Voriconazole and Itraconazole/Posaconazole. Antimicrob Agents Chemother. 2011 doi: 10.1128/AAC.05502-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan DP, Ohvo-Rekila H, Baumann NA, Beh CT, Menon AK. Sterol trafficking between the endoplasmic reticulum and plasma membrane in yeast. Biochem Soc Trans. 2006;34:356–358. doi: 10.1042/BST0340356. [DOI] [PubMed] [Google Scholar]
- Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Varma A, Kwon-Chung KJ. Heteroresistance of Cryptococcus gattii to fluconazole. Antimicrob Agents Chemother. 2010;54:2303–2311. doi: 10.1128/AAC.00153-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- Washton H. Review of fluconazole: a new triazole antifungal agent. Diagn Microbiol Infect Dis. 1989;12:229S–233S. doi: 10.1016/0732-8893(89)90141-7. [DOI] [PubMed] [Google Scholar]
- Yoneda A, Doering TL. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol Biol Cell. 2006;17:5131–5140. doi: 10.1091/mbc.E06-08-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]