Abstract
Although the genomic pattern of nucleosome positioning is broadly conserved, quantitative aspects vary over evolutionary timescales. We identify the cis and trans determinants of nucleosome positioning using a functional evolutionary approach involving S. cerevisiae strains containing large genomic regions from other yeast species. In a foreign species, nucleosome depletion at promoters is maintained over poly(dA:dT) tracts, whereas internucleosome spacing and all other aspects of nucleosome positioning tested are not. Interestingly, the locations of the +1 nucleosome and RNA start sites shift in concert. Strikingly, in a foreign species, nucleosome-depleted regions occur fortuitously in coding regions, and they often act as promoters that are associated with a positioned nucleosome array linked to the length of the transcription unit. We suggest a three-step model, in which nucleosome remodelers, general transcription factors, and the transcriptional elongation machinery are primarily involved in generating the nucleosome positioning pattern in vivo.
INTRODUCTION
In living cells, nucleosome positions are influenced by intrinsic DNA sequence preferences due to the thermodynamic costs associated with wrapping stiff DNA around the histone octamer (Drew and Travers, 1985; Jiang and Pugh, 2009; Radman-Livaja and Rando, 2010). In addition, a wide variety of proteins can affect nucleosome positions and occupancy, most notably ATP-dependent chromatin remodeling complexes. The relative importance of DNA sequence and protein factors in determining nucleosome positioning has been subject to considerable debate. In vitro reconstitution of genomic DNA into nucleosomes by salt dialysis recapitulates gross variation in nucleosome occupancy in yeast and in humans – AT-rich sequences such as those found at yeast promoters are intrinsically nucleosome-depleted (Kaplan et al., 2009; Sekinger et al., 2005; Zhang et al., 2009), whereas the GC-rich sequences prevalent at human promoters are intrinsically nucleosome-enriched (Valouev et al., 2011). These studies typically find little role for intrinsic preferences in precise nucleosome positioning, although the enrichment of particular sequence features (10 bp periodicity of AA/AT/TA dinucleotides) at the +1 position in budding yeast has nonetheless led to forceful (Kaplan et al., 2010; Kaplan et al., 2009; Segal et al., 2006), but disputed (Fan et al., 2010; Stein et al., 2009; Weiner et al., 2010; Zhang et al., 2009, 2010) claims that intrinsic DNA sequence preferences play a major determining role in nucleosome positioning.
Conversely, several experimental approaches, largely in budding yeast, have revealed a key role for proteins in establishing nucleosome positions in vivo. While in vitro reconstitution of DNA into nucleosomes does not properly establish nucleosome positions at PHO5, addition of yeast whole cell extract enables more accurate assembly of nucleosomes at this locus (Korber and Horz, 2004). Genome-wide analysis subsequently showed that one or more ATP-dependent activities in yeast whole cell extract can assemble nucleosomes in positions that resemble, but do not completely coincide with, in vivo positioning (Zhang et al., 2011), thereby demonstrating a major role for nucleosome-remodeling complexes in nucleosome positioning. Decades of biochemical studies have identified many specific proteins and protein complexes capable of altering nucleosome positions on DNA in vitro (Clapier and Cairns, 2009), and increasingly these factors are being implicated in proper nucleosome positioning in vivo (Gkikopoulos et al., 2011; Whitehouse et al., 2007; Whitehouse and Tsukiyama, 2006). For example, the ATP-dependent remodeling enzymes Chd1, Isw1, and Isw2 globally affect nucleosome positioning in vivo, as their deletion in yeast leads to nearly complete loss of nucleosome positioning downstream of the +2 nucleosome of coding regions (Gkikopoulos et al., 2011).
In a genetic approach to this problem, diploid hybrids between the closely-related species, S. cerevisiae and S. paradoxus, have been used to determine to what extent divergent nucleosome positioning on specific orthologous genes can be attributed to cis or trans factors, with the majority of chromatin changes between these species being attributed to poly(dA:dT) elements at promoters (Tirosh et al., 2010). However, S. cerevisiae and S. paradoxus differ very little in bulk aspects of chromatin architecture. In contrast, chromatin structure exhibits far greater differences between more divergent species: for example, average nucleosome spacing differs by ~15–20 bp between S. cerevisiae and K. lactis (last common ancestor ~150 MYA) (Heus et al., 1993; Tsankov et al., 2010).
Here, we describe a functional evolutionary approach to systematically dissect the contributions of DNA sequence and the nuclear environment to nucleosome positioning in vivo. This approach relies on the finding that there are species-specific differences in parameters of nucleosome positioning in a variety of yeast species, even though the general pattern is highly conserved (Tsankov et al., 2010). Specifically, we compare nucleosome maps of artificial chromosomes (YACs) containing large, heterologous genomic regions from different yeast species in S. cerevisiae with maps of the same regions in their native organism (Figure 1A). In principle, features that change in the context of S. cerevisiae are determined by protein factors that are functionally distinct in the two species, whereas features that are retained when the foreign yeast DNA is present in S. cerevisiae are either due to intrinsic DNA sequence or to conserved trans-acting regulators. For example, when the S. cerevisiaeHIS3-PET56 region is introduced into S. pombe, it retains the nucleosome-depleted promoter region, but not the positions of nucleosomes in the coding region (Sekinger et al., 2005). In addition, the generation of fortuitous functional elements arising from heterologous genomic sequences makes it possible to address mechanistic issues that are presumably free of evolutionary constraints. Here, we show that nucleosome spacing is established in trans, and that promoter nucleosome depletion can be established either by intrinsic sequence cues or by trans-acting factors. Further, we find that +1 nucleosome positioning is most likely established by some aspect of the transcriptional machinery, and positioning of more downstream nucleosomes in the mRNA coding region is linked to Pol II elongation. Based on results presented here and elsewhere, we propose a unifying, three-step model for how nucleosome positions are established in vivo.
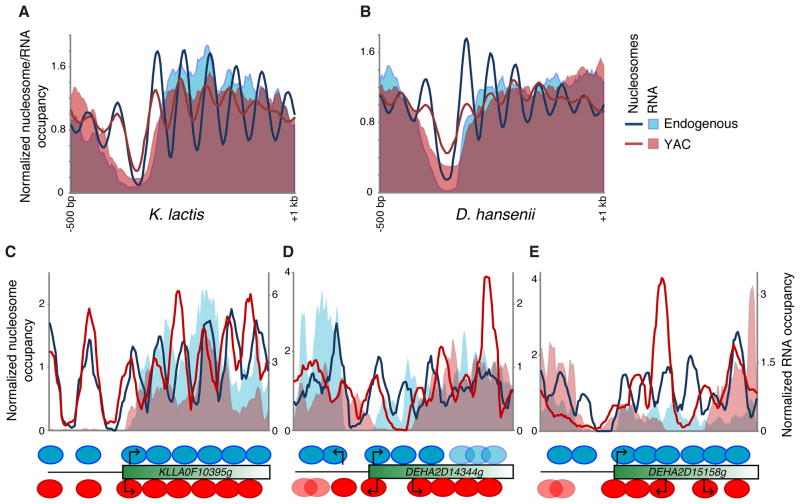
Figure 1. Functional evolutionary dissection of chromatin establishment mechanisms.
(A) Schematic of experimental design. Yeast Artificial Chromosomes are constructed carrying sequence from species such as K. lactis, and introduced into S. cerevisiae. Comparison of nucleosome mapping data between the same sequence in two different environments (its endogenous genome, and in S. cerevisiae) can be used to disentangle DNA-driven from trans-mediated aspects of chromatin organization.
(B) Chromosomal complement of parental S. cerevisiae (AB1380) and 3 different YAC-bearing strains. Pulsed field gel electrophoresis of YAC-bearing strains, as indicated.
(C–D) Examples of nucleosome mapping data from two genes. Blue line indicates nucleosome mapping data from wild-type K. lactis (Tsankov et al., 2010), red line shows data from the same sequence carried on a YAC in S. cerevisiae.
(E–F) Data for all K. lactis genes on all 3 YACs. (E) shows data for all genes from wild-type K. lactis, with genes sorted by NDR width, while (F) shows data from these genes on YACs, sorted identically. Black indicates no sequencing reads, yellow intensity indicates number of sequencing reads. C and D indicate the example genes shown above.
RESULTS
Generation of S. cerevisiae strains harboring artificial chromosomes with large segments of foreign yeast DNA
To generate yeast artificial chromosomes (YACs, Figure 1A), genomic DNA from K. lactis, K. waltii, and D. hansenii was sheared to ~100–200 kb average size, and ligated to the pYAC4 vector carrying sequences for S. cerevisiae telomeres, centromere, and origin of replication, as well as two selectable markers. YACs were transformed into wild-type S. cerevisiae and confirmed by pulsed-field gel electrophoresis (Figure 1B). Furthermore, both ends of YACs containing foreign yeast DNA inserts were validated by DNA sequencing. In total, we generated seven strains carrying distinct YACs from the three species, with an average insert length of ~140 kb (Table S1). YAC strains were grown in identical conditions to those previously used for mapping nucleosomes in these four species (Tsankov et al., 2010), and formaldehyde cross-linked chromatin was digested to ~80% mononucleosomes using micrococcal nuclease (Yuan et al., 2005). Mononucleosomal DNA was analyzed by deep sequencing as previously described (Shivaswamy et al., 2008; Tsankov et al., 2010; Weiner et al., 2010). Figures 1C–D show nucleosome mapping data for two genes from K. lactis, with data from wild-type K. lactis in blue (“endogenous”), and data for these same genes in the context of a YAC-carrying S. cerevisiae strain in red (“YAC”). Notable in these views are a number of well-described aspects of fungal chromatin structure – in the endogenous context, nucleosomes are generally well-positioned (nucleosome peaks are well separated and exhibit high peak to trough ratios), and both genes have a nucleosome-depleted region (NDR) that contains the gene’s promoter.
Promoter NDRs are largely maintained in a foreign species in a manner strongly correlated with poly(dA:dT) tracts
The endogenous positions of promoter NDRs were largely maintained in the YACs (Figures 1E–F) – 50% and 56% of D. hansenii and K. lactis NDRs, respectively, were located within 50 bp of their endogenous position, and for both species’ sets of YACs only 13% of NDRs did not overlap the endogenous NDR at all. Furthermore, the extent of the NDR, which varies considerably among genes, is largely maintained in the YAC-containing strains. These data are consistent with the view that nucleosome depletion at fungal promoters is largely programmed by genomic sequence. However, the average extent of depletion over promoters is not as great in YACs as in wild-type (Figures 1C–F, and Figures S1A–B), potentially as a consequence of the reduced expression of YAC genes (see below). This observation suggests that some of the depletion at promoters is not intrinsically determined by DNA sequence, consistent with the previous observation that nucleosome depletion at promoters is more pronounced in vivo than in vitro (Kaplan et al., 2009; Zhang et al., 2009; Zhang et al., 2011).
To further investigate the role for intrinsic sequence cues in establishing nucleosome depletion, we sorted genes by the difference in nucleosome occupancy over the proximal NDR between endogenous genes and YACs (Figure 2A, Figures S1C–D). Notably, in both K. lactis and D. hansenii we observed very few genes with lower nucleosome occupancy in the YAC context, with the majority of promoters showing a range from little change to substantially increased nucleosome occupancy. Genes that maintained the same level of nucleosome depletion in the YAC as in wild-type were characterized by promoter sequences with greater numbers of long poly(dA:dT) elements (Figures 2B–C, see Methods), consistent with the idea that these sequences intrinsically program nucleosome depletion in any context. Genes from D. hansenii tended to exhibit much less dramatic nucleosome depletion at promoters in the YAC context than in their endogenous context (Figure S1B). This is consistent with our prior observation (Tsankov et al., 2011; Tsankov et al., 2010) that D. hansenii promoters have fewer poly(dA:dT) sequences than most other organisms in the Hemiascomycota phylogeny (potentially due to their ecological niche in high salt environments) and with the hypothesis that D. hansenii promoters are more often established by trans-acting proteins such as General Regulatory Factors (GRFs). In this regard, promoters that gained substantial nucleosome occupancy when carried in the YAC sometimes, but not always, contained known binding motifs for transcription factors we previously (Tsankov et al., 2011; Tsankov et al., 2010) inferred to be GRFs in D. hansenii but not in S. cerevisiae (Figure 2D). Together, these results indicate that intrinsic sequence determinants (or conserved trans-acting factors) play a major role in generating nucleosome depletion at fungal promoters, and that poly(dA:dT) tracts are the primary DNA sequence determinant.
Figure 2. Promoter nucleosome depletion is maintained over poly(dA:dT) elements.
(A) D. hansenii genes sorted by the extent of change in nucleosome occupancy over the NDR. Left panel shows differences in nucleosome occupancy between D. hansenii and YACs for 114 genes – blue indicates increased nucleosome occupancy in the YAC relative to endogenous context. Middle and right panels show nucleosome mapping data for endogenous D. hansenii sequences and for YACs, as indicated.
(B) Strength of poly(dA:dT) element (Field et al., 2008; Tsankov et al., 2010) for genes, ordered as in (A). 40 gene running window average is shown.
(C) An example of a gene with little change in nucleosome depletion between endogenous and YAC contexts. Sequence from this stable NDR contains multiple poly(dA:dT) elements, as indicated in red.
(D) An example of a gene exhibiting dramatically increased nucleosome occupancy at the native NDR when carried on YAC. Here, this NDR includes few polyA elements, and carries a binding site for Cbf1, which has nucleosome-evicting activity in D. hansenii but not in S. cerevisiae (Tsankov et al., 2011; Tsankov et al., 2010).
Nucleosome positions differ markedly in the endogenous and YAC-containing strains
In contrast to the widespread but not universal conservation of NDRs, a given DNA sequence is generally packaged differently when carried in the endogenous species or in S. cerevisiae. Nucleosome positions change markedly in the YAC strains – the +1 nucleosome can be found near to (Figure 1C), upstream (see below), or downstream (Figure 1D) of its endogenous location, while nucleosomes farther downstream of the +1 occur farther and farther away from their endogenous locations. By definition, differences in nucleosome positioning of a given genomic region in the endogenous organism or in S. cerevisiae cannot be due to intrinsic DNA sequence, but rather trans-acting factor(s). These measured differences are not secondary to technical artifacts such as differences in MNase digestion, as we observe remarkably consistent results for S. cerevisiae genes for the various YAC datasets (Figure S2). Interestingly, the average deviation in chromatin structure between genes in their endogenous context and in the YAC was quite different for the three species studied – K. lactis genes appeared closest to their native structure when in YACs, whereas chromatin structure of K. waltii sequences in YACs appeared random with respect to genic structure (data not shown).
Nucleosome spacing is determined by protein factors in the host organism, not DNA sequence
As observed by MNase cleavage of bulk chromatin, most nucleosomes in any given species are found in arrays in which the linker regions between adjacent nucleosomes are similar in size. Interestingly, nucleosome spacing is substantially different between S. cerevisiae, with an average internucleosomal distance of ~165 bp, and K. lactis, with an average spacing of ~178 bp (Heus et al., 1993; Tsankov et al., 2010). We therefore used our YAC dataset to assess whether nucleosome spacing over K. lactis genes is established by DNA sequence, or by the nuclear environment. As can be appreciated in Figures 1C–D, nucleosome spacing appears shorter over K. lactis genes when carried in S. cerevisiae, relative to the endogenous spacing. Figure 3A shows the average nucleosome data for all K. lactis genes present on one of the 3 YACs, aligned by the endogenous location of the +1 nucleosome. Average nucleosome spacing decreases when these genes are carried in YACs. The distribution underlying this average trend is quantified in Figure 3B. Here, we called nucleosome positions (Weiner et al., 2010), then plotted the distribution of all internucleosomal distances as indicated. K. lactis genomic sequence in its endogenous context is packaged with nucleosomes occurring every 178 bp, whereas the same sequence in the S. cerevisiae trans environment exhibits ~165 bp nucleosome spacing, precisely the same spacing observed over native S. cerevisiae genes. Importantly, we observed no change in the spacing of S. cerevisiae genes between wild type yeast and our YAC strains, indicating no artifactual effects on nucleosome spacing from, for example, MNase titration level (Figure 3B, Figure S2). The primary role for proteins in determining internucleosomal spacing is not surprising, as different cell types in multicellular organisms (sharing identical genomes) can exhibit different nucleosome spacing (Van Holde, 1989). Importantly, the observation that internucleosomal spacing depends on protein factors means that the precise positions for the vast majority of nucleosomes are not determined by intrinsic DNA sequence.
Figure 3. Nucleosome spacing is set in trans.
(A) Averaged data for all K. lactis genes on YACs 1–3. Genes are aligned by the +1 nucleosome position as defined in Tsankov et al., and data from either wild-type K. lactis or from the YAC strains are averaged for 184 genes, as indicated.
(B) K. lactis sequences adopt S. cerevisiae spacing when carried in S. cerevisiae. Nucleosome positions were called, and the distribution of all internucleosomal distances (center to center) is shown for 184 K. lactis genes from wild-type or in the YACs. Similar distributions for S. cerevisiae nucleosome positioning from wild-type and YAC-containing strains indicates that YACs do not perturb host chromatin state (See also Figure S2).
The position of the +1 nucleosome is not determined by DNA sequence, but rather is mechanistically linked to transcriptional initiation
The claim that the +1 nucleosome is positioned by DNA sequence (Segal et al., 2006) has been subject to debate, not least because in vitro reconstitution experiments reveal no significant recovery of +1 nucleosome positioning. Alternatively, it has been proposed that the +1 nucleosome is positioned by either transcription factors such as Rap1, Abf1, and Reb1 (Kornberg and Stryer, 1988; Zhang et al., 2009; Zhang et al., 2011), NDRs (Mavrich et al., 2008; Yuan et al., 2005), or the preinitiation complex (Zhang et al., 2009).
Interestingly, although the average position of the +1 nucleosome is similar between YACs and the endogenous context for both K. lactis and D. hansenii genes (see Figure 3A, Figure S1), examination of individual genes shows that +1 positioning is highly variable for the same sequence in two different nuclear environments. The distribution of +1 nucleosome shifts for genes carried on YACs was far more variable than the experimental variability measured using the background of S. cerevisiae genes – while only 17% (14%–21% in different YAC strains) of S. cerevisiae +1 nucleosomes appeared > 20 bp apart between strains, 45% of K. lactis and 57% of D. hansenii +1 nucleosomes in the YAC strains shifted at least 20 bp from their endogenous location (Figure S3). +1 nucleosomes could shift in either direction on YACs (Figure 4, Figure S3), although in K. lactis YACs these shifts were biased towards upstream shifts (Figure 3A). Thus, our observations demonstrate that pronucleosomal sequences do not “program” the position of the +1 nucleosome in vivo.
Figure 4. +1 nucleosome shifts associated with transcriptional changes.
(A–B) Nucleosome data and RNA-Seq data are shown for K. lactis and D. hansenii genes in wild-type and YACs, as indicated. RNA-Seq data for YAC-derived transcripts are normalized independently from S. cerevisiae transcripts here – see Figures S4B–C for data normalized genome-wide.
(C–E) Examples of +1 nucleosome shifts associated with changes in transcription. (C) shows a moderate upstream shift in a +1 nucleosome with a similar change in transcript length, while (DE) show large scale NDR gain/loss with associated changes in transcription. Schematic interpretation of the nucleosome positioning for the endogenous gene is shown in blue above the rectangle, nucleosome positioning in the YAC is shown in red below the rectangle. Arrows indicate inferred TSSs (note that RNA-sequencing data are not strand-specific, but TFIIB mapping data support our inferred TSSs) – the furthest 5′ RNA in (E), for example, derives from the upstream gene as opposed to a divergent promoter.
The strong correspondence between +1 nucleosome positioning and transcriptional start sites in many species (Jiang and Pugh, 2009) led us to consider the hypothesis that changes in transcriptional activity might underlie the repositioning of the +1 nucleosomes (Zhang et al., 2009). We therefore carried out deep sequencing of RNA isolated from D. hansenii, K. lactis, and the S. cerevisiae YAC strains in this study, and carried out ChIP-Seq for TFIIB localization in the YAC-containing strains (a full analysis of these data will be published separately). Alignment of RNA-Seq data from wild-type strains with nucleosome mapping data confirmed prior predictions that the positioning of +1 nucleosomes with respect to a gene’s transcription start site (TSS) varies between these species (Tirosh et al., 2007; Tsankov et al., 2010) – transcription begins further inside the +1 nucleosome in K. lactis than in D. hansenii (Figure S4A).
Comparing endogenous to YAC-based gene expression, we found on average that genes on YACs were expressed at lower levels than host genes – average sequencing reads per kilobase of coding sequence for YACs was ~40% of the average value for endogenous RNAs – consistent with extensive promoter sequence divergence between species resulting in widespread misinterpretation of exogenous regulatory information by the S. cerevisiae transcriptional machinery (Figures S4B–C and E). In general, we found a good correlation between expression levels for genes in their endogenous genome versus expression from the YACs (Figure S4D) – genes expressed at high levels in K. lactis remained the most highly-expressed genes when carried on YACs, but were expressed at lower levels relative to S. cerevisiae genes. In D. hansenii, we also observed increased expression of intergenic regions in the YACs (Figure S4C and see below), again indicating evolutionary divergence in transcriptional control sequences (e.g. loss of transcriptional termination signals and/or gain of cryptic promoters).
Consistent with a relationship between +1 nucleosome positioning and transcription start sites, we found that the 5′ ends of RNAs in YACs shifted on average towards a S. cerevisiae-like location relative to the +1 nucleosome (Figures 4A–B) – K. lactis RNAs started farther upstream in the YAC, whereas D. hansenii RNAs started farther downstream. Furthermore, +1 nucleosome shifts largely were accompanied by coherent shifts in inferred transcription start sites. These include ~90 examples such as that seen in Figure 4C, in which both RNA-Seq data and the +1 nucleosome for a given gene shift in the same direction. This is visualized in Figure S5A – RNA-Seq data for K. lactis genes exhibiting no +1 nucleosome shift, and for those exhibiting upstream +1 nucleosome shifts, is averaged for both endogenous RNA expression and YAC-based expression. Despite no average difference in 5′ ends of transcripts between these two classes in the endogenous case, we find that genes exhibiting upstream shifts in the +1 nucleosome also showed more strongly 5′-shifted transcripts relative to genes without a +1 nucleosome shift, consistent with the idea that there is a mechanistic coupling between +1 nucleosome positioning and transcriptional initiation. Furthermore, we also used 5′ RACE to more precisely map TSSs for 4 K. lactis genes in their endogenous context and in the YAC, confirming that +1 nucleosome shifts corresponded to shifts in the location of the TSS (Figures S5B–E). These observations provide functional evidence for a mechanistic linkage between nucleosome positioning and transcriptional initiation, although they do not establish the cause-and-effect relationship between these two processes.
Generation of NDRs in foreign coding regions via fortuitous interactions of S. cerevisiae activator proteins
More dramatic cases of non-conserved nucleosome positioning are observed, particularly in D. hansenii -derived YACs, in which many fortuitous NDRs arise in coding regions (Figures 4D–E, Figures 5A–B, Figures S6A–B). The sequences underlying these NDRs are not associated with poly(dA:dT) elements (analysis not shown), as expected given that they do not intrinsically form NDRs in their host genomic context (Figure 5B, blue line). Interestingly, these NDRs are associated with TFIIB binding (Figures 5C–D) and concomitant changes in RNA abundance (Figures 4D–E, Figure 5A, Figure S6A) indicating a wholesale functional change in which a coding sequence from one species (D. hansenii) is used as a promoter in a foreign species (S. cerevisiae). These NDRs are most likely determined by S. cerevisiae transcription factors that fortuitously recognize and functionally act on foreign DNA sequences that do not act as promoters in the native organism. In other words, DNA-binding transcriptional activator proteins recruit nucleosome-remodeling complexes to these fortuitously recognized sites, thereby evicting histones and generating an NDR. These NDRs are associated with varying levels of TFIIB and RNA transcripts, presumably depending on the quality of TATA elements and other core promoter sequences in the vicinity.
Figure 5. Characterization of fortuitousNDRs in YACs.
(A) Example of a fortuitous NDR that occurs only in the YAC but not in the native genome, and is associated with transcription. This fortuitous NDR occurs in the middle of a D. hansenii coding region, and is associated with two shorter, divergent transcripts in the YAC context (data cover 2.2 kb of sequence). Note that nucleosome organization correlates with transcript length – rightmost transcript shows greater nucleosome positioning at the 5′ end than at the 3′ end of the transcript.
(B) Fortuitous NDRs are generally associated with well-positioned +/−1 nucleosomes. Averaged data for 120 NDRs observed in D. hansenii YACs but not in the endogenous context, as indicated.
(C–D) Fortuitous NDRs represent functional promoters. (C) shows TFIIB ChIP-Seq data from YAC-bearing strain for the genomic locus shown in (A), while (D) shows averaged data for all fortuitous NDRs. Note that TFIIB localization in the endogenous context could not be obtained as our anti-TFIIB antibody does not recognize TFIIB from D. hansenii.
Fortuitous coding region NDRs are associated with typical nucleosome patterns
The existence of fortuitous and presumably evolutionarily meaningless promoters in D. hansenii coding regions in the context of S. cerevisiae cells makes it possible to determine the role of transcription in establishing the nucleosome positioning pattern. Strikingly, these coding region NDRs are associated with a typical nucleosome pattern of highly positioned +1 and −1 nucleosomes as well as progressively less positioned downstream nucleosomes (Figure 5B). Thus in the absence of any intrinsic nucleosome-destabilizing sequences, transcription factors and associated co-factors are sufficient to generate a nucleosome positioning pattern that is very similar to the standard pattern at endogenous promoters. Furthermore, at such fortuitous NDRs, the extent of the positioned array is linked to the length of the RNA transcript (Figure 5A, Figure S6C), strongly suggesting a role for transcriptional elongation in the generation of the nucleosomal pattern. These results demonstrating a functional role for transcription-related events appear to conflict with the conclusion that nucleosome-remodeling complexes are sufficient to establish aspects of the nucleosome positioning pattern in the absence of transcription (Zhang et al., 2011). However, these observations are not mutually exclusive, and indeed are complementary as both mechanisms are likely to contribute to establishing the nucleosome pattern.
DISCUSSION
A functional evolutionary approach to address the determinants of molecular phenomena in vivo
Here, we used a functional evolutionary approach to systematically dissect the role for cis-acting sequence elements and trans-acting proteins in establishment of nucleosome positioning in fungi. This approach, which is based on species-specific differences in parameters of nucleosome positioning in a variety of yeast species (Tsankov et al., 2010), involves placing large segments of foreign yeast DNA in S. cerevisiae and comparing molecular properties in such strains with those in the native organism. In principle, non-conserved properties are determined by protein factors that are functionally distinct in the two species, whereas conserved properties are due either to DNA sequence or to conserved trans-acting regulators. The use of yeast artificial chromosomes to carry the foreign yeast DNA makes it possible to examine many genes at once, and hence to obtain information that is both statistically robust and permits one to identify many examples of new phenomena. Furthermore, the ability to generate fortuitous functional events (e.g. the NDRs in D. hansenii coding regions) that do not occur in the native organisms makes it possible to address mechanistic questions in a manner that is, most likely, independent of evolutionary history. An extension of this approach should also permit one to identify factors responsible for the species-specific behavior, specifically by replacing a candidate factor by its homolog in the foreign species and examining whether the pattern resembles that of the foreign species.
More generally, this functional evolutionary approach should allow for elucidating the determinants of other molecular phenomena that are broadly conserved but show species-specific differences. For example, more detailed analysis of the RNA generated in the YAC-containing strains with the corresponding endogenous yeast species should be informative of determinants of 5′ and 3′ end formation, splicing, and half-lives. As such, this approach combines the virtues of evolutionary comparison and classic functional genetic analysis.
A three-step model for establishing the nucleosome positioning pattern in vivo
Based on results presented here and elsewhere, we propose a three-step model (Figure 6) for how nucleosome positioning is established in eukaryotic organisms. The first step involves the generation of an NDR, which can occur either by transcription factors and their recruited nucleosome remodeling complexes and/or by poly(dA:dT) sequences that intrinsically disfavor nucleosome formation. Even at poly(dA:dT)-containing promoters, it is likely that transcriptional machinery contributes to nucleosome depletion, as nucleosome depletion is more pronounced in vivo than in vitro (Kaplan et al., 2009; Zhang et al., 2009), and nucleosome-remodeling complexes enhance the depletion in vitro (Zhang et al., 2011). In this sense, intrinsic programming of NDRs represents a specialized mechanism that is used frequently (S. cerevisiae), moderately (D. hansenii), or rarely (D. melanogaster), depending on the species.
Figure 6. Three-step model for establishment of nucleosome positioning in vivo.
A unifying three-step model for how nucleosome positioning pattern is generated in eukaryotic organisms. The first step is the generation of an NDR, either by poly(dA:dT) elements and/or by transcription factors and their recruited nucleosome remodeling complexes. In the second step, nucleosome-remodeling complexes recognize the NDRs and generate highly positioned nucleosomes flanking the NDR; and the RNA polymerase II preinitiation complex fine-tunes the position of the +1 nucleosome. In the final step, positioning of the more downstream nucleosomes depends on transcriptional elongation, and the recruitment of nucleosome-remodeling activities and histone chaperones by the elongating RNA polymerase II machinery.
In the second step, nucleosome-remodeling complexes recognize the NDRs and generate highly positioned nucleosomes flanking the NDR. Strong positioning could, in principle, arise simply from the boundary of the NDR and/or from sequence preferences of the nucleosome remodelers. Indeed, it has been argued that this step does not require transcription factors or transcription per se (Zhang et al., 2011), although it is important to note that there is overall poor correspondence between +1 nucleosome positioning observed in vivo and that recapitulated using ATP-dependent extracts in the absence of transcription (Zhang et al., 2011) (Figure S7). In this regard, Zhang et al. compared nucleosome positioning generated by ATP-dependent extracts with the nucleosome positions measured from yeast lysed without crosslinking and allowed to redistribute prior to crosslinking. Indeed, we find mediocre correspondence between the “native” nucleosome positions from Zhang et al. and true in vivo nucleosome positions generated from crosslinked yeast (Figure S7), so the ability of whole cell extracts to recover these “native” positions in the absence of transcription does not have any bearing on the question of whether in vivo positioning is influenced by transcription prior to lysis of cells. Although nucleosome remodelers can generate somewhat positioned nucleosomes flanking the NDR and unquestionably perform far better than salt dialysis, they apparently are insufficient to generate the precise in vivo nucleosome positions, particularly for the +1 nucleosome (Figure S7).
Here, the strong, and species-specific, spacing relationship between the +1 nucleosome and mRNA start site that is observed both in the native and YAC strains indicates that there is a mechanistic connection between transcriptional initiation and the location of the +1 nucleosome. Given the strong in vivo positioning of both the preinitiation complex and the +1 nucleosome, a spacing relationship between these two entities requires that at least one of these is anchored to a specific location, thereby permitting a defined location for the second entity. As discussed above, nucleosome remodeling complexes alone are insufficient to generate proper positioning of the +1 nucleosome, and hence sequence and nucleosome remodelers are insufficient to provide an anchor. In contrast, preinitiation complexes bound at core promoters are clearly sufficient to provide an anchor, with the location of the TBP bound to the TATA element or TATA-related sequence being the major determinant of the anchor point. From these considerations, and our finding that the TSS to +1 distance in YACs shifts to the S. cerevisiae spacing (Figure 5 and Figure S5), we suggest that the preinitiation complex plays a role in fine-tuning the position of the +1 nucleosome.
In the third step, positioning of downstream nucleosomes, with progressively less positioned nucleosomes downstream within the gene, depends on transcriptional elongation, and hence recruitment of nucleosome-remodeling activities and histone chaperones by the elongating RNA polymerase II machinery. This elongation-dependent step explains why nucleosome-remodeling complexes, though capable of weakly positioning nucleosomes flanking the NDR, are unable to position more downstream nucleosomes (Zhang et al., 2011). Conversely, yeast mutant strains lacking nucleosome-remodeling complexes (Chd1 and Isw1) that are recruited to coding regions by elongating RNA polymerase show drastically reduced positioning of downstream nucleosomes, but relatively normal positioning of the +1 and +2 nucleosomes (Gkikopoulos et al., 2011). Finally, a transcription-based step nicely helps to explain why nucleosome arrays occur largely in the transcribed direction even though highly positioned nucleosomes can occur both at the +1 and −1 position, as well as the curious observation that the decay of nucleosome positioning towards the center of genes displays a 5′/3′ asymmetry (Vaillant et al., 2010); both of these observations are inconsistent with a pure packing-based model.
The above model can explain why the general pattern of nucleosome positioning is highly conserved among eukaryotes, yet shows species-specific differences in various aspects of chromatin structure. These species-specific differences reflect the relative utilization of poly(dA:dT) sequences and hence intrinsic histone-DNA interactions, as well as differences in the enzymatic and recruitment properties of the nucleosome remodelers.
METHODS
Growth Conditions
All cultures were grown in medium containing: SC – Tryptophan – Uracil (Sunrise Sciences) (0.2%), Yeast extract (1.5%), Peptone (1%), Dextrose (2%), and Adenine (0.01%), as previously described (Tsankov et al., 2010).
Preparation of YACs
Yeast chromosomal DNA was prepared in InCert agarose blocks (LONZA), with a final cell concentration of 2 × 109 cells/ml. Agarose blocks with intact chromosomal DNA were subjected to EcoRI partial digestion with a titrated Mg2+ concentration, followed by size fractionation using pulsed field gel electrophoresis (PFGE). ~100–200 kb partially-digested DNA fragments were excised from the gel. YAC vector pYAC4 was purified by successive CsCl gradient ultracentrifugation and digested with BamHI and EcoRI, followed by calf intestine alkaline phosphatase treatment. Digested pYAC4 and partially-digested yeast chromosomal fragments were ligated by T4 DNA ligase in agarose blocks. Prior to YAC transformation, ligated DNA was size-fractionated again by PFGE and DNA larger than 100 kb was excised from the gel. The excised gel slice was further digested with β-agarase and ligated DNA was transformed into S. cerevisiae host cells (AB1380), using either spheroplast transformation protocol or standard yeast LiCl transformation method. Further details of YAC construction are available upon request.
Validation of YAC-bearing strains
Transformants with red color, which survived double selection (Ura+/Trp+) on AHC plates, were collected for validation. Chromosomal DNA of candidate strains was prepared in agarose blocks and resolved by PFGE (Figure 1B). Strains with desired YAC bands were selected, and terminal sequences from selected YAC clones were isolated and confirmed with DNA sequencing analysis (Riley et al., 1990).
Nucleosome Isolation and Illumina Deep Sequencing
Micrococcal nuclease (MNase) digestions were performed as previously described (Yuan et al., 2005). Briefly, 450mL cultures were grown to OD600 of ~0.5 at 30°C, 220rpm. Cultures were fixed for 30 minutes at 30°C with 1.85% formaldehyde, then spheroplasted with 10 mg zymolyase (Cape Cod Associates) for 45 minutes at 30°C. Spheroplasts were subjected to 20 minutes of MNase digestion, and DNA was purified. MNase titrations were selected to obtain largely mononucleosomal DNA with some di- and tri-nucleosomal DNA apparent. Mononucleosomal DNA was gel purified (BioRad Freeze N′ Squeeze) and used to create a library for deep sequencing on the Solexa 1G Genome Analyzer, as previously performed (Shivaswamy et al., 2008; Tsankov et al., 2010; Weiner et al., 2010). Briefly, DNA was blunt ended, A-tailed, and ligated to Illumina genomic adapters, followed by a final PCR with a size-selecting gel purification.
Data Normalization and Nucleosome TSS Alignments
Reads from deep sequencing were mapped back to the relevant hybrid genome (S. cerevisiae plus the relevant species’ chromosome), using blat. Uniquely mapping reads that had fewer than three mismatches were kept for analysis. Reads were extended by the cross correlations between those from the Watson and Crick strands, to create nucleosome peaks. Read count numbers were normalized to one by dividing each base read count by the genome-wide average read count per base. Gene alignments were carried out using the endogenous boundary of the +1 nucleosome (Tsankov et al., 2010). RNA-seq data was treated similarly but without extending reads. RNA abundance for YAC-based transcripts was, on average, ~30–40% (in reads per kb per million reads) of the RNA abundance of endogenous S. cerevisiae transcripts.
Nucleosome Calls
Template Filtering (Weiner et al., 2010) was used to call the locations of nucleosomes.
5′ RACE
Trizol (Invitrogen) extracted RNA was enriched for mRNA on polyT magnetic beads (NEB). Calf Intestinal Phosphatase removed all phosphates prior to the hydrolysis of the mRNA cap to a phosphate with Tobacco Acid Pyrophosphatase (Epicentre). An oligo was ligated to the 5′ end of the mRNA (T4 RNA Ligase) and the RNA was reverse transcribed (SuperScript III Reverse Transcriptase, Invitrogen) with a tailed random hexamer. The cDNA was amplified with a low cycle PCR (Phusion, NEB) using primers matching the sequences added in the ligation and reverse transcription. A gene specific PCR amplified the transcription start site sequence, which was cloned (StrataClone, Agilent) and sequenced.
Supplementary Material
Figure S1. NDRs are generally better-maintained over sequence from K. lactis than over D. hansenii sequence
(A–B) Nucleosome mapping data for all genes from K. lactis (A) or D. hansenii (B) are shown for wild-type and YACs, as indicated. Genes are sorted by wild-type NDR width.
(C) NDR maintenance correlates with poly(dA:dT) elements. As in Figures 2A–B, but for K. lactis.
(D) Identical to Figures 2A–B, reproduced here for comparison between D. hansenii and K. lactis.
Figure S2. Bulk chromatin is not affected in YAC-bearing strains
(A) Nucleosome sequencing data for all strains in this study was mapped to the S. cerevisiae genome, and data are averaged for all genes aligned by the +1 nucleosome.
(B) Data for all S. cerevisiae genes are shown for wild-type (BY4741) and 5 YAC-bearing strains. Genes are sorted by K means clustering (K=4) of BY4741 dataset.
Figure S3. +1 nucleosome shifts associated with transcription.
(A) Distribution of shifts for +1 nucleosomes. Distributions are shown for the shifts between +1 positions in wild-type and YAC-bearing strains. The three S. cerevisiae distributions show changes in +1 positioning for S. cerevisiae genes in the indicated YACs, showing that technical variability or analytical variability do not account for nucleosome position changes for YAC-associated genes.
(B) Data for all K. lactis and D. hansenii genes, from wild-type and YACs. Data are shown from −100 to +300 bp relative to the +1 nucleosome upstream border (+1 nucleosome center indicated as a red line). Green box for K. lactis genes indicates a set of genes with low +1 nucleosome occupancy in wild-type, where the downstream shift in the YAC is likely due to failure to correctly call the low occupancy +1 nucleosome in the YAC.
(C–E) Examples of K. lactis genes exhibiting different +1 nucleosome shifts in the YAC context, including an upstream shift (C), an unchanged +1 (D), and a downstream shift (E).
Figure S4. Comparison of RNA-Seq and MNase-Seq datasets.
(A) TSS positioning relative to +1 nucleosome. Averaged nucleosome data (solid lines) or RNA-Seq data (shaded area) for K. lactis and D. hansenii wild type cells, as indicated. All genes are aligned by +1 nucleosome position.
(B–C) Lower expression of genes on YACs relative to endogenous expression. For K. lactis (B) or D. hansenii (C), nucleosome data and RNA-Seq data are plotted as indicated for YAC-associated genes. For YAC-associated genes, RNA-Seq data are normalized to whole-genome RNA data (e.g. including S. cerevisiae genes, in contrast to the normalization in Figure 4), with lower normalized abundance indicating that YAC-associated genes are expressed at lower levels than are endogenous genes, assuming similar RNA content of the various species.
(D) RNA abundance for K. lactis genes. Normalized RNA-Seq data from K. lactis (x axis) or YAC-bearing strains (y axis) is scatter-plotted with each point representing a single gene. mRNA abundance data are shown as reads per kb per million reads. Note good correlation between endogenous and YAC-based expression, indicating that differences between poorly and highly-expressed genes are maintained in a foreign nuclear environment.
(E) ChIP-Seq was carried out for TFIIB in the YAC-bearing strains. TSS-aligned data are shown for all S. cerevisiae genes in each strain, or for all YAC-based genes for either K. lactis or D. hansenii. TFIIB ChIP could not be carried out for the other organisms due to the limited cross-reactivity of our antibody. Note that foreign promoters continue to recruit TFIIB in S. cerevisiae, but that for both organisms TFIIB exhibits worse localization in the YAC than expected, due to divergence of regulatory information between species. This is especially true for D. hansenii.
Figure S5. Nucleosome positioning shifts are associated with shifts in TSS.
(A) Averaged nucleosome mapping and RNA-Seq data are shown for all K. lactis genes exhibiting either no +1 nucleosome shift (less than 20 bp in either direction) in the YAC, or a 20 bp or more upstream shift in the YAC. Note that RNA-Seq data shift 5′ in both cases when K. lactis genes are expressed in S. cerevisiae, but that genes with 5′ nucleosome shifts exhibit a greater 5′ shift, consistent with a constant distance being maintained between TSS and +1 nucleosome positioning. Note that genes with 3′ shifts in the +1 nucleosome are not included, as these largely represent genes where the +1 nucleosome decreases occupancy and hence is miscalled (Figure S3B).
(B-E) TSS mapping for 4 individual genes. TSSs were mapped by 5′ RACE (Methods), and individual clone locations are shown as indicated below the nucleosome mapping data. Note that for two genes with little +1 nucleosome position shift (B, E) there is little change in TSS, whereas the two genes with 5′ shifts in the +1 nucleosome (C, D) also show upstream shifts in the TSS in the YAC.
Figure S6. Examples of fortuitousNDRs in D. hanseniiYACs.
(A–B) As in Figure 5, another example of a fortuitous NDR that occurs only in YACs, and is associated with transcription. (A) shows nucleosome and RNA data for 4 kb surrounding a YAC-specific NDR, with (B) showing nucleosome data only for the indicated region. Note increasing nucleosome fuzziness in the YAC nucleosome data at the 3′ end of the transcript.
(C) Extent of positioned nucleosome array is linked to RNA transcript length. Schematic interpretation of the nucleosome positioning for RNA transcripts with different lengths, derived from fortuitous coding region NDRs. RNA transcript is shown in green rectangle and nucleosome positioning is shown in solid red (well positioned nucleosome) and transparent light red (less positioned nucleosome) above the rectangle. Black arrows indicate inferred TSSs.
Figure S7. Yeast whole cell extracts poorly position nucleosomes
(A) Nucleosome mapping data from intact yeast (“in vivo”), from lysed yeast equilibrated prior to crosslinking (“native”), or from yeast genomic DNA incubated with yeast whole cell extract and ATP (WCE+ATP) in vitro (Zhang et al., 2011). Yeast whole cell extract performs significantly better than salt gradient dialysis, as reported, but the nucleosome positions recovered nonetheless do not precisely match nucleosome positions recovered from intact yeast – compare “native” and in vivo positioning.
(B) +1 nucleosome positioning in “native” yeast extracts exhibits systematic deviation from in vivo positioning. +1 nucleosome positions were called, and distance from +1 nucleosome positions in vivo to the positions recovered in various datasets is shown as a histogram. As a comparison for technical variation, we show data for +1 positioning variability in our YAC strains (to keep datasets on the same y axis we used a 5 bp bin size for YACs rather than the 10 bp used for other datasets).
Supplementary Table 1. Strain list
Strains used in this study.
Highlights.
Systematic dissection of cis and trans contributions to nucleosome positioning
Nucleosome positioning is generally established by proteins, not DNA sequence
Fortuitous promoters establish canonical promoter chromatin architecture
3 step model for establishment of in vivo nucleosome positioning
Acknowledgments
This paper is dedicated to the memory of Jonathan Widom. His experimental and conceptual contributions were a major influence on this work, and we will miss our numerous memorable and enjoyable discussions. We thank Yajie Niu for assistance in CsCl gradient ultracentrifugation, Caitlin Reavey and Matthew Pipkin for helpful discussions on pulsed field gel electrophoresis, and members of the Rando and Struhl labs for helpful discussions and comments. OJR is funded by NIH grant GM079205, KS is funded by GM030186. OJR and KS conceived the experiments, and YJ and AH designed the experiments. YJ generated YACs and YAC-bearing strains, and AH carried out nucleosome isolation and deep sequencing, and 5′ RACE. YJ carried out all RNA sequencing and TFIIB ChIP sequencing. AH and YJ carried out data analysis with assistance from OJR and KS. AH, YJ, OJR, and KS wrote the manuscript.
Footnotes
This paper is dedicated to the memory of Jonathan Widom, our friend and colleague who was passionately interested in nucleosome positioning and many other aspects of chromatin structure, and enjoyed vigorous scientific debate
Accession numbers
The GEO accession number for the datasets reported in this paper is GSE39011.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Drew HR, Travers AA. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985;186:773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Fan X, Moqtaderi Z, Jin Y, Zhang Y, Liu XS, Struhl K. Nucleosome depletion at yeast terminators is not intrinsic and can occur by a transcriptional mechanism linked to 3′-end formation. Proc Natl Acad Sci USA. 2010;107:17945–17950. doi: 10.1073/pnas.1012674107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkikopoulos T, Schofield P, Singh V, Pinskaya M, Mellor J, Smolle M, Workman JL, Barton GJ, Owen-Hughes T. A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science. 2011;333:1758–1760. doi: 10.1126/science.1206097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heus JJ, Zonneveld BJ, Bloom KS, de Steensma HY, van den Berg JA. The nucleosome repeat length of Kluyveromyces lactis is 16 bp longer than that of Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2247–2248. doi: 10.1093/nar/21.9.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore I, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, Hughes TR, Lieb JD, Widom J, Segal E. Nucleosome sequence preferences influence in vivo nucleosome organization. Nat Struct Mol Biol. 2010;17:918–920. doi: 10.1038/nsmb0810-918. author reply 920–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber P, Horz W. In vitro assembly of the characteristic chromatin organization at the yeast PHO5 promoter by a replication-independent extract system. J Biol Chem. 2004;279:35113–35120. doi: 10.1074/jbc.M405446200. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Stryer L. Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res. 1988;16:6677–6690. doi: 10.1093/nar/16.14.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman-Livaja M, Rando OJ. Nucleosome positioning: How is it established, and why does it matter? Dev Biol. 2010;339:258–266. doi: 10.1016/j.ydbio.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley J, Butler R, Ogilvie D, Finniear R, Jenner D, Powell S, Anand R, Smith JC, Markham AF. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 1990;18:2887–2890. doi: 10.1093/nar/18.10.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A, Takasuka TE, Collings CK. Are nucleosome positions in vivo primarily determined by histone-DNA sequence preferences? Nucleic Acids Res. 2009;38:709–719. doi: 10.1093/nar/gkp1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Berman J, Barkai N. The pattern and evolution of yeast promoter bendability. Trends Genet. 2007;23:318–321. doi: 10.1016/j.tig.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Sigal N, Barkai N. Divergence of nucleosome positioning between two closely related yeast species: genetic basis and functional consequences. Mol Syst Biol. 2010;6:365. doi: 10.1038/msb.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov A, Yanagisawa Y, Rhind N, Regev A, Rando OJ. Evolutionary divergence of intrinsic and trans-regulated nucleosome positioning sequences reveals plastic rules for chromatin organization. Genome Res. 2011;21:1851–1862. doi: 10.1101/gr.122267.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov AM, Thompson DA, Socha A, Regev A, Rando OJ. The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol. 2010;8:e1000414. doi: 10.1371/journal.pbio.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant C, Palmeira L, Chevereau G, Audit B, d’Aubenton-Carafa Y, Thermes C, Arneodo A. A novel strategy of transcription regulation by intragenic nucleosome ordering. Genome Res. 2010;20:59–67. doi: 10.1101/gr.096644.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Holde KE. Chromatin. New York: Springer-Verlag; 1989. [Google Scholar]
- Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010;20:90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Tsukiyama T. Antagonistic forces that position nucleosomes in vivo. Nat Struct Mol Biol. 2006;13:633–640. doi: 10.1038/nsmb1111. [DOI] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol. 2009;16:847–852. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K. Evidence against a genomic code for nucleosome positioning. Nat Struct Mol Biol. 2010;17:920–923. doi: 10.1038/nsmb0810-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wippo CJ, Wal M, Ward E, Korber P, Pugh BF. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science. 2011;332:977–980. doi: 10.1126/science.1200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. NDRs are generally better-maintained over sequence from K. lactis than over D. hansenii sequence
(A–B) Nucleosome mapping data for all genes from K. lactis (A) or D. hansenii (B) are shown for wild-type and YACs, as indicated. Genes are sorted by wild-type NDR width.
(C) NDR maintenance correlates with poly(dA:dT) elements. As in Figures 2A–B, but for K. lactis.
(D) Identical to Figures 2A–B, reproduced here for comparison between D. hansenii and K. lactis.
Figure S2. Bulk chromatin is not affected in YAC-bearing strains
(A) Nucleosome sequencing data for all strains in this study was mapped to the S. cerevisiae genome, and data are averaged for all genes aligned by the +1 nucleosome.
(B) Data for all S. cerevisiae genes are shown for wild-type (BY4741) and 5 YAC-bearing strains. Genes are sorted by K means clustering (K=4) of BY4741 dataset.
Figure S3. +1 nucleosome shifts associated with transcription.
(A) Distribution of shifts for +1 nucleosomes. Distributions are shown for the shifts between +1 positions in wild-type and YAC-bearing strains. The three S. cerevisiae distributions show changes in +1 positioning for S. cerevisiae genes in the indicated YACs, showing that technical variability or analytical variability do not account for nucleosome position changes for YAC-associated genes.
(B) Data for all K. lactis and D. hansenii genes, from wild-type and YACs. Data are shown from −100 to +300 bp relative to the +1 nucleosome upstream border (+1 nucleosome center indicated as a red line). Green box for K. lactis genes indicates a set of genes with low +1 nucleosome occupancy in wild-type, where the downstream shift in the YAC is likely due to failure to correctly call the low occupancy +1 nucleosome in the YAC.
(C–E) Examples of K. lactis genes exhibiting different +1 nucleosome shifts in the YAC context, including an upstream shift (C), an unchanged +1 (D), and a downstream shift (E).
Figure S4. Comparison of RNA-Seq and MNase-Seq datasets.
(A) TSS positioning relative to +1 nucleosome. Averaged nucleosome data (solid lines) or RNA-Seq data (shaded area) for K. lactis and D. hansenii wild type cells, as indicated. All genes are aligned by +1 nucleosome position.
(B–C) Lower expression of genes on YACs relative to endogenous expression. For K. lactis (B) or D. hansenii (C), nucleosome data and RNA-Seq data are plotted as indicated for YAC-associated genes. For YAC-associated genes, RNA-Seq data are normalized to whole-genome RNA data (e.g. including S. cerevisiae genes, in contrast to the normalization in Figure 4), with lower normalized abundance indicating that YAC-associated genes are expressed at lower levels than are endogenous genes, assuming similar RNA content of the various species.
(D) RNA abundance for K. lactis genes. Normalized RNA-Seq data from K. lactis (x axis) or YAC-bearing strains (y axis) is scatter-plotted with each point representing a single gene. mRNA abundance data are shown as reads per kb per million reads. Note good correlation between endogenous and YAC-based expression, indicating that differences between poorly and highly-expressed genes are maintained in a foreign nuclear environment.
(E) ChIP-Seq was carried out for TFIIB in the YAC-bearing strains. TSS-aligned data are shown for all S. cerevisiae genes in each strain, or for all YAC-based genes for either K. lactis or D. hansenii. TFIIB ChIP could not be carried out for the other organisms due to the limited cross-reactivity of our antibody. Note that foreign promoters continue to recruit TFIIB in S. cerevisiae, but that for both organisms TFIIB exhibits worse localization in the YAC than expected, due to divergence of regulatory information between species. This is especially true for D. hansenii.
Figure S5. Nucleosome positioning shifts are associated with shifts in TSS.
(A) Averaged nucleosome mapping and RNA-Seq data are shown for all K. lactis genes exhibiting either no +1 nucleosome shift (less than 20 bp in either direction) in the YAC, or a 20 bp or more upstream shift in the YAC. Note that RNA-Seq data shift 5′ in both cases when K. lactis genes are expressed in S. cerevisiae, but that genes with 5′ nucleosome shifts exhibit a greater 5′ shift, consistent with a constant distance being maintained between TSS and +1 nucleosome positioning. Note that genes with 3′ shifts in the +1 nucleosome are not included, as these largely represent genes where the +1 nucleosome decreases occupancy and hence is miscalled (Figure S3B).
(B-E) TSS mapping for 4 individual genes. TSSs were mapped by 5′ RACE (Methods), and individual clone locations are shown as indicated below the nucleosome mapping data. Note that for two genes with little +1 nucleosome position shift (B, E) there is little change in TSS, whereas the two genes with 5′ shifts in the +1 nucleosome (C, D) also show upstream shifts in the TSS in the YAC.
Figure S6. Examples of fortuitousNDRs in D. hanseniiYACs.
(A–B) As in Figure 5, another example of a fortuitous NDR that occurs only in YACs, and is associated with transcription. (A) shows nucleosome and RNA data for 4 kb surrounding a YAC-specific NDR, with (B) showing nucleosome data only for the indicated region. Note increasing nucleosome fuzziness in the YAC nucleosome data at the 3′ end of the transcript.
(C) Extent of positioned nucleosome array is linked to RNA transcript length. Schematic interpretation of the nucleosome positioning for RNA transcripts with different lengths, derived from fortuitous coding region NDRs. RNA transcript is shown in green rectangle and nucleosome positioning is shown in solid red (well positioned nucleosome) and transparent light red (less positioned nucleosome) above the rectangle. Black arrows indicate inferred TSSs.
Figure S7. Yeast whole cell extracts poorly position nucleosomes
(A) Nucleosome mapping data from intact yeast (“in vivo”), from lysed yeast equilibrated prior to crosslinking (“native”), or from yeast genomic DNA incubated with yeast whole cell extract and ATP (WCE+ATP) in vitro (Zhang et al., 2011). Yeast whole cell extract performs significantly better than salt gradient dialysis, as reported, but the nucleosome positions recovered nonetheless do not precisely match nucleosome positions recovered from intact yeast – compare “native” and in vivo positioning.
(B) +1 nucleosome positioning in “native” yeast extracts exhibits systematic deviation from in vivo positioning. +1 nucleosome positions were called, and distance from +1 nucleosome positions in vivo to the positions recovered in various datasets is shown as a histogram. As a comparison for technical variation, we show data for +1 positioning variability in our YAC strains (to keep datasets on the same y axis we used a 5 bp bin size for YACs rather than the 10 bp used for other datasets).
Supplementary Table 1. Strain list
Strains used in this study.