Abstract
Trisomies 18 and 21 are the two most common live born autosomal aneuploidies in humans. While the anatomic abnormalities in affected fetuses are well documented, the dysregulated biological pathways associated with the development of the aneuploid phenotype are less clear. Amniotic fluid (AF) cell-free RNA is a valuable source of biological information obtainable from live fetuses. In this study, we mined gene expression data previously produced by our group from mid-trimester AF supernatant samples. We identified the euploid, trisomy 18 and trisomy 21 AF transcriptomes, and analyzed them with a particular focus on the nervous system. We used multiple bioinformatics resources, including DAVID, Ingenuity Pathway Analysis, and the BioGPS Gene Expression Atlas. Our analyses confirmed that AF supernatant from aneuploid fetuses is enriched for nervous system gene expression and neurological disease pathways. Tissue analysis showed that fetal brain cortex and Cajal–Retzius cells were significantly enriched for genes contained in the AF transcriptomes. We also examined AF transcripts known to be dysregulated in aneuploid fetuses compared with euploid controls and identified several brain-specific transcripts among them. Many of these genes play critical roles in nervous system development. NEUROD2, which was downregulated in trisomy 18, induces neurogenic differentiation. SOX11, downregulated in trisomy 21, is a transcription factor that is essential for pan-neuronal protein expression and axonal growth of sensory neurons. Our results show that whole transcriptome analysis of cell-free RNA in AF from live pregnancies permits discovery of biomarkers of abnormal human neurodevelopment and advances our understanding of the pathophysiology of aneuploidy.
Introduction
Trisomies 18 and 21 are the two most common live born autosomal aneuploidies in humans. While the anatomic abnormalities in affected fetuses have been well documented with prenatal sonography (Bianchi et al. 2010), the dysregulated biological pathways associated with the aneuploid phenotypes are less clear. To study human fetal development, gene expression studies have largely relied on post-partum or post-mortem tissue samples. Samples from live fetuses have almost exclusively been performed using cultured amniocytes or chorionic villi (Altug-Teber et al. 2007; Chung et al. 2005; Chou et al. 2008). These sample sources are all inherently limited in their ability to reflect in vivo physiology. Mid-trimester amniotic fluid (AF) is valuable because it is a pure fetal biofluid that reflects ongoing in utero physiology. The residual cell-free RNA that is present after amniocytes have been removed for clinical diagnosis is an excellent source of biological information (Hui and Bianchi 2011). Prior studies from our group have shown that cell-free RNA from AF can be successfully isolated and used for global gene expression studies of abnormal pregnancies (Larrabee et al. 2005; Slonim et al. 2009; Koide et al. 2011).
In recent work, we analyzed the AF transcriptome in euploid fetuses by mining gene expression data from 12 mid-trimester AF supernatants (Hui et al. 2012). Our results suggested that the cell-free RNA in AF derives from multiple tissues. More specifically, functional analysis indicated that nervous system development and function is enriched in the euploid AF transcriptome. Despite our lack of knowledge about their precise route of entry and their in vivo half-life, brain-derived transcripts may be a novel source of biomarkers of nervous system development that are uniquely accessible from living fetuses. These findings prompted us to establish whether AF supernatant could be a useful tool for studying fetal neurodevelopment in aneuploid pregnancies.
In this study, we mined gene expression data previously produced by our group from aneuploid AF supernatant samples (Koide et al. 2011; Slonim et al. 2009), focusing on the transcripts associated with nervous system development. We newly created the trisomy 18 and 21 transcriptomes, which consist of the universally detected transcripts in AF from mid-trimester trisomy 18 and trisomy 21 fetuses, respectively. Our aims were to determine if nervous system gene expression is enriched in AF supernatant from aneuploid fetuses, and to identify differentially regulated nervous system-specific transcripts in AF that may represent potential biomarkers of abnormal neurodevelopment.
Materials and methods
The euploid, trisomy 21 and trisomy 18 transcriptomes
We newly analyzed gene expression datasets from mid-trimester euploid and aneuploid AF samples produced in prior microarray studies from our group. These data are publicly available at http://www.ncbi.nlm.nih.gov/geo/ (GSE25634, GSE16176, and GSE33168). Institutional review board approval was obtained for the initial collection and analysis of all samples. The methods of sample collection, RNA extraction, fragmentation, labeling, and hybridization have been previously described (Slonim et al. 2009). In brief, RNA was extracted from 10 ml of AF supernatant from women undergoing fetal testing for clinical indications. The trisomy 21 transcriptome was derived from five male and two female fetuses with trisomy 21 (16–21 weeks), the trisomy 18 transcriptome from five female fetuses with trisomy 18 (17–20 weeks) and the euploid transcriptome from six male and six female euploid fetuses (16–21 weeks). There was no pooling of samples. The final amplified cDNA products were hybridized to Affymetrix U133 Plus 2.0 microarrays (Affymetrix, Santa Clara, CA). Each AF sample was anonymized before being received in the research laboratory. Only gestational age and karyotype were available. No data were available for the presence of structural anomalies at the time of amniocentesis.
Data were normalized using the threestep function from the affyPLM package in Bioconductor (version 2.8.1) (Gentleman et al. 2004), with ideal-mismatch background/signal adjustment, quantile normalization, and the Tukey biweight summary method (Bolstad 2004). This summary method includes a logarithmic transformation that improves the normality of the data. To obtain detection calls consistent with those produced by Affymetrix® 5.0 software, we used the mas5calls function from the Bioconductor affy package.
We identified the euploid, trisomy 18 and 21 transcriptomes by selecting those transcripts present in 100 % of euploid, trisomy 18 and trisomy 21 AF supernatant samples, respectively. The enrichment of specific tissue expression was explored using multiple tools. See Table 1 for summary of study methods.
Table 1.
Summary of methods
| Identification of the euploid, trisomy 21 and 18 AF transcriptomes |
| Trisomy 21 AF transcriptome compared with Down syndrome meta analysisa |
| DAVIDb tissue expression analysis of euploid, trisomy 21 and 18 AF transcriptomes |
| BioGPS analysis for tissue-specific genes in the trisomy 21 and 18 AF transcriptomesc |
| IPAd functional analysis of trisomy 21 and 18 AF transcriptomesc |
| Genes differentially regulated in trisomy 21 and 18 compared with euploid fetusese |
| Identification of nervous system-specific genes that were up or down regulated in aneuploid compared with euploid fetuses |
Database for annotation, visualization and integrated discovery
We have recently reported these results for the euploid AF transcriptome (Hui et al. 2012)
Ingenuity Pathway Analysis™
As previously published by our group in Koide et al. (2011), Slonim et al. (2009)
Comparison of the trisomy 21 transcriptome with a Down syndrome gene expression meta analysis
To determine if our transcriptomic approach produced biologically relevant data, we compared the trisomy 21 AF transcriptome with a meta analysis of 45 trisomy 21 gene expression studies that included many different tissue samples (Vilardell et al. 2011). This meta analysis by Vilardell et al. identified 324 genes with significant genomewide dosage effects in Down syndrome. The authors noted that their meta analysis included many brain experiments and was therefore able to detect a high fraction of genes related to neuron development, synapsis and neurodegeneration. We identified the individual genes and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in common between the trisomy 21 AF transcriptome and the meta analysis results. The KEGG pathways were identified using the functional annotation cluster tool in DAVID and a Benjamini–Hochberg corrected P value with a cutoff value <0.05.
Tissue expression analyses
We used the tissue expression function in the Database for Annotation, Visualization and Integrated Discovery (DAVID) to identify tissues that most highly expressed the genes contained in each AF transcriptome (http://david.abcc.ncifcrf.gov/) (Dennis et al. 2003). This tool is designed to facilitate biomarker identification and gene expression pattern discovery. DAVID integrates tissue expression databases from a publicly available Gene Expression Atlas (Su et al. 2004) (http://biogps.org/index.html), the Cancer Genome Anatomy Project (Boon et al. 2002) (http://cgap.nci.nih.gov/), Unigene (ftp://ftp.ncbi.nih.gov/repository/UniGene/Homo_sapiens/) and UniProt (Consortium 2011) (http://www.uniprot.org) to allow the identification of the most enriched gene expression patterns across hundreds of normal and diseased tissues for any given gene list. For each resource, the expression values for a given gene in all tissues are ranked from greatest to least. All tissues with an expression value in the top quartile are associated with that gene and an enrichment P value called the EASE score (a modified Fisher exact P value) is calculated for that tissue (Hosack et al. 2003). DAVID also calculates a more conservative P value using the Benjamini–Hochberg correction to control the false discovery rate of family-wise enriched terms. We defined results as significant if the tissue term had a Benjamini–Hochberg corrected P value <0.05. We uploaded the euploid, trisomy 21 and trisomy 18 transcriptome gene lists to DAVID. We reported the top ranked tissues for each database and karyotype, as well as the individual neurological tissues that met statistical significance.
Identification of tissue-specific genes in the aneuploid transcriptomes
The BioGPS Gene Expression Atlas at http://biogps.org identified genes with tissue-specific expression patterns in the trisomy 21 and 18 transcriptomes. This atlas of the human protein-encoding transcriptome used Affymetrix Human Genome-U133A and custom human arrays to map gene expression profiles in 78 normal human tissues (Su et al. 2004). We chose this resource because of its Affymetrix microarray platform, coverage of normal adult and fetal tissues, high reproducibility and good correlation between transcript levels and protein abundance (Kislinger et al. 2006). The BioGPS Gene Expression Atlas allowed us to assess the gene expression patterns of individual Affymetrix probe sets in the trisomy 18 and 21 transcriptomes. We categorized probe sets as highly organ specific if they mapped to a single organ with an expression value >30 multiples of the median (MoM) and had no unrelated tissue expression greater than one-third of the maximum expression level.
Functional analyses of the trisomy 18 and 21 transcriptomes
The web-based software tool Ingenuity Pathway Analysis Version 9.0 (IPA) (Content Version 11904312) was used for the biological interpretation of the trisomy 18 and trisomy 21 AF transcriptome gene lists (Ingenuity Systems, Redwood City, CA, USA; http://www.ingenuity.com). Functional analysis of the euploid transcriptome has been previously reported by our group and was not repeated here (Hui et al. 2012). IPA uses a manually curated repository of biological interactions and functional annotations to identify the most statistically significant signaling pathways and biological processes represented in a given gene set. The right-tailed Fisher’s exact test is used by IPA to calculate a P value representing the probability that a biological function not really relevant to the AF transcriptome is reported as relevant. Controlling for multiple pathway testing is performed with the Benjamini–Hochberg correction. Diseases and disorder pathways that contained at least one functional annotation with a Benjamini–Hochberg corrected P value <0.05 were considered statistically significant.
Identification of differentially regulated tissue-specific genes in aneuploid fetuses
Genes that were significantly differentially expressed in trisomies 18 or 21 AF supernatants compared with euploid controls matched for gestational age and fetal sex were obtained from prior published work from our group (Slonim et al. 2009; Koide et al. 2011). We used the BioGPS Gene Expression Atlas to identify the differentially regulated genes that were specifically expressed by the nervous system according to the method described above for the aneuploid transcriptomes.
Results
In the trisomy 21 dataset, 1,644 probe sets received a present call in all seven samples, corresponding to 1,184 individual genes in the transcriptome (Supplemental Table 1). In the trisomy 18 dataset, 1,076 probe sets were present in all five samples, corresponding to 746 genes (Supplemental Table 2). The euploid dataset contained 806 probe sets that were present in all 12 samples, corresponding to 536 genes. Nervous system gene expression was consistently represented in our analyses of these datasets.
Comparison of the trisomy 21 AF transcriptome to the Down syndrome gene expression meta analysis
Results from the statistical meta analysis of 45 heterogeneous trisomy 21 data sets were compared to the trisomy 21 AF transcriptome (Vilardell et al. 2011). Fifty of 324 (15.4 %) genes identified in the meta analysis were present in trisomy 21 AF transcriptome, including genes physically located on chromosome 21 (APP, SOD1, DYRK1A, and RCAN1) (Supplemental Table 3). In contrast, only 17 (5.2 %) of the meta analysis genes were universally detectable in the euploid AF samples.
Analysis of KEGG pathways in the trisomy 21 AF transcriptome using the functional annotation tool in DAVID showed one statistically significant functional cluster (Enrichment score >1.3). This single cluster contained three of the four neuropathological KEGG pathways present in the meta analysis gene list (Table 2).
Table 2.
Significant functional cluster of KEGG pathways in the trisomy 21 AF transcriptome
| Annotation Cluster 1 Category |
Enrichment score: 6.520a Term |
Count | P value* |
|---|---|---|---|
| KEGG_PATHWAY | hsa05016: Huntington’s disease | 41 | 1.46E–06 |
| KEGG_PATHWAY | hsa05012: Parkinson’s disease | 32 | 1.03E–06 |
| KEGG_PATHWAY | hsa00190: Oxidative phosphorylation | 29 | 9.79E–05 |
| KEGG_PATHWAY | hsa05010: Alzheimer’s disease | 33 | 3.46E–04 |
Benjamini–Hochberg corrected P value
Group Enrichment Score is the geometric mean (in −log scale) of member’s P values in a corresponding annotation cluster, and is used to rank their biological significance
Tissue expression analyses
The DAVID tissue expression analysis showed that transcripts from multiple organs are enriched in AF, including the fetal nervous system, embryonic stem cells, pooled cell lines, umbilical cord, and epithelia (Table 3). Fetal brain cortex and Cajal–Retzius cells consistently ranked in the top four tissues in the UniProt database (Table 4). A Unigene expressed sequence tag library derived primarily from fetal cochlea was also highly ranked. Many other nervous system tissues met statistical significance across all DAVID databases, including adult caudate nucleus, whole brain, whole fetal brain, spinal cord, retina and many neurological malignancies (Supplemental Table 4).
Table 3.
DAVID tissue expression results for AF transcriptomes: top ranking tissues by database and karyotype
| Database | Euploid | Trisomy 21 | Trisomy 18 |
|---|---|---|---|
| BioGPS Gene Expression Atlas | Caudate nucleus | Caudate nucleus | Caudate nucleus |
| UniProt tissue | Fetal brain cortex | Epithelium | Epithelium |
| CGAP ESTa | Pooled tissue normalc | Pooled tissue normalc | Embryonic tissue |
| CGAP SAGEb | Embryonic stem cell | Embryonic stem cell | Embryonic stem cell |
| Unigene EST | Umbilical cord | Eard | Eard |
Cancer Genome Anatomy Project expressed sequence tag library
Cancer Genome Anatomy Project serial analysis of gene expression library
Library 10550 (dbEST ID) pooled from 40 cell lines including brain
Only two Unigene EST ear libraries contained >1,000 sequences. Both were derived from fetal cochlea: Lib.371 Morton Fetal Cochlea (14805) and Lib.18222 Human Fetal Cochlea cDNA Library (3880)
Table 4.
Top five terms in UniProt tissue expression chart by karyotype
| Karyotype | UniProt KB term | No. of genes | P value* |
|---|---|---|---|
| Euploid | Fetal brain cortex | 41 | 6.34E–18 |
| Cajal–Retzius cella | 35 | 2.83E–14 | |
| Epithelium | 154 | 1.23E–13 | |
| B-cell lymphoma | 24 | 5.86E–10 | |
| Skin | 102 | 1.71E–07 | |
| Trisomy 21 | Epithelium | 342 | 1.21E–35 |
| Placenta | 377 | 6.06E–27 | |
| Fetal brain cortex | 63 | 4.24E–22 | |
| Cajal–Retzius cella | 59 | 3.20E–21 | |
| Skin | 216 | 1.26E–16 | |
| Trisomy 18 | Epithelium | 224 | 1.11E–23 |
| Fetal brain cortex | 50 | 1.37E–20 | |
| Cajal–Retzius cella | 44 | 2.39E–17 | |
| Skin | 140 | 2.05E–10 | |
| Placenta | 220 | 2.24E–10 |
Benjamini–Hochberg corrected modified Fisher exact P value
See Supplemental Table 5 for genes expressed by Cajal–Retzius cells that were detected in the euploid, trisomy 21 and trisomy 18 AF transcriptomes
Tissue-specific genes in the aneuploid transcriptomes
The search for tissue-specific genes using the Gene Expression Atlas at BioGPS identified genes in the trisomy 21 and 18 AF transcriptomes that were specifically expressed by the nervous system. A total of 61 organ-specific genes were identified in the trisomy 21 AF transcriptome gene list. The most highly represented tissues were hematopoetic/immune cells (19 genes), epithelial cells from skin, oral cavity and respiratory tract (14 genes) and nervous system (12 genes). These results are consistent with the tissues identified in the DAVID tissue expression analysis. The trisomy 18 transcriptome contained six nervous system-specific genes among the total of 36 tissue-specific genes. The complete lists of tissue-specific genes are available in Supplemental Tables 6 and 7.
Functional analyses of trisomy 21 and trisomy 18 AF transcriptomes
IPA identified neurological disease as the first and second most highly enriched disease pathway in the trisomy 21 and 18 datasets, respectively (Tables 5, 6). There was a considerable overlap of specific functional annotations within the neurological disease category between the two aneuploid groups (Supplemental Table 8). The infectious diseases annotations that were highly ranked in both groups related primarily to immune mechanisms against infection or cell surface proteins involved in the mechanisms of viral infection. We specifically excluded the biological category of cancer because cell proliferation functions that are normal in the developing human cause the IPA disease category “cancer” to be highly significant in fetal datasets (unpublished observations).
Table 5.
Trisomy 21 IPA significant diseases and disorders
| Category | P value range* | No. of genes |
|---|---|---|
| Neurological disease | 2.76E–12 to 4.61E–02 | 201 |
| Dermatological diseases and conditions | 7.05E–06 to 3.06E–02 | 114 |
| Infectious disease | 6.52E–05 to 4.20E–02 | 177 |
| Reproductive system disease | 1.97E–04 to 2.51E–04 | 96 |
| Connective tissue disorders | 7.38E–03 to 4.61E–02 | 6 |
| Respiratory disease | 4.61E–02 | 6 |
| Cardiovascular disease | 4.20E–02 | 4 |
Benjamini–Hochberg corrected right-tailed Fisher exact P value ranges for individual functional annotations within each category
Table 6.
Trisomy 18 IPA significant diseases and disorders
| Category | P value range* | No. of genes |
|---|---|---|
| Infectious disease | 1.61E–05 to 2.82E–02 | 119 |
| Neurological disease | 3.80E–05 to 3.76E–02 | 107 |
| Dermatological diseases and conditions | 1.67E–03 to 3.79E–03 | 45 |
| Skeletal and muscular disorders | 8.49E–03 to 3.45E–02 | 9 |
| Connective tissue disorders | 2.00E–02 to 4.64E–02 | 5 |
Benjamini–Hochberg corrected right-tailed Fisher exact P value ranges for individual functional annotations within each category
Identification of differentially regulated tissue-specific genes in aneuploid fetuses
There was significant differential regulation of several nervous system-specific genes in trisomy 21 and 18 fetuses, including transcription factors with critical roles in neuronal differentiation. These genes represent potential biomarkers of abnormal neurodevelopment (Table 7). Of the 318 genes that were differentially regulated in trisomy 21 fetuses compared with matched euploid controls, 22 were tissue specific (Supplemental Table 9). Four genes of these genes were specifically expressed by the nervous system: SOX11 and DAAM2 were downregulated, and MEF2C and CELSR2 were upregulated in trisomy 21 fetuses, respectively.
Table 7.
Differentially regulated nervous system-specific genes detected in trisomy 18 and 21 AF supernatants
| Differential regulationa |
Gene | Gene name | Tissues expressing gene >30 MoMb |
Gene function in nervous systemc |
|---|---|---|---|---|
| Downregulated in trisomy 18 | PTPRD | Protein tyrosine phosphatase, receptor type, D | Fetal brain, prefrontal cortex, hypothalamus, amygdala, spinal cord | Implicated in axonal growth and guidance of motor neurons in mammals |
| SOBP | Sine oculis binding protein homolog (Drosophila) | Fetal brain, amygdala, prefrontal cortex, whole brain | The protein encoded by this gene is a nuclear zinc finger protein that is involved in development of the cochlea. Defects in this gene linked to intellectual disability | |
| NEUROD2 | Neurogenic differentiation 2 | Cerebellum peduncles, fetal brain, cerebellum | Expression of this gene can induce transcription from neuron-specific promoters. The product of the human gene can induce neurogenic differentiation in non-neuronal cells in Xenopus embryos, and is thought to play a role in the determination and maintenance of neuronal cell fates | |
| Upregulated in trisomy 18 | PLEKHA4 | Pleckstrin homology domain containing, family A (phosphoinositide binding-specific) member 4 | Olfactory bulb | The second messenger phospatidylinositol 3-phosphate regulates a plethora of cellular processes |
| PRPH2 | Peripherin 2 (retinal degeneration, slow) | Pineal (day), pineal (night) | The proteins mediate signal transduction events that play a role in the regulation of cell development, activation, growth and motility. This encoded protein is a cell surface glycoprotein found in the outer segment of both rod and cone photoreceptor cells. Defects in this gene are associated with both central and peripheral retinal degenerations | |
| GPM6A | Glycoprotein M6A | Amygdala, fetal brain, prefrontal cortex, occipital lobe, whole brain | An abundant cell surface protein on neurons in the central nervous system associated with differentiation and neuronal migration (Michibata et al. 2009). Downregulated in the brains of transgenic mice that overexpress tau, the neuronal phosphoprotein associated with Alzheimer’s disease (Woo et al. 2010) | |
| Downregulated in trisomy 21 | SOX11 | SRY (sex determining region Y)-box 11 | Fetal brain | A transcription factor involved in the regulation of embryonic development and in the determination of the cell fate; promotes neuronal maturation in the developing neural tube and is essential for establishment of pan-neuronal protein expression (Bergsland et al. 2006) |
| DAAM2 | Disheveled associated activator of morphogenesis 2 | Spinal cord, hypothalamus, prefrontal cortex, retina, thalamus, caudate nucleus | Disheveled-binding protein transducing WNT signals to the PCP pathway. Involved in pivotal steps in neuronal cell differentiation and movement (Kida et al. 2004) | |
| Upregulated in trisomy 21 | MEF2C | Myocyte enhancer factor 2C | Fetal brain | Crucial for normal neuronal development, distribution, and electrical activity in the neocortex. May also be involved in neurogenesis and in the development of cortical architecture |
| CELSR2 | Cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila) | Whole brain, prefrontal cortex, amygdala, medulla oblongata, cingulate cortex, parietal lobe | Receptor involved in cell/cell signaling during nervous system formation |
As identified from Slonim et al. 2009, Koide et al. 2011
BioGPS Gene Expression Atlas
Entrez gene or UniProt summaries unless references given
Of the 264 genes that were differentially regulated in trisomy 18 fetuses compared with matched euploid controls, there were 13 organ-specific genes (Supplemental Table 10). Three neurological tissue-specific genes were upregulated (PLEKHA4, PRPH2, and GPM6Z) and three were downregulated (NEUROD2, SOBP and PTPRD). There was no overlap between the trisomy 21 and trisomy 18 differentially regulated nervous system-specific genes.
Discussion
We applied multiple bioinformatic tools to explore the gene expression information contained in AF cell-free RNA from euploid, trisomy 21 and trisomy 18 fetuses. In this study, we focused on the neurodevelopmental data in the aneuploid groups because of the importance of developmental disability to the phenotypes of trisomy 21 and 18. We showed that the transcripts consistently present in AF are enriched for nervous system tissue expression and identified nervous-specific genes that are differentially regulated in aneuploid fetuses compared with euploid controls.
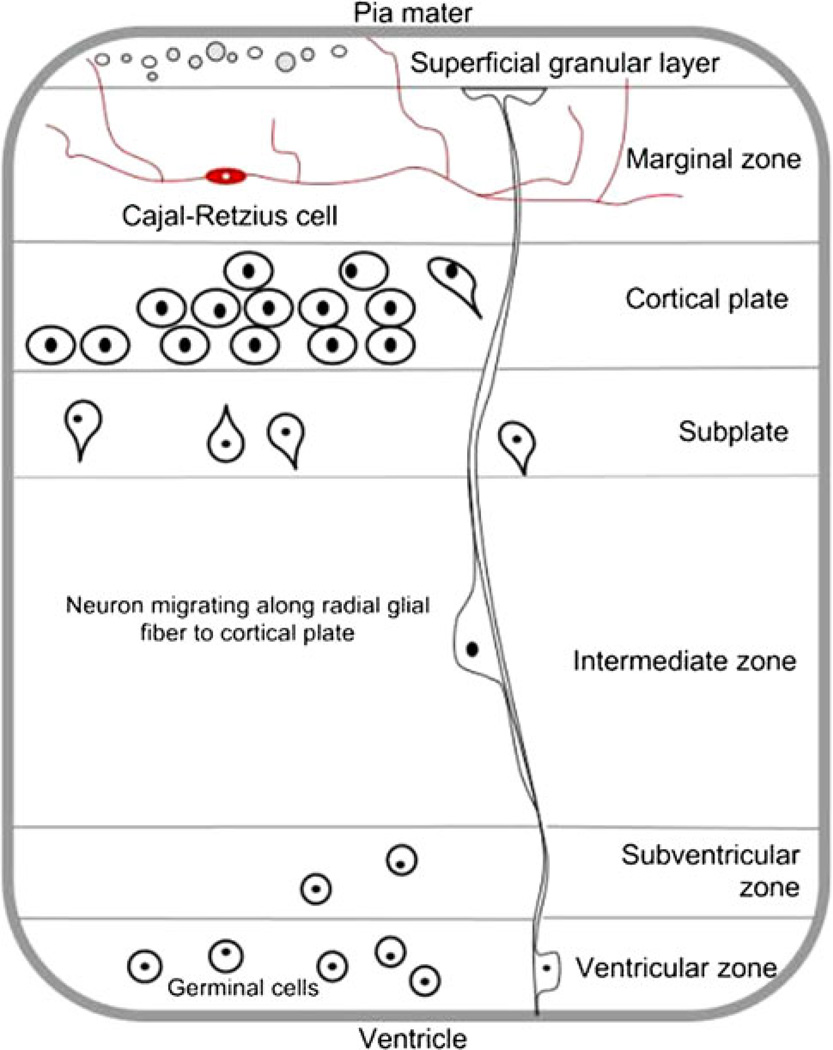
The significant result for the KEGG pathway for Alzheimer’s disease in both the trisomy 21 AF transcriptome and the Down syndrome meta analysis is consistent with the known neuropathology of Down syndrome. This suggests that AF supernatant reflects real tissue biology and is therefore valid for studying human neurodevelopment. Of particular interest was the finding from the DAVID tissue expression analysis that Cajal–Retzius cells and fetal cortex had highly enriched expression of genes in the AF transcriptomes. Cajal–Retzius cells are reelin-producing neurons found in the marginal zone of the fetal brain that play an essential role in neuronal migration and cortical development (Fig. 1 Cajal–Retzius cell in the developing human fetal cerebral cortex at 13 weeks’ gestation). The fact that nervous system tissues were enriched in AF across multiple independent databases gives us greater confidence that these findings reflect actual fetal biology and are not the result of noise inherent in high throughput gene expression data.
Fig. 1.
Cajal–Retzius cell in the developing human fetal cerebral cortex at 13 weeks of gestation
The significant neurological disease annotations obtained in the IPA functional analysis of the aneuploid transcriptomes provided additional confirmation that nervous system pathophysiology can be reflected in AF. However, it is important to note that the functional annotation of diseases within IPA is largely based on adult studies. We do not propose that all the features of specific disorders such as Huntington’s disease or Parkinson’s disease occur in aneuploid fetuses. Rather, we believe that the significant results for these categories are a result of the fact that many of the genes involved in these adult diseases are essential for normal neurodevelopment and are dysregulated in aneuploid fetuses.
We identified brain-specific genes that were dysregulated in aneuploid fetuses compared with euploid controls. Many of these genes play critical roles in nervous system development, as summarized in Table 7. NEUROD2, which was downregulated in trisomy 18, is most highly expressed in the marginal zone in week 19 of human gestation (Franklin et al. 2001) and induces neurogenic differentiation (Ravanpay et al. 2010; Mattar et al. 2008). SOBP, also downregulated in Trisomy 18, is involved in development of the cochlea, and defects in this gene have been linked to developmental disability (Birk et al. 2010; Chen et al. 2008). Both hearing and intellectual impairment are consistent features of the trisomy 18 phenotype. SOX11, downregulated in trisomy 21, is a transcription factor expressed in the embryonic central and peripheral nervous systems and is of critical importance for the establishment of pan-neuronal protein expression (Bergsland et al. 2006) and axonal growth of embryonic sensory neurons (Lin et al. 2011). These findings, uniquely derived from live human fetuses, provide novel information about the altered neurodevelopmental gene expression in trisomies 18 and 21.
There is still a general paucity of information on the biology of cell-free RNA in AF (Hui and Bianchi 2011). The mechanisms by which fetal transcripts enter the AF are still unknown. It is possible that differential entry or degradation of cell-free transcripts could influence the results of our analysis. Furthermore, cell-free RNA is inherently more degraded and generally produces lower hybridization rates than cellular RNA. Fresh tissue would be an ideal source of gene expression information, but given the practical issues associated with human studies, AF provides an important window on fetal development.
Conclusions
Multiple independent bioinformatics analyses confirm that AF supernatant of aneuploid fetuses is enriched for nervous system gene expression and neurological disease pathways. We identified nervous system-specific transcription factors with key roles in brain development that are differentially regulated in aneuploid fetuses compared with euploid controls. Whole transcriptome analysis of cell-free RNA in AF from live pregnancies permits discovery of biomarkers of abnormal human neurodevelopment and advances understanding of the pathophysiology of human aneuploidy.
Supplementary Material
Acknowledgments
The authors thank Janet Cowan, PhD, and Uma Tantravahi, PhD, who provided the AF supernatant samples. Financial support was provided by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (R01 HD 42053-09 to Dr Bianchi and R01 HD 058880 to Dr Slonim); the University of Sydney Medical School (Albert S. McKern Research Scholarship to Dr Hui); and the Royal Australian and New Zealand College of Obstetricians and Gynaecologists Research Foundation (Fotheringham Fellowship to Dr Hui).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00439-012-1195-x) contains supplementary material, which is available to authorized users.
The authors declare that they have no conflicts of interest to disclose. This study complies with current laws of the United States of America.
Contributor Information
Lisa Hui, Email: lisahui77@gmail.com, Mother Infant Research Institute and the Division of Genetics, Department of Pediatrics, The Floating Hospital for Children at Tufts Medical Center, 800 Washington St, Boston, MA 02111, USA; The Department of Obstetrics, Gynecology and Neonatalogy, University of Sydney, Sydney, NSW, Australia.
Donna K. Slonim, Department of Computer Science, Tufts University, Medford, MA, USA Department of Pathology, Tufts University School of Medicine, Boston, MA, USA.
Heather C. Wick, Department of Computer Science, Tufts University, Medford, MA, USA
Kirby L. Johnson, Mother Infant Research Institute and the Division of Genetics, Department of Pediatrics, The Floating Hospital for Children at Tufts Medical Center, 800 Washington St, Boston, MA 02111, USA
Keiko Koide, Mother Infant Research Institute and the Division of Genetics, Department of Pediatrics, The Floating Hospital for Children at Tufts Medical Center, 800 Washington St, Boston, MA 02111, USA.
Diana W. Bianchi, Mother Infant Research Institute and the Division of Genetics, Department of Pediatrics, The Floating Hospital for Children at Tufts Medical Center, 800 Washington St, Boston, MA 02111, USA
References
- Altug-Teber O, Bonin M, Walter M, Mau-Holzmann UA, Dufke A, Stappert H, Tekesin I, Heilbronner H, Nieselt K, Riess O. Specific transcriptional changes in human fetuses with autosomal trisomies. Cytogenet Genome Res. 2007;119(3–4):171–184. doi: 10.1159/000112058. [DOI] [PubMed] [Google Scholar]
- Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Gene Dev. 2006;20(24):3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi DW, Crombleholme TM, D’Alton ME, Malone FD. Fetology. 2nd edn. USA: McGraw-Hill Companies, Inc; 2010. pp. 910–925. [Google Scholar]
- Birk E, Har-Zahav A, Manzini CM, Pasmanik-Chor M, Kornreich L, Walsh CA, Noben-Trauth K, Albin A, Simon AJ, Colleaux L, Morad Y, Rainshtein L, Tischfield DJ, Wang P, Magal N, Maya I, Shoshani N, Rechavi G, Gothelf D, Maydan G, Shohat M, Basel-Vanagaite L. SOBP is mutated in syndromic and nonsyndromic intellectual disability and is highly expressed in the brain limbic system. Am J Hum Genet. 2010;87(5):694–700. doi: 10.1016/j.ajhg.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM. Low level analysis of high-density oligonucleotide array data: background, normalization and summarization. Berkeley: University of California; 2004. [Google Scholar]
- Boon K, Osorio EC, Greenhut SF, Schaefer CF, Shoemaker J, Polyak K, Morin PJ, Buetow KH, Strausberg RL, De Souza SJ, Riggins GJ. An anatomy of normal and malignant gene expression. Proc Natl Acad Sci USA. 2002;99(17):11287–11292. doi: 10.1073/pnas.152324199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Montcouquiol M, Calderon R, Jenkins NA, Copeland NG, Kelley MW, Noben-Trauth K. Jxc1/Sobp, encoding a nuclear zinc finger protein, is critical for cochlear growth, cell fate, and patterning of the organ of corti. J Neurosci. 2008;28(26):6633–6641. doi: 10.1523/JNEUROSCI.1280-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CY, Liu LY, Chen CY, Tsai CH, Hwa HL, Chang LY, Lin YS, Hsieh FJ. Gene expression variation increase in trisomy 21 tissues. Mamm Genome. 2008;19(6):398–405. doi: 10.1007/s00335-008-9121-1. [DOI] [PubMed] [Google Scholar]
- Chung IH, Lee SH, Lee KW, Park SH, Cha KY, Kim NS, Yoo HS, Kim YS, Lee S. Gene expression analysis of cultured amniotic fluid cell with Down syndrome by DNA microarray. J Korean Med Sci. 2005;20(1):82–87. doi: 10.3346/jkms.2005.20.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium U. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. ((Database issue)) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- Franklin A, Kao A, Tapscott S, Unis A. NeuroD homologue expression during cortical development in the human brain. J Child Neurol. 2001;16(11):849–853. doi: 10.1177/08830738010160111201. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4(10):R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui L, Bianchi DW. Cell-free fetal nucleic acids in amniotic fluid. Hum Reprod Update. 2011;17(3):362–371. doi: 10.1093/humupd/dmq049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui L, Slonim DK, Wick HC, Johnson KL, Bianchi DW. The amniotic fluid transcriptome: a source of novel information about human fetal development. Obstet Gynecol. 2012;119(1):111–118. doi: 10.1097/AOG.0b013e31823d4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida Y, Shiraishi T, Ogura T. Identification of chick and mouse Daam1 and Daam2 genes and their expression patterns in the central nervous system. Brain Res Dev Brain Res. 2004;153(1):143–150. doi: 10.1016/j.devbrainres.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Kislinger T, Cox B, Kannan A, Chung C, Hu P, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, Rossant J, Hughes TR, Frey B, Emili A. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125(1):173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- Koide K, Slonim DK, Johnson KL, Tantravahi U, Cowan JM, Bianchi DW. Transcriptomic analysis of cell-free fetal RNA suggests a specific molecular phenotype in trisomy 18. Hum Genet. 2011;129(3):295–305. doi: 10.1007/s00439-010-0923-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrabee PB, Johnson KL, Lai C, Ordovas J, Cowan JM, Tantravahi U, Bianchi DW. Global gene expression analysis of the living human fetus using cell-free messenger RNA in amniotic fluid. JAMA. 2005;293(7):836–842. doi: 10.1001/jama.293.7.836. [DOI] [PubMed] [Google Scholar]
- Lin L, Lee VM, Wang Y, Lin JS, Sock E, Wegner M, Lei L. Sox11 regulates survival and axonal growth of embryonic sensory neurons. Dev Dyn. 2011;240(1):52–64. doi: 10.1002/dvdy.22489. [DOI] [PubMed] [Google Scholar]
- Mattar P, Langevin LM, Markham K, Klenin N, Shivji S, Zinyk D, Schuurmans C. Basic helix-loop-helix transcription factors cooperate to specify a cortical projection neuron identity. Mol Cell Biol. 2008;28(5):1456–1469. doi: 10.1128/MCB.01510-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michibata H, Okuno T, Konishi N, Kyono K, Wakimoto K, Aoki K, Kondo Y, Takata K, Kitamura Y, Taniguchi T. Human GPM6A is associated with differentiation and neuronal migration of neurons derived from human embryonic stem cells. Stem Cells & Dev. 2009;18(4):629–639. doi: 10.1089/scd.2008.0215. [DOI] [PubMed] [Google Scholar]
- Ravanpay AC, Hansen SJ, Olson JM. Transcriptional inhibition of REST by NeuroD2 during neuronal differentiation. Mol Cell Neurosci. 2010;44(2):178–189. doi: 10.1016/j.mcn.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slonim DK, Koide K, Johnson KL, Tantravahi U, Cowan JM, Jarrah Z, Bianchi DW. Functional genomic analysis of amniotic fluid cell-free mRNA suggests that oxidative stress is significant in Down syndrome fetuses. Proc Natl Acad Sci USA. 2009;106(23):9425–9429. doi: 10.1073/pnas.0903909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101(16):6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardell M, Rasche A, Thormann A, Maschke-Dutz E, Perez-Jurado LA, Lehrach H, Herwig R. Meta-analysis of heterogeneous Down syndrome data reveals consistent genome-wide dosage effects related to neurological processes. BMC Genomics. 2011;12:229. doi: 10.1186/1471-2164-12-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J-M, Park SJ, Kang HI, Kim BG, Shim SB, Jee SW, Lee SH, Sin JS, Bae CJ, Jang MK, Cho C, Hwang DY, Kim CK. Characterization of changes in global gene expression in the brain of neuron-specific enolase/human Tau23 transgenic mice in response to overexpression of Tau protein. Int J Mol Med. 2010;25(5):667–675. doi: 10.3892/ijmm_00000390. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.