Abstract
14C-labeled nicotinamide cofactors are widely employed in biomedical investigations, e.g. to delineate metabolic pathways, to elucidate enzymatic mechanisms, or as substrates in kinetic isotope effect (KIE) experiments. The 14C-label has generally been located remote from the reactive position, frequently at the adenine ring. Rising costs of commercial precursors, and disruptions in the availability of enzymes required for established syntheses, have recently made the preparation of labeled nicotinamides such as [Ad-14C]-NADPH inviable. Here, we report the syntheses and characterization of several alternatives: [carbonyl-14C]-NADPH, 4R-[carbonyl-14C, 4-2H]-NADPH, and [carbonyl-14C, 4-2H2]-NADPH. The new procedures utilize [carbonyl-14C]-nicotinamide as starting material, as it is significantly cheaper than other commercial 14C-precursors of NADPH, and require only one commercially available enzyme to prepare NAD(P)+ and NAD(P)H. The proximity of carbonyl-14C to the reactive center raises the risk of an inopportune 14C-isotope effect. This concern has been alleviated via competitive KIE measurements with Escherichia coli dihydrofolate reductase (EcDHFR), that use this specific carbonyl-14C NADPH. A combination of binding isotope effect and KIE measurements yielded no significant 12C/14C isotope effect at the amide carbonyl (KIE = 1.003 ± 0.004). The reported procedure provides a high-yield, high-purity and cost-effective alternative to labeled nicotinamide cofactors synthesized by previously published routes.
Keywords: radioactive labeling, kinetic isotope effect, nicotinamide cofactor, mechanistic enzymology, NAD+ nucleosidase
Introduction
An enormous number of metabolic enzymes depend on the nicotinamide cofactors, viz. NAD+, NADP+, or their reduced forms, NADH and NADPH [1]. Chemical and isotopic modification of nicotinamide cofactors have enabled enzymological studies for several decades now (see refs.[2], [3], [4] and [5] for selected examples, and [6] for a comprehensive review of these cofactors). The development of new biophysical techniques drives the continued quest for novel nicotinamide derivatives prepared by rapid, cost-effective means [7]. When investigating enzymes that utilize one of these molecules as a redox cofactor, the fate of the labeled or modified cofactor may thus be followed to determine the mechanistic or kinetic course of the enzyme in question. While this can be achieved by a number of methods (e.g., following the change in 340 nm absorbance of the reduced forms through out the reaction), radiolabeling offers higher sensitivity than most other techniques [8] and also facilitates the measurement of isotope effects.
Among the many uses of isotopically labeled nicotinamide cofactors is the measurement of kinetic isotope effects (KIEs)1 for H-transfer enzymatic reactions. This is exemplified below by KIE experiments with the Escherichia coli dihydrofolate reductase (EcDHFR) enzyme. EcDHFR is an extensively studied system and thus an excellent benchmark with which to test new nicotinamide labeling patterns. EcDHFR uses NADPH as a hydride source in the reduction of 7,8-dihydrofolate (H2F) to S-5,6,7,8-tetrahydrofolate (H4F), transferring the pro-R hydrogen from C4 on the nicotinamide ring of NADPH to the si-face of C6 of the pteridine ring of H2F (Scheme 1). Several previous works have used NADPH labeled with deuterium (2H) or tritium (3H) at C4 to determine the KIE on the hydride transfer step for EcDHFR and its mutants [9; 10; 11; 12; 13]. When such KIEs are measured competitively, both isotopologues are allowed to compete simultaneously for the enzyme. In these experiments the ratio of the reaction rates between the isotopologues is measured by following the fractionation of the isotopes at the product or the reactant, instead of determining the absolute rates [14]. For example, in competitive H/T and D/T KIE measurements [denoted also by T(V/K)H,obs and T(V/K)D,obs, respectively] it is imperative that the non-radioactive isotopologues (1H- or 2H-labeled) are tracked by a radioactive labeling other than 3H at a position on the NADPH molecule remote from the transferred hydrogen. This remote tracer (commonly 14C) in the product thus represents the H (or 2H) that has been transferred and so permits analytical examination of the 3H depletion relative to H (or 2H) in the product by following the 3H/14C ratio at different fractional conversions (as explained in detail in Materials and Methods). The same remote tracer can also be used to measure competitive H/D KIEs [denoted also by D(V/K)H,obs], where a remote 3H label tracks one isotope (e.g., 2H) and a remote 14C label identifies the other (e.g., H). Consequently, in such experiments it is important that the remote tracer has no measurable effect on the reaction rate, and thus no effect on the KIEs under study. If the labeling is too close to the reactive center, isotope effects due to the tracer isotope may lead to inflation or deflation of the KIEs being measured. As an extreme example, the use of [4-14C]-NADPH as a labeled cofactor in the EcDHFR reaction would add a primary (1°, see below) 14C KIE to the measured hydrogen KIE. While such an unwanted isotope effect would have a minor effect on 1° hydrogen KIEs (due to their large size), it could have a more detrimental impact on the much smaller secondary (2°, see below) KIEs, or on KIEs of heavy isotopes.
Scheme 1.
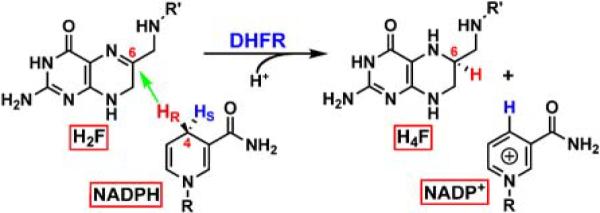
The reaction catalyzed by DHFR. R = adenine dinucleotide 2' phosphate and R' = (p-aminobenzoyl) glutamate.
Using such a remote-labeling dual-isotope method lends itself very well to a sequential HPLC/liquid scintillation counting (LSC) analysis, as demonstrated by a wealth of studies over the years [15; 16]. For example, in the case of EcDHFR, we previously performed competitive measurements of H/T KIEs using [Ad-14C]-NADPH in competition with 4R-[4-3H]-NADPH (see Figure 1 for radiolabeling details): here, the 14C label on the adenine ring is far from the point of hydride transfer and cannot affect the rate of this step. The H/T KIEs are measured by simultaneously following the conversion of [Ad-14C]-NADPH to [Ad-14C]-NADP+, and of 4R-[4-3H]-NADPH to tritiated H4F and its oxidized derivatives [17]. The measured values can be used together with D/T KIEs (measured in a similar fashion) to extract the intrinsic KIEs for the reaction, thus providing direct insight into the nature of the chemical step in the enzymatic reaction under consideration [18; 19].
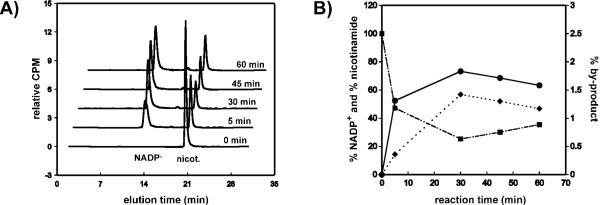
Figure 1.
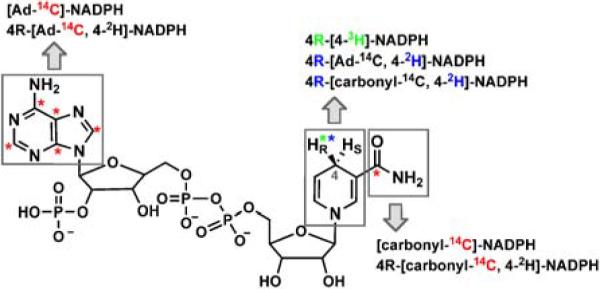
Labeled NADPH derivatives. Asterixes indicate the position of isotopic labeling with 14C (red), 2H (blue) or 3H (green). HR=1H for NADPH, [Ad-14C]-NADPH and [carbonyl-14C]-NADPH; HR=2H for 4R-[Ad-14C, 4-2H]-NADPH and 4R-[carbonyl-14C, 4-2H]-NADPH; HR=3H for 4R-[4-3H]-NADPH. For the sake of clarity, 3H-labeling on [Ad-3H]-NADPH and its derivatives is not shown.
The isotopic labeling pattern must be modified according to the type of KIE being measured - i.e., either a 1 KIE, which results when the isotopic difference is in an atom directly involved in making or breaking bonds during the reaction, or a 2 KIE, which arises when the nature of the bonding to a labeled atom changes without formation or cleavage of bonds to that atom. Moreover, the labeling pattern should take into consideration whether H/T, D/T or H/D KIEs are being measured, as well as the interaction of the labeled nicotinamide cofactor with the enzyme. Indeed, the degree of interaction between the remotely-labeled cofactor and the enzyme can be determined by the measurement of binding isotope effects (BIEs), which arise due to a difference in the binding constants of isotopologic reactants [20]. An inverse (<1) or normal (>1) BIE is indicative of differential binding constants for the labeled versus unlabeled cofactor. While binding effects are commonly determined by direct comparison of binding parameters (e.g. κd), BIEs for enzymatic reactions can be sensitively measured by competitive KIE measurements of the labeled vs. the unlabeled cofactors as substrates (assuming that here KIEs represent the fractionation of the isotopes between the bound and the free state) [20]. In other words, location of the isotopic labeling is such that measured V/K KIEs only represent the fractionation of the bound substrate since the labeling is on the adenine ring i.e., remote from the reaction site.
The [Ad-14C]-NADPH cofactors used in the aforementioned studies were all synthesized using the expensive (~$2,380/50 μCi) and periodically unavailable [Ad-14C]-NAD+ as the starting material, which significantly limits their use [5]. Here, we present the synthesis of [carbonyl-14C]-NADPH, 4R-[carbonyl-14C, 4-2H]-NADPH and [carbonyl-14C, 4-2H2]-NADPH from the less expensive (~$750/250 μCi) and readily available [carbonyl-14C]-nicotinamide starting material. The synthesized cofactors have been examined as cost-effective alternatives to [Ad-14C]-NADPH and its derivatives as reagents in KIE studies of enzymatic systems, with special emphasis on possible artifacts arising from the 14C labeling at the amide position that might affect hydrogen KIEs of interest.
The remote location of the 14C-label on the adenine ring in [Ad-14C]-NADPH and other [Ad-14C]-NAD+ cofactors used previously is not likely to have any measurable effect on either binding or the chemistry of the aforementioned cofactors. The remoteness of the heavy-atom label from the reactive C4 hydrogen of the nicotinamide cofactor ensures no isotope effect due to the 14C label on the rate of the H- or D- transfer being considered. However, in the case of the [carbonyl-14C]-NADPH described here, the 14C-label is located at the amide carbonyl, γ to the reactive C4 hydrogen. This proximity to the reactive center gives rise to the question: does labeling the amide carbonyl (rather than the adenine ring) with 14C lead to a significant isotope effect on H-transfer reactions occurring at the C4 position? In other words, is there a measurable isotope effect when [carbonyl-14C]-NADPH is used instead of [Ad-14C]-NADPH?
Since direct measurement of the isotope effect between [carbonyl-14C]-NADPH and [Ad-14C]-NADPH is not practical, we first measured the BIE at the adenine ring on the EcDHFR reaction, using [Ad-3H]-NADPH and [Ad-14C]-NADPH, and then the KIE between [Ad-3H]-NADPH and [carbonyl-14C]-NADPH. Since [Ad-3H]-NADPH is a common reference between the BIE and KIE experiments, the ratio between the measured BIE to the KIE allowed us to establish the −2 14C KIE of interest. In other words, since it is not possible to directly compare the reaction rate of the [carbonyl 14C]-NADPH to that using unlabeled NADPH or [Ad-14C]-NADPH, we have instead used multiple KIE and BIE measurements to compare the rates of [Ad-14C]-NADPH to that of [carbonyl-14C]-NADPH using [Ad−3H]-NADPH as a common reference. The measured value after standard error propagation is 1.003 ± 0.004 for the −2 14C KIE, which suggests that [carbonyl-14C]-NADPH and its derivatives are viable substitutes for [Ad-14C]-NADPH and other labeled cofactors synthesized in the past from [Ad-14C]-NAD+.
Materials and Methods
Materials
All reagents were purchased from Sigma-Aldrich, unless otherwise noted. NADP+-dependent glucose dehydrogenase from Bacillus megaterium (bmGDH, 218 units/mg solid) was purchased from USB-Affymetrix, while NAD+/NADP+-dependent nucleosidase (NADase) from porcine brain (0.007 un/mg solid) was purchased from Sigma-Aldrich. EcDHFR was expressed and purified according to the procedure of Cameron et al. [21]. [carbonyl-14C]-nicotinamide (55 mCi/mmol, 100 μCi/ml) was obtained from Moravek, [Ad-14sC]-NAD+ (>220 mCi/mmol, 25 μCi/ml) and nicotinamide-[2,5',8-3H]-adenine nucleotide ([Ad-3H]-NAD+, 24 Ci/mmol, 0.1 mCi/ml) were from Amersham (now available from Perkin-Elmer). H2F was synthesized from folic acid by the method of Blakley [22]. [1-2H]-glucose was synthesized with > 99.9% deuteration (as characterized by 1H-NMR) by reduction of -gluconolactone with sodium amalgam in 99.96% D2O [23].
Analytical methods
All analytical separations were made by HPLC on a Beckman-Coulter System Gold instrument (model 126), using a Supelco Discovery C-18 column (250 × 4.6 mm i.d., 5 m particle size) and the gradient elution method described in Table 1, but with an altered flow-rate of 0.8 ml/min suited for use with the analytical HPLC column. Eluted peaks were analyzed using an online Beckman UV-Vis detector (model 168) and a Packard 500TR Series flow scintillation detector. NADP+ synthesized in the first step (Scheme 2) was isolated in a single broad peak at ~ 12 min using a Supelco Discovery semipreparative C-18 column (250 × 10 mm i.d., 5 m particle size) following the elution gradient shown in Table 1; a representative elution profile is depicted in Figure 2a. The semipreparative HPLC method described previously [24] was used to purify the final NADPH product, and radioactive product yields were quantified using a Packard TriCarb 2900 TR liquid scintillation counter.
Table 1.
Semi-preparative HPLC method using reverse-phase C-18 column and gradient elution. Solvent A is 0.2 M NaCl/1 mM Tris-HCl (pH 8.2) and solvent B is MeOH.
| Time (min) | % solvent A | % solvent B | Flow rate (ml/min) |
|---|---|---|---|
| 0 | 100 | 0 | 3.2 |
| 11 | 100 | 0 | 3.2 |
| 11.10 | 92.5 | 7.5 | 3.2 |
| 21 | 92.5 | 7.5 | 3.2 |
| 21.10 | 50 | 50 | 3.2 |
| 23 | 50 | 50 | 3.2 |
| 23.10 | 0 | 100 | 3.2 |
| 30 | 0 | 100 | 3.2 |
Scheme 2.
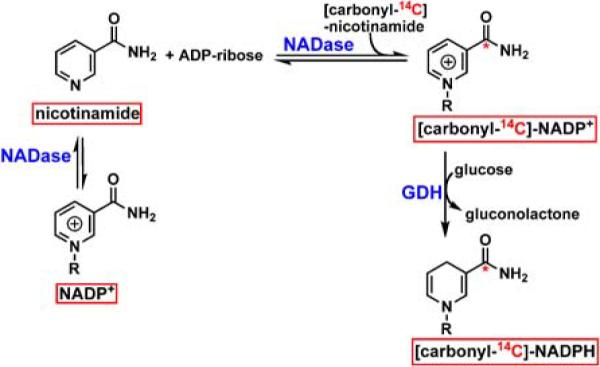
Synthesis of [carbonyl-14C]-NADPH from [carbonyl-14C]-nicotinamide. R = 2'-monophosphoadenosine-5'-diphosphate ribose, where the asterixes denote the position of the 14C-label.
Figure 2.
Summary of the results for the first synthetic step - synthesis of [carbonyl-14C]-NADP+. A) Reverse-phase HPLC-flow radiogram of the reaction mixture at different time points. The main peaks correspond to [carbonyl-14C]-NADP+, ~12.5 min, and [carbonyl-14C] nicotinamide, ~20.5 min. The middle peak represents an unidentified by-product, which accounts for <1.6% of total radioactivity, and was easily separated from the NADP+. B) Percent of each compound (from panel A) during the course of 60 min reaction, where [carbonyl-14C]-NADP+ is presented by closed circles, [carbonyl-14C]-nicotinamide by squares, and the 14C-labeled by-product by diamonds.
Preparation of porcine brain NADase
Prior to the synthesis of the radiolabeled cofactor, NADase solutions were prepared in 0.1 M KH2PO4 buffer (pH 7.5) using a modified version of the procedure described by Hixson et al. [2]. In short, 1 g of lyophilized NADase (~7 units) was dissolved in 0.1 M KH2PO4 buffer (pH 7.5) to a final concentration of 0.5 units/ml. The buffered solution was sonicated for 45 min at 4 C in the dark, then 6 mg of trypsin (≥10,000 units/mg) was added and the reaction mixture was incubated at 37 C for 40 min. 3.6 mmol of phenylmethylsulfonate (PMSF) was then added, and incubation continued for an additional 5 min at 37 C. The reaction mixture was centrifuged at 4 C and 4000 to 5000 × g to remove insoluble debris. The supernatant was removed and stored at 4 C in the dark until used. Under these conditions, NADase solution was found to be stable for at least 2 months. Prior to every synthesis, an aliquot of NADase solution corresponding to 0.175 units was spun down at 10,000 × g to remove any denatured or precipitated protein that may have accumulated during storage.
Synthesis of [carbonyl-14C]-NADPH
Scheme 2 illustrates the general synthetic strategy employed for the preparation of [carbonyl-14C]-NADPH in two steps: first, the exchange reaction of NADP+ and [carbonyl-14C]-nicotinamide to yield [carbonyl-14C]-NADP+, and the subsequent reduction with bmGDH and glucose to yield [carbony-14C]-NADPH.
Synthesis of [carbonyl-14C]-NADP+ (Scheme 2, first step)
125 L of the ethanolic solution of [carbonyl-14C]-nicotinamide (12.5 Ci, 227 nmol) was blown to dryness with N2 or Ar gas, and combined with 0.175 units NADase, 2.3 μmol NADP+ and 0.1 M KH2PO4 (pH 7.5) buffer to a final volume of 400 L. The reaction was allowed to proceed at 35 C in the dark for 60 min. Reaction progress was periodically monitored by HPLC/flow scintillation detector analysis. It is important to note that, since NADase catalyzes the reversible hydrolysis of NADP+ to nicotinamide and 2'-monophosphoadenosine-5'-diphosphoribose, the reaction mixture for the first step should contain a 10:1 excess of NADP+ over [carbonyl-14C]-nicotinamide. Different ratios lead to lower yields or lower specific radioactivities of the radiolabeled NADP+ (see discussion). It is also important to note that in the synthesis of radiolabeled material, the yield is measured in terms of the radiolabeled starting material ([carbonyl-14C]-nicotinamide in this case), which is the limiting-reagent in the synthesis.
After completion of the exchange reaction (generally after 30 min), NADase was removed from the reaction mixture by filtration through an Amicon Ultra Ultracel-10K (10,000 MWCO) regenerated cellulose centrifugal filter. LSC analysis of the filtrate indicated minimal loss of radioactivity due to filtration. [carbonyl-14C]-NADP+ was then isolated from the filtrate by semipreparative HPLC purification as described in the Methods section, and lyophilized overnight to remove all traces of solvent. The lyophilized NADP+ was stable at −80 C for at least a year. Before continuing with further synthetic steps, it was critical that the large amounts of salt (the result of lyophilization) be removed, as high salt concentrations can inhibit certain enzymatic reactions. Therefore, lyophilized [carbonyl-14C]-NADP+ was dissolved in 1 ml of doubly de-ionized (DDI) H2O and loaded onto a 3 ml Sephadex C-18 desalting column. The loaded column was washed through with 3 ml DDI H2O, and [carbonyl-14C] NADP+ was then eluted using 3 ml of 15% MeOH/0.1 M KH2PO4 buffer. This solution was lyophilized overnight.
Synthesis of [carbonyl-14C]-NADPH (Scheme 2, second step)
The lyophilized [carbonyl-14C]-NADP+ was re-suspended in 450 l 0.1 M KH2PO4 (pH 7.5) buffer immediately before its reduction to [carbonyl-14C]-NADPH. 1.25 mol D-glucose and 1 mg (~218 units) bmGDH were added to the [carbonyl-14C]-NADP+ solution and the pH was adjusted to 8.0 at 37 C, with a final volume of 500 l. The reaction was allowed to proceed at 37 C for 1 hour, when HPLC/flow scintillation analysis showed >99% conversion of [carbonyl-14C]-NADP+ to [carbonyl-14C]-NADPH (Figure 3). Upon completion of the reaction, the reaction mixture was centrifuged at room temperature and 13,000 × g for 5 min, and the products were purified by semipreparative HPLC as described in the Methods section. The purified [carbonyl-14C]-NADPH was quantified by LSC analysis, and divided into 3 million DPM aliquots. These aliquots were immediately lyophilized and stored in the dark at −80 °C. Previously published data indicates that NADPH stored under these conditions remains viable for more than 24 months [24]. After purification and lyophilization, the NADPH product was found to be more than 99.5% pure by HPLC/radiographic flow-scintillation analysis (Figure 3).
Figure 3.

Reverse-phase HPLC-flow radiogram of the reaction mixture after the [carbonyl-14C]-NADP+ reduction. Radiogram indicates a complete conversion (>99.5%) of NADP+ to NADPH.
Synthesis of 4R-[carbonyl-14C,4-2H]-NADPH from [carbonyl-14C]-NADP+
4R-[carbonyl-14C, 4-2H]-NADPH was synthesized from [carbonyl-14C]-NADP+ using the one-step reaction catalyzed by Thermoanaerobium brockii alcohol dehydrogenase (tbADH) [25]. The final product was purified, characterized and stored in the same manner as [carbonyl-14C]-NADPH described in the previous section.
Synthesis of [carbonyl-14C,4-2H2]-NADPH from [carbonyl-14C]-NADP+
[carbonyl-14C, 4-2H2]-NADPH was synthesized from [carbonyl-14C]-NADP+ following the 3-step procedure involving bmGDH, tbADH and 1-2H-glucose as described in refs. [26] and [17]. Purification, characterization and storage were the same as for [carbonyl-14C]-NADPH.
Determination of 1 H/T and D/T KIEs for EcDHFR using synthesized [carbonyl-14C] derivatives
For the measurement of H/T KIEs, 4R-[4-3H]-NADPH was prepared according to previously published procedure [27], and co-purified in a 6:1 ratio with [carbonyl-14C]-NADPH to compensate for the lower efficiency in LSC counting of tritium [24]. For the measurement of D/T KIEs, 4R-[4-3H]-NADPH was co-purified in a 6:1 ratio with 4R-[carbonyl-14C, 4-2H]-NADPH. The co-purified materials in both cases were divided into 300,000 DPM aliquots (of 14C) and frozen at −80 C for short-term storage (up to 15 days). KIE experiments were performed according to the procedure used by Wang et al.[17] in 1X MTEN buffer (100 mM NaCl, 50 mM MES, 25 mM Tris and 25 mM ethanolamine, pH 9.0 at 25 C). As per the previous procedure, EcDHFR was allowed to react with co-purified NADPH and H2F (final concentration of 0.85 mM) at 25 C in 1X MTEN (pH 9.0), and aliquots were removed from the reaction mixture and quenched with methotrexate to yield samples with fractional conversions (f; determined from the ratio of 14C in the product to the total amount of 14C in products and reactants) ranging from 20–80%. Observed KIEs were calculated using the following equation [14]:
| (1) |
where Rt and R∞ are ratios of 3H to 14C in the products at various fractional conversions and at 100% conversion, respectively. Intrinsic H/T KIEs were calculated using the Northrop method, as elaborated upon in previous publications [17] and [28]. All KIE results are displayed in Table 2.
Table 2.
Observed and intrinsic 1° KIEs measured using [carbonyl-14C]-NADPH and 4R-[4-3H]-NADPH (for H/T experiments) and 4R-[carbonyl-14C, 4-2H]-NADPH with 4R-[4-3H]-NADPH (for D/T experiments). The results are compared to previously published values obtained using [Ad-14C] NADPH and [Ad-14C, 4-2H2] NADPH (with 4R-[4-3H]-NADPH and 4S-[4,4-3H,2H]-NADPH, respectively) [28].
| 14C-labeling pattern | H/Tobs | D/Tobs | H/Tint | D/Tint | H/Dint |
|---|---|---|---|---|---|
| [carbonyl-14C] | 4.86 ± 0.06 | 1.66 ± 0.01 | 6.13 ± 0.18 | 1.72 ± 0.02 | 3.56 ± 0.07 |
| [Ad-14C] a | 4.85 ± 0.09 | 1.66 ± 0.03 | 6.10 ± 0.42 | 1.72 ± 0.04 | 3.55 ± 0.17 |
Measurement of - 2 14C KIE due to 14C isotopic labeling at the amide carbonyl of NADPH
[Ad-3H]-NADPH and [carbonyl-14C]-NADPH for KIE measurement, and [Ad-3H]-NADPH and [Ad-14C]-NADPH for the BIE measurement, were combined and co-purified in a DPM ratio of 5:1 (3H/14C). These two mixtures were used in the kinetic measurements following the same procedure as described for 1° KIEs above. As mentioned in the introduction, while the experimental procedure is the same for BIE and KIE experiments, the value calculated in the BIE measurement using equation 1 corresponds to a BIE, while KIE obtained by [Ad-3H]-NADPH and [carbonyl-14C]-NADPH reflects the product of the BIE and the - 2 14C KIE of interest. Dividing the second by the first yields a value indicative of the carbonyl - 2 14C KIE.
Results and Discussion
Synthesis of [carbonyl-14C]-NADPH, 4R- [carbonyl-14C, 4-2H]-NADPH and [carbonyl-14C, 4-2H2]-NADPH
NADase catalyzes the conversion of NAD(P)+ to nicotinamide and 2'-monophosphoadenosine-5'-diphosphoribose. The hydrolysis is reversible in both cases, and the reaction equilibrium is governed by the ratio of NAD(P+) to free nicotinamide, and the amount of ADP-ribose or 2'-monophosphoadenosine-5'-diphosphoribose in solution. It was found that the reaction could be optimized for minimal reaction times (< 1 hour) and maximal yields of radioactive cofactor by using a 10:1 ratio of NADP+ to [carbonyl-14C]-nicotinamide in the initial reaction mixture. Deviations from this ratio produced deleterious effects on the time required for maximal yield, and the yield itself. Moreover, dissimilar ratios resulted in different specific radioactivities in the product, and in some cases led to the formation of large amounts of unidentified by-products (as determined by HPLC analysis). For example, a lower ratio (3:1) of NADP+ to labeled nicotinamide required longer reaction times and resulted in maximum yields of 37% after 1 hour, while ratios higher than 10:1 led to final products with low specific radioactivity, thus limiting their use in KIE experiments. When a ratio of 10:1 was used, yields reached a maximum value of 70–75% (measured as the percentage of total radioactivity in the desired product) after ~30 minutes (Figure 2B). It is important to establish a point at which the amount of formed product is at its maximum, as a slow steady decline occurs at longer reaction times. This decline could be result of several different processes, and not simply a reverse reaction catalyzed by NADase (as this is an equilibrium reaction). Moreover, we observed a small amount of unidentified by-product (max ~1.6% of total radioactivity; Figure2B) that was easily separated from the NADP+, and therefore did not present a major obstacle in either total yield or purity of the product. Since a 10:1 ratio produced satisfactory yields of [carbonyl-14C]-NADP+, the reaction was not further optimized. However, if lower specific radioactivities are acceptable in the product, it is possible to obtain up to 90% yields in 1 hr using higher ratios of NADP+/nicotinamide. The relatively low price of the radioactive [carbonyl-14C]-nicotinamide starting material was also a factor in the experimental design described above. Since the cost of this starting material is the major contributor to the overall cost of each synthesis, the price difference makes the [carbonyl-14C]-nicotinamide more appealing as a reagent.
The [carbonyl-14C]-NADP+ produced must be separated from the reaction mixture that also contains [carbonyl-14C]-nicotinamide and 2'-monophosphoadenosine-5'-diphosphoribose. Otherwise, it is possible that subsequent kinetic experiments with the final [carbonyl-14C]-NADPH product will show artifacts due to nicotinamide contamination. In addition, 2'-monophosphoadenosine-5'-diphosphoribose has been shown to inhibit certain NADP+-dependent dehydrogenases, including GDH [29]. This makes isolating the [carbonyl-14C]-NADP+ from the exchange reaction mixture crucial for successful downstream steps. A previously published method for the purification of NADPH [5] was found to be inefficient in separating NADP+ from nicotinamide. Therefore, that method was modified by reducing the percentage of methanol from 15 to 7.5% during the isocratic elution that starts at 11 min, consequently increasing separation between NADP+ and nicotinamide peaks to 8 min. Thus, in the modified method, NADP+ elutes at 12.5 min, and nicotinamide at 20.5 min. The [carbonyl-14C] NADP+ elutes in a broad peak over 5 min, as depicted in Figure 2, and is collected in at least 6 ml of 7.5% MeOH /0.1 M KH2PO4 solution. Further reduction in the methanol percentage caused significant peak broadening that rendered greater separation in retention times meaningless.
The lyophilized product from the first exchange step contains [carbonyl-14C]-NADP+ as well as significant amounts of KH2PO4. When this solid was simply resuspended in 500 l water (to a final concentration of ~ 1.2 M KH2PO4) and used for the subsequent GDH reaction, a dramatic decrease in the rate of the reduction to NADPH was observed (80% of the NADP+ still remained unreacted at 1 hour, in contrast to the 99% conversion to NADPH usually seen at 37 C and pH 7.0 for the same reaction in the presence of 0.1 M KH2PO4). We hypothesized that this was due to the high salt concentration in the resuspension solution, which was either causing active GDH to precipitate out of the solution (a `salting out' effect) or directly inhibiting the GDH reaction. To test this hypothesis, we used a Sephadex 1 ml C-18 column to remove the excess salt from the lyophilized product, and [carbonyl-14C]-NADP+ was eluted from the column using a 15% MeOH/0.1 M KH2PO4 (pH 7.5) solution. The losses in yield due to the addition of this step were minor (<2%). Addition of this step restored the reaction rate of subsequent GDH-catalyzed reductions, providing support to the above hypothesis.
Our procedure describes the reduction of [carbonyl-14C]-NADP+ to [carbonyl-14C]-NADPH, 4R-[carbonyl-14C, 4-2H]-NADPH, or [carbonyl-14C, 4-2H2]-NADPH. If desired, 4S-[carbonyl-14C-4-2H] NADPH could be produced by simply truncating the procedure and reducing [carbonyl-14C]-NADP+ with [1-2H]-glucose and GDH. Also, since NADase can accept both NAD+ and NADP+ as substrates, experimental procedures similar to the ones described here can be envisioned for the preparation of [carbonyl-14C]-NAD+/NADH and their deuterated derivatives for kinetic and mechanistic studies of NAD(H) specific enzymes.
14C-labeling at the nicotinamide carbonyl carbon of NADPH does not result in a significant -2 14C KIE
Due to the location of the new label near the reaction center, more specifically at the amide carbonyl of NADPH, the possible issue of a γ-2° KIE artifact on hydride transfer KIEs to be measured using this compound had to be addressed. To examine the extent of that effect on cofactor oxidation, we designed experiments using [Ad-3H]-NADPH in competition with either [Ad-14C]-NADPH or [carbonyl-14C]-NADPH as substrates of the EcDHFR catalyzed reaction. The first combination ([Ad-14C]-NADPH/[Ad-3H]-NADPH) resulted in a BIE on the adenine ring of 1.002 0.002 and the second ([carbonyl-14C]-NADPH/[Ad-3H]-NADPH) in a mixed KIE of 1.005 0.002. The ratio of these values yield a γ-2° 14C KIE ([carbonyl-14C]-NADPH/[Ad-14C]-NADPH) of 1.003 ± 0.004. This value is practically unity (no KIE) as the errors usually obtained on either 1° and even 2° hydrogen KIEs are larger than 0.01. When measuring small KIEs (e.g. 2° KIEs) with other enzymes than EcDHFR, or when using other carbonyl-labeled nicotinamide cofactors, we recommend that researchers test for such possible artifacts for their specific system.
Testing the viability of synthesized materials using KIEs: Comparison of intrinsic KIEs measured using [carbonyl-14C]-NADPH vs. [Ad-14C]-NADPH
To validate the use of the new [carbonyl-14C]-NADPH and its deuterated derivatives in kinetic experiments, we have compared the outcome of KIE experiments that used the new synthesized materials, to previous KIE measurements that utilized adenine labeled cofactors as substrates of EcDHFR. We conducted 1° H/T experiments using [carbonyl-14C]-NADPH and 4R-3H-NADPH as substrates, and 1° D/T KIEs using 4R-[carbonyl-14C, 4-2H]-NADPH and 4R-3HNADPH as substrates, for the EcDHFR reaction. All experiments were performed at 25 C and pH 9.0. The results, along with the calculated intrinsic KIEs, are compared to previously published values in Table 2. All previous measurements were done using [Ad-14C]-NADPH and its deuterated analogs as substrates. The close agreement between our measured and intrinsic values and all prior results indicates that with this enzyme the [carbonyl-14C]-NADPH materials are excellent substitutes for the [Ad-14C]-NADPH materials in KIE experiments.
Comparison of cost, effort, and yield of [Ad-14C]-NADPH [5] vs. [carbonyl-14C]-NADPH syntheses
The new synthesis of [carbonyl-14C]-NADPH presented here is advantageous because: (i) the [carbonyl-14C]-nicotinamide precursor is substantially cheaper starting material than the commercially available [Ad-14C]-NAD+ precursor used to make [Ad-14C]-NADPH (by a factor of >12 based on $/Ci and synthetic yields); (ii) the new synthesis is much faster (e.g., 30 min glycosidic exchange vs. 3 hours 2'-phosphorilation); (iii) it requires less enzymes and reagents (e.g., ca. $1 NADase vs. ca. $70 NAD-kinase and phosphocreatine kinase); and (iv) has better yield than that of [Ad-14C]-NADPH (e.g., 60–75% vs. 40–60%). In summary, the new synthetic procedure is a cost- and effort-effective alternative to previously published procedures”.
Conclusion
The development of a new labeling pattern for 14C-labled nicotinamide cofactors is presented along with some applications and the examination of possible experimental artifacts. We have shown that [carbonyl-14C]-NADP+, [carbonyl-14C]-NADPH, 4R-[carbonyl-14C, 4-2H]-NADPH and [carbonyl-14C, 4-2H2]-NADPH can be synthesized quickly, in reasonable yields, and at relatively low costs, using a simple chemoenzymatic procedure. We have utilized [carbonyl-14C]-NADPH to demonstrate that NADPH labeled with 14C at the nicotinamide carbonyl has no significant effects on the hydrogen KIEs measured using those isotopologues as substrates in the EcDHFR reaction. 1 H/T and D/T KIEs measured using the newly synthesized materials are in complete agreement with KIEs previously measured with NADPH labeled at the adenine ring, and confirm the utility of this material. Finally, the synthesized material can easily be adapted for use in kinetic experiments with other NADP+/NADPH/NAD+/NADH-dependent enzymes, and as these are ubiquitous in nature the synthetic methods described herein could potentially be of broad utility.
Acknowledgments
This work was supported by NSF CHE-1149023, NIH R01 GM065368, and BSF-2007256.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations used: KIE, kinetic isotope effect; EcDHFR, Escherichia coli dihydrofolate reductase; H2F, dihydrofolate; H4F, tetrahydrofolate; T(V/K)H,obs and H/Tobs, observed tritium KIE; D(V/K)H,obs and H/Dobs, observed deuterium KIE; H/Tint, intrinsic tritium KIE; H/Dint, intrinsic deuterium KIE; BIE, binding isotope effect; NADase, DPN nucleosidase/NAD+ glycohydrolase; PMSF, phenylmethylsulfonate; bmGDH, Bacillus megaterium glucose dehydrogenase; tbADH, Thermoanaerobium brockii alcohol dehydrogenase; DPM, disintegrations per minute
References
- [1].Voet D, Voet JG. Biochemistry. Wiley; 1995. [Google Scholar]
- [2].Hixson SS, Hixson SH. Photochemical labeleing of yeast alcohol dehydrogenase with an azide analog of NAD+ Photochem. Photobiol. 1973;18:135–138. doi: 10.1111/j.1751-1097.1973.tb06403.x. [DOI] [PubMed] [Google Scholar]
- [3].Cook PF, Oppenheimer NJ, Cleland WW. Secondary deuterium and nitrogen-15 isotope effects in enzyme catalyzed reactions. Chemical mechanism of liver alcohol dehydrogenase. Biochemistry. 1981;20:1817–1825. doi: 10.1021/bi00510a016. [DOI] [PubMed] [Google Scholar]
- [4].Stone SR, Mark A, Morrison JF. Interaction of analogues of nicotinamide adenine dinucleotide phosphate with dihydrofolate reductase from Escherichia coli. Biochemistry. 1984;23:4340–4346. [Google Scholar]
- [5].Markham KA, Sikorski RS, Kohen A. Synthesis and utility of 14C Labeled nicotinamide cofactors. Anal. Biochem. 2004;325:62–67. doi: 10.1016/j.ab.2003.10.027. [DOI] [PubMed] [Google Scholar]
- [6].Woenckhaus C, Jeck R. In: Pyridine nucleotide coenzymes: chemical, biochemical, and medical aspects. Dolphin D, Poulson R, Avramovic O, editors. John Wiley & Sons Inc.; New York: 1987. pp. 449–568. [Google Scholar]
- [7].Dutta S, Cook RJ, Houtman JCD, Kohen A, Cheatum CM. Characterization of azo-NAD to assess its potential as a two-dimensional infrared probe of enzyme dynamics. Anal. Biochem. 2010;407:241–246. doi: 10.1016/j.ab.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Markham KA, Kohen A. Analytical procedures for the preparation, isolation, analysis and preservation of reduced nicotinamides. Curr. Anal. Chem. 2006;2:379–388. [Google Scholar]
- [9].Sikorski RS, Wang L, Markham KA, Rajagopalan PTR, Benkovic SJ, Kohen A. Tunneling and coupled motion in the Escherichia coli dihydrofolate reductase catalysis. J. Am. Chem. Soc. 2004;126:4778–4779. doi: 10.1021/ja031683w. [DOI] [PubMed] [Google Scholar]
- [10].Antikainen NM, Smiley RD, Benkovic SJ, Hammes GG. Conformation coupled enzyme catalysis: single-molecule and transient kinetics investigation of dihydrofolate reductase. Biochemistry. 2005;44:16835–16843. doi: 10.1021/bi051378i. [DOI] [PubMed] [Google Scholar]
- [11].Fierke CA, Johnson KA, Benkovic SJ. Construction and evaluation of the kinetic scheme associated with dihydrofolate reductase from Escherichia coli. Biochemistry. 1987;26:4085–4092. doi: 10.1021/bi00387a052. [DOI] [PubMed] [Google Scholar]
- [12].Stojkovic V, Perissinotti L, Willmer D, Benkovic S, Kohen A. Effects of the donor acceptor distance and dynamics on hydride tunneling in the dihydrofolate reductase catalyzed reaction. J. Am. Chem. Soc. 2012;134:1738–1745. doi: 10.1021/ja209425w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang L, Goodey NM, Benkovic SJ, Kohen A. Coordinated effects of distal mutations on environmentally coupled tunneling in dihydrofolate reductase. Proc. Natl. Acad. Sci. USA. 2006;103:15753–15758. doi: 10.1073/pnas.0606976103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Melander L, Saunders WH. Reaction rates of isotopic molecules. Krieger, R.E.; Malabar, FL: 1987. [Google Scholar]
- [15].Cook PF. Enzyme Mechanism from Isotope Effects. CRC Press; Boca Raton, Fl.: 1991. [Google Scholar]
- [16].Cook PF, Cleland WW. Enzyme kinetics and mechanism. Taylor & Francis Group LLC; New York, NY: 2007. [Google Scholar]
- [17].Wang L, Tharp S, Selzer T, Benkovic SJ, Kohen A. Effects of a distal mutation on active site chemistry. Biochemistry. 2006;45:1383–1392. doi: 10.1021/bi0518242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Northrop DB. Intrinsic isotope effects in enzyme catalyzed reactions. In: Cook PF, editor. Enzyme mechanism from isotope effects. CRC Press; Boca Raton, Fl.: 1991. pp. 181–202. [Google Scholar]
- [19].Sen A, Yahashiri A, Kohen A. Triple isotopic labeling and kinetic isotope effects: a sensitive and accurate method forexposing H-transfer steps in enzymatic systems. Biochemistry. 2011;50:6462–6468. doi: 10.1021/bi2003873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Schramm VL. Binding isotope effects: boon and bane. Curr. Opin. Chem. Biol. 2007;11:529–536. doi: 10.1016/j.cbpa.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cameron CE, Benkovic SJ. Evidence for a functional role of the dynamics of glycine-121 of Escherichia coli dihydrofolate reductase obtained from kinetic analysis of a site-directed mutant. Biochemistry. 1997;36:15792–15800. doi: 10.1021/bi9716231. [DOI] [PubMed] [Google Scholar]
- [22].Blakley RL. Crystalline dihydropteroylglutamic acid. Nature. 1960;188:231–232. [Google Scholar]
- [23].Kohen A, Jonsson T, Klinman JP. Effect of protein glycosylation on catalysis: changes in hydrogen tunneling and enthalpy of activation in the glucose oxidase reaction. Biochemistry. 1997;36:2603–2611. doi: 10.1021/bi962492r. [DOI] [PubMed] [Google Scholar]
- [24].Markham KA, Sikorski RS, Kohen A. Purification, analysis, and preservation of reduced nicotinamide adenine dinucleotide 2'-phosphate. Anal. Biochem. 2003;322:26–32. doi: 10.1016/j.ab.2003.07.010. [DOI] [PubMed] [Google Scholar]
- [25].Yahashiri A, Sen A, Kohen A. Microscale synthesis and kinetic isotope effect analysis of (4R)-[Ad-14C, 4-2H] NADPH and (4R)-[Ad-3H,4-2H] NADPH. J. Labelled Compd. Radiopharm. 2009;52:463–466. doi: 10.1002/jlcr.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].McCracken JA, Wang L, Kohen A. Synthesis of R and S tritiated reduced β-nicotinamide adenine dinucleotide 2' phosphate. Anal. Biochem. 2003;324:131–136. doi: 10.1016/j.ab.2003.09.025. [DOI] [PubMed] [Google Scholar]
- [27].Agrawal N, Kohen A. Microscale synthesis of 2-tritiated isopropanol and 4R-tritiated reduced nicotinamide adenine dinucleotide phosphate. Anal. Biochem. 2003;322:179–184. doi: 10.1016/j.ab.2003.08.012. [DOI] [PubMed] [Google Scholar]
- [28].Sikorski RS, Wang L, Markham KA, Rajagopalan PTR, Benkovic SJ, Kohen A. Tunneling and coupled motion in the E. coli dihydrofolate reductase catalysis. J. Am. Chem. Soc. 2004;126:4778–4779. doi: 10.1021/ja031683w. [DOI] [PubMed] [Google Scholar]
- [29].Ben-Hayyim G, Hochman A, Avron M. Phosphoadenosine diphosphate ribose, a specific inhibitor of nicotinamide adenine dinucleotide phosphate enzymes. J. Biol. Chem. 1967;242:2837–2839. [PubMed] [Google Scholar]