Summary
The RNA exosome is an essential multi-subunit ribonuclease (RNase) that contributes to cytoplasmic and nuclear RNA decay and quality control. The nine-subunit exosome core (Exo9) features a prominent central channel formed by stacked asymmetric rings of six RNase PH-like proteins and three S1/KH domain proteins. Exo9 is catalytically inert but associates with Rrp44, an endoribonuclease and processive 3’→5’ exoribonuclease, and Rrp6, a distributive 3’→5’ exoribonuclease. We show that Exo9 and its central channel modulate all three yeast exosome RNase activities because channel occlusion attenuates RNA binding and RNase activities in vitro and fails to complement exosome functions in vivo. We find that Rrp6 stimulates Rrp44 RNase activities and that Rrp6 is inhibited by a mutation in the Rrp44 exoribonuclease active site in eleven-subunit nuclear exosomes. These results suggest the exosome core and central channel is essential because it modulates each of the known RNase activities of the yeast RNA exosome.
Introduction
The eukaryotic RNA exosome plays an essential role in RNA decay, quality control, and maturation. Messenger RNAs can be deadenylated, decapped and degraded either 5’→3’ via the exoribonuclease Xrn1 (Larimer and Stevens, 1990), or 3’→5’ by the cytoplasmic exosome (Anderson and Parker, 1998; Houseley et al., 2006). In addition to normal mRNA turnover, the exosome functions in rapid decay of CUTs, or cryptic unstable transcripts (Davis and Ares, 2006; Neil et al., 2009), transcripts endonucleolytically cleaved by the RNA Induced Silencing Complex (Orban and Izaurralde, 2005), RNAi-induced heterochromatin spreading at centromeres (Buhler et al., 2007), microRNA processing (Flynt et al., 2010) and in targeting activation-induced cytidine deaminase for optimal class switch recombination (Basu et al., 2011). Quality control functions for the RNA exosome include degradation of misfolded RNAs, including tRNAs, rRNAs, snRNAs, snoRNAs, and pre-mRNAs (Allmang et al., 1999; van Hoof et al., 2000), and degradation of aberrant mRNAs that feed into nonsense-mediated decay, nonstop decay, and no-go decay (van Hoof et al., 2002; Doma and Parker, 2006).
The eukaryotic exosome core (Exo9) contains nine distinct gene products that are essential for growth in budding yeast. Six belong to the RNase PH-like protein family (Rrp41, Rrp42, Rrp46, Rrp43, Mtr3 and Rrp45) and three (Rrp4, Rrp40 and Csl4) belong to a family of S1/KH proteins. These proteins form an asymmetric double-stacked ring with a six-membered RNase PH-like trimer of dimers that is capped by a three-membered S1/KH protein ring (Liu et al., 2006). Although devoid of catalytic activity, the architecture of eukaryotic Exo9 core bears striking structural resemblance to bacterial and archaeal 3’→5’ phosphorolytic exoribonucleases with respect to domain composition and a prominent central channel that is large enough to accommodate single stranded RNA (Symmons et al., 2000; Liu et al., 2006; Lorentzen and Conti, 2006; Buttner et al., 2006; Januszyk and Lima, 2010).
Ribonuclease activities of the eukaryotic exosome are attributable to two subunits that interact with Exo9, Rrp44 and Rrp6. Rrp44 interacts with Exo9 in the cytoplasm and nucleus and includes an N-terminal PIN domain, two cold-shock domains, an S1 domain and a ribonuclease domain (RNB) (Lorentzen et al., 2008). The RNB contains an active site that catalyzes processive 3’→5’ hydrolytic exoribonuclease activity while the N-terminal PIN domain catalyzes endoribonuclease activity and interacts directly with Rrp41 and Rrp45 (Lebreton et al., 2008; Schaeffer et al., 2009; Schneider et al., 2009; Bonneau et al., 2009). While RRP44 is essential, a point mutation in the RNB domain that disrupts exoribonuclease activity (D551N) is viable although it exhibits a severe growth phenotype (Dziembowski et al., 2007). A point mutation that disrupts endoribonuclease activity (D171N) has no discernable phenotype but a double mutant disrupting both RNB and PIN activities fails to support growth (Lebreton et al., 2008; Schaeffer et al., 2009).
The nuclear exosome includes Rrp6, Rrp44 and Exo9. Rrp6 is a distributive 3’→5’ exoribonuclease and contains an N-terminal domain important for binding to Rrp47 and the TRAMP complex (Stead et al., 2007; Callahan and Butler, 2010; Callahan and Butler, 2008; Butler and Mitchell, 2010), a central exoribonucleolytic domain and a C-terminal HRDC domain (Midtgaard et al., 2006; Januszyk et al., 2011). RRP6 is not essential, but rrp6Δ phenotypes include slow growth and buildup of 3’ extended forms of 5.8S rRNA, snRNAs and snoRNAs (Allmang et al., 1999; Allmang et al., 2000). Furthermore, combining rrp6Δ with rrp44exo- results in synthetic lethality and combining rrp6Δ with rrp44endo- results in a synthetic growth defect (Lebreton et al., 2008).
The function of the eukaryotic Exo9 core in regulating the RNA decay activities of the exosome has been a subject of interest given that each of its nine subunits are essential for viability although devoid of catalytic activity (Liu et al., 2006; Dziembowski et al., 2007). Analysis of the human exosome structure and comparison to bacterial and archaeal counterparts revealed the presence of conserved residues in the central channel that could contribute to interaction with RNA substrates (Liu et al., 2006). Furthermore, co-incubation of Rrp44 with Exo9 harboring channel-facing charge reversal point mutations diminished Rrp44 exoribonuclease activity in vitro (Bonneau et al., 2009) and electron microscopy showed density at the entry and exit points of the channel in RNA bound complexes of Exo9 and Rrp44 (Malet et al., 2010). Although these data are highly suggestive that Exo9 functions to regulate Rrp44 exoribonuclease activities, the contribution of the RNA exosome core and central channel to each RNase activity of the exosome or for function in vivo remains unclear.
We report here that Exo9 modulates Rrp6 exoribonuclease, Rrp44 exoribonuclease and endoribonuclease activities and that mutations designed to physically occlude the PH-like ring and central channel inhibit exosome activities in vitro and fail to support the essential functions of the RNA exosome in vivo. Our data invokes an RNA path that passes through the S1/KH and PH-like rings to Rrp44, one that partially overlaps with a path used by RNA to reach Rrp6 that entails the S1/KH cap and upper portions of PH-like ring. We show that Rrp6 stimulates Rrp44 activities independent of Rrp6 catalytic activity and that Rrp6 is potently inhibited by a mutation in the Rrp44 exoribonuclease active site. Taken together, these data suggest that the RNA exosome core coordinates RNA binding to regulate substrate access to the endo- and exoribonuclease activities of the RNA exosome.
Results and Discussion
The exosome core modulates Rrp44 and Rrp6 activities in ten-subunit exosomes
The contributions of Exo9 to Rrp44 and Rrp6 RNase activities were analyzed by reconstituting 34 distinct exosomes and comparing their ability to bind and degrade RNA (Table S1). Exosome reconstitutions used recombinant proteins as described (Liu et al., 2006; Greimann and Lima, 2008) with notable exceptions including removal of N-terminal hexa-histidine tags from Csl4, Rrp4, Rrp40, Rrp42, Rrp46 and Rrp41. We isolated a ten-subunit complex containing Rrp44 and Exo9 (Exo1044), a ten-subunit complex containing Rrp6 and Exo9 (Exo106), and an eleven-subunit complex containing Exo9, Rrp44 and Rrp6 (Exo1144/6) (Figure S1A). Although Exo106 has not been reported in yeast, recent work suggests its presence in nucleoli of human cells (Tomecki et al., 2010). Exo106 and Exo1144/6 solubility was increased by removing 128 N-terminal residues of Rrp6 (NΔ128Rrp6) that are not required for association with exosomes (Callahan and Butler, 2008; Assenholt et al., 2008). Differences in catalytic activity are not observed in vitro when comparing exosomes containing NΔ128Rrp6 or Rrp6 (Figure S1B).
Ribonuclease activities of Rrp44, Rrp6 and reconstituted exosomes were assayed using synthetic 5’-fluorescein labeled 49 nucleotide RNAs as models for cytoplasmic (AU-rich) and nuclear (polyA) substrates of the RNA exosome. Products were detected by monitoring fluorescein fluorescence as described (Liu et al,. 2006; Greimann and Lima, 2008). AU-rich and polyA RNA are single stranded and small enough to penetrate the central channel; however biophysical data suggest that AU-rich RNA is highly unstructured and readily deformable while polyA RNA forms helical domains with base-stacking interactions that make it more rigid and less deformable (Seol et al., 2004; 2007). RNase activities were assayed under multiple turnover conditions (10-fold substrate excess) to facilitate initial rate measurements, as rates were too fast to measure using single turnover conditions for AU-rich RNA. It is important to note that degradation patterns for polyA RNA were similar using single (enzyme excess) or multiple turnover conditions (Figure S1C). Apparent dissociation constants (Kd) for RNA binding to catalytically dead versions of Rrp6 and Rrp44 (exo-) in free and exosome-associated states were derived using fluorescence polarization.
Comparative analysis of Rrp44 exoribonuclease activity shows Exo1044 is 8-fold and 42-fold less active than free Rrp44 on AU-rich and polyA RNA, respectively (Figure 1A, 1B, Table S2). These results are consistent with trends observed previously (Liu et al., 2006, Bonneau et al., 2009). Exo1044exo- binds AU-rich RNA 3-fold weaker than free Rrp44exo- and more than 30-fold weaker for polyA RNA although binding can not be saturated for Exo1044exo- with polyA RNA indicating a Kd >10 μM (Figures 1A, 1B; Table S4; Figure S2). Thus, the magnitude of defects for RNA binding with Exo1044exo- mirror catalytic deficiencies observed for Exo1044 RNase activity when compared to Rrp44.
Figure 1. The Exo9 core alters exoribonuclease, endoribonuclease and RNA binding activities of Rrp44.
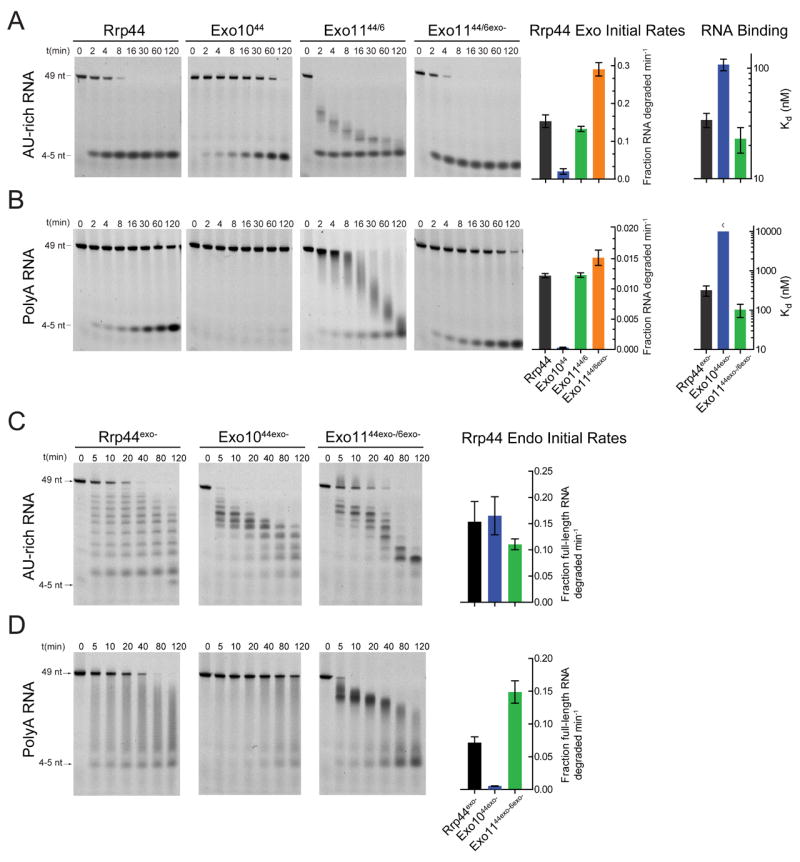
Association of Rrp44 with Exo9 (Exo1044) diminishes the exoribonuclease and RNA binding activities on (A) AU-rich and (B) polyA RNAs. Rrp6 stimulates Rrp44 activity in Exo1144/6 independent of Rrp6 catalytic activity. Multiple turnover RNA decay assays performed using 5’ fluorescein labeled RNAs and reaction products separated by Urea-PAGE. Representative gels and quantitation. Bar graphs depicting initial rates of Rrp44-mediated exoribonuclease activity determined from data obtained in the linear range. Bar graphs depicting dissociation constants (Kd) were derived by fluorescence polarization of catalytically dead variants of free and Exo9 core-associated Rrp44 assayed using 5’ fluorescein labeled (A) AU-rich or (B) polyA RNA. The pattern of intermediates generated by Rrp44exo- changes after association with Exo9 (Exo1044exo-) or Exo9 and Rrp6exo- (Exo1144exo-/6exo-) for (C) AU-rich and (D) polyA RNA (See Figure S3C for assays using unlabeled RNA). The distributive pattern of intermediates observed for Rrp44exo- (short and long 5’ labeled products accumulate simultaneously) is altered to patterns that appear to be generated 3’ to 5’ in Exo1044exo- and Exo1144exo-/6exo- for AU-rich RNA because longer 5’ labeled products appear before appearance of shorter products. Exo1044exo- has weaker endonuclease activity on polyA RNA but is stimulated by addition of Rrp6exo- in Exo1144exo-/6exo-. A stoichiometric ratio of enzymes and 5’-fluorescein RNA (10 nM) was incubated in reaction buffer in the presence of 3 mM MnCl2 (See Experimental Procedures). Error bars represent ± 1 standard deviation as calculated from three independent experiments. Bar graphs color coded according to Table S1.
Endoribonuclease activity of free Rrp44 has been shown to be dependent on manganese and is most readily observed when Rrp44 exoribonuclease activity is disrupted in Rrp44exo- (Lebreton et al., 2008) while inactivating mutations in both exoribonuclease and endoribonuclease active sites (Rrp44exo-/endo-) result in no ribonuclease activity (Figure S3A). Products of free Rrp44exo- endoribonuclease activity differ dramatically in length and distribution when compared to those generated by Exo1044exo- (Figures 1C, 1D). While free Rrp44exo- generates a distributive pattern of 5’ labeled RNA intermediates (from 5 nt to 49 nt) at the earliest time points for AU-rich and polyA RNAs, Exo1044exo- generates intermediates with the AU-rich RNA with apparent 3’ to 5’ directionality because longer 5’ labeled products appear before shorter products and intermediates accumulate around 23-26 nt prior to complete degradation (Figure 1C). Patterns are similar using unlabeled RNA and are consistent with distributive endoribonuclease activities for Rrp44exo- and 3’ to 5’ directionality for Exo1044exo- (Figure S3C). Endonuclease activity of Rrp44 is attenuated upon association with the Exo9 core. For polyA, the initial rate of substrate degradation, as measured by loss of the 49 nt substrate over time, is diminished 13-fold in Exo1044exo- compared to Rrp44exo- (Figure 1D; Figure S3B; Table S3).
Comparing Rrp6 and Exo106 activities reveals similar initial rates as determined by measuring the decrease in median RNA intermediate lengths over time (Table S4); however, Exo106 and Rrp6 products differ with respect to their pattern and distribution (Figure 2). Rrp6 generates RNA products that range in size between 49 nt and 20 nt as early as 0.5 minutes while Exo106 produces intermediates that cluster closer together for both AU-rich and polyA RNA substrates (Figures 2A, 2B; Figure S3D). Rrp6exo- and Exo106exo- bind AU-rich RNA with similar affinities (23 nM and 19 nM, respectively), but Rrp6exo- binds polyA RNA 13-fold better than Exo106exo- (121 nM and 1619 nM, respectively) (Figure 2B; Table S5). Taken together, RNase and binding data for Rrp44, Rrp6, Exo1044 and Exo106 suggest that the non-catalytic Exo9 core modulates Rrp44 and Rrp6 exoribonuclease and Rrp44 endoribonuclease activities, especially for polyA, and that defects in RNase activity can be explained in large part by a diminished capacity to bind RNA.
Figure 2. The Exo9 core alters Rrp6 exoribonuclease activity and RNA binding.
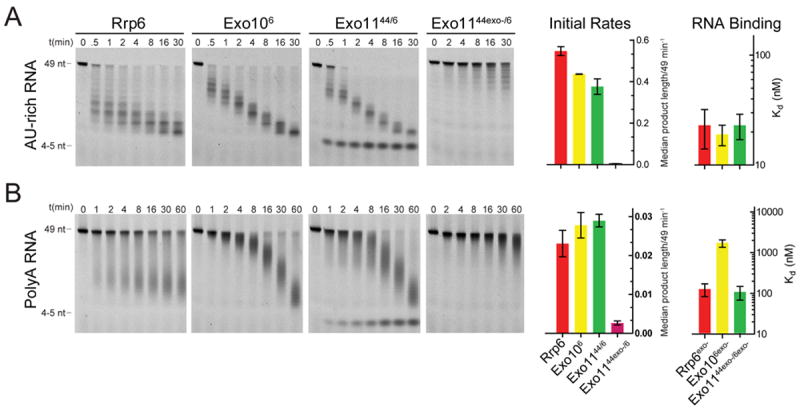
Association of Rrp6 with Exo9 (Exo106 or Exo1144/6) results in formation of a unique pattern of (A) AU-rich or (B) polyA RNA intermediates compared to Rrp6. Bar graphs representing initial rates of Rrp6-mediated exoribonuclease activity calculated by determining the median length of products generated over time and dividing the median length by substrate length (49 nt). Bar graphs depicting dissociation constants (Kd) derived by fluorescence polarization of catalytically dead variants of free and Exo9 core-associated Rrp6 using 5’ fluorescein labeled (A) AU-rich or (B) polyA RNA. Error bars represent ± 1 standard deviation as calculated from three independent experiments. Bar graphs color coded according to Table S1.
Rrp6 and Rrp44 activities in the eleven-subunit exosome
Rrp6 and Rrp44 activities were assessed next in the context of the eleven-subunit exosome (Exo1144/6). The presence of Rrp44 does not noticeably alter Rrp6 activities (Figure 2), but Rrp6 confers a 7-fold or 43-fold increase in Rrp44 exoribonuclease activity compared to Exo1044 using AU-rich or polyA RNA, respectively (Figures 1A, 1B). This is not due to Rrp6 generating additional RNA substrates for Rrp44 as Rrp6exo- is also able to stimulate Rrp44 in Exo1144/6. In agreement with these biochemical trends, the Kd for polyA RNA is the lowest when all eleven components are present in Exo1144/6 (Figure 1B).
The initial rates of RNA decay and intermediate patterns generated by endonuclease activity in Exo1144exo-/6exo- are similar to Exo1044exo-; however, degradation products are reduced to smaller sizes more quickly for the AU-rich RNA for Exo1144exo-/6exo- (Figure 1C). The stimulatory effect of having Rrp6 in the complex is more evident for polyA RNA when comparing initial rates and products as Exo1144exo-/6exo- is 28-fold more active than Exo1044exo- (Figure 1D). Furthermore, the distributive pattern of intermediates generated by Rrp44exo- with polyA RNA is compressed in reactions with Exo1144exo-/6exo-, more closely resembling the pattern and accumulation of 23-26 nt intermediates generated by Exo1044exo- and Exo1144exo-/6exo- with AU-rich RNA. These data suggest that Rrp6 stimulates both Rrp44 endo- and exoribonuclease activities within the context of Exo1144/6.
Differences are not detected for Rrp6 activity in Exo1144/6 when compared to Exo106 but it is noteworthy that Rrp6 is inhibited 100-fold in Exo1144exo-/6 (Figure 2). This suggests that RNA binding to Rrp44exo- in Exo1144exo-/6 precludes Rrp6 from accessing RNA despite a 10-fold excess of RNA substrate in these reactions. These data also suggest that apparent Kd values for Exo1144exo-/6exo- (23 nM for AU-rich and 103 nM for polyA) report on RNA binding to Rrp44 rather than Rrp6 (Table S5). Why does Rrp44 outcompete Rrp6 for binding? We believe this is likely due to differences in catalytic mechanism between Rrp44, a processive enzyme that binds and holds onto the substrate until it is completely degraded, and Rrp6, a distributive enzyme that catalyzes successive rounds of degradation and product release during decay. Data presented thus far suggests that Exo9 modulates RNA binding and RNase activities of both Rrp6 and Rrp44, albeit to differing extents. Furthermore, exosome-associated activities of Rrp6 and Rrp44 appear interdependent because Rrp6 confers a gain of function on Rrp44 activities and Rrp44exo- severely inhibits Rrp6 activity in Exo1144exo-/6.
RNA-protein interactions in exosomes containing Rrp6 and Rrp44
To identify exosome subunits that contact RNA we employed UV cross-linking in reactions containing Rrp6exo- or Rrp44exo- and their respective complexes with Exo9. It is worth noting that appearance of UV cross-linked species only suggests and by no means proves that interactions are on pathway to a productive substrate/enzyme complex. UV cross-linking efficiency was enhanced by using AU-rich RNA, and protein-RNA adducts were resolved by SDS-PAGE. The identities of core subunits were determined by reconstituting exosomes containing unique combinations of N-terminal hexa-histidine tagged subunits to generate tag-dependent mobility shifts of RNA-protein adducts upon UV-crosslinking (Figure S4).
Very weak cross-linking is observed for Exo9 (Figure 3), consistent with fluorescence polarization data (Figure S2) and RNase foot-printing (Bonneau et al., 2009) that fail to detect a specific RNA bound complex. In contrast, UV cross-linking produces several protein-RNA adducts unique to Exo1044exo-, Exo106exo- and Exo1144exo-/6exo- (Figure 3). Specifically, adducts are observed to Rrp44exo-, the S1/KH cap proteins (Csl4, Rrp4, and Rrp40) and half of the six PH-like proteins (Rrp43, Rrp45, and Rrp41) using Exo1044exo-. Adducts are observed to Rrp6exo- and more weakly to the S1/KH cap proteins and to lesser extent Rrp41 using Exo106exo-. Cross-linking patterns of Exo1144exo-/6exo- are similar to Exo1044exo-, namely adducts are observed to Rrp44exo- and the respective Exo9 subunits with only minimal cross-linking to Rrp6exo-. The observation that Rrp44exo- inhibits RNA cross-linking to Rrp6exo- in Exo1144exo-/6exo- and Rrp6 catalytic activity in Exo1144exo-/6, suggests that RNA, when bound to Rrp44exo-, blocks a path used by RNA to engage Rrp6 in respective Exo1144/6 complexes. Because UV cross-linking to the S1/KH-proteins is observed in Exo1044exo-, Exo106exo- and Exo1144exo-/6exo-, and since UV cross-linking to the RNase PH-like proteins is only observed in Exo1044exo- and Exo1144exo-/6exo- we posit that RNA enters a channel formed by the S1/KH cap proteins on its way to Rrp6 or Rrp44 and that RNA passes through or by the RNase PH-like ring to engage Rrp44.
Figure 3. UV-RNA cross-linking reveals exosome-RNA contacts.
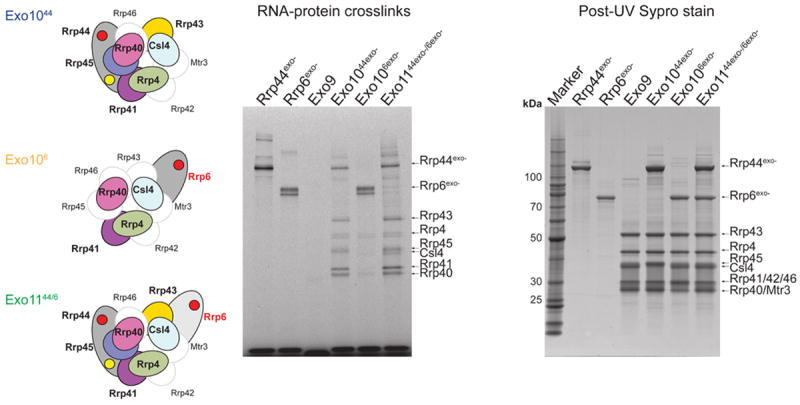
Schematics of Exo1044exo- (top), Exo106exo- (middle) and Exo1144exo-/6exo- (bottom) shown on left. Subunits for which no cross-linking was observed are outlined, labeled and shown in white while subunits for which RNA-protein cross-links were detected are outlined, labeled and colored. Exosome subunits and exosomes were incubated with 5’ fluorescein AU-rich RNA and illuminated by UV to induce cross-linking. UV RNA-protein cross-linked products were separated by SDS-PAGE and gels were scanned to detect 5’-fluorescein RNA-protein adducts (middle), then stained with Sypro Ruby and scanned to visualize total protein (right). Subunit positions in respective gels indicated by labels and arrows. Molecular weight markers shown in left lane for the Sypro Ruby stained gel.
The exosome central channel is essential in vivo
Previous in vitro studies indicated that Rrp44 exoribonuclease activity was compromised when Rrp44 was incubated in the presence of exosome cores that contained channel-lining charge-swap mutations in the PH-like protein Rrp41 (Bonneau et al., 2009). Consistent with earlier complementation assays that incorporated three channel-lining point mutations (Dziembowski et al., 2007), we observe no growth phenotype for charge-swap point mutations in Rrp41 in vivo (Figure S5A). If the central channel plays an important role in exosome function one might expect phenotypes such as growth defects and aberrant RNA processing as observed in strains lacking RRP6 or in strains harboring a mutation in the Rrp44 exoribonuclease active site (Dziembowski et al., 2007; Schaeffer et al., 2009; Schneider et al., 2009). These apparent discrepancies suggest that interior channel surfaces are not important, that RNA can bypass the central channel to engage Rrp44 or Rrp6, or that mutations on the channel surface are not sufficient to physically occlude the channel in vivo.
To physically and electrostatically occlude the channel that is formed by the PH-like ring we constructed insertions based on the structure of human Exo9 (Liu et al., 2006) in two loops that project into the channel. Insertions include acidic side chains and range between 2 (EL), 7 (ELGESEG) and 11 (ELGESEGESEG) amino acid residues. We refer to these insertions as Small [S], Medium [M], and Large [L] and rough calculations suggest they could project into the channel by 3.5 Å, 9 Å and 15 Å, respectively. Given the dimensions of the RNA exosome central channel and position of the loops we posit that the flexible, extended loops in Rrp41 and Rrp45 would pose steric impediments to RNAs entering near the top and middle of the PH-like ring, respectively (Figure 4A).
Figure 4. The exosome central channel is essential in vivo.
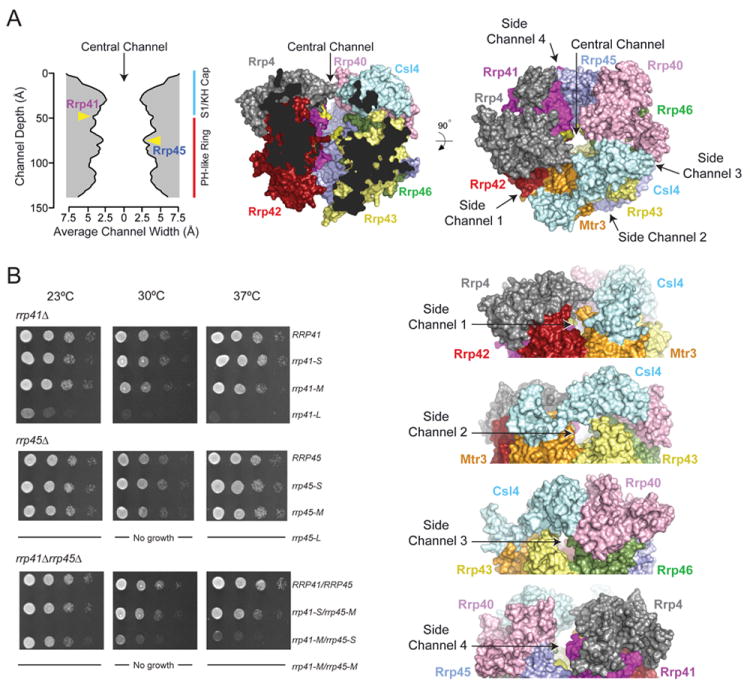
A) The width and dimensions of the channel (left) were calculated using the dimensional analysis program, HOLE (Smart et al., 1996). Axes are labeled. The middle panel shows a view of the human Exo9 core (PDB 2NN6) in surface representation. The central channel is made apparent by removing Mtr3 and cutting away remaining surfaces of Csl4, Rrp43 and Rrp42. The positions of insertions in Rrp41 and Rrp45 are colored yellow in the structure and indicated by labels and yellow arrows in the graph depicting channel dimensions. Right panel depicts an orthogonal view of the Exo9 core in surface representation showing all nine subunits. A label and black arrow indicates the positions of the central channel and four putative side channels. Subunits colored and labeled. Below this panel are four side views of the S1/KH cap and top portion of the PH-like ring highlighting four potential side channels. B) Serial 10-fold dilutions of S. cerevisiae rrp41Δ (top), rrp45Δ (middle) or rrp41Δrrp45Δ (bottom) bearing RRP41 or RRP45 or mutant rrp41 or rrp45 alleles containing insertions ranging between 2 [S], 7 [M] and 11 [L] amino acids spotted on YPD agar grown at 23°C (left), 30°C (middle), and 37°C (right).
The rrp41 or rrp45 alleles containing S, M, or L insertions were assessed in complementation assays in S. cerevisiae using rrp41Δ, rrp45Δ, and rrp41Δrrp45Δ strains (Figure 4B). While S and M insertions had little impact on growth in isogenic rrp41Δ and rrp45Δ strains, L insertions in rrp41 or rrp45 (rrp41-L or rrp45-L) resulted in a severe temperature sensitive growth defect or lethality, respectively (Figure S5B). In the rrp41Δrrp45Δ strain, rrp41-S/rrp45-M exhibited a slight growth defect at 37°C, while rrp41-M/rrp45-S grew slowly at 30°C and 37°C. Although individual rrp41-M and rrp45-M mutants had no discernable growth phenotype in rrp41Δrrp45Δ, combining these alleles resulted in synthetic lethality at all temperatures tested. TAP-purification of Rrp44 from the viable rrp41-M/rrp45-S strain confirms that channel occluded exosomes remain associated in vivo (Figure S5C). These results suggest that mutations predicted to physically occlude the central channel fail to support the essential functions of the RNA exosome in vivo.
Channel occluding mutations inhibit Rrp44 and Rrp6 activities in vitro
Our data suggest that Exo9 modulates Rrp44 and Rrp6 RNase activities, and binding and cross-linking data suggest that RNAs interact with and potentially pass through the S1/KH-protein ring to access Rrp6 and through the S1/KH-protein and PH-like ring to access Rrp44. To test this model we combined medium length insertions (Rrp41-M/Rrp45-M) to reconstitute Exo1144/6/channel- because this combination failed to complement the essential functions of the exosome in vivo (Figure S5B). Exo1144/6/channel- behaves similarly in vitro to Exo1144/6 during reconstitution and purification suggesting that loop insertions do not interfere with exosome assembly (Figure S5D).
Rrp44 exoribonuclease activity on AU-rich and polyA RNA is inhibited 45-fold and 100-fold, respectively, by channel occlusion in Exo1144/6/channel- relative to WT Exo1144/6 (Rrp41-WT/Rrp45-WT) (Figures 5A, 5B; Figure S6A). This effect is independent of Rrp6, as we observe similar trends in Exo1044 (Figure S6B). Occluding the channel also diminishes Rrp44 endonuclease activities in Exo1144exo-/6exo-/channel- by 20-fold on polyA in comparison to Exo1144exo-/6exo- (Figures 5A, 5B), a defect that is further exacerbated using exosomes reconstituted with large insertions in both Rrp41 and Rrp45 (Figure S6D).
Figure 5. Channel occlusion inhibits RNA binding and RNase activities of Rrp44 and Rrp6.
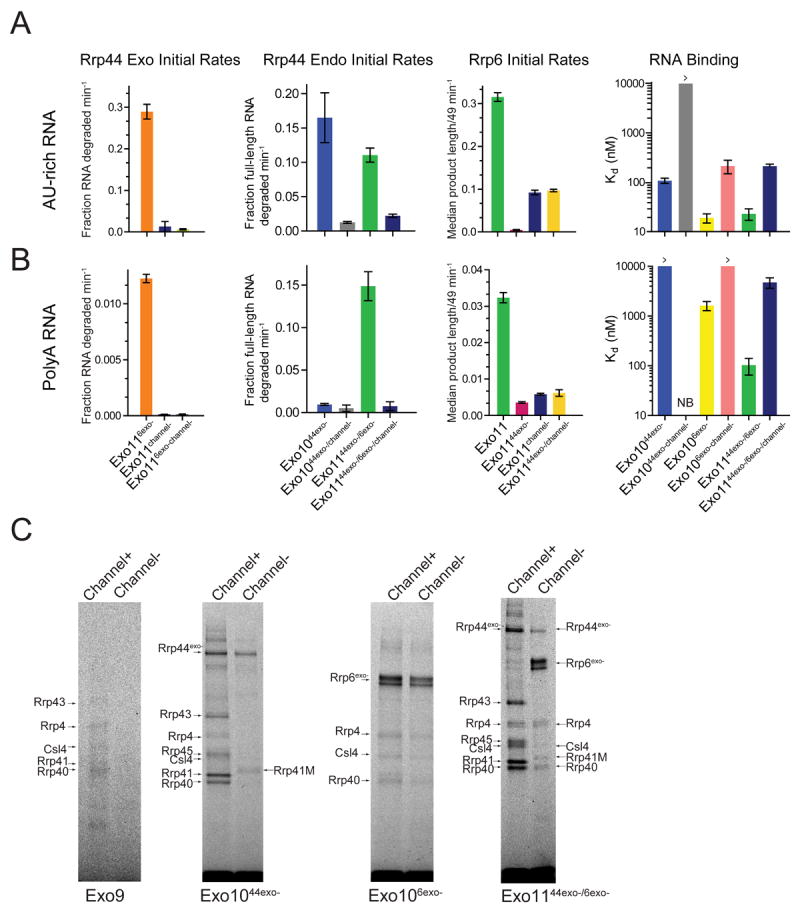
Bar graphs depicting initial rates calculated for Rrp6 and Rrp44 RNase on A) AU-rich and B) polyA RNA. Occluding the central channel of Exo1144/6 (Exo1144/6/channel-) with loop insertions (Rrp41-M/Rrp45-M) inhibits both Rrp44 activities while Exo1144exo-/6/channel- diminishes Rrp6 exoribonuclease activities and alleviates Rrp6 inhibition by Rrp44exo-. Rrp44 endonuclease activities are inhibited by channel occlusion in Exo1044exo- and Exo1144exo-/6exo-. Assays performed in triplicate and initial rates calculated from data obtained in the linear range (Figures S6A and S6C). Bar graphs on the right depict apparent Kd values for various exosome complexes with RNA. Error bars are ± 1 standard deviation. C) Channel occlusion in Exo9 leads to diminished cross-linking (left). Panel to the right shows that channel occlusion in Exo1044exo- leads to loss of most RNA-protein adducts to core subunits. Next panel shows that channel occlusion in Exo106exo- slightly weakens cross-linking to the S1/KH cap proteins and Rrp6. On the right, channel occlusion in Exo1144exo-/6exo- shifts the cross-linking pattern from one involving Rrp44, the PH-like and S1/KH rings to one involving Rrp6 and the S1/KH ring. Products separated by SDS-PAGE and imaged by detecting the fluorescence of the 5’ labeled AU-rich RNA. Major cross-linked species are labeled and indicated by arrows.
If the central channel is used to guide RNA to Rrp44 then channel occlusion in the PH-like ring should impair RNA binding and UV cross-linking and this is exactly what is observed. Binding assays confirm that Exo1044exo-/channel- is >100-fold less able to interact with AU-rich RNA (Figure 5A) while binding to polyA RNA is further weakened to nearly undetectable levels compared to Exo1044exo- (Figure 5B; Figure S2). Consistent with binding data, UV cross-linking patterns to Exo9 are weakened further when its channel is occluded and RNA adducts to Rrp44exo- and Exo9 are greatly diminished in Exo1044exo-/channel- relative to Exo1044exo- although some cross-linking to Rrp41 and Rrp44exo- is still observed (Figure 5C).
Channel occlusion in the PH-like ring was not predicted to impact Rrp6 to the same degree as Rrp44 because strong cross-linking was not observed to PH-like ring subunits in Exo106exo-; however we observe that PH-ring channel occlusion attenuates Rrp6 activity in Exo1144/6/channel-, particularly with the polyA RNA, resulting in a relatively modest 5-fold reduction in initial rate relative to Exo1144/6 (Figure 5B). Perhaps more interesting is that PH-ring channel occlusion prevents Rrp6 from stimulating Rrp44 activities in Exo1144/6 and partially alleviates Rrp6 inhibition by Rrp44exo- as evidenced by comparing activities of Exo1144exo-/6, Exo1144exo-/6/channel- and Exo1144/6/channel- (Figure 5A; Figure S6A). Similar changes in activities were observed for Exo106/channel- (Figure S6B).
Consistent with inhibition of Rrp6 RNase activities in Exo106/channel-, Exo106exo-/channel- binds AU-rich and polyA RNAs 10- and 40-fold less well, respectively, compared to Exo106exo- (Figures 5A, 5B) and exhibits slightly diminished cross-linking patterns to the S1/KH cap proteins, Rrp41 and Rrp6exo- (Figure 5C). Similarly, channel occlusion in Exo1144exo-/6exo-/channel- attenuates binding to AU-rich and polyA RNA by 20- and 50-fold, respectively, relative to Exo1144exo-/6exo- (Figure 5A). While cross-linking is diminished to PH-like ring subunits and Rrp44exo- (Figure 5C) it is interesting that cross-linking is enhanced to Rrp6exo- in Exo1144exo-/6exo-/channel- compared to Exo1144exo-/6exo-. Because PH-like ring channel occlusion inhibits RNA from gaining access to Rrp44 and partially alleviates inhibition of Rrp6 activity in exosomes containing Rrp44exo-, this cross-linking pattern is consistent with PH-like ring channel occlusion diverting RNA to Rrp6. This also suggests that RNA binding to Exo1144exo-/6exo-/channel- reports on RNA interactions with the S1/KH-proteins and Rrp6exo- because apparent Kd values are similar to those measured for Exo106exo-/channel- (Table S5).
If RNA passes by or through the S1/KH cap and PH-like rings to engage Rrp44, and by or through the S1/KH cap and top portion of the PH-like ring to bind Rrp6, we might expect that a larger insertion at the top of the PH-ring would impact Rrp6 more than a large insertion in the middle of the PH-ring. As predicted, Rrp6 activity is diminished 4-fold further in an Exo1144/6/channel- variant that contains a large insertion near the top of the channel (Rrp41-L) compared to one where a large insertion is placed in the middle (Rrp45-L) (Figure 6A). Furthermore, UV cross-linking patterns between RNA, the S1/KH cap proteins and Rrp6 is enhanced slightly in Exo106exo- complexes containing a large insertion in the middle of the channel, rather than the top (Figure S7A). Importantly, Rrp44 activity is similarly inhibited independent of insertion placement (Figure 6A).
Figure 6. Rrp6 is more dependent on the upper portion of the channel to access RNA.
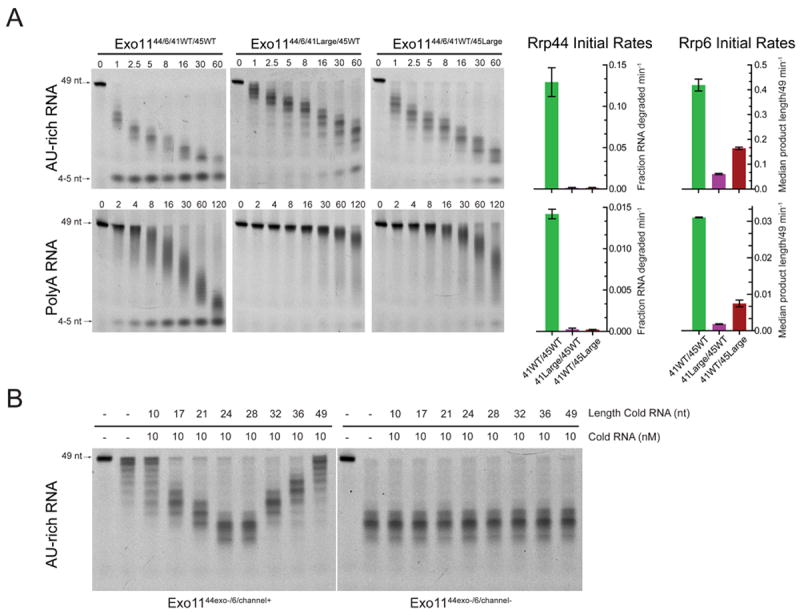
A) RNA decay assays using AU-rich or polyA RNA for WT exosomes and exosomes reconstituted with large loop insertions at the top (41Large) or middle (45Large) of the channel. Bar graphs on right indicate initial rates of decay mediated by Rrp44 or Rrp6. Rrp44 activity is inhibited by insertions at either position while the insertion at the top of the channel (Exo1144/6 41Large/45WT) impedes Rrp6 activity more than insertion in the middle (Exo1144/6 41WT/45Large). Decay assays performed in triplicate. Error bars are ± 1 standard deviation. B) Inhibition of Rrp6 activity in Exo1144exo-/6 by 49 nt AU-rich RNA is alleviated by short competing RNAs that mimic channel occlusion in the PH-like ring. End point assay (15 minutes) of reactions containing 10 nM unlabeled AU-rich RNAs (10, 17, 21, 24, 28, 32, 36, 49 nts) that were pre-incubated with 1 nM Exo1144exo-/6 or Exo1144exo-/6/channel-. Reactions were then challenged with 10 nM 49 nt 5-fluorescein AU-rich RNA and reaction intermediates detected by denaturing PAGE. Reactions containing unlabeled 24 or 28 nt mimic channel occlusion and alleviate Rrp6 inhibition in Exo1144exo-/6 to similar levels observed in Exo1144exo-/6/channel-.
To test this model by other means we took advantage of the observation that Rrp6 is potently inhibited (100-fold) in Exo11 complexes containing Rrp44exo- when a 49 nt AU-rich RNA is used (Figure 2A). The human Exo9 channel extends 125 Å and is long enough to accommodate approximately 30 nts of single stranded RNA if extended (~4 Å per base; Figure 4A). Based on a model of Rrp44 bound to Rrp41/Rrp45 (Bonneau et a., 2009) and the observation that at least 9 nts are required for binding to Rrp44exo- (Lorentzen et al., 2008), we posit that Rrp6 is inhibited when a 49 nt RNA substrate is bound to Exo1144exo-/6 because the RNA is long enough to span the entirety of the channel through the S1/KH and PH-like rings thereby competing with the portion of the S1/KH ring channel that is required by Rrp6. If true, competing 23-30 nt single-stranded RNAs should bind Rrp44exo- (9 nt) and engage the PH-like ring (14-23 nt). Because these RNAs are too short to reach the S1/KH ring channel, they should mimic PH-like ring channel occlusion in Exo1144exo-/6 and alleviate inhibition of Rrp6 (Figure 4A). We tested this by incubating Exo1144exo-/6 and Exo1144exo-/6/channel- with unlabeled AU-rich RNAs of 10, 17, 21, 24, 28, 32, 36, and 49 nts and then challenged these mixtures with 5’ labeled 49 nt AU-rich RNA. Indeed, Rrp6 inhibition is alleviated in Exo1144exo-/6 in a manner dependent on RNA length – 17 and 21 nt RNAs partially restore Rrp6 activity while 24 and 28 nt RNAs mimic PH-like ring channel occlusion by loop insertions. Further increase in RNA length beyond 32 nt recapitulates Rrp6 inhibition (Figure 6B). This effect is mediated by RNA binding to Rrp44exo- and the PH-like ring central channel because competing RNAs have no discernable effect on Rrp6 activity when assayed with Exo1144exo-/6/channel- (Figure 6B). It is worth nothing that inhibition is only partially alleviated by the 10 nt RNA, an RNA too short to simultaneously bind Rrp44exo- and engage the PH-like ring channel.
Functional consequences of channel occlusion in vivo
The data presented thus far suggest that the central channel mediates RNA decay because channel occluding mutations inhibit RNA binding and RNase activities in vitro and fail to complement the essential functions of the exosome in vivo. To test if partial occlusion of the central channel alters exosome functions in viable strains containing rrp41-S/rrp45-M and rrp41-M/rrp45-S, we analyzed the abundance of several RNA substrates previously identified as exosome substrates by quantitative PCR and compared them to strains containing RRP41-WT/RRP45-WT or lacking RRP6. RNA targets included a pre-ribosomal RNA, ITS2-1 (Allmang et al., 2000), the U4 snRNA (Allmang et al., 1999; van Hoof et al., 2000), and several CUTs, including NEL025C (Wyers et al., 2005), CUT638, and CUT273 (Xu et al., 2009). We also analyzed the open reading frames (ORFs) APM2 and YLR356W that are located adjacent to CUT638 and CUT273, respectively (Xu et al., 2009).
Defects could be observed for many of the substrates in rrp41-S/rrp45-M and rrp41-M/rrp45-S when compared to WT (Figure 7). The rrp41-M/rrp45-S strain exhibits similar deficiencies in early pre-rRNA processing and reduced levels of ITS2-1, corresponding to the 27SA2 pre-rRNA, as observed in rrp6Δ and reported in previous studies (Allmang et al, 2000). While rrp6Δ exhibits greater defects for all but CUT638, strains including channel occlusions accumulate a 3’ extended form of the U4 snRNA (3-fold), and the CUTs NEL025C (3-fold), CUT638 (5-fold) and CUT273 (4-fold) when compared to WT.
Figure 7. Channel occlusion results in aberrant RNA processing in vivo.
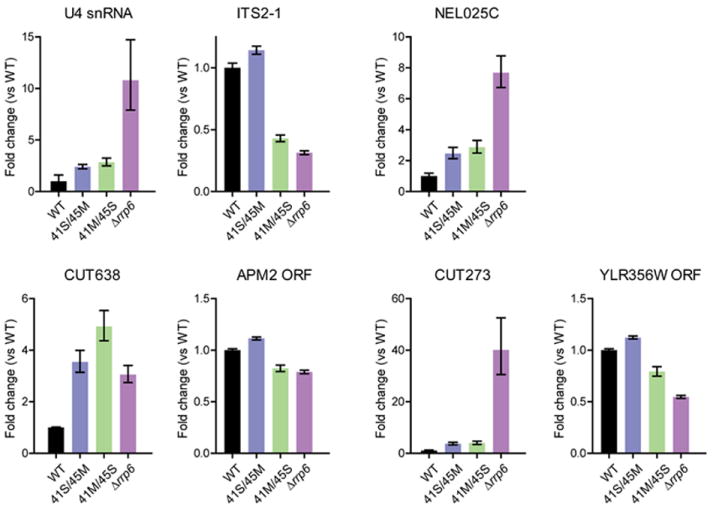
Previously identified substrates of the RNA exosome were analyzed by qPCR from total RNA obtained from rrp41Δrrp45Δ strains bearing WT RRP41/RRP45 or mutant alleles containing small/medium (rrp41-S/rrp45-M), and medium/small (rrp41-M/rrp45-S) insertions and compared to levels in an rrp6Δ strain. Bar graphs show quantitation of results obtained from triplicate experiments. Error bars represent ± 1 standard deviation. Targets were normalized to Scr1 mRNA. APM2 and YLR356W ORFs serve as controls for CUT638 and CUT273, respectively.
Conclusion
The results presented here underscore the importance of the Exo9 core and central channel in RNA processing and decay because Exo9 modulates the endo- and exoribonuclease activities between Rrp44 and Rrp6 and because mutations that occlude the PH-ring channel inhibit Rrp44 and Rrp6 RNase activities in vitro and fail to complement essential functions of the exosome in vivo. Our data suggest that RNA passes through a channel formed by the S1/KH cap and PH-like ring to reach Rrp44 because PH-like ring channel occlusion inhibits Rrp44 RNase activities, weakens binding and UV cross-linking and alleviates Rrp44exo- mediated inhibition of Rrp6 with RNAs longer than 32 nt (Figures S7B, S7C). This latter observation, along with RNA binding and UV cross-linking, supports the hypothesis that RNA passes through the S1/KH ring to access Rrp6. How RNA exits the upper portion of the S1/KH channel to access Rrp6 remains unknown, but four channels that are large enough to accommodate single stranded RNA exist below the S1/KH cap and above the PH-like ring in the human Exo9 structure (Figure 4A).
Another unresolved issue is how the exosome core selects a path for RNA to Rrp6 or Rrp44 although it appears stochastic based on the two substrates used in this study, as both Rrp6 and Rrp44 activities are evident in Exo1144/6 (Figure 1). Once through the S1/KH and PH-like rings RNA can be directed to either the Rrp44 endoribonuclease or exoribonuclease site because unlike Rrp6, which is potently inhibited by Rrp44exo-, Rrp44 endoribonuclease activity is clearly evident in exosomes containing Rrp44exo-. These in vitro data are perhaps consistent with the genetic interplay reported between endo- and exoribonuclease active sites in vivo (Lebreton et al., 2008; Schneider et al., 2009; Schaeffer et al., 2009; Schaeffer and van Hoof, 2011). Details regarding how a particular RNA is directed down a distinct path to one or more of the three RNase activities of the nuclear exosome will remain a focus of investigation for some time.
Our studies may shed some light on genetic and functional analysis of exosome activities in strains harboring rrp44-D551N (exo-) or in strains lacking RRP6. The near lethal growth phenotype observed for strains harboring rrp44-D551N and shared RNA processing defects in rrp44-D551N (exo-) and rrp6Δ indicated partially overlapping functions for Rrp6 and Rrp44 in vivo (Dziembowski et al., 2007). Our data suggest that strains harboring rrp44-D551N may also experience a partial loss of Rrp6 function because Exo1144exo-/6 inhibits Rrp6 activity by 100-fold over Exo1144/6. With respect to Rrp6, phenotypes observed in rrp6Δ strains could be due in part to partial loss of Rrp44 activities since we observe that Rrp6 can stimulate Rrp44 activities in vitro, especially for polyA RNA. In support of this hypothesis, strains harboring rrp6-D238A (exo-) grow better than rrp6Δ and accumulate less nucleolar polyA(+) than rrp6Δ (Assenholt et al., 2008).
Rrp44 activities are severely compromised by channel occlusion and Rrp6 is potently inhibited by RNA bound Rrp44exo- in Exo1144exo-/6. These observations suggest that each active site is sequestered from solvent when associated with the exosome core. The structural basis for this remains unclear because Exo1044 models derived from human Exo9 and S. cerevisiae Rrp44-Rrp41-Rrp45 structures show Rrp44 exo- and endoribonuclease active sites to be solvent exposed (Liu et al., 2006; Wang et al., 2007; Bonneau et al., 2009). Sequestration of catalytic activities within a chamber is a regulatory mechanism and organization shared among other RNA and protein degrading complexes as noted previously (Lorentzen and Conti, 2006) and our data suggest that the exosome core functions in this regard to modulate or limit access to all three identified RNase activities of the RNA exosome.
No structural models exist yet for Exo1144/6, but the observation that Rrp6 can stimulate RNA binding and Rrp44 activities in Exo1144/6 is particularly intriguing as evidenced by the 40-fold activation in polyA degradation when comparing Exo1044 and Exo1144/6. As noted earlier, single stranded polyA differs from the AU-rich substrate because it has secondary structure that makes it more rigid compared to the more flexible AU-rich RNA. As such, we posit that polyA is more susceptible to exclusion from the central channel because it is less able to navigate through steric impediments, including those presented by loop insertions. Furthermore, our data suggest that polyA is restricted from binding and entering the central channel in Exo1044, an effect that is alleviated by Rrp6 in Exo1144/6. This suggests that Rrp6 serves to gate the channel, activate unwinding of the polyA structure and/or activate Rrp44 through some other mechanism. These features are particularly attractive as they provide opportunities to regulate the activities of the exosome through direct targeting of RNA to the central channel or by gating the channel with other RNA or RNA-protein complexes.
Experimental Procedures
Cloning, expression, purification and reconstitution
Purification of recombinant exosome subunits and exosome reconstitutions were performed using methods published previously (Liu et al., 2006; Greimann and Lima, 2008). Cloning, expression, and purification of subunits with cleavable N-terminal His6 tags are described in Supplementary Experimental Procedures.
Ribonuclease assays
Synthetic 49 nt RNAs bearing a 5’ fluorescein label (Invitrogen) or unlabeled RNAs (Invitrogen or IDT) were used to assay exoribonuclease activities of Rrp44 and Rrp6, and endoribonuclease activity of Rrp44. Reactions assaying exoribonuclease activity were performed at 30°C in 100 μL under multiple turnover conditions (1 nM enzyme, 10 nM RNA). Endonuclease assays were performed at 30°C in 100 μL with 10 nM enzyme and 10 nM RNA. Details of reaction conditions and quantitation are described in Supplementary Experimental Procedures.
UV-RNA crosslinking
700 nM of exosome subunits or complexes with mutations in the exoribonuclease active site were incubated on ice for 20 minutes with 50 nM of a 36 nt 5’ fluorescein AU-rich RNA in a 70 μL reaction volume. The binding reaction buffer consisted of 50 mM KCl, 20 mM Tris (pH 8.0), 10 mM DTT, 0.5 mM MgCl2. Samples were then placed in a UV-Stratalinker (Stratagene) and subjected to 400,000 or 700,000 μJ of short-wave UV radiation (254 nM). 15 μL aliquots were taken, added to 4X LDS loading buffer, and separated by SDS-PAGE on a 4-12% Bis-Tris gel (Invitrogen). RNA-protein adducts were visualized immediately with a fluoroimager (FITC filter); protein integrity was assessed after initial imaging by staining with Sypro Ruby (Invitrogen) and visualized with a fluoroimager (LPB filter).
Fluorescence Polarization
Full description is provided in Supplementary Experimental Procedures.
Yeast strains and complementation by plasmid shuffle
Yeast strain construction and details of complementation assays are in Supplementary Experimental Procedures.
RNA analysis by quantitative PCR
Full methods describing qPCR and oligonucleotides used can be found in Supplementary Experimental Procedures.
Supplementary Material
Highlights.
The exosome core and central channel modulate Rrp44 and Rrp6 RNase activities
Rrp6 stimulates Rrp44 activities in the eleven-subunit exosome
Insertions that occlude the central channel inhibit exosome RNase activities
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award numbers F31GM097910 (E.V.W) and R01GM079196 (C.D.L). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1684–91. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JS, Parker RP. The 3’ to 5’ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3’ to 5’ exonucleases of the exosome complex. EMBO J. 1998;17:1497–506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenholt J, Mouaikel J, Andersen KR, Brodersen DE, Libri D, Jensen TH. Exonucleolysis is required for nuclear mRNA quality control in yeast THO mutants. RNA. 2008;14:2305–13. doi: 10.1261/rna.1108008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu U, Meng FL, Keim C, Grinstein V, Pefanis E, Eccleston J, Zhang T, Myers D, Wasserman CR, Wesemann DR, Januszyk K, Gregory RI, Deng H, Lima CD, Alt FW. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011;144:353–63. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–59. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- Bühler M, Haas W, Gygi SP, Moazed D. RNAi-dependent and – independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–21. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- Butler JS, Mitchell P. Rrp6, Rrp47 and cofactors of the nuclear exosome. In RNA Exosome. In: Jensen TH, editor. Adv Exp Med Biol. Vol. 702. New York: Landes Bioscience and Springer Science; 2010. pp. 91–104. [PubMed] [Google Scholar]
- Büttner K, Wenig K, Hopfner KP. The exosome: a macromolecular cage for controlled RNA degradation. Mol Microbiol. 2006;61:1372–9. doi: 10.1111/j.1365-2958.2006.05331.x. [DOI] [PubMed] [Google Scholar]
- Callahan KP, Butler JS. Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res. 2008;36:6645–55. doi: 10.1093/nar/gkn743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan KP, Butler JS. TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. J Biol Chem. 2010;285:3540–7. doi: 10.1074/jbc.M109.058396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Ares M., Jr Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2006;103:3262–7. doi: 10.1073/pnas.0507783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–4. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Séraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- Flynt AS, Greimann JC, Chung WJ, Lima CD, Lai EC. MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol Cell. 2010;38:900–7. doi: 10.1016/j.molcel.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greimann JC, Lima CD. Reconstitution of RNA exosomes from human and Saccharomyces cerevisiae cloning, expression, purification, and activity assays. Methods Enzymol. 2008;448:185–210. doi: 10.1016/S0076-6879(08)02610-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–39. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Januszyk K, Lima CD. Structural components and architectures of RNA exosomes. In RNA Exosome. In: Jensen TH, editor. Adv Exp Med Biol. Vol. 702. New York: Landes Bioscience and Springer Science; 2010. pp. 9–28. [PMC free article] [PubMed] [Google Scholar]
- Januszyk K, Liu Q, Lima CD. Activities of human RRP6 and structure of the human RRP6 catalytic domain. RNA. 2011;17:1566–77. doi: 10.1261/rna.2763111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimer FW, Stevens A. Disruption of the gene XRN1, coding for a 5’ to 3’ exoribonuclease, restricts yeast cell growth. Gene. 1990;95:85–90. doi: 10.1016/0378-1119(90)90417-p. [DOI] [PubMed] [Google Scholar]
- Lebreton A, Tomecki R, Dziembowski A, Séraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–6. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–37. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Conti E. The exosome and the proteasome: nano-compartments for degradation. Cell. 2006;125:651–4. doi: 10.1016/j.cell.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell. 2008;29:717–28. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- Malet H, Topf M, Clare DK, Ebert J, Bonneau F, Basquin J, Drazkowska K, Tomecki R, Dziembowski A, Conti E, Saibil HR, Lorentzen E. RNA channelling by the eukaryotic exosome. EMBO Rep. 2010;11:936–42. doi: 10.1038/embor.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midtgaard SF, Assenholt J, Jonstrup AT, Van LB, Jensen TH, Brodersen DE. Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain. Proc Natl Acad Sci USA. 2006;103:11898–903. doi: 10.1073/pnas.0604731103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil H, Malabat C, d’Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–42. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- Orban TI, Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 2005;11:459–69. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer D, van Hoof A. Different nuclease requirements for the exosome-mediated degradation of normal and nonstop mRNAs. Proc Natl Acad Sci USA. 2011;108:2366–71. doi: 10.1073/pnas.1013180108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Leung E, Brown J, Tollervey D. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009;37:1127–40. doi: 10.1093/nar/gkn1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol Y, Skinner GM, Visscher K. Elastic properties of a single-stranded charged homopolymeric ribonucleotide. Phys Rev Lett. 2004;93:118102. doi: 10.1103/PhysRevLett.93.118102. [DOI] [PubMed] [Google Scholar]
- Seol Y, Skinner GM, Visscher K, Buhot A, Halperin A. Stretching of homopolymeric RNA reveals single-stranded helices and base-stacking. Phys Rev Lett. 2007;98:158103. doi: 10.1103/PhysRevLett.98.158103. [DOI] [PubMed] [Google Scholar]
- Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–60. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- Stead JA, Costello JL, Livingstone MJ, Mitchell P. The PMC2NT domain of the catalytic exosome subunit Rrp6p provides the interface for binding with its cofactor Rrp47p, a nucleic acid-binding protein. Nucleic Acids Res. 2007;35:5556–67. doi: 10.1093/nar/gkm614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons MF, Jones GH, Luisi BF. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure. 2000;8:1215–26. doi: 10.1016/s0969-2126(00)00521-9. [DOI] [PubMed] [Google Scholar]
- Tollervey D, Mattaj IW. Fungal small nuclear ribonucleoproteins share properties with plant and vertebrate U-snRNPs. EMBO J. 1987;6:469–76. doi: 10.1002/j.1460-2075.1987.tb04777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, Dziembowski A, Jensen TH. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010;29:2342–57. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3’-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol. 2000;20:441–52. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–4. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- Wang HW, Wang J, Ding F, Callahan K, Bratkowski MA, Butler JS, Ke A. Architecture of the yeast Rrp44 exosome complex suggests routes of RNA recruitment for 3’ end processing. Proc Natl Acad Sci USA. 2007;104:16844–9. doi: 10.1073/pnas.0705526104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, Libri D, Jacquier A. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–37. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–7. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


