Abstract
All cells have developed various mechanisms to respond and adapt to a variety of environmental challenges, including stresses that damage cellular proteins. One such response, the heat shock response (HSR), leads to the transcriptional activation of a family of molecular chaperone proteins that promote proper folding or clearance of damaged proteins within the cytosol. In addition to its role in protection against acute insults, the HSR also regulates lifespan and protects against protein misfolding that is associated with degenerative diseases of aging. As a result, identifying pharmacological regulators of the HSR has become an active area of research in recent years. Here, we review progress made in identifying small molecule activators of the HSR, what cellular targets these compounds interact with to drive response activation, and how such molecules may ultimately be employed to delay or reverse protein misfolding events that contribute to a number of diseases.
1. Cellular Stress Responses to Adverse Environmental Conditions
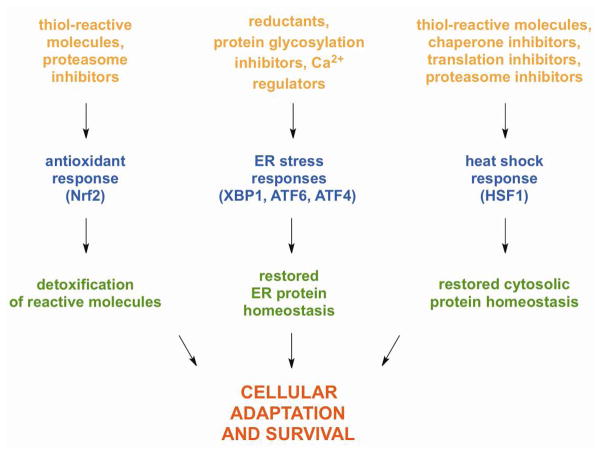
The environment in which organisms exist is not static, and most biological systems can adapt to maintain homeostasis, an ideal steady state that promotes growth, replication, and/or survival. However, a wide range of physiological and environmental stimuli continuously disrupt this equilibrium, resulting in constant flux around a homeostatic point. Unfavorable conditions such as elevated temperature, oxidant accumulation, abrupt alterations in osmolarity, and nutrient deprivation change the intracellular environment, restrict cell growth and development, and can lead to cell death. To restore homeostasis under such unfavorable conditions, organisms have developed sophisticated stress response mechanisms that increase expression of protective genes within a cell. A cell’s response to acute stress is generally transient and can involve a number of transcriptional regulators, some of which are activated simultaneously by particular stressors. Many of these stress responses are in place to prevent, repair, or clear damage to proteins caused by harsh environmental conditions, thereby influencing protein homeostasis. Examples of transcriptional responses to stress that are often simultaneously induced include the antioxidant response, ER stress responses, and the heat shock response (HSR) (Fig. 1).
Figure 1. Parallel Stress Pathways.
The antioxidant response, ER stress response, and HSR pathways help maintain cellular homeostasis following exposure to different environmental stresses.
1.1. Antioxidant Response
In response to certain stresses (e.g., oxidative stress, proteasome inhibition), many organisms induce the expression of genes involved in detoxification. In mammalian cells, the antioxidant response is mediated by the transcription factor Nrf2 and its negative regulator, Keap1. Normally, Nrf2 undergoes rapid degradation in cells, a process that is facilitated by Keap1; upon exposure to stress, Nrf2 accumulates and migrates into the nucleus to promote the expression of its target genes, including many involved glutathione metabolism and scavenging of free radicals.1 Transcription factors related to mammalian Nrf2 regulate similar responses in multicellular eukaryotes.2 However, in unicellular eukaryotes, the response is controlled by unrelated transcription factors with different mechanisms of activation.3 Despite the lack of conserved antioxidant response transcription factors in all eukaryotes, the genes that are induced by these disparate factors universally protect cells against excessive protein and DNA damage caused by reactive species (e.g., oxidants, electrophiles) and are commonly induced by sub-cytotoxic concentrations of these molecules.4
1.2. ER Stress Responses
Eukaryotic cells have one or more transcriptional regulators that respond to protein misfolding in the endoplasmic reticulum.5 In mammalian cells, the transcription factors XBP1, ATF6, and ATF4 are activated following treatment with protein reductants, protein glycosylation inhibitors, molecules that influence Ca2+ stores, and other conditions that perturb ER homeostasis.6,7 Collectively, XBP1, ATF6, and ATF4 control the expression of multiple protective proteins including molecular chaperones that reside within the ER.7 These chaperone proteins, in turn, allow for proper folding and function of many membrane and secreted proteins. In addition, the ER chaperones can assist with targeting misfolded or damaged ER proteins for export into the cytosol where they are degraded by the proteasome.8
1.3. Heat Shock Response
In contrast to ER stress responses, which predominantly serve the secretory pathway, the heat shock response (HSR) primarily responds to protein misfolding conditions in the cytoplasm.9,10 It results in the transcriptional activation of genes encoding cytosolic molecular chaperones, proteases, and other proteins essential for protection and recovery from cellular damage. Under stress conditions, many heat shock proteins (Hsps) (e.g., Hsp70) hold and refold denatured proteins, prevent them from aggregating, and/or regulate their degradation. Some Hsps (e.g., Hsp90) play integral roles in signal transduction, immunity, and apoptosis.11–16 Modulation of the HSR, therefore, may be directly beneficial for a variety of human diseases associated with cell growth (e.g., cancer) and accumulation of damaged and misfolded proteins (e.g., neurodegenerative diseases).
2. HSF1: A Key Regulator of Heat Shock Genes
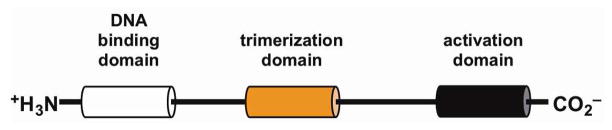
In all eukaryotes, the HSR is primarily mediated by heat shock transcription factors (HSFs), which bind to heat shock elements (HSEs) in the promoters of target genes.17,18 A functional HSE contains a series of pentameric units arranged as inverted adjacent arrays of the sequence 5′-nGAAn. At least three pentamers are required for optimal interaction between HSF and HSE.19,20 Whereas a single HSF has been identified in Saccharomyces cerevisiae, Caenorhabditis elegans, and Drosophila melanogaster, multiple HSF isoforms are encoded in the genomes of vertebrates and many plants.21–24 Among these HSFs, HSF1 is the principal activator of heat shock gene transcription in response to elevated temperature and other environmental stresses. Despite variation in size and homology, HSFs share several conserved features: a conserved DNA-binding domain composed of a winged helix-turn-helix motif located near the N-terminus, three leucine zipper repeats that regulate trimerization of the factor, and an activation domain that undergoes many inducible post-translational modifications during stress (Fig. 2).24–28
Figure 2. HSF1 Domain Structure.
HSF1 has three principal domains that facilitate its regulation of Hsp genes in response to stress. The DNA binding domain can bind inverted 5′-nGAA-3′ repeats upon trimerization of monomeric HSF1 through its trimerization domain. The activation domain near the C-terminus is the site of many inducible post-translational modification following exposure to various stresses.
In many organisms, HSF1 activation is a multi-step process that includes nuclear accumulation, trimerization, acquisition of DNA binding activity, and transactivation of genes encoding Hsps (Fig. 3).29 The stress-dependent conversion of HSF1 into its active form implies that HSF1 is negatively regulated under normal conditions.22,30 The Hsps themselves are proposed to inhibit heat shock gene expression via an autoregulatory loop.24,30,31 Under normal growth conditions, monomeric HSF1 is purportedly stabilized by cytosolic Hsps such as Hsp70 and Hsp90.32 During heat shock, accumulated unfolded proteins may titrate Hsps away from HSF1 and relieve HSF1 repression. Ultimately, HSF1 trimerizes and acquires DNA binding ability, resulting in an increased amount of Hsps in the stressed cells.
Figure 3. Steps in HSF1 Activation.

HSF1 activation following exposure to protein-damaging stresses often involves its dissociation from various negative regulators (including Hsp70 and Hsp90), its trimerization, and its inducible post-translational modification. Post-transalational modifications to HSF1 include sumoylation (SUMO), acetylation (Ac), and phosphorylation (P).
2.1. Subcellular Localization
HSF1 undergoes changes in its cellular localization in response to stress in mammalian cells. Under homeostatic conditions, it is distributed in a diffuse pattern in both the cytoplasm and/or the nucleus, depending on the cell type. However, in heat-shocked cells, HSF1 is found primarily in the nucleus in punctate granules that form in various cell lines and are correlated with the active form of HSF1.33–35 During attenuation and recovery, the granules disappear and HSF1 redistributes to the cytoplasm, thus suggesting a role for HSF1 granules as a temporal nuclear compartment for spatial regulation of HSF1 activity.35
2.2. Trimerization
In vertebrate and Drosophila cells, HSF1 is expressed as an inert monomer with both DNA binding and transcription activity repressed through intramolecular interaction and constitutive phosphorylation.31,36 Upon activation, HSF1 trimerizes to acquire DNA binding activity.37 The trimerization domain in HSF1 contains high α-helical content and can be divided into two subdomains, each containing an amphipathic helix with hydrophobic heptad repeats HR-A and HR-B.28,38 In trimeric HSF1, HR-A and HR-B possibly form a highly elongated structure. Intermolecular hydrophobic interactions between HR-A and HR-B mediate the formation of a triple-stranded coiled-coil configuration.38 Spontaneous trimerization of HSF1 is inhibited by another hydrophobic heptad repeat HR-C between the trans-activation and regulatory domains near the C-terminus. It is thought that HR-C interacts with the HR-A and HR-B domains and maintains HSF1 in monomeric form under normal growth conditions.36
2.3. Post-Translational Modifications
HSF1 can undergo numerous post-translational modifications, including phosphorylation, sumoylation, and acetylation. Of these, phosphorylation has been the most extensively studied. For human HSF1, phosphorylation occurs on at least 15 Ser residues (S121, S216, S230, S292, S303, S307, S314, S319, S320, S326, S344, S363, S368, S419 and S444) and four Thr residues (T142, T323, T367 and T369) and has both positive and negative effects on HSF1 activity.39–45 Inducible phosphorylation of S230 by the calcium/calmodulin-dependent kinase CaMKII induces HSF1 activation.46 In addition, phosphorylation of S326 facilitates the association between HSF1 and the coactivator DAXX.47 However, other phosphorylation events repress the activity of HSF1. For instance, mutation of S303 and S307 to Ala derepresses mammalian HSF1 activity under non-stress conditions.48,49
Phosphorylation at certain sites may influence additional post-translational modification events. In support of this, phosphorylation at S303 correlates with sumoylation of HSF1 on K298. Phosphorylation mapping and analysis further established that HSF1 is modified by SUMO-1 and SUMO-2 in a stress-inducible and phosphorylation-dependent manner at S303.50,51 Similar to S303 phosphorylation, sumoylation of HSF1 is thought to repress HSF1 transcriptional activity. HSF1 also undergoes stress-inducible acetylation on several different lysine residues. Activation of the deacetylase sirtuin 1 (SIRT1) promotes maintenance of HSF1 in a DNA-binding competent state.52 Unlike phosphorylation events that primarily occur in the transactivation domain, lysines prone to acetylation are found in the DNA binding, oligomerization, and transactivation domains of HSF1, suggesting potentially broader regulation of acetylation upon its transcriptional activity.43
2.4. Binding to Negative Regulators
The heat shock response is self-regulated under thermal stress.53,54 In yeast, deletion of the two constitutively expressed cytosolic Hsp70 genes results in activation of the HSR at normal growth temperature.55,56 Hsp70 associates with the HSF1 transactivation domain in vivo and in vitro.31,57–59 However, Hsp70 alone is not sufficient to deactivate HSF1 in vitro.59 Regulation of HSF1 by Hsp90 and associated co-chaperones represents another plausible means of repressing HSF1 activity, as Hsp90 associates with HSF1 in vivo and in vitro.60 In support of this, direct injection of anti-Hsp90 in a Xenopus oocytes induces the HSR.61 A role of Hsp90 in HSF1 repression is also supported by the finding that Hsp90 inhibitors (e.g., geldanamycin, radicicol, and celastrol) also activate HSF1.16,62–66 Therefore, the likely model is that HSF1 remains inactive due to binding with Hsp70 and Hsp90 under normal growth conditions.
3. Small Molecular Activators of the Heat Shock Response
In recent years, there has been growing interest in identifying small molecule regulators of the HSR. These molecules have a variety of chemical features–some are highly reactive with a diverse array of cellular proteins, whereas others have specific targets that impair protein homeostasis. Below, we will highlight recent studies aimed at identifying small molecule HSF1 activators and their mechanisms of action.
3.1. Pharmacological Activators with Known Targets in the Protein Homeostasis Network
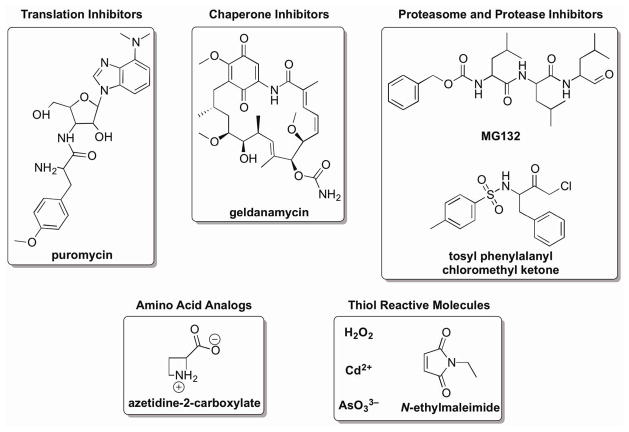
Many known HSR activators influence parts of the protein homeostasis network, a collection of proteins that provide surveillance for the synthesis, maturation, and turnover of proteins. Known activators of the HSR that play roles in this capacity include protein translation inhibitors (e.g., puromycin), amino acid analogs that prevent proper protein folding (e.g., azetidine 2-carboxylate, canavanine), several different pharmacological inhibitors of molecular chaperones (e.g., geldanamycin and radicicol), and proteasome and protease inhibitors (e.g., MG132).67,68 A defining feature for these molecules is that their molecular targets are integral parts of the protein homeostasis network.12,69 Drastically altering the function of the protein synthesis, folding, and clearance machineries imbalances cellular homeostasis and is likely to lead to accumulation of misfolded polypeptides that oligomerize and aggregate in the cytosol,70,71 thereby necessitating an increase in molecular chaperone expression.
3.2. Thiol-Reactive Molecules Activating the Heat Shock Response
For other HSF1 activators, the molecular targets that are linked with HSR activation are less clearly defined. Three distinct classes of reactive molecules–oxidants, certain transition metals/metalloids, and organic electrophiles–fall into this category (Fig. 4).32,67,72 Each class of molecules has the propensity to damage proteins directly, often on the side chains of highly reactive Cys residues in target proteins (Fig. 5).73–75 Below, we explore the reactions that each of these classes of thiol-reactive HSF1 activators undergo with proteins.
Figure 4. Families of Molecules That Activate HSF1.
HSF1 activators can perturb various aspects of protein homeostasis including translation, protein folding, processing, and degradation.
Figure 5. Thiol-Modifications by Oxidants, Transition Metals, and Organic Electrophiles.
Oxidants, metals and metalloids, and organic electrophiles carry out different modifications on reactive Cys residues in target proteins.
3.2.1. Oxidants
A variety of oxidants are capable of activating HSF1 in mammalian cells, including hydrogen peroxide, hypochlorous acid, and diamide.76–78 Depending on its strength, an oxidant can cause breaks within the peptide backbone or react with the side chains on several amino acids, including Cys, His, Met, Phe, and Tyr.79 Of these residues, highly reactive Cys residues (i.e., those with lower pKa values) in specific target proteins are often the most susceptible to oxidation (Fig. 5), with the residue’s thiol group initially forming a sulfenic acid (Fig. 5).74,80 There are several possible fates of the sulfenic acid form of Cys. It can undergo a condensation reaction with a nearby thiol (intramolecularly, with a Cys residue in another protein, or with free thiols within the cell (e.g., GSH)) to form a disulfide bond.74,80 Alternatively, the Cys sulfenic acid can undergo further oxidation to a sulfinic or sulfonic acid.74,80 Oxidative modifications of Cys can play a regulatory role in protein function, as Cys residues are often incorporated into sensor proteins to detect the accumulation of oxidants and promote expression of various oxidant defense genes.74,81–83 Such sensory Cys residues may be present in HSF1 or particular Hsps which are implicated in HSF1 regulation, as will be described later.
3.2.2. Transition metals and metalloids
Exposure to many metal ions (e.g., Zn2+, Cd2+, Hg2+, Pb2+, Co2+) and some metalloid-containing anions (e.g., AsO33−) also promotes HSF1 activation.84–86 In terms of their reactions with proteins, these molecules function as Lewis acids and form coordinate covalent bonds with amino acid side chains bearing readily accessible lone pairs of electrons.87 Clusters of nearby Cys, His, Asp, or Glu residues in proteins commonly chelate the metal or metalloid ions (Fig. 5), and such reactions may perturb protein structure and function by displacing normal metal centers or by forming non-natural metal binding sites.75,87 These molecules typically are slower to activate the HSR compared with other thiol-reactive compounds or acute heat stress,33,88 which may be because they require increased time to cross the cell membrane through a transporter and/or that they react more slowly with protein targets responsible for bringing about the response. Nonetheless, increased levels of metal chelators (e.g., GSH, N-acetyl Cys) decrease overall Hsp expression by these molecules,89–91 suggesting that their reaction with protein targets is required for HSR activation.
3.2.3. Organic electrophiles
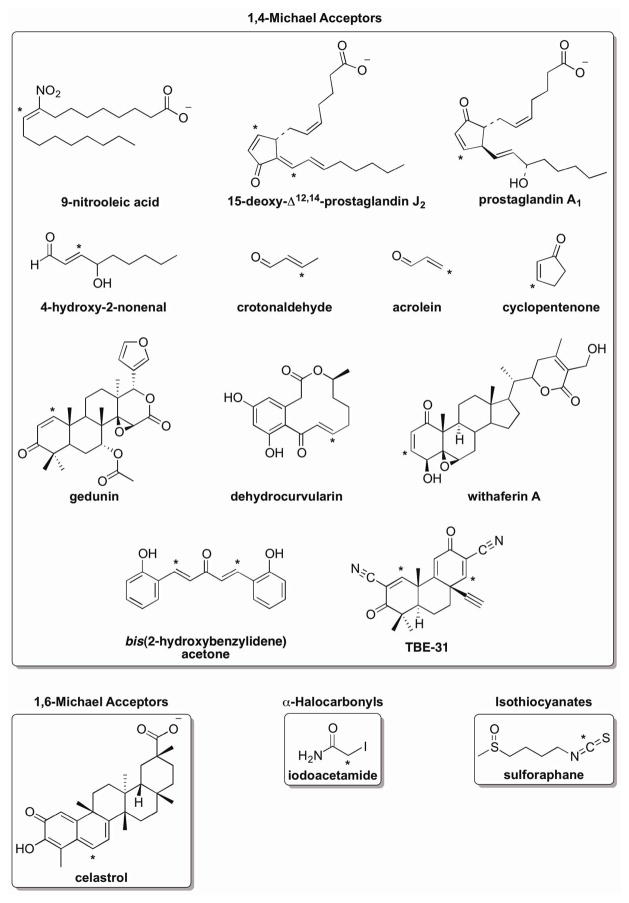
Many organic electrophiles induce Hsp expression, including the cyclopentenone prostaglandins (PGs) 15-deoxy-Δ12,14-PGJ2 (15d-PGJ2) and PGA1,92–95 the lipid peroxidation products 4-hydroxy-2-nonenal (HNE), acrolein, and crotonaldehyde,96–99 the lipid nitration product nitrooleic acid,100 the phytochemicals celastrol, withaferin A, and sulforaphane,16,101,102 and numerous synthetic chemicals including iodoacetamide and cyclopentenone.103–105 All of these molecules contain one (or more) electrophilic center(s) (Fig. 6), but they vary widely in their physical and chemical properties (Fig. 1, Table 1). Below, we will highlight several features within these molecules that may contribute to their potency as HSF1 activators.
Figure 6. Classes of Organic Electrophiles That Activate HSF1.
Electrophiles that activate HSF1 are grouped according to the type of addition reaction that they carry out with thiol groups. An asterisk (*) indicates where thiol groups are likely to add within these molecules. More information about the physicochemical properties of these molecules and their thiol adducts is provided in Table 1.
Table 1.
Electrophilic Activators of the Heat Shock Response in Cultured Mammalian Cells.
| Class | Electrophile | Origin | Active Concentrationsa | Predicted logPb | Adduct Properties |
|---|---|---|---|---|---|
| 1,4-Michael Acceptors | |||||
| nitrooleic acid | lipid nitration | 1–5 μM100 | 6.6 | reversibility111 | |
| 15d-PGJ2 | lipid metabolism | ~5–15 μM94,95 | 4.1 | cross-linking113,114 | |
| PGA2 | lipid metabolism | 20–100 μM92,93 | 2.6 | –c | |
| HNE | lipid peroxidation | 20–60 μM97,132 | 1.5 | hemiacetal formation; cross-linking116–118 | |
| crotonaldehyde | lipid peroxidation; combustion | 40–80 μM99 | 0.5 | cross-linkingd | |
| acrolein | lipid peroxidation; combustion | ~100 μM98 | 0.1 | cross-linking115 | |
| cyclopentenone | synthetic | 250–1000 μM104 | 0.3 | – | |
| gedunin | natural product | 1–5 μM102 | 2.9 | – | |
| dehydrocurvularin | natural product | 1–5 μM102 | 2.0 | – | |
| withaferin A | natural product | 1–5 μM102 | 2.6 | – | |
| bis (2-hydroxybenzylidene) acetone | synthetic | 1–5 μM119 | 3.2 | cross-linking120,d | |
| TBE-31 | synthetic | 0.1–0.5 μM119 | 2.5 | cross-linking,d reversible121,d | |
| 1,6-Michael Acceptors | celastrol | natural product | 1–10 μM16 | 6.9 | reversibility112,d |
| α-Halocarbonyls | iodoacetamide | synthetic | 50–100 μM103 | 0.1 | – |
| Isothiocyanates | sulforaphane | natural product | 5–10 μM101 | −0.5 | reversibility110 |
Values are estimated from literature reports cited in various mammalian cell lines.
logP values were predicted using ChemDraw.
Dashes (−) indicate predicted lack of reversibility, cross-linking, or other adduct rearrangement.
Predicted based on structure.
Like oxidants and transition metals, electrophilic molecules react predominantly with the thiol group in Cys, although the nucleophilic amine groups of Lys and His residues may become modified in some target proteins.73,82,106–108 Since nucleophilicity (and hence, overall reactivity) of Cys residues is increased upon thiol deprotonation to the thiolate anion, Cys residues with low pKa values are particularly susceptible to modification by electrophilic molecules in target proteins.109 Reactivity of these molecules with proteins is closely linked with HSF1 activation. Analogs of cyclopentenone PGs, cyclopentenone, withaferin A, gedunin, and dehydrocurvularin that are saturated at their electrophilic centers cannot react with proteins and no longer activate HSF1.92,93,102,104 Similarly, the addition of DTT to celastrol or iodoacetamide prevents these electrophiles from increasing the expression of HSR genes.66,103 The overall differences in potency between these molecules may be due, in part, to differences in their specificity and rates of reactivity with target proteins, but a comprehensive study to explore these possibilities has not been performed.
Other properties of these electrophiles, including their hydrophobicity, adduct reversibility, and the ability to form intermolecular cross-links between target proteins, may contribute to HSF1 activation as well. Studies on the cyclopentenone PGs suggest that hydrophobicity alters the potency of HSR activation. 15d-PGJ2 is considerably more hydrophobic (estimated logP ~ 4.1) and more potent than cyclopentenone (estimated logP ~ 0.3).94,95,104 PGA1, which has an estimated logP value between that of 15d-PGJ2 and cyclopentenone, activates HSF1 at doses between that of the other molecules.92,93,104 Conversely, converting the natural product celastrol into more hydrophobic derivatives by esterifying its carboxylate group decreases its potency,16 perhaps due to poor solubility in aqueous solution.
In addition to an electrophile’s hydrophobicity, reversibility of the thiol adducts they form may contribute to HSR activation by these molecules. Several of the most potent HSF1 activators–nitrooleic acid, celastrol, and sulforaphane–carry out reversible reactions with thiol-containing molecules, with each molecule activating HSF1 in the low micromolar range.110–112 The equilibrium between the adducted form and the free form of the electrophile may be influenced by the relative concentrations of the electrophile and various target proteins (and nucleophilic small molecules), by pH changes, and by the degree of protein turnover. Therefore, it is likely that protein damage by these molecules is transient in treated cells.
The ability of an electrophile to form protein cross-links may also enhance HSR activation. Consistent with this idea, 15d-PGJ2 can cause cross-links to form between two proteins,113,114 suggesting that both its hydrophobicity and ability to form intermolecular cross-links may contribute to its increased potency in comparison with analogous electrophiles. In addition, HNE, acrolein, and crotonaldehyde are heterobifunctional electrophiles that are capable of causing unstable intermolecular cross-links between Cys and Lys residues in proteins,115–118 although the extent to which protein cross-linking contributes to HSF1 activation has not been explored with these molecules. Likewise, two recently identified and relatively potent HSF1 activators, TBE-31 and bis(2-hydroxybenzylidene) acetone, contain two Michael acceptor sites,119 through which they presumably cause protein cross-linking.120,121 Protein cross-linking may greatly enhance activation of cytoprotective response genes, as our recent studies indicate that protein cross-linkers impact cellular physiology at much lower concentrations than corresponding monofunctional alkylating agents.122,123
4. Models for HSF1 Activation by Pharmacological Activators
Over the past two decades, much has been learned about the properties and targets of the molecules that activate HSF1 and the specific molecular events occurring most proximal to HSF1 activation. However, the mechanism through which individual small molecule stressors are initially sensed to drive activation of the heat shock response, whether multiple sensors exert influence on different regulatory steps in HSF1 activation, and whether these mechanisms are common for all stresses remain largely unresolved. Below, we highlight several potential models that have been proposed for HSF1 activation by the pharmacological agents described earlier. Our focus will be on how certain stresses are sensed to promote activation of the response.
4.1. HSF1 as a Direct Stress Sensor
HSF1 may directly sense multiple types of stress, including elevated temperature, high levels of oxidants, altered pH, and exposure to hydrophobic molecules.24,76,124–126 This causes HSF1 to trimerize and acquire DNA binding activity. Consistent with these observations, HSF1 undergoes secondary and tertiary structural changes in its C-terminal regulatory domain, thereby promoting its trimerization in vitro.125 Structural changes that occur in HSF1 following exposure to increased temperature are readily reversible when the stress is removed, causing the protein return to its inactive, monomeric state.124 The nature of the reversibility for other stressors and whether this sensing mechanism occurs in a cellular context remain unresolved.
Several groups have carried out additional in vitro studies on the oxidation of mammalian HSF1 to explore if and how it senses peroxides. Following exposure to peroxides, HSF1 is oxidized in vitro on a number of Cys residues located within the DNA binding and/or the trimerization domains of the protein, leading to the formation of disulfide bonds.76,126–129 Conversely, exposure of HSF1 to dithiothreitol (DTT) prevents activation in these instances, an effect that mirrors the inhibitory effects of DTT on endogenous HSF1 activation.103 While these reports indicate that direct redox regulation of HSF1 is possible, there is some discrepancy regarding the Cys residues in HSF1 that are involved in oxidant sensing and whether disulfides are formed intra- or inter-molecularly. In addition, a limitation of this work is that disulfide formation has not been definitively shown to occur in endogenous HSF1 exposed to oxidants. Moreover, HSF1 in certain organisms (including S. cerevisiae) lacks Cys residues at proposed sites of disulfide formation,128 implying that direct oxidation may be a contributing factor but is not a conserved mechanism of HSF1 activation.
4.2. Chaperones as Stress Sensors
In the absence of stress, the chaperones Hsp70 and Hsp90 repress HSF1 trimerization and/or activation directly by binding to it in an inactive form.24,57,60,130 In the case of Hsp70, these binding interactions are mediated through its C-terminal substrate binding domain and the activation domain of HSF1.57 Following exposure to some stresses, interactions between HSF1 and these chaperones decrease.131,132 In addition, when chaperone function is inhibited through either pharmacological or genetic means,67,133 HSF1 activity increases significantly in a variety of organisms. There are at least two potential ways that HSF1 may be released from repression by these chaperones, depending on the type of pharmacological agent being added. One of these models involves titration of chaperones away from HSF1 to deal with protein folding problems elsewhere, whereas the other model posits that Hsp70 and Hsp90 function as direct sensors of many thiol-reactive HSR inducers.
4.2.1. Chaperones as Sensors of Cytosolic Protein Misfolding
Chaperones monitor all aspects of a protein’s ‘life cycle,’ from its emergence as a nascent peptide from the ribosome to its degradation within the cell.12,69 Many different pharmacological agents that activate HSF1 have been implicated in disrupting translation, post-translational protein processing and modification, protein folding, and protein degradation (Fig. 4).67 The end result of perturbing these points in the protein homeostasis network is the generation of peptide fragments or polypeptides that are prone to self-association.70,71 Indeed, several studies using genetic and pharmacological approaches have shown that inhibition of protein synthesis by the ribosome or protein degradation by the proteasome leads to enhanced protein aggregation in the cytosol.71,134–136 Such misfolding events are likely to place increased demands on the cytosolic chaperones, recruiting them away from HSF1 (Fig. 8).
Figure 8. Stress-Induced Derepression of HSF1 Activity.
Molecular chaperones (e.g., Hsp70 and Hsp90) that bind to and repress HSF1 activity are recruited to sites of misfolded polypetides and aggregated proteins that accumulate upon exposure to many stresses, allowing HSF1 to trimerize and bind DNA. For simplicity, only Hsp70 is shown as a negative regulator in complex with HSF1.
Likewise, protein modification by oxidants, metals, and electrophiles may cause misfolding of some target proteins, leading to their self-association and aggregation.137–139 Oligomerization of aggregation-prone proteins is accelerated by thiol-reactive molecules in many instances,140,141 perhaps through the molecules having a ‘seeding effect’ to promote further oligomerization. Since many different proteins are susceptible to modification by thiol-reactive molecules, protein folding problems may occur when cellular concentrations exceed levels that can be readily detoxified. In support of this, proteins form aggregates in cells treated with high concentrations of the oxidants hydrogen peroxide and hypochlorous acid.142,143 Taken together, these results indicate that protein damage caused by cellular exposure to thiol-reactive molecules may perturb the structures of proteins in their native state, creating a burden on the chaperones that normally repress HSF1 activity (Fig. 8).
In addition, several studies have highlighted that thiol-reactive molecules can impair efficient protein degradation by the proteasome. This inhibition may involve the covalent inactivation of multiple enzymatic activities within the proteasome, including those involved in ubiquitin chain removal and proteolysis of the targeted substrate.144–147 Similar to the effects of specific proteasome inhibitors,71 thiol-reactive molecules may cause increased levels of misfolded oligomers and protein aggregates in the cytosol through their inhibitory effects on proteasome activity and may thereby promote HSF1 activation.
4.2.2. Chaperones as Direct Sensors of Thiol-Reactive Molecules
Thiol-reactive molecules can modify the chaperones that repress HSF1 by directly reacting with key Cys residues within these proteins. Specifically, both Hsp70 and Hsp90 have been identified as potential targets of electrophiles and oxidants in multiple proteome-wide profiling studies.148–152 Hsp70 association with HSF1 decreases upon treatment with electrophiles,132 implying that alkylation may alter its ability to bind HSF1. Indeed, in vitro chaperone activity assays reveal that Hsp70, Hsp90, and several of their co-chaperones are inactivated by thiol-reactive molecules.112,139,148,149,153,154 Taken together, these results imply that chaperones are targets for inactivation by thiol-reactive molecules.
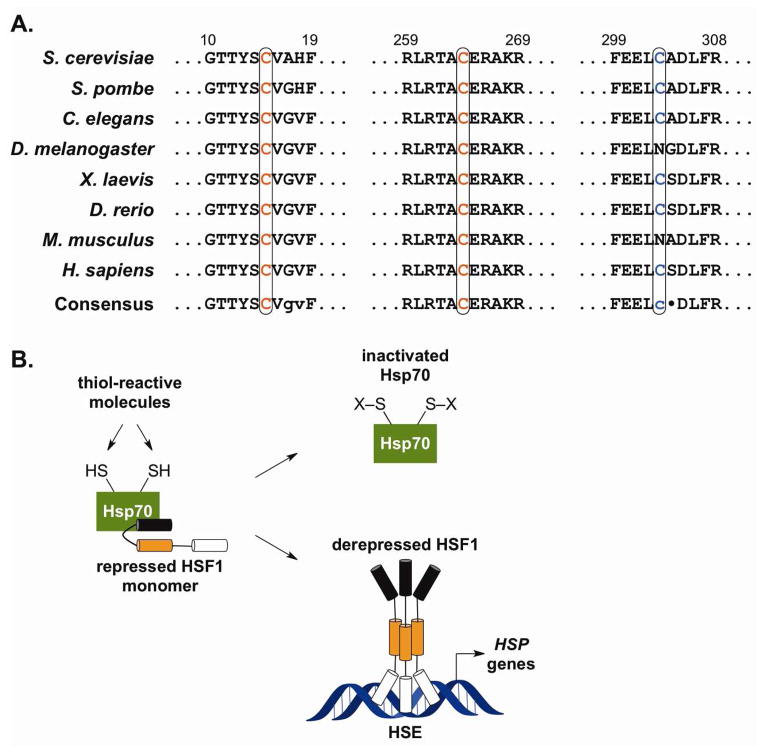
Using a yeast model system, we have found that HSR activation by thiol-reactive molecules depends on the presence of Cys residues 264 and 303 in the Hsp70 Ssa1.155 Several lines of evidence suggest that modification of these residues in Hsp70 may be a universal sensing mechanism for activation of the HSR by thiol-reactive molecules. First, one of these Cys residues (C264 in S. cerevisiae) is highly conserved among eukaryotic Hsp70s (Fig. 9A), whereas other potential sites of modification in Hsp70 and Hsp90 are not as highly conserved. The conserved Cys residues in Hsp70 are alkylated by several different electrophiles in vitro, leading to inactivation of the chaperone function of the protein.148,153,156 Conversely, DnaK, an Hsp70 from E. coli that lacks C264, is not sensitive to inactivation by the electrophile HNE.148 Mutation of C264 to Ser in Ssa1 significantly decreases the magnitude of the HSR in response to thiol-reactive molecules, but not heat.155 Since it is widely thought that Hsp70 forms a repressive, multichaperone complex with HSF1, then it is likely that modification of this key thiol group at C264 disrupts the interactions between these proteins, thereby derepressing HSF1 activity (Fig. 9B). A similar mechanism of derepression has been proposed for electrophilic modification of Hsp90;119,157 however, the yeast Hsp90 lacks conservation of reactive amino acids most likely to be modified by thiol-modifying agents in mammalian Hsp90.149,158
Figure 9. Direct Sensing of Thiol-Reactive Compounds by Hsp70.
(A) Conservation of Cys residues in Hsp70 homologs among various eukaryotic species. C264 and C303 are sites of alkylation in the S. cerevisiae Hsp70 Ssa1. (B) Hypothetical model for activation of HSF1 by thiol-reactive molecules. Hsp70 is modified on conserved Cys residues by reactive molecules, leading to its dissociation from HSF1 and subsequent steps in HSF1 activation.
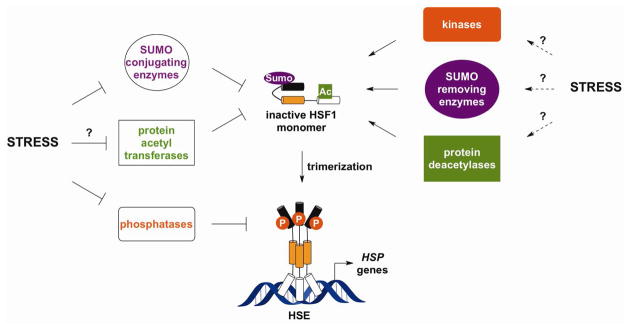
4.3. HSF1-Modifying Enzymes as Potential Sensors of Stress
As described earlier, HSF1 undergoes numerous constitutive and inducible post-translational modifications.43 Many of the enzymes that attach or remove these modifications may exhibit altered cellular activities in response to cellular stress. If so, these HSF1-regulating enzymes or upstream proteins may function as stress sensors and thereby influence the magnitude or duration of the HSR (Fig. 10). In support of this, treatment of cells with Ser/Thr phosphatase inhibitors or overexpression of specific phosphatases prolongs HSF1 phosphorylation and DNA binding,159–162 whereas enhancing acetylation of HSF1 through inhibition of deacetylase activity or increased expression of acetyl transferases results in reduced HSF1 DNA binding.52 Likewise, stress-induced alterations in HSF1 sumoylation may play a key role in altering its activity,50,163,164 with prolonged sumoylation presumably blocking HSF1 DNA binding. While the sensory mechanisms that allow for altered post-translational modification of HSF1 are still being investigated, several of the enzymes that play a direct role in carrying out in post-translational modifications are modified by thiol-reactive molecules.165–168 Accordingly, stress-responsive modification of these enzymatic activities may significantly influence the overall HSR in stressed cells.
Figure 10. Sensing by Enzymes Involved in Post-Translational Modification of HSF1.
Stresses that activate the HSR may influence the activities of multiple enzymes that facilitate HSF1 post-translational modification.
4.4. Future Work to Explore the Models of HSF1 Activation
Because HSF1 regulation is a complex, multi-step process, determining the upstream molecular events leading to its activation and the order of steps in its inactivation has represented a challenge, leading to a variety of models for how the process occurs in cells. While many of these models are referenced herein, it is important to note that none of them fully addresses the complexity of HSF1 regulation. For instance, we still know relatively little about the enzymes responsible for post-translational modification of HSF1, the timing of these events in relation to transcription of Hsp-encoding genes, and their overall effects on HSF1 activity. To assess the importance of these and other events, validating each effect in a cellular context (to complement in vitro biochemical work), performing detailed time-course experiments to tease out the timing of specific molecular events, and utilizing new biological and chemical tools to explore these problems will be essential in moving the field forward.
5. Ramifications of Influencing the Heat Shock Response
With the recent advances in understanding activation of the HSR at a biological and chemical level, we are approaching the point where pharmacological manipulation of this cytoprotective response is a distinct possibility. A number of pathophysiological conditions have been strongly linked to dysregulation of the HSR, including several debilitating diseases. For many of these disorders, there are few, if any, therapies available, making the possibility of HSR-targeted approaches highly attractive.
5.1. Acute Stress Tolerance
Defense against acute insult, most notably thermal stress, is perhaps the most appreciated cellular role of the HSR. The ability to tolerate severe stress is directly dependent on the capacity to produce Hsps in all organisms studied. This is true for survival of an immediate acute stress, dependent on sufficient basal Hsp expression, as well as acquired stress tolerance, the phenomenon whereby challenge with a sub-lethal stress potentiates survival of a subsequent lethal stress. Yeast cells lacking the Hsp70 and Hsp104 protein chaperones, or deficient in HSF1 function, are hypersensitive to stress.169–171 Correspondingly, HSF1−/− mice are compromised in their ability to withstand environmental insult.172 While severe heat stress (45–55°C) would be expected to perturb cellular ultrastructure, including membranes and cytoskeleton, the elevated expression of Hsp104 in yeast is sufficient to confer enhanced thermotolerance, suggesting that proteotoxicity is the primary cause of cell death.173 In contrast to the situation in unicellular eukaryotes, a careful balance is struck between Hsp-mediated cell survival and apoptosis in mammalian cells.
Because the regulation of apoptosis involves Hsps, it is possible that they play a major role in a cell’s decision to repair damaged proteins or undergo cell death. Hsp70, Hsp90, and Hsp27 inhibit apoptosome formation and caspase activation.174,175 In addition, Hsp90 is required for activity of a number of pro-survival proteins, including Akt and proteins in the Ras-Raf cascade.176 It is currently thought that when challenged with lethal stress, HSF1-mediated HSR induction is the first line of response. If protein homeostasis is restored, growth may resume. If not, an irreversible commitment to apoptosis is triggered to eliminate the terminally damaged cell. In support of this conjecture, pharmacological inhibition of Hsp90 enhances apoptosis, and this effect is amplified when combined with the compound KNK437, which inhibits the HSR through an unknown mechanism.177 Conversely, activation of the HSR can delay or block the cell death response. Thus, pharmacological induction of the HSR provides protection against cell death in tissues immediately after cytotoxic stresses. Indeed, thermal induction of Hsps correlates with an increase in the success rate of organ transplant, a prime example of physiological stress.178
5.2. The HSR and Human Disease
There is growing evidence that the HSR may play both positive and negative roles in initiation and progression of human disease. At the heart of this involvement is the ability of Hsps to fold and promote maturation of substrate proteins. Because Hsps are generally considered non-selective in their targets and interact with a number of proteins, abrogation of their function can alter fundamental aspects of cell physiology, leading to pathology and disease. In contrast, the known dependence of select signaling and regulatory molecules such as cyclin dependent kinases, Akt, growth-promoting tyrosine kinases, and telomerase as client of the Hsp90 chaperone system means that upregulation of the HSR favors cancer progression and metastasis. In addition to the HSR, the degradative capacity of the cell provides the counterweight to protein repair functions, together defining protein homeostasis from cradle to grave. We examine these features and their relevance to human health below.
5.2.1. Aging
All cells must replenish and recycle their components over time to maintain viability. This is especially true of many terminally differentiated cells, which are not renewed as organisms age. Mounting evidence suggests that the ability of cells to maintain proper protein homeostasis also deteriorates over time. Aged rats (20+ months) display marked reduction in their capacity to mount a functional cardiac HSR when subjected to thermal stress relative to adults (~ 6 months), which correlates with reduced tolerance to ischemia.179 HSF1 levels remain unchanged in aged versus younger rats, but the transcription factor exhibits poor DNA binding activity that may account for diminished HSR and stress sensitivity.180 This phenomenon extends to other mammalian cell types, as human lymphocytes and fibroblasts from aged donors exhibit the same loss of HSF1 activity, as do serially passaged cells of both types from younger donors.180,181
Non-mammalian model systems have been instrumental in probing the molecular details of the links between aging and the HSR. Longevity in C. elegans is dependent on HSF-1, an effect more pronounced in worms with a genetic deficiency in the insulin-like growth factor pathway.182 The insulin-like growth factor pathway also regulates lifespan in mice and other organisms, underscoring its central role in aging. In addition to HSF-1, the FOXO-like transcription factor DAF-16 also plays a role in slowing aging.183 Both factors contribute to the expression of chaperones of the small Hsp (sHsp) family, whose deletion (namely Hsp22) leads to decreased lifespan in Drosophila.184 In contrast, overexpression of Hsp22 in motor neurons extends lifespan, with specific benefits in tolerating oxidative and thermal stresses.185 Gradual loss of protein homeostasis in early adulthood occurs in C. elegans, and this can be slowed by enhancing cytoplasmic- and endoplasmic reticulum-based stress response pathways.186 These results are consistent with the observation that soluble polyglutamine (polyQ)-fluorescent protein chimeras aggregate within muscle tissue in aged, but not young, worms.187
Further studies have yielded mechanistic explanations for the observed decline in protein quality control with age. Caloric restriction extends lifespan in many organisms, and this effect requires HSF1 in the worm model.188 The age-related decline in HSF1 DNA binding affinity and accompanying loss in Hsp70 production is also reversed by caloric restriction in rat hepatocytes.189 Many of the benefits of caloric restriction are purportedly mediated by the sirtuin family of protein deacetylases, and both HSF1 and FOXO are regulated by SIRT1 in mammalian cells.52,190 In the case of HSF1, SIRT1 promotes prolonged DNA binding and Hsp gene expression, providing a possible direct link between nutritional status, the HSR, and protein homeostasis in the aged cell.52 Pharmacological SIRT1 activators are under development, based on initial indications that the natural product resveratrol mimics caloric restriction in a SIRT1-dependent manner. However, recent findings suggest the mechanisms favoring prolonged lifespan may be more complex than anticipated.191
5.2.2. Neurodegenerative disease
A major advance in our understanding of a wide range of neurodegenerative disorders including Alzheimer’s, Parkinson’s, Huntington’s, and Lou Gehrig’s diseases is the realization that all are fundamentally pathologies caused by the misfolding or altered processing/localization of key proteins. For example, the etiology of Alzheimer’s disease is strongly linked with production of the Aβ1–42 peptide and aggregation of the cytoskeletal protein tau. Many of these maladies are diseases associated with aging, and their inception may correlate with the aforementioned decline in protein quality control. Genetic factors also play a role, with familial forms of some neurodegenerative diseases linked to polymorphisms in the causal protein, such as Cu/Zn-superoxide dismutase (SOD-1) mutants that predispose individuals to Lou Gehrig’s disease (amyotrophic lateral sclerosis).192 The preponderance of known protein misfolding pathologies are neuronal in nature, prompting the speculation that neurons are relatively deficient in protein quality control. This hypothesis is supported by evidence that HSF1 is inactive or weakly active in cultured motor neurons, resulting in diminished Hsp production.193,194 This phenomenon is ameliorated by introduction of constitutively active HSF1, demonstrating that these cells are competent to mount an HSF1-mediated response but fail to do so for reasons that have yet to be determined.193
A growing body of evidence indicates that the HSR may play a role in preventing or delaying the onset of age-associated degenerative diseases. Cultured HSF1−/− cells accumulate ubiquitinated proteins and polyQ-fluorescent protein fusions, demonstrating a requirement for mediating clearance of damaged proteins.195 A recent human genetics study identified an allele of the HspB1 gene that produces lower levels of the sHsp Hsp27 as a contributing factor in a form of Charcot-Marie-Tooth disease, a hereditary neuromuscular disorder.196 Aβ toxicity was also decreased in an Alzheimer’s disease mouse model when lifespan was extended through inhibition of the IGF signaling pathway, which correlates with heightened HSR activity.197 Balance in the protein homeostasis network appears to be delicately maintained, as the inclusion of metastable folding variants of select proteins enhances the aggregation of SOD-1 and aggregation-prone polyQ fusion proteins in C. elegans muscle cells.198 The converse is also true; overexpression of polyQ proteins causes aggregation and loss of function of the same metastable proteins.199
5.2.3. Cancer development and progression
Cells regulate growth and proliferation at a sustainable rate that does not exceed their biosynthetic or degradative capacity. Transformation into the malignant state deregulates this control and inhibits apoptosis, resulting in a highly active, proliferative and physiologically robust cell. Among other diagnostic and prognostic indicators, elevated Hsp levels have been observed in a variety of cancers including those of the prostate and breast.200 Hsp90, Hsp70, and Hsp27 levels are routinely found to be two to fourfold higher in tumor tissue and still higher in metastatic variants.201 Hsps may be limiting for cancer transformation, as overexpression of the kinase chaperone Cdc37 is sufficient to induce mammary gland tumors in a transgenic mouse model.202 Indeed, Hsp90 and Cdc37 promote the activity of a number of pro-growth kinases such as the cyclin-dependent kinases, receptor tyrosine kinases, and Akt.203 Hsp90 is also required for signaling by steroid hormone receptors, and a chemical genomic search for novel compounds capable of blocking elevated androgen receptor activity linked to prostate cancer identified celastrol and the structurally related compound gedunin as potent inhibitors, highlighting this chaperone as a primary pharmacological target.65
The observations that chaperones promote cancer growth and progression extend to global control of the HSR. A recent study revealed elevated levels and constitutive nuclear localization of HSF1 in in situ and invasive breast carcinomas, and statistical analysis supports a strong correlation between these characteristics and poor prognosis.204 These and other results have led to the “Hsp addiction” hypothesis, wherein cancer cells depend on elevated Hsp levels to support their rapid growth and metabolism. This idea is bolstered by the finding that HSF1 is required for tumor formation. HSF1−/− mice exhibit a marked reduction in tumor formation induced by either oncogenic Ras or p53 mutations in mice, and HSF1 knockdown by siRNA technology decreases proliferation of human cancer cell lines.205 Chaperone-mediated autophagy is an important aspect of cellular recycling and degradation, and was recently shown to be required for tumorigenesis and metastasis, underscoring the degree to which Hsps and chaperones are involved in cancer.206
A corollary to the apparently constitutive activation of the HSR is that the resultant Hsp production levels render cancer cells highly resistant to cytotoxic compounds typically used in chemotherapy. Overexpression of Hsp27 increased resistance of a bladder cancer cell line to paclitaxel, while siRNA knockdown had the opposite effect, leading to enhanced apoptosis upon chemotherapeutic treatment.207,208 Pancreatic cancers are highly resistant to known chemotherapies and are thus extremely difficult to treat. Hsp27 knockdown in a pancreatic cell line resulted in sensitivity to one of the few partially effective drugs, gemcitabine.209 Similarly, pharmacologic inhibition of Hsp90 with 17-allyamino 17-demethoxygeldanamycin synergizes with suberanoylanilide hydroxamic acid (a histone deacetylase inhibitor) to promote apoptosis in leukemia cell lines.210 Together these findings suggest that inhibiting HSR activation at the level of HSF1, or selective inhibition of one or more Hsps, may be a promising avenue to explore in the fight against cancer.
5.2.4. Other protein folding maladies
In contrast to the involvement of the HSR in cancer, which appears to be a “gain-of-function” relationship, many human diseases are caused by deficiencies in critical metabolic enzymes due to misfolding and/or degradation caused by spontaneous or inherited polymorphisms. Two lysosomal storage disorders, Gaucher’s and Tay-Sachs disease, result from ER-associated degradation of glucocerebrosidase and β-hexosaminidase, respectively. Lysosomal accumulation of glucosylceramide in Gaucher’s and GM2 gangliosides in Tay-Sachs leads to the accompanying pathologies, fostering the idea that disease symptoms could be reversed by increasing the amount of enzyme that escapes degradation to be successfully delivered to the lysosome.211 This has been achieved at the cellular level through the combined use of the proteasome inhibitor MG132 and celastrol to upregulate the HSR and ER stress responses in parallel.212 A similar effect on multiple lysosomal storage disorders has been observed using Ca2+ channel blockers. These agents presumably enhance ER chaperone production through ER stress response induction and activation of Ca2+-dependent chaperones.213,214
The role of the protein quality control machinery in regulating the abundance and targeting of the cystic fibrosis transmembrane conductance regulator (CFTR) and known disease-inducing mutants has been heavily investigated. The CFTRΔ508 mutant is a naturally occurring variant that is misfolded within the ER and selected for ER-associated degradation, greatly reducing the amount of functional channel at the cell surface, which ultimately leads to altered chloride and water transport across the airway epithelium. The synthesis of CFTR is regulated by ER and cytosolic chaperones as well as the ER-associated degradation and proteasomal machinery, providing multiple opportunities for pharmacologic intervention. In support of this notion, S-nitrosation of the Hsp70/Hsp90 organizing protein (Hop) increases cell surface expression of CFTRΔ508, as does downregulation of other Hsp90 co-chaperones including Aha1.215,216 Induction of Hsp70 expression via treatment with 4-phenylbutyrate rescues CFTRΔ508 processing, raising the possibility that the next generation of Hsp70 activators may have significant therapeutic potential in this and other diseases.217
5.3. Efficacy of HSR Activators in Disease Models
Pharmacological HSR activators show promise for slowing the onset of age-related phenotypes and degenerative diseases in various animal models. For instance, Hsp90 inhibitors that activate HSF1 are capable of ameliorating cytotoxicity in an Alzheimer’s disease model with only moderate induction of Hsps.218 Another recently described HSR activator, the benzyl pyrazole derivative HSF1A, abrogates polyQ-dependent toxicity in neuronal cells.68,219 Similarly, several HSR activators with Michael acceptor properties decrease overall protein aggregation and motility defects in worms expressing expanded polyQ proteins.105 In addition, administration of geranylgeranylacetone, an HSR activator that interacts with the substrate binding domain of Hsp70, protects against degenerative cell death in mouse models of spinal and bulbar muscular atrophy, age-related hearing loss, and ischemia-reperfusion tissue injury.220–223 Collectively, these findings suggest that pharmacological manipulation of the HSR will be of benefit in a variety of disorders of both an acute and chronic nature.
However, several issues must be resolved before HSR activators may be of clinical use in humans. A number of current HSR activators show moderate to significant cytotoxicity, including hepatotoxicity, at the higher concentrations that would be required in clinical trials. In addition, many of these compounds are reactive and therefore have numerous targets and signaling effects that occur independently of HSR activation. Another challenge to consider is that prolonged HSR activation may not always be desirable, as it could lead to the onset of other conditions (e.g., cancer) later on. Therefore, current efforts in many laboratories are focused on generating derivatives of HSR activators with increased potency and specificity while limiting toxicity and additional complications that may arise from long-term administration.
5.4. Future Work
Because of their ability to fundamentally alter the stability, processing, or localization of multiple substrates, compounds that modulate aspects of protein quality control including the HSR, ER stress responses, and the degradation machinery have been collectively dubbed “proteostasis regulators.” As described earlier in this review, these compounds, both natural and synthetic, have the capacity to significantly impact human disease. Most exciting is the apparent synergy with pharmacologic chaperones, small molecules that stabilize protein folding states or otherwise promote the activity of targets expressed at low levels. Current work has revealed a number of relatively broad-acting compounds whose therapeutic utility is compromised by frequent cytotoxicity or bioavailability issues. Therefore, an important goal for the immediate future will be the rational design of molecules that build on these findings to provide a much higher degree of specificity, while retaining efficacy, either alone or as adjuvants to increase the success of traditional therapies. An additional area that needs exploration is determining whether altering the HSR to treat one disorder leads to the onset of others, given that the HSR can play opposing roles in specific diseases (Fig. 11). Determining appropriate dosing for HSR activators and inhibitors will be key in validating their efficacy in treating a given disorder without triggering another. Nonetheless, by pharmacologically targeting the HSR, we can envision enhancing the protein homeostasis network as a general means to ameliorate loss of function problems in proteins in aging, metabolic disease, and neurodegenerative disease or selectively interrupting it to slow the development and spread of cancer (Fig. 11). Given the recent advances and success of in vitro models, it is conceivable that treatments of protein folding disorders may be among the first to realize the potential of personalized medicine.
Figure 11. Model for Therapeutic Utility of Pharmacological HSR Regulators.
The HSR is altered in aging and several disease states. As a possible way of intervening in each of these physiological states, pharmacological regulators of HSF1 activity may be used to restore the HSR to normal activity levels.
Figure 7. Direct Regulation of HSF1 Under Stress Conditions.

Upon exposure to several stress conditions in vitro, HSF1 undergoes trimerization and shows increased DNA binding.
Acknowledgments
Funding Support. Work in the authors’ laboratories is sponsored by a Cottrell College Science Award from Research Corporation for Science Advancement (to J.D.W.), the Luce Fund for Distinguished Scholarship from The College of Wooster (to J.D.W.), and NIH grant GM074696 from NIGMS (to K.A.M.). Y.W. was supported by the Cameron Foundation.
We thank Sandy Westerheide and Aaron Jacobs for critically reading the manuscript.
Abbreviations
- HSR
heat shock response
- HSF1
heat shock factor 1
- HSE
heat shock element
- Hsp
heat shock protein
- UPR
unfolded protein response
- SUMO
small ubiquitin-like modifier
- HR
heptad repeat
- SIRT1
sirtuin 1
- ROS
reactive oxygen species
- PG
prostaglandin
- 15d-PGJ2
15-deoxy-Δ12,14-PGJ2
- HNE
4-hydroxy-2-nonenal
- sHsp
small Hsp
- CFTR
cystic fibrosis transmembrane conductance regulator
- Hop
Hsp70/Hsp90 organizing protein
References
- 1.Osburn WO, Kensler TW. Nrf2 signaling: an adaptive response pathway for protection against environmental toxic insults. Mutat Res. 2008;659:31–39. doi: 10.1016/j.mrrev.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sykiotis GP, Bohmann D. Stress-activated cap’n’collar transcription factors in aging and human disease. Sci Signal. 2010;3:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Autreaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 4.Hur W, Gray NS. Small molecule modulators of antioxidant response pathway. Curr Opin Chem Biol. 2011;15:162–173. doi: 10.1016/j.cbpa.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Mori K. Signalling pathways in the unfolded protein response: development from yeast to mammals. J Biochem. 2009;146:743–750. doi: 10.1093/jb/mvp166. [DOI] [PubMed] [Google Scholar]
- 6.Wu J, Kaufman RJ. From acute ER stress to physiological roles of the unfolded protein response. Cell Death Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- 7.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 8.Brodsky JL, Skach WR. Protein folding and quality control in the endoplasmic reticulum: recent lessons from yeast and mammalian cell systems. Curr Opin Cell Biol. 2011;23:464–475. doi: 10.1016/j.ceb.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mager WH, Ferreira PM. Stress response of yeast. Biochem J. 1993;290:1–13. doi: 10.1042/bj2900001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Chang A. Heat shock response relieves ER stress. EMBO J. 2008;27:1049–1059. doi: 10.1038/emboj.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartl FU. Heat shock proteins in protein folding and membrane translocation. Semin Immunol. 1991;3:5–16. [PubMed] [Google Scholar]
- 12.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 13.Csermely P, Schnaider T, Soti C, Prohaszka Z, Nardai G. The 90-kDa molecular chaperone family: structure, function, and clinical applications. Pharmacol Ther. 1998;79:129–168. doi: 10.1016/s0163-7258(98)00013-8. [DOI] [PubMed] [Google Scholar]
- 14.Morano KA, Liu PC, Thiele DJ. Protein chaperones and the heat shock response in Saccharomyces cerevisiae. Curr Opin Microbiol. 1998;1:197–203. doi: 10.1016/s1369-5274(98)80011-8. [DOI] [PubMed] [Google Scholar]
- 15.Neckers L, Ivy SP. Heat shock protein 90. Curr Opin Oncol. 2003;15:419–424. doi: 10.1097/00001622-200311000-00003. [DOI] [PubMed] [Google Scholar]
- 16.Westerheide SD, Bosman JD, Mbadugha BN, Kawahara TL, Matsumoto G, Kim S, Gu W, Devlin JP, Silverman RB, Morimoto RI. Celastrols as inducers of the heat shock response and cytoprotection. J Biol Chem. 2004;279:56053–56060. doi: 10.1074/jbc.M409267200. [DOI] [PubMed] [Google Scholar]
- 17.Morimoto RI, Kroeger PE, Cotto JJ. The transcriptional regulation of heat shock genes: a plethora of heat shock factors and regulatory conditions. EXS. 1996;77:139–163. doi: 10.1007/978-3-0348-9088-5_10. [DOI] [PubMed] [Google Scholar]
- 18.Bienz M, Pelham HR. Heat shock regulatory elements function as an inducible enhancer in the Xenopus hsp70 gene and when linked to a heterologous promoter. Cell. 1986;45:753–760. doi: 10.1016/0092-8674(86)90789-0. [DOI] [PubMed] [Google Scholar]
- 19.Amin J, Ananthan J, Voellmy R. Key features of heat shock regulatory elements. Mol Cell Biol. 1988;8:3761–3769. doi: 10.1128/mcb.8.9.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perisic O, Xiao H, Lis JT. Stable binding of Drosophila heat shock factor to head-to-head and tail-to-tail repeats of a conserved 5 bp recognition unit. Cell. 1989;59:797–806. doi: 10.1016/0092-8674(89)90603-x. [DOI] [PubMed] [Google Scholar]
- 21.Nakai A, Tanabe M, Kawazoe Y, Inazawa J, Morimoto RI, Nagata K. HSF4, a new member of the human heat shock factor family which lacks properties of a transcriptional activator. Mol Cell Biol. 1997;17:469–481. doi: 10.1128/mcb.17.1.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santoro MG. Heat shock factors and the control of the stress response. Biochem Pharmacol. 2000;59:55–63. doi: 10.1016/s0006-2952(99)00299-3. [DOI] [PubMed] [Google Scholar]
- 23.Tanabe M, Nakai A, Kawazoe Y, Nagata K. Different thresholds in the responses of two heat shock transcription factors, HSF1 and HSF3. J Biol Chem. 1997;272:15389–15395. doi: 10.1074/jbc.272.24.15389. [DOI] [PubMed] [Google Scholar]
- 24.Abravaya K, Myers MP, Murphy SP, Morimoto RI. The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev. 1992;6:1153–1164. doi: 10.1101/gad.6.7.1153. [DOI] [PubMed] [Google Scholar]
- 25.Jakobsen BK, Pelham HR. Constitutive binding of yeast heat shock factor to DNA in vivo. Mol Cell Biol. 1988;8:5040–5042. doi: 10.1128/mcb.8.11.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jakobsen BK, Pelham HR. A conserved heptapeptide restrains the activity of the yeast heat shock transcription factor. EMBO J. 1991;10:369–375. doi: 10.1002/j.1460-2075.1991.tb07958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison CJ, Bohm AA, Nelson HC. Crystal structure of the DNA binding domain of the heat shock transcription factor. Science. 1994;263:224–227. doi: 10.1126/science.8284672. [DOI] [PubMed] [Google Scholar]
- 28.Peteranderl R, Nelson HC. Trimerization of the heat shock transcription factor by a triple-stranded α-helical coiled-coil. Biochemistry. 1992;31:12272–12276. doi: 10.1021/bi00163a042. [DOI] [PubMed] [Google Scholar]
- 29.Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 30.Schlesinger MJ, Ryan C. An ATP- and hsc70-dependent oligomerization of nascent heat-shock factor (HSF) polypeptide suggests that HSF itself could be a “sensor” for the cellular stress response. Protein Sci. 1993;2:1356–1360. doi: 10.1002/pro.5560020819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baler R, Welch WJ, Voellmy R. Heat shock gene regulation by nascent polypeptides and denatured proteins: hsp70 as a potential autoregulatory factor. J Cell Biol. 1992;117:1151–1159. doi: 10.1083/jcb.117.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 33.Sarge KD, Murphy SP, Morimoto RI. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cotto J, Fox S, Morimoto R. HSF1 granules: a novel stress-induced nuclear compartment of human cells. J Cell Sci. 1997;110:2925–2934. doi: 10.1242/jcs.110.23.2925. [DOI] [PubMed] [Google Scholar]
- 35.Jolly C, Morimoto R, Robert-Nicoud M, Vourc’h C. HSF1 transcription factor concentrates in nuclear foci during heat shock: relationship with transcription sites. J Cell Sci. 1997;110:2935–2941. doi: 10.1242/jcs.110.23.2935. [DOI] [PubMed] [Google Scholar]
- 36.Rabindran SK, Haroun RI, Clos J, Wisniewski J, Wu C. Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science. 1993;259:230–234. doi: 10.1126/science.8421783. [DOI] [PubMed] [Google Scholar]
- 37.Westwood JT, Wu C. Activation of Drosophila heat shock factor: conformational change associated with a monomer-to-trimer transition. Mol Cell Biol. 1993;13:3481–3486. doi: 10.1128/mcb.13.6.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peteranderl R, Rabenstein M, Shin YK, Liu CW, Wemmer DE, King DS, Nelson HC. Biochemical and biophysical characterization of the trimerization domain from the heat shock transcription factor. Biochemistry. 1999;38:3559–3569. doi: 10.1021/bi981774j. [DOI] [PubMed] [Google Scholar]
- 39.Guettouche T, Boellmann F, Lane WS, Voellmy R. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem. 2005;6:4. doi: 10.1186/1471-2091-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soncin F, Zhang X, Chu B, Wang X, Asea A, Ann Stevenson M, Sacks DB, Calderwood SK. Transcriptional activity and DNA binding of heat shock factor-1 involve phosphorylation on threonine 142 by CK2. Biochem Biophys Res Commun. 2003;303:700–706. doi: 10.1016/s0006-291x(03)00398-x. [DOI] [PubMed] [Google Scholar]
- 41.Lee P, Cho BR, Joo HS, Hahn JS. Yeast Yak1 kinase, a bridge between PKA and stress-responsive transcription factors, Hsf1 and Msn2/Msn4. Mol Microbiol. 2008;70:882–895. doi: 10.1111/j.1365-2958.2008.06450.x. [DOI] [PubMed] [Google Scholar]
- 42.Mayya V, Han DK. Phosphoproteomics by mass spectrometry: insights, implications, applications and limitations. Expert Rev Proteomics. 2009;6:605–618. doi: 10.1586/epr.09.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anckar J, Sistonen L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem. 2011;80:1089–1115. doi: 10.1146/annurev-biochem-060809-095203. [DOI] [PubMed] [Google Scholar]
- 44.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 45.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 46.Holmberg CI, Hietakangas V, Mikhailov A, Rantanen JO, Kallio M, Meinander A, Hellman J, Morrice N, MacKintosh C, Morimoto RI, Eriksson JE, Sistonen L. Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. EMBO J. 2001;20:3800–3810. doi: 10.1093/emboj/20.14.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boellmann F, Guettouche T, Guo Y, Fenna M, Mnayer L, Voellmy R. DAXX interacts with heat shock factor 1 during stress activation and enhances its transcriptional activity. Proc Natl Acad Sci USA. 2004;101:4100–4105. doi: 10.1073/pnas.0304768101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kline MP, Morimoto RI. Repression of the heat shock factor 1 transcriptional activation domain is modulated by constitutive phosphorylation. Mol Cell Biol. 1997;17:2107–2115. doi: 10.1128/mcb.17.4.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knauf U, Newton EM, Kyriakis J, Kingston RE. Repression of human heat shock factor 1 activity at control temperature by phosphorylation. Genes Dev. 1996;10:2782–2793. doi: 10.1101/gad.10.21.2782. [DOI] [PubMed] [Google Scholar]
- 50.Hietakangas V, Ahlskog JK, Jakobsson AM, Hellesuo M, Sahlberg NM, Holmberg CI, Mikhailov A, Palvimo JJ, Pirkkala L, Sistonen L. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol Cell Biol. 2003;23:2953–2968. doi: 10.1128/MCB.23.8.2953-2968.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci USA. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Westerheide SD, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindquist S. Varying patterns of protein synthesis in Drosophila during heat shock: implications for regulation. Dev Biol. 1980;77:463–479. doi: 10.1016/0012-1606(80)90488-1. [DOI] [PubMed] [Google Scholar]
- 54.Voellmy R, Boellmann F. Chaperone regulation of the heat shock protein response. Adv Exp Med Biol. 2007;594:89–99. doi: 10.1007/978-0-387-39975-1_9. [DOI] [PubMed] [Google Scholar]
- 55.Craig EA, Jacobsen K. Mutations of the heat inducible 70 kilodalton genes of yeast confer temperature sensitive growth. Cell. 1984;38:841–849. doi: 10.1016/0092-8674(84)90279-4. [DOI] [PubMed] [Google Scholar]
- 56.Craig EA, Gross CA. Is hsp70 the cellular thermometer? Trends Biochem Sci. 1991;16:135–140. doi: 10.1016/0968-0004(91)90055-z. [DOI] [PubMed] [Google Scholar]
- 57.Shi Y, Mosser DD, Morimoto RI. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baler R, Zou J, Voellmy R. Evidence for a role of Hsp70 in the regulation of the heat shock response in mammalian cells. Cell Stress Chaperones. 1996;1:33–39. doi: 10.1379/1466-1268(1996)001<0033:efaroh>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rabindran SK, Wisniewski J, Li L, Li GC, Wu C. Interaction between heat shock factor and hsp70 is insufficient to suppress induction of DNA-binding activity in vivo. Mol Cell Biol. 1994;14:6552–6560. doi: 10.1128/mcb.14.10.6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- 61.Ali A, Bharadwaj S, O’Carroll R, Ovsenek N. HSP90 interacts with and regulates the activity of heat shock factor 1 in Xenopus oocytes. Mol Cell Biol. 1998;18:4949–4960. doi: 10.1128/mcb.18.9.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Conde R, Belak ZR, Nair M, O’Carroll RF, Ovsenek N. Modulation of Hsf1 activity by novobiocin and geldanamycin. Biochem Cell Biol. 2009;87:845–851. doi: 10.1139/o09-049. [DOI] [PubMed] [Google Scholar]
- 63.Lawson B, Brewer JW, Hendershot LM. Geldanamycin, an hsp90/GRP94-binding drug, induces increased transcription of endoplasmic reticulum (ER) chaperones via the ER stress pathway. J Cell Physiol. 1998;174:170–178. doi: 10.1002/(SICI)1097-4652(199802)174:2<170::AID-JCP4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 64.Schulte TW, Akinaga S, Soga S, Sullivan W, Stensgard B, Toft D, Neckers LM. Antibiotic radicicol binds to the N-terminal domain of Hsp90 and shares important biologic activities with geldanamycin. Cell Stress Chaperones. 1998;3:100–108. doi: 10.1379/1466-1268(1998)003<0100:arbttn>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hieronymus H, Lamb J, Ross KN, Peng XP, Clement C, Rodina A, Nieto M, Du J, Stegmaier K, Raj SM, Maloney KN, Clardy J, Hahn WC, Chiosis G, Golub TR. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10:321–330. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 66.Trott A, West JD, Klaic L, Westerheide SD, Silverman RB, Morimoto RI, Morano KA. Activation of heat shock and antioxidant responses by the natural product celastrol: transcriptional signatures of a thiol-targeted molecule. Mol Biol Cell. 2008;19:1104–1112. doi: 10.1091/mbc.E07-10-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Westerheide SD, Morimoto RI. Heat shock response modulators as therapeutic tools for diseases of protein conformation. J Biol Chem. 2005;280:33097–33100. doi: 10.1074/jbc.R500010200. [DOI] [PubMed] [Google Scholar]
- 68.Neef DW, Jaeger AM, Thiele DJ. Heat shock transcription factor 1 as a therapeutic target in neurodegenerative diseases. Nat Rev Drug Discov. 2011;10:930–944. doi: 10.1038/nrd3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 70.Salomons FA, Menendez-Benito V, Bottcher C, McCray BA, Taylor JP, Dantuma NP. Selective accumulation of aggregation-prone proteasome substrates in response to proteotoxic stress. Mol Cell Biol. 2009;29:1774–1785. doi: 10.1128/MCB.01485-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilde IB, Brack M, Winget JM, Mayor T. Proteomic characterization of aggregating proteins after the inhibition of the ubiquitin proteasome system. J Proteome Res. 2011;10:1062–1072. doi: 10.1021/pr1008543. [DOI] [PubMed] [Google Scholar]
- 72.West JD, Marnett LJ. Endogenous reactive intermediates as modulators of cell signaling and cell death. Chem Res Toxicol. 2006;19:173–194. doi: 10.1021/tx050321u. [DOI] [PubMed] [Google Scholar]
- 73.Marnett LJ, Riggins JN, West JD. Endogenous generation of reactive oxidants and electrophiles and their reactions with DNA and protein. J Clin Invest. 2003;111:583–593. doi: 10.1172/JCI18022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paulsen CE, Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem Biol. 2010;5:47–62. doi: 10.1021/cb900258z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Casini A, Scozzafava A, Supuran CT. Cysteine-modifying agents: a possible approach for effective anticancer and antiviral drugs. Environ Health Perspect. 2002;110:801–806. doi: 10.1289/ehp.02110s5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ahn SG, Thiele DJ. Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev. 2003;17:516–528. doi: 10.1101/gad.1044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Woods CG, Fu J, Xue P, Hou Y, Pluta LJ, Yang L, Zhang Q, Thomas RS, Andersen ME, Pi J. Dose-dependent transitions in Nrf2-mediated adaptive response and related stress responses to hypochlorous acid in mouse macrophages. Toxicol Appl Pharmacol. 2009;238:27–36. doi: 10.1016/j.taap.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Borrelli MJ, Stafford DM, Rausch CM, Bernock LJ, Freeman ML, Lepock JR, Corry PM. Diamide-induced cytotoxicity and thermotolerance in CHO cells. J Cell Physiol. 1998;177:483–492. doi: 10.1002/(SICI)1097-4652(199812)177:3<483::AID-JCP11>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 79.Davies MJ. The oxidative environment and protein damage. Biochim Biophys Acta. 2005;1703:93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 80.Winterbourn CC, Hampton MB. Thiol chemistry and specificity in redox signaling. Free Radic Biol Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 81.Claiborne A, Yeh JI, Mallett TC, Luba J, Crane EJ, 3rd, Charrier V, Parsonage D. Protein-sulfenic acids: diverse roles for an unlikely player in enzyme catalysis and redox regulation. Biochemistry. 1999;38:15407–15416. doi: 10.1021/bi992025k. [DOI] [PubMed] [Google Scholar]
- 82.Rudolph TK, Freeman BA. Transduction of redox signaling by electrophile-protein reactions. Sci Signal. 2009;2:re7. doi: 10.1126/scisignal.290re7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dickinson BC, Chang CJ. Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat Chem Biol. 2011;7:504–511. doi: 10.1038/nchembio.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Levinson W, Oppermann H, Jackson J. Transition series metals and sulfhydryl reagents induce the synthesis of four proteins in eukaryotic cells. Biochim Biophys Acta. 1980;606:170–180. doi: 10.1016/0005-2787(80)90108-2. [DOI] [PubMed] [Google Scholar]
- 85.Murata M, Gong P, Suzuki K, Koizumi S. Differential metal response and regulation of human heavy metal-inducible genes. J Cell Physiol. 1999;180:105–113. doi: 10.1002/(SICI)1097-4652(199907)180:1<105::AID-JCP12>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 86.Zou J, Salminen WF, Roberts SM, Voellmy R. Correlation between glutathione oxidation and trimerization of heat shock factor 1, an early step in stress induction of the Hsp response. Cell Stress Chaperones. 1998;3:130–141. doi: 10.1379/1466-1268(1998)003<0130:cbgoat>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moulis JM. Cellular mechanisms of cadmium toxicity related to the homeostasis of essential metals. Biometals. 2010;23:877–896. doi: 10.1007/s10534-010-9336-y. [DOI] [PubMed] [Google Scholar]
- 88.Shi Y, Kroeger PE, Morimoto RI. The carboxyl-terminal transactivation domain of heat shock factor 1 is negatively regulated and stress responsive. Mol Cell Biol. 1995;15:4309–4318. doi: 10.1128/mcb.15.8.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Abe T, Yamamura K, Gotoh S, Kashimura M, Higashi K. Concentration-dependent differential effects of N-acetyl-L-cysteine on the expression of HSP70 and metallothionein genes induced by cadmium in human amniotic cells. Biochim Biophys Acta. 1998;1380:123–132. doi: 10.1016/s0304-4165(97)00144-x. [DOI] [PubMed] [Google Scholar]
- 90.Abe T, Gotoh S, Higashi K. Attenuation by glutathione of hsp72 gene expression induced by cadmium in cisplatin-resistant human ovarian cancer cells. Biochem Pharmacol. 1999;58:69–76. doi: 10.1016/s0006-2952(99)00049-0. [DOI] [PubMed] [Google Scholar]
- 91.Chao PL, Fan SF, Chou YH, Lin AM. N-Acetylcysteine attenuates arsenite-induced oxidative injury in dorsal root ganglion explants. Ann New York Acad Sci. 2007;1122:276–288. doi: 10.1196/annals.1403.020. [DOI] [PubMed] [Google Scholar]
- 92.Amici C, Sistonen L, Santoro MG, Morimoto RI. Antiproliferative prostaglandins activate heat shock transcription factor. Proc Natl Acad Sci USA. 1992;89:6227–6231. doi: 10.1073/pnas.89.14.6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Holbrook NJ, Carlson SG, Choi AM, Fargnoli J. Induction of HSP70 gene expression by the antiproliferative prostaglandin PGA2: a growth-dependent response mediated by activation of heat shock transcription factor. Mol Cell Biol. 1992;12:1528–1534. doi: 10.1128/mcb.12.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Clay CE, Atsumi GI, High KP, Chilton FH. Early de novo gene expression is required for 15-deoxy-Δ12,14-prostaglandin J2-induced apoptosis in breast cancer cells. J Biol Chem. 2001;276:47131–47135. doi: 10.1074/jbc.C100339200. [DOI] [PubMed] [Google Scholar]
- 95.Kondo M, Shibata T, Kumagai T, Osawa T, Shibata N, Kobayashi M, Sasaki S, Iwata M, Noguchi N, Uchida K. 15-Deoxy-Δ12,14-prostaglandin J2: the endogenous electrophile that induces neuronal apoptosis. Proc Natl Acad Sci USA. 2002;99:7367–7372. doi: 10.1073/pnas.112212599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cajone F, Salina M, Benelli-Zazzera A. 4-Hydroxynonenal induces a DNA-binding protein similar to the heat-shock factor. Biochem J. 1989;262:977–979. doi: 10.1042/bj2620977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.West JD, Marnett LJ. Alterations in gene expression induced by the lipid peroxidation product, 4-hydroxy-2-nonenal. Chem Res Toxicol. 2005;18:1642–1653. doi: 10.1021/tx050211n. [DOI] [PubMed] [Google Scholar]
- 98.Thompson CA, Burcham PC. Genome-wide transcriptional responses to acrolein. Chem Res Toxicol. 2008;21:2245–2256. doi: 10.1021/tx8001934. [DOI] [PubMed] [Google Scholar]
- 99.Liu XY, Yang ZH, Pan XJ, Zhu MX, Xie JP. Gene expression profile and cytotoxicity of human bronchial epithelial cells exposed to crotonaldehyde. Toxicol Lett. 2010;197:113–122. doi: 10.1016/j.toxlet.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 100.Kansanen E, Jyrkkanen HK, Volger OL, Leinonen H, Kivela AM, Hakkinen SK, Woodcock SR, Schopfer FJ, Horrevoets AJ, Yla-Herttuala S, Freeman BA, Levonen AL. Nrf2-dependent and -independent responses to nitro-fatty acids in human endothelial cells: identification of heat shock response as the major pathway activated by nitro-oleic acid. J Biol Chem. 2009;284:33233–33241. doi: 10.1074/jbc.M109.064873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gan N, Wu YC, Brunet M, Garrido C, Chung FL, Dai C, Mi L. Sulforaphane activates heat shock response and enhances proteasome activity through up-regulation of Hsp27. J Biol Chem. 2010;285:35528–35536. doi: 10.1074/jbc.M110.152686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Santagata S, Xu YM, Wijeratne K, Kontnik R, Rooney C, Perley C, Kwon H, Clardy J, Kesari S, Whitesell L, Lindquist S, Gunatilaka L. Using the heat-shock response to discover compounds that target the malignant phenotype by disrupting protein pomeostasis. ACS Chem Biol. 2011;7:340–349. doi: 10.1021/cb200353m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu H, Lightfoot R, Stevens JL. Activation of heat shock factor by alkylating agents is triggered by glutathione depletion and oxidation of protein thiols. J Biol Chem. 1996;271:4805–4812. [PubMed] [Google Scholar]
- 104.Rossi A, Elia G, Santoro MG. 2-Cyclopenten-1-one, a new inducer of heat shock protein 70 with antiviral activity. J Biol Chem. 1996;271:32192–32196. doi: 10.1074/jbc.271.50.32192. [DOI] [PubMed] [Google Scholar]
- 105.Calamini B, Silva MC, Madoux F, Hutt DM, Khanna S, Chalfant MA, Saldanha SA, Hodder P, Tait BD, Garza D, Balch WE, Morimoto RI. Small-molecule proteostasis regulators for protein conformational diseases. Nat Chem Biol. 2012;8:185–196. doi: 10.1038/nchembio.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liebler DC. Protein damage by reactive electrophiles: targets and consequences. Chem Res Toxicol. 2008;21:117–128. doi: 10.1021/tx700235t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brown KK, Hampton MB. Biological targets of isothiocyanates. Biochim Biophys Acta. 2011;1810:888–894. doi: 10.1016/j.bbagen.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 108.Klaic L, Trippier PC, Mishra RK, Morimoto RI, Silverman RB. Remarkable stereospecific conjugate additions to the Hsp90 inhibitor celastrol. J Am Chem Soc. 2011;133:19634–19637. doi: 10.1021/ja208359a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang Y, Kolm RH, Mannervik B, Talalay P. Reversible conjugation of isothiocyanates with glutathione catalyzed by human glutathione transferases. Biochem Biophys Res Commun. 1995;206:748–755. doi: 10.1006/bbrc.1995.1106. [DOI] [PubMed] [Google Scholar]
- 111.Batthyany C, Schopfer FJ, Baker PR, Duran R, Baker LM, Huang Y, Cervenansky C, Branchaud BP, Freeman BA. Reversible post-translational modification of proteins by nitrated fatty acids in vivo. J Biol Chem. 2006;281:20450–20463. doi: 10.1074/jbc.M602814200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sreeramulu S, Gande SL, Gobel M, Schwalbe H. Molecular mechanism of inhibition of the human protein complex Hsp90-Cdc37, a kinome chaperone-cochaperone, by triterpene celastrol. Angew Chem Int Ed Engl. 2009;48:5853–5855. doi: 10.1002/anie.200900929. [DOI] [PubMed] [Google Scholar]
- 113.Perez-Sala D, Cernuda-Morollon E, Canada FJ. Molecular basis for the direct inhibition of AP-1 DNA binding by 15-deoxy-Δ12,14-prostaglandin J2. J Biol Chem. 2003;278:51251–51260. doi: 10.1074/jbc.M309409200. [DOI] [PubMed] [Google Scholar]
- 114.Sanchez-Gomez FJ, Diez-Dacal B, Pajares MA, Llorca O, Perez-Sala D. Cyclopentenone prostaglandins with dienone structure promote cross-linking of the chemoresistance-inducing enzyme glutathione transferase P1–1. Mol Pharmacol. 2010;78:723–733. doi: 10.1124/mol.110.065391. [DOI] [PubMed] [Google Scholar]
- 115.Burcham PC, Raso A, Thompson C, Tan D. Intermolecular protein cross-linking during acrolein toxicity: efficacy of carbonyl scavengers as inhibitors of heat shock protein-90 cross-linking in A549 cells. Chem Res Toxicol. 2007;20:1629–1637. doi: 10.1021/tx700192e. [DOI] [PubMed] [Google Scholar]
- 116.Uchida K, Stadtman ER. Covalent attachment of 4-hydroxynonenal to glyceraldehyde-3-phosphate dehydrogenase. A possible involvement of intra- and intermolecular cross-linking reaction. J Biol Chem. 1993;268:6388–6393. [PubMed] [Google Scholar]
- 117.Nadkarni DV, Sayre LM. Structural definition of early lysine and histidine adduction chemistry of 4-hydroxynonenal. Chem Res Toxicol. 1995;8:284–291. doi: 10.1021/tx00044a014. [DOI] [PubMed] [Google Scholar]
- 118.Liu Z, Minkler PE, Sayre LM. Mass spectroscopic characterization of protein modification by 4-hydroxy-2-(E)-nonenal and 4-oxo-2-(E)-nonenal. Chem Res Toxicol. 2003;16:901–911. doi: 10.1021/tx0300030. [DOI] [PubMed] [Google Scholar]
- 119.Zhang Y, Ahn YH, Benjamin IJ, Honda T, Hicks RJ, Calabrese V, Cole PA, Dinkova-Kostova AT. HSF1-dependent upregulation of Hsp70 by sulfhydryl-reactive inducers of the KEAP1/NRF2/ARE pathway. Chem Biol. 2011;18:1355–1361. doi: 10.1016/j.chembiol.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dinkova-Kostova AT, Cory AH, Bozak RE, Hicks RJ, Cory JG. Bis(2-hydroxybenzylidene)acetone, a potent inducer of the phase 2 response, causes apoptosis in mouse leukemia cells through a p53-independent, caspase-mediated pathway. Cancer Lett. 2007;245:341–349. doi: 10.1016/j.canlet.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 121.Dinkova-Kostova AT, Talalay P, Sharkey J, Zhang Y, Holtzclaw WD, Wang XJ, David E, Schiavoni KH, Finlayson S, Mierke DF, Honda T. An exceptionally potent inducer of cytoprotective enzymes: elucidation of the structural features that determine inducer potency and reactivity with Keap1. J Biol Chem. 2010;285:33747–33755. doi: 10.1074/jbc.M110.163485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.West JD, Stamm CE, Brown HA, Justice SL, Morano KA. Enhanced toxicity of the protein cross-linkers divinyl sulfone and diethyl acetylenedicarboxylate in comparison to related monofunctional electrophiles. Chem Res Toxicol. 2011;24:1457–1459. doi: 10.1021/tx200302w. [DOI] [PubMed] [Google Scholar]
- 123.West JD, Stamm CE, Kingsley PJ. Structure-activity comparison of the cytotoxic properties of diethyl maleate and related molecules: identification of diethyl acetylenedicarboxylate as a thiol cross-linking agent. Chem Res Toxicol. 2011;24:81–88. doi: 10.1021/tx100292n. [DOI] [PubMed] [Google Scholar]
- 124.Zhong M, Orosz A, Wu C. Direct sensing of heat and oxidation by Drosophila heat shock transcription factor. Mol Cell. 1998;2:101–108. doi: 10.1016/s1097-2765(00)80118-5. [DOI] [PubMed] [Google Scholar]
- 125.Pattaramanon N, Sangha N, Gafni A. The carboxy-terminal domain of heat-shock factor 1 is largely unfolded but can be induced to collapse into a compact, partially structured state. Biochemistry. 2007;46:3405–3415. doi: 10.1021/bi061124c. [DOI] [PubMed] [Google Scholar]
- 126.Lu M, Lee YJ, Park SM, Kang HS, Kang SW, Kim S, Park JS. Aromatic-participant interactions are essential for disulfide-bond-based trimerization in human heat shock transcription factor 1. Biochemistry. 2009;48:3795–3797. doi: 10.1021/bi802255c. [DOI] [PubMed] [Google Scholar]
- 127.Manalo DJ, Liu AY. Resolution, detection, and characterization of redox conformers of human HSF1. J Biol Chem. 2001;276:23554–23561. doi: 10.1074/jbc.M011300200. [DOI] [PubMed] [Google Scholar]
- 128.Manalo DJ, Lin Z, Liu AY. Redox-dependent regulation of the conformation and function of human heat shock factor 1. Biochemistry. 2002;41:2580–2588. doi: 10.1021/bi0159682. [DOI] [PubMed] [Google Scholar]
- 129.Lu M, Kim HE, Li CR, Kim S, Kwak IJ, Lee YJ, Kim SS, Moon JY, Kim CH, Kim DK, Kang HS, Park JS. Two distinct disulfide bonds formed in human heat shock transcription factor 1 act in opposition to regulate its DNA binding activity. Biochemistry. 2008;47:6007–6015. doi: 10.1021/bi702185u. [DOI] [PubMed] [Google Scholar]
- 130.Bonner JJ, Carlson T, Fackenthal DL, Paddock D, Storey K, Lea K. Complex regulation of the yeast heat shock transcription factor. Mol Biol Cell. 2000;11:1739–1751. doi: 10.1091/mbc.11.5.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Guo Y, Guettouche T, Fenna M, Boellmann F, Pratt WB, Toft DO, Smith DF, Voellmy R. Evidence for a mechanism of repression of heat shock factor 1 transcriptional activity by a multichaperone complex. J Biol Chem. 2001;276:45791–45799. doi: 10.1074/jbc.M105931200. [DOI] [PubMed] [Google Scholar]
- 132.Jacobs AT, Marnett LJ. Heat shock factor 1 attenuates 4-Hydroxynonenal-mediated apoptosis: critical role for heat shock protein 70 induction and stabilization of Bcl-XL. J Biol Chem. 2007;282:33412–33420. doi: 10.1074/jbc.M706799200. [DOI] [PubMed] [Google Scholar]
- 133.Stone DE, Craig EA. Self-regulation of 70-kilodalton heat shock proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:1622–1632. doi: 10.1128/mcb.10.4.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chan HY, Warrick JM, Andriola I, Merry D, Bonini NM. Genetic modulation of polyglutamine toxicity by protein conjugation pathways in Drosophila. Hum Mol Genet. 2002;11:2895–2904. doi: 10.1093/hmg/11.23.2895. [DOI] [PubMed] [Google Scholar]
- 135.Nollen EA, Garcia SM, van Haaften G, Kim S, Chavez A, Morimoto RI, Plasterk RH. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci USA. 2004;101:6403–6408. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hyun DH, Lee M, Halliwell B, Jenner P. Proteasomal inhibition causes the formation of protein aggregates containing a wide range of proteins, including nitrated proteins. J Neurochem. 2003;86:363–373. doi: 10.1046/j.1471-4159.2003.01841.x. [DOI] [PubMed] [Google Scholar]
- 137.Chapman AL, Winterbourn CC, Brennan SO, Jordan TW, Kettle AJ. Characterization of non-covalent oligomers of proteins treated with hypochlorous acid. Biochem J. 2003;375:33–40. doi: 10.1042/BJ20030685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bieschke J, Zhang Q, Bosco DA, Lerner RA, Powers ET, Wentworth P, Jr, Kelly JW. Small molecule oxidation products trigger disease-associated protein misfolding. Acc Chem Res. 2006;39:611–619. doi: 10.1021/ar0500766. [DOI] [PubMed] [Google Scholar]
- 139.Sharma SK, Goloubinoff P, Christen P. Heavy metal ions are potent inhibitors of protein folding. Biochem Biophys Res Commun. 2008;372:341–345. doi: 10.1016/j.bbrc.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 140.Zhang Q, Powers ET, Nieva J, Huff ME, Dendle MA, Bieschke J, Glabe CG, Eschenmoser A, Wentworth P, Jr, Lerner RA, Kelly JW. Metabolite-initiated protein misfolding may trigger Alzheimer’s disease. Proc Natl Acad Sci USA. 2004;101:4752–4757. doi: 10.1073/pnas.0400924101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Siegel SJ, Bieschke J, Powers ET, Kelly JW. The oxidative stress metabolite 4-hydroxynonenal promotes Alzheimer protofibril formation. Biochemistry. 2007;46:1503–1510. doi: 10.1021/bi061853s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Winter J, Ilbert M, Graf PC, Ozcelik D, Jakob U. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell. 2008;135:691–701. doi: 10.1016/j.cell.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rand JD, Grant CM. The thioredoxin system protects ribosomes against stress-induced aggregation. Mol Biol Cell. 2006;17:387–401. doi: 10.1091/mbc.E05-06-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mullally JE, Moos PJ, Edes K, Fitzpatrick FA. Cyclopentenone prostaglandins of the J series inhibit the ubiquitin isopeptidase activity of the proteasome pathway. J Biol Chem. 2001;276:30366–30373. doi: 10.1074/jbc.M102198200. [DOI] [PubMed] [Google Scholar]
- 145.Bulteau AL, Lundberg KC, Humphries KM, Sadek HA, Szweda PA, Friguet B, Szweda LI. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J Biol Chem. 2001;276:30057–30063. doi: 10.1074/jbc.M100142200. [DOI] [PubMed] [Google Scholar]
- 146.Yang H, Chen D, Cui QC, Yuan X, Dou QP. Celastrol, a triterpene extracted from the Chinese “Thunder of God Vine,” is a potent proteasome inhibitor and suppresses human prostate cancer growth in nude mice. Cancer Res. 2006;66:4758–4765. doi: 10.1158/0008-5472.CAN-05-4529. [DOI] [PubMed] [Google Scholar]
- 147.Yang H, Shi G, Dou QP. The tumor proteasome is a primary target for the natural anticancer compound Withaferin A isolated from “Indian winter cherry. Mol Pharmacol. 2007;71:426–437. doi: 10.1124/mol.106.030015. [DOI] [PubMed] [Google Scholar]
- 148.Carbone DL, Doorn JA, Kiebler Z, Sampey BP, Petersen DR. Inhibition of Hsp72-mediated protein refolding by 4-hydroxy-2-nonenal. Chem Res Toxicol. 2004;17:1459–1467. doi: 10.1021/tx049838g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Carbone DL, Doorn JA, Kiebler Z, Ickes BR, Petersen DR. Modification of heat shock protein 90 by 4-hydroxynonenal in a rat model of chronic alcoholic liver disease. J Pharmacol Exp Ther. 2005;315:8–15. doi: 10.1124/jpet.105.088088. [DOI] [PubMed] [Google Scholar]
- 150.Le Moan N, Clement G, Le Maout S, Tacnet F, Toledano MB. The Saccharomyces cerevisiae proteome of oxidized protein thiols: contrasted functions for the thioredoxin and glutathione pathways. J Biol Chem. 2006;281:10420–10430. doi: 10.1074/jbc.M513346200. [DOI] [PubMed] [Google Scholar]
- 151.Vila A, Tallman KA, Jacobs AT, Liebler DC, Porter NA, Marnett LJ. Identification of protein targets of 4-hydroxynonenal using click chemistry for ex vivo biotinylation of azido and alkynyl derivatives. Chem Res Toxicol. 2008;21:432–444. doi: 10.1021/tx700347w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Leonard SE, Reddie KG, Carroll KS. Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem Biol. 2009;4:783–799. doi: 10.1021/cb900105q. [DOI] [PubMed] [Google Scholar]
- 153.Liu Q, Levy EJ, Chirico WJ. N-Ethylmaleimide inactivates a nucleotide-free Hsp70 molecular chaperone. J Biol Chem. 1996;271:29937–29944. doi: 10.1074/jbc.271.47.29937. [DOI] [PubMed] [Google Scholar]
- 154.Chadli A, Felts SJ, Wang Q, Sullivan WP, Botuyan MV, Fauq A, Ramirez-Alvarado M, Mer G. Celastrol inhibits Hsp90 chaperoning of steroid receptors by inducing fibrillization of the co-chaperone p23. J Biol Chem. 2010;285:4224–4231. doi: 10.1074/jbc.M109.081018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang Y, Gibney PA, West JD, Morano KA. The yeast Hsp70 Ssa1 is a sensor for heat shock response activation by thiol-reactive compounds. Mol Biol Cell. 2012 doi: 10.1091/mbc.E12-06-0447. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hermawan A, Chirico WJ. N-Ethylmaleimide-modified Hsp70 inhibits protein folding. Arch Biochem Biophys. 1999;369:157–162. doi: 10.1006/abbi.1999.1354. [DOI] [PubMed] [Google Scholar]
- 157.Shibata T, Kimura Y, Mukai A, Mori H, Ito S, Asaka Y, Oe S, Tanaka H, Takahashi T, Uchida K. Transthiocarbamoylation of proteins by thiolated isothiocyanates. J Biol Chem. 2011;286:42150–42161. doi: 10.1074/jbc.M111.308049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Connor RE, Marnett LJ, Liebler DC. Protein-selective capture to analyze electrophile adduction of hsp90 by 4-hydroxynonenal. Chem Res Toxicol. 2011;24:1275–1282. doi: 10.1021/tx200157t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kim D, Ouyang H, Li GC. Heat shock protein hsp70 accelerates the recovery of heat-shocked mammalian cells through its modulation of heat shock transcription factor HSF1. Proc Natl Acad Sci USA. 1995;92:2126–2130. doi: 10.1073/pnas.92.6.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Xia W, Voellmy R. Hyperphosphorylation of heat shock transcription factor 1 is correlated with transcriptional competence and slow dissociation of active factor trimers. J Biol Chem. 1997;272:4094–4102. doi: 10.1074/jbc.272.7.4094. [DOI] [PubMed] [Google Scholar]
- 161.Schett G, Steiner CW, Xu Q, Smolen JS, Steiner G. TNFα mediates susceptibility to heat-induced apoptosis by protein phosphatase-mediated inhibition of the HSF1/hsp70 stress response. Cell Death Differ. 2003;10:1126–1136. doi: 10.1038/sj.cdd.4401276. [DOI] [PubMed] [Google Scholar]
- 162.Conde R, Xavier J, McLoughlin C, Chinkers M, Ovsenek N. Protein phosphatase 5 is a negative modulator of heat shock factor 1. J Biol Chem. 2005;280:28989–28996. doi: 10.1074/jbc.M503594200. [DOI] [PubMed] [Google Scholar]
- 163.Hong Y, Rogers R, Matunis MJ, Mayhew CN, Goodson ML, Park-Sarge OK, Sarge KD. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J Biol Chem. 2001;276:40263–40267. doi: 10.1074/jbc.M104714200. [DOI] [PubMed] [Google Scholar]
- 164.Hilgarth RS, Hong Y, Park-Sarge OK, Sarge KD. Insights into the regulation of heat shock transcription factor 1 SUMO-1 modification. Biochem Biophys Res Commun. 2003;303:196–200. doi: 10.1016/s0006-291x(03)00312-7. [DOI] [PubMed] [Google Scholar]
- 165.Nemani R, Lee EY. Reactivity of sulfhydryl groups of the catalytic subunits of rabbit skeletal muscle protein phosphatases 1 and 2A. Arch Biochem Biophys. 1993;300:24–29. doi: 10.1006/abbi.1993.1004. [DOI] [PubMed] [Google Scholar]
- 166.Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell. 2006;21:349–357. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 167.Codreanu SG, Adams DG, Dawson ES, Wadzinski BE, Liebler DC. Inhibition of protein phosphatase 2A activity by selective electrophile alkylation damage. Biochemistry. 2006;45:10020–10029. doi: 10.1021/bi060551n. [DOI] [PubMed] [Google Scholar]
- 168.Caito S, Rajendrasozhan S, Cook S, Chung S, Yao H, Friedman AE, Brookes PS, Rahman I. SIRT1 is a redox-sensitive deacetylase that is post-translationally modified by oxidants and carbonyl stress. FASEB J. 2010;24:3145–3159. doi: 10.1096/fj.09-151308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sanchez Y, Lindquist SL. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 170.Morano KA, Santoro N, Kock KA, Thiele DJ. A trans-activation domain in yeast heat shock transcription factor is essential for cell cycle progression during stress. Mol Cell Biol. 1999;19:402–411. doi: 10.1128/mcb.19.1.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Becker J, Walter W, Yan W, Craig EA. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.McMillan DR, Xiao X, Shao L, Graves K, Benjamin IJ. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J Biol Chem. 1998;273:7523–7528. doi: 10.1074/jbc.273.13.7523. [DOI] [PubMed] [Google Scholar]
- 173.Sanchez Y, Taulien J, Borkovich KA, Lindquist S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 1992;11:2357–2364. doi: 10.1002/j.1460-2075.1992.tb05295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Beere HM. Stressed to death: regulation of apoptotic signaling pathways by the heat shock proteins. Sci STKE. 2001:re1. doi: 10.1126/stke.2001.93.re1. [DOI] [PubMed] [Google Scholar]
- 175.Beere HM, Green DR. Stress management: heat shock protein-70 and the regulation of apoptosis. Trends Cell Biol. 2001;11:6–10. doi: 10.1016/s0962-8924(00)01874-2. [DOI] [PubMed] [Google Scholar]
- 176.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 177.Khong T, Spencer A. Targeting HSP90 induces apoptosis and inhibits critical survival and proliferation pathways in multiple myeloma. Mol Cancer Ther. 2011;10:1909–1917. doi: 10.1158/1535-7163.MCT-11-0174. [DOI] [PubMed] [Google Scholar]
- 178.Perdrizet GA. Heat shock and tissue protection. New Horiz. 1995;3:312–320. [PubMed] [Google Scholar]
- 179.Locke M, Tanguay RM. Diminished heat shock response in the aged myocardium. Cell Stress Chaperones. 1996;1:251–260. doi: 10.1379/1466-1268(1996)001<0251:dhsrit>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Fawcett TW, Sylvester SL, Sarge KD, Morimoto RI, Holbrook NJ. Effects of neurohormonal stress and aging on the activation of mammalian heat shock factor 1. J Biol Chem. 1994;269:32272–32278. [PubMed] [Google Scholar]
- 181.Jurivich DA, Qiu L, Welk JF. Attenuated stress responses in young and old human lymphocytes. Mech Ageing Dev. 1997;94:233–249. doi: 10.1016/s0047-6374(96)01856-8. [DOI] [PubMed] [Google Scholar]
- 182.Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 184.Morrow G, Battistini S, Zhang P, Tanguay RM. Decreased lifespan in the absence of expression of the mitochondrial small heat shock protein Hsp22 in Drosophila. J Biol Chem. 2004;279:43382–43385. doi: 10.1074/jbc.C400357200. [DOI] [PubMed] [Google Scholar]
- 185.Morrow G, Samson M, Michaud S, Tanguay RM. Overexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. FASEB J. 2004;18:598–599. doi: 10.1096/fj.03-0860fje. [DOI] [PubMed] [Google Scholar]
- 186.Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci USA. 2009;106:14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Steinkraus KA, Smith ED, Davis C, Carr D, Pendergrass WR, Sutphin GL, Kennedy BK, Kaeberlein M. Dietary restriction suppresses proteotoxicity and enhances longevity by an hsf-1-dependent mechanism in Caenorhabditis elegans. Aging Cell. 2008;7:394–404. doi: 10.1111/j.1474-9726.2008.00385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Heydari AR, You S, Takahashi R, Gutsmann A, Sarge KD, Richardson A. Effect of caloric restriction on the expression of heat shock protein 70 and the activation of heat shock transcription factor 1. Dev Genet. 1996;18:114–124. doi: 10.1002/(SICI)1520-6408(1996)18:2<114::AID-DVG4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 190.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 191.Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, Cohen HY. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 192.Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat Rev Neurosci. 2003;4:49–60. doi: 10.1038/nrn1007. [DOI] [PubMed] [Google Scholar]
- 193.Batulan Z, Shinder GA, Minotti S, He BP, Doroudchi MM, Nalbantoglu J, Strong MJ, Durham HD. High threshold for induction of the stress response in motor neurons is associated with failure to activate HSF1. J Neurosci. 2003;23:5789–5798. doi: 10.1523/JNEUROSCI.23-13-05789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kaarniranta K, Oksala N, Karjalainen HM, Suuronen T, Sistonen L, Helminen HJ, Salminen A, Lammi MJ. Neuronal cells show regulatory differences in the hsp70 gene response. Brain Res Mol Brain Res. 2002;101:136–140. doi: 10.1016/s0169-328x(02)00179-1. [DOI] [PubMed] [Google Scholar]
- 195.Homma S, Jin X, Wang G, Tu N, Min J, Yanasak N, Mivechi NF. Demyelination, astrogliosis, and accumulation of ubiquitinated proteins, hallmarks of CNS disease in hsf1-deficient mice. J Neurosci. 2007;27:7974–7986. doi: 10.1523/JNEUROSCI.0006-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Evgrafov OV, Mersiyanova I, Irobi J, Van DBL, Dierick I, Leung CL, Schagina O, Verpoorten N, Van IK, Fedotov V, Dadali E, Auer-Grumbach M, Windpassinger C, Wagner K, Mitrovic Z, Hilton-Jones D, Talbot K, Martin JJ, Vasserman N, Tverskaya S, Polyakov A, Liem RK, Gettemans J, Robberecht W, De JP, Timmerman V. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat Genet. 2004;36:602–606. doi: 10.1038/ng1354. [DOI] [PubMed] [Google Scholar]
- 197.Cohen E, Paulsson JF, Blinder P, Burstyn-Cohen T, Du D, Estepa G, Adame A, Pham HM, Holzenberger M, Kelly JW, Masliah E, Dillin A. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell. 2009;139:1157–1169. doi: 10.1016/j.cell.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Gidalevitz T, Krupinski T, Garcia S, Morimoto RI. Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLoS Genet. 2009;5:e1000399. doi: 10.1371/journal.pgen.1000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- 200.Tang D, Khaleque MA, Jones EL, Theriault JR, Li C, Wong WH, Stevenson MA, Calderwood SK. Expression of heat shock proteins and heat shock protein messenger ribonucleic acid in human prostate carcinoma in vitro and in tumors in vivo. Cell Stress Chaperones. 2005;10:46–58. doi: 10.1379/CSC-44R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem Sci. 2006;31:164–172. doi: 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 202.Stepanova L, Finegold M, DeMayo F, Schmidt EV, Harper JW. The oncoprotein kinase chaperone CDC37 functions as an oncogene in mice and collaborates with both c-myc and cyclin D1 in transformation of multiple tissues. Mol Cell Biol. 2000;20:4462–4473. doi: 10.1128/mcb.20.12.4462-4473.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wandinger SK, Richter K, Buchner J. The Hsp90 chaperone machinery. J Biol Chem. 2008;283:18473–18477. doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- 204.Santagata S, Hu R, Lin NU, Mendillo ML, Collins LC, Hankinson SE, Schnitt SJ, Whitesell L, Tamimi RM, Lindquist S, Ince TA. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc Natl Acad Sci USA. 2011;108:18378–18383. doi: 10.1073/pnas.1115031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kon M, Kiffin R, Koga H, Chapochnick J, Macian F, Varticovski L, Cuervo AM. Chaperone-mediated autophagy is required for tumor growth. Sci Transl Med. 2011;3:109ra117. doi: 10.1126/scitranslmed.3003182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kamada M, So A, Muramaki M, Rocchi P, Beraldi E, Gleave M. Hsp27 knockdown using nucleotide-based therapies inhibit tumor growth and enhance chemotherapy in human bladder cancer cells. Mol Cancer Ther. 2007;6:299–308. doi: 10.1158/1535-7163.MCT-06-0417. [DOI] [PubMed] [Google Scholar]
- 208.Vargas-Roig LM, Gago FE, Tello O, Aznar JC, Ciocca DR. Heat shock protein expression and drug resistance in breast cancer patients treated with induction chemotherapy. Int J Cancer. 1998;79:468–475. doi: 10.1002/(sici)1097-0215(19981023)79:5<468::aid-ijc4>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 209.Mori-Iwamoto S, Kuramitsu Y, Ryozawa S, Mikuria K, Fujimoto M, Maehara S, Maehara Y, Okita K, Nakamura K, Sakaida I. Proteomics finding heat shock protein 27 as a biomarker for resistance of pancreatic cancer cells to gemcitabine. Int J Oncol. 2007;31:1345–1350. [PubMed] [Google Scholar]
- 210.Rahmani M, Reese E, Dai Y, Bauer C, Kramer LB, Huang M, Jove R, Dent P, Grant S. Cotreatment with suberanoylanilide hydroxamic acid and 17-allylamino 17-demethoxygeldanamycin synergistically induces apoptosis in Bcr-Abl+ cells sensitive and resistant to STI571 (imatinib mesylate) in association with down-regulation of Bcr-Abl, abrogation of signal transducer and activator of transcription 5 activity, and Bax conformational change. Mol Pharmacol. 2005;67:1166–1176. doi: 10.1124/mol.104.007831. [DOI] [PubMed] [Google Scholar]
- 211.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 212.Mu TW, Ong DS, Wang YJ, Balch WE, Yates JR, 3rd, Segatori L, Kelly JW. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell. 2008;134:769–781. doi: 10.1016/j.cell.2008.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ong DS, Mu TW, Palmer AE, Kelly JW. Endoplasmic reticulum Ca2+ increases enhance mutant glucocerebrosidase proteostasis. Nat Chem Biol. 2010;6:424–432. doi: 10.1038/nchembio.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Mu TW, Fowler DM, Kelly JW. Partial restoration of mutant enzyme homeostasis in three distinct lysosomal storage disease cell lines by altering calcium homeostasis. PLoS Biol. 2008;6:e26. doi: 10.1371/journal.pbio.0060026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Marozkina NV, Yemen S, Borowitz M, Liu L, Plapp M, Sun F, Islam R, Erdmann-Gilmore P, Townsend RR, Lichti CF, Mantri S, Clapp PW, Randell SH, Gaston B, Zaman K. Hsp 70/Hsp 90 organizing protein as a nitrosylation target in cystic fibrosis therapy. Proc Natl Acad Sci USA. 2010;107:11393–11398. doi: 10.1073/pnas.0909128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Wang X, Venable J, LaPointe P, Hutt DM, Koulov AV, Coppinger J, Gurkan C, Kellner W, Matteson J, Plutner H, Riordan JR, Kelly JW, Yates JR, 3rd, Balch WE. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- 217.Singh OV, Pollard HB, Zeitlin PL. Chemical rescue of deltaF508-CFTR mimics genetic repair in cystic fibrosis bronchial epithelial cells. Mol Cell Proteomics. 2008;7:1099–1110. doi: 10.1074/mcp.M700303-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Allison AC, Cacabelos R, Lombardi VR, Alvarez XA, Vigo C. Celastrol, a potent antioxidant and anti-inflammatory drug, as a possible treatment for Alzheimer’s disease. Prog Neuropsychopharmacol Biol Psychiatry. 2001;25:1341–1357. doi: 10.1016/s0278-5846(01)00192-0. [DOI] [PubMed] [Google Scholar]
- 219.Neef DW, Turski ML, Thiele DJ. Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease. PLoS Biol. 2010;8:e1000291. doi: 10.1371/journal.pbio.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Otaka M, Yamamoto S, Ogasawara K, Takaoka Y, Noguchi S, Miyazaki T, Nakai A, Odashima M, Matsuhashi T, Watanabe S, Itoh H. The induction mechanism of the molecular chaperone HSP70 in the gastric mucosa by Geranylgeranylacetone (HSP-inducer) Biochem Biophys Res Commun. 2007;353:399–404. doi: 10.1016/j.bbrc.2006.12.031. [DOI] [PubMed] [Google Scholar]
- 221.Katsuno M, Sang C, Adachi H, Minamiyama M, Waza M, Tanaka F, Doyu M, Sobue G. Pharmacological induction of heat-shock proteins alleviates polyglutamine-mediated motor neuron disease. Proc Natl Acad Sci USA. 2005;102:16801–16806. doi: 10.1073/pnas.0506249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Mikuriya T, Sugahara K, Sugimoto K, Fujimoto M, Takemoto T, Hashimoto M, Hirose Y, Shimogori H, Hayashida N, Inouye S, Nakai A, Yamashita H. Attenuation of progressive hearing loss in a model of age-related hearing loss by a heat shock protein inducer, geranylgeranylacetone. Brain Res. 2008;1212:9–17. doi: 10.1016/j.brainres.2008.03.031. [DOI] [PubMed] [Google Scholar]
- 223.Yasuda H, Shichinohe H, Kuroda S, Ishikawa T, Iwasaki Y. Neuroprotective effect of a heat shock protein inducer, geranylgeranylacetone in permanent focal cerebral ischemia. Brain Res. 2005;1032:176–182. doi: 10.1016/j.brainres.2004.11.009. [DOI] [PubMed] [Google Scholar]