SUMMARY
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder characterized by memory and cognitive impairment and is the leading cause of dementia in the elderly. A number of genome wide association studies and subsequent replication studies have been published recently on late onset AD (LOAD). These studies identified several new susceptibility genes including phosphatidylinositol-binding clathrin assembly protein (PICALM) on chromosome 11. The aim of our study was to examine the entire coding sequence of PICALM to determine if the association could be explained by any previously undetected sequence variation. Therefore, we sequenced 48 cases and 48 controls homozygous for the risk allele in the signal SNP rs3851179. We did not find any new variants; however, rs592297, a known coding synonymous SNP that is part of an exonic splice enhancer region in exon 5, is in strong linkage disequilibrium with rs3851179 and should be examined for functional significance in Alzheimer pathophysiology.
Keywords: Alzheimer, Neurodegenerative disease, PICALM, Sequencing, Exonic splicing
Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder characterized by memory and cognitive impairment, and is the leading cause of dementia in the elderly. Mutations in three genes encoding β-amyloid (APP), presenilin 1 (PS1), and presenilin 2 (PS2) cause rare Mendelian early onset AD (EOAD) (Brouwers et al. 2008). One common allele (ε4) in the apolipoprotein E gene (APOE) has been unequivocally accepted as an important risk factor for late onset Alzheimer’s disease (LOAD) (Corder et al. 1993), while another allele (ε2) decreases risk (Corder et al. 1994). A number of genome wide association studies (GWAS) and subsequent replication studies have been recently published on LOAD (Carrasquillo et al. 2010; Corneveaux et al. 2010; Harold et al. 2009; Hollingworth et al. 2011; Jun et al. 2010; Lambert et al. 2009; Naj et al. 2011; Seshadri et al. 2010). These studies identified several new susceptibility genes including phosphatidylinositol-binding clathrin assembly protein (PICALM). Because GWAS associations rarely identify the actual susceptibility variation, additional detailed studies of the implicated genes are warranted. The aim of this study was to examine the coding sequence of PICALM to determine if the observed association could be explained by any previously undetected sequence variation.
Methods
Ascertainment and Samples
Our sample set is derived from the Collaborative Alzheimer Project (CAP: The Hussman Institute for Human Genomics at the Miller School of Medicine, University of Miami and The Center for Human Genetics Research at the Vanderbilt University School of Medicine). After a complete description of the study to the subjects, written informed consent was obtained from all participants in agreement with protocols approved by the institutional review board at each contributing center. For inclusion, each LOAD affected individual met the NINCDS/ADRDA criteria for definite or probable AD and had an age at onset greater than 60 years of age. Age at onset (AAO) for LOAD was determined from specific probe questions within the clinical history provided by a reliable family informant or documented significant impairment in the medical record. Cognitive controls were spouses, friends, and other biologically unrelated individuals who were frequency age and gender matched to the cases, and all were from within the same clinical catchment areas. All cognitive controls were examined and none showed signs of dementia by history and upon interview. Additionally, cognitive controls each have a documented Mini-Mental State Exam (MMSE) ≥ 27 or Modified Mini-Mental State Exam (3MS) ≥ 87 (Teng & Chui, 1987). The sample set consisted of 48 cases and 48 controls who were T/T homozygotes for the protective allele at rs3851179, the signal SNP previously associated to LOAD found at the 5’ end of the PICALM gene (Harold et al. 2009; Jun et al. 2010; Seshadri et al. 2010).
Sequencing
Twenty one amplicons for the twenty four different exons and UTRs were optimized for amplification (see Supplementary Information for details) and the products were sequenced using ABI’s 3730xl DNA analyzer (Life Technologies Corporation, Carlsbad, CA, 92008, USA) in the Vanderbilt University DNA Sequencing Core Facility. Both forward and reverse strands were sequenced to provide confirmation of any changes. Sequencher (Gene Codes Corporation, Ann Arbor, MI) was used to align the sequences; both the forward and the reverse fragment were aligned to confirm all possible variants.
ESEfinder
The program ESEfinder (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home) was used to determine the predicted effects of variation at rs592297 on the binding of exon splicing enhancers. ESEfinder 2.0 was used with a matrix library “SR proteins” and the default (thresholds) settings (Cartegni et al. 2003). We added ~20bp on each side of exon 5 as input for ESEfinder, in total 138bp was submitted to ESEfinder.
Results and Discussion
PICALM is one of the four currently accepted associated genes for LOAD (along with APOE, CLU, and CR1). Recent studies have identified rs3851179, a SNP at the 5’ end of the PICALM gene and thought to be part of a regulatory sequence, as being associated to LOAD with a combined odds ratio = 0.88 (95% CI, 0.86–0.91) for the T allele across all the studies (http://www.alzgene.org/meta.asp?geneid=636&polyID=2656) (Bertram et al. 2007). In the GWAS published by Harold et al. (Harold et al. 2009), several functional SNPs were identified at the PICALM locus. Two SNPs, rs561655 (meta analysis, p=1−10−7), which is within a putative transcription factor binding site, and rs592297 (meta analysis, p=2×10−7), which is a synonymous SNP in exon 5 that may influence a predicted exon splicing enhancer (ESE) sequence, showed evidence for association. The association for rs561655 was confirmed in the Alzheimer Disease Genetics Consortium (ADGC) GWAS (Hollingworth et al. 2011; Jun et al. 2010; Naj et al. 2011). In addition, two longitudinal studies suggested that rs7110631 in PICALM, a SNP in high LD with rs3851179, is associated with age-related cognitive decline (Chibnik et al. 2011).
Further studies performed on other populations have revealed a strong architectural complexity for PICALM (Greene et al. 2009). A Caribbean Hispanic cohort study used to identify new LOAD associated loci and to replicate existing findings found that only rs17159904 was marginally associated in population stratification-adjusted and APOE-adjusted analyses (Lee et al. 2010). This SNP is situated between the 5’ end of PICALM and rs3851179. A mixed Hispanic, African American, and Wadi Ari population was also examined but no association to PICALM was detected. It was hypothesized that the lack of association was the result of the small sample size (Jun et al. 2010). In a Han Chinese population the association between rs3851179 and LOAD could not be replicated (Yu et al. 2010). The authors concluded that rs381179 might not play a major role in the development of LOAD in the Han Chinese population. They pointed out that their study had only 266 cases and 343 controls and might be underpowered, or that population specificity might play a role, and finally that their results required further confirmation as rs3851179 was the only SNP to be genotyped (Yu et al. 2010). Another study by Li et al. confirmed the lack of association between PICALM and isolated Alzheimer’s disease in Chinese (Li et al. 2011). No association was found between PICALM rs3851179 and Italian LOAD subjects, however a possible role of PICALM in longevity was suggested (Piaceri et al. 2011).
PICALM was first described in U937 cell lines as the clathrin assembly lymphoid myeloid leukemia (CALM) gene (Dreyling et al. 1996). Its N-terminus binds to phosphatidylinositol-4,5-biphosphate present in the plasma membrane and its C-terminus to clathrin and adaptor protein 2 (AP-2), recruiting both to the membrane (Baig et al. 2010). PICALM is important in clathrin-mediated endocytosis and intracellular trafficking of the synaptic vesicle protein VAMP2 (Harel et al. 2008). VAMP2 is a soluble N-ethylmaleimide-sensitive attachment protein receptor (SNARE) protein that is involved in neurotransmitter release at the presynaptic membrane, which is important for neuronal function and memory formation (Harold et al. 2009; Sleegers et al. 2010). Furthermore, PICALM may also be involved in the regulation of β-amyloid levels through the degradation of APP upon entry into endosomes and Aβ production (Kyriazis et al. 2008). PICALM is expressed in all tissue types with prominent expression in neurons (Harold et al. 2009) and is present in synapses (Harel et al. 2008). PICALM is a large gene (112kb, http://www.ensembl.org, ENSG00000073921, GRCh37, May 2011) on 11q23, with 28 alternative transcripts, 19 of them encoding for a protein, and ranging in size from 134 residues to 660 residues. PICALM, like most eukaryotic genes, is comprised of multiple relatively short exons that are separated by longer introns (Fig. 1).
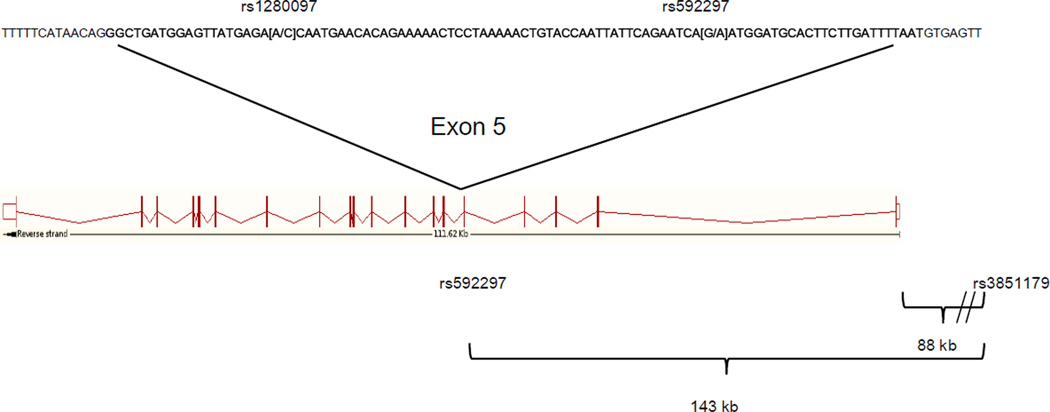
Figure 1.
A PICALM representation from http://www.ensembl.org. The full sequence of exon 5 is represented in bold and the location of the SNPs and base pair change can be seen on the sequence. Rs3851179, has a combined OR= 0.88 for all studies (http://www.AlzGene.org, April 2011), and is 88kb from the 5’ end of the gene, and 143kb from rs592297 that has a p value of 2×10−7 in theHarold et al. (2009) meta analysis.
Point mutations in the coding regions of genes are commonly assumed to exert their effects by altering single amino acids in the encoded proteins. However, there is increasing evidence that many human disease genes harbor exonic mutations that affect pre-mRNA splicing (Cartegni et al. 2002). The mechanism of splicing is complex due to the presence of numerous intronic pseudo exons; therefore, the additional information required for “true” splicing is contained in cis-acting regulatory enhancer and silencer sequences (Cartegniet al. 2002).
For our sequencing of the exonic regions of PICALM, we picked 48 cases and 48 controls that were homozygous for the protective allele for rs3851179 and that were of mixed APOE status (see Supplementary Information for details on primer sequences and PCR protocol). Our study was designed to detect rare variants with a minor allele frequency of 0.01, and even though we selected for homozygotes at rs3851179 to provide a more consistent background for the search of new variants, we did not find any new exonic polymorphisms or rare variants; only heterozygotes for the previously known SNPs rs76719109 and rs592297 were detected (see supplementary material, Fig. 1 for details). Thus the strong association in this region is unlikely to be explained by a novel coding variant in PICALM.
We saw altered, but non-significant (when corrected for multiple comparison), allele frequencies for rs592297 in exon 5 (see Table S2). Even though this SNP is approximately 143 kb from rs3851179, they are in strong linkage disequilibrium (LD). A slight increase in LD is observed between the ADGC GWAS data (Jun et al. 2010) and the HapMap data (GWAS D’=0.937, r2=0.40 and HapMap D’=0.923, r2=0.28). The bias in sampling, only T/T homozygotes, and the high correlation between the two SNPs likely explains the altered genotype frequencies that we see in our sample set.
Alteration of the allele for rs592297 from a G to an A changes the predicted exon enhancer binding at this site from an SF2/ASF protein to SRp40 (see Fig. S2). This alteration of the ESE could change the splicing pattern at exon 5. However, there are limitations in using ESEfinder; the presence of a high-score motif in a sequence, as seen in Figure S2, does not necessarily identify that sequence as a functional ESE, and in general, there is not a very strict quantitative correlation between numerical scores and ESE activity (Cartegni et al. 2003). Direct experimental evidence is necessary before we can reach a final conclusion. Reverse transcriptase PCR experiments could be designed for the different cell lines, with the different genotypes, to detect possible splicing differences. The lack of additional variation within the coding sequence of the gene suggests that novel variation in PICALM does not explain the association results and that the known variation should be explored further. Expression studies could be designed using the known coding variant in order to see if the variation could indeed change the expression of PICALM.
Supplementary Material
Acknowledgements
This research was funded by NIH grant 5R01AG027944.
Footnotes
The authors have no conflict of interest to declare. The authors would like to thank Marie Boutaud for her contributions to the project.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article:
Amplification protocol
Table S1: primer sequences
Table S2: Allele frequencies for rs592297
Figure S1: PICALM exon 5 sequence
Figure S2: Representation of the PICALM gene. All the coding SNPs from dbSNP (list obtained April 5th 2011) were added to the figure next to their corresponding coding region.
Figure S3: ESE results for exon 5 (http://rulai.cshl.edu/tools/ESE2/)
Figure S4: Linkage disequilibrium schematic for PICALM
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organised for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Baig S, Joseph SA, Tayler H, Abraham R, Owen MJ, Williams J, Kehoe PG, Love S. Distribution and expression of picalm in Alzheimer disease. J Neuropathol Exp Neurol. 2010;69:1071–1077. doi: 10.1097/NEN.0b013e3181f52e01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Brouwers N, Sleegers K, van Broeckhoven C. Molecular genetics of Alzheimer's disease: an update. Ann Med. 2008;40:562–583. doi: 10.1080/07853890802186905. [DOI] [PubMed] [Google Scholar]
- Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou F, Crook JE, Pankratz VS, Dickson DW, Graff-Radford NR, Petersen RC, Morgan K, Younkin SG. Replication of CLU, CR1, and PICALM associations with alzheimer disease. Arch Neurol. 2010;67:961–964. doi: 10.1001/archneurol.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibnik LB, Shulman JM, Leurgans SE, Schneider JA, Wilson RS, Tran D, Aubin C, Buchman AS, Heward CB, Myers AJ, Hardy JA, Huentelman MJ, Corneveaux JJ, Reiman EM, Evans DA, Bennett DA, De Jager PL. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Ann Neurol. 2011;69:560–569. doi: 10.1002/ana.22277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC, Jr, Rimmler JB, Locke PA, Conneally PM, Schmader KE, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994;7:180–184. doi: 10.1038/ng0694-180. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Corneveaux JJ, Myers AJ, Allen AN, Pruzin JJ, Ramirez M, Engel A, Nalls MA, Chen K, Lee W, Chewning K, Villa SE, Meechoovet HB, Gerber JD, Frost D, Benson HL, O'Reilly S, Chibnik LB, Shulman JM, Singleton AB, Craig DW, Van Keuren-Jensen KR, Dunckley T, Bennett DA, De Jager PL, Heward C, Hardy J, Reiman EM, Huentelman MJ. Association of CR1, CLU and PICALM with Alzheimer's disease in a cohort of clinically characterized and neuropathologically verified individuals. Hum Mol Genet. 2010;19:3295–3301. doi: 10.1093/hmg/ddq221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyling MH, Martinez-Climent JA, Zheng M, Mao J, Rowley JD, Bohlander SK. The t(10;11)(p13;q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family. Proc Natl Acad Sci U S A. 1996;93:4804–4809. doi: 10.1073/pnas.93.10.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene CS, Penrod NM, Williams SM, Moore JH. Failure to replicate a genetic association may provide important clues about genetic architecture. PLoS.One. 2009;4:e5639. doi: 10.1371/journal.pone.0005639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A, Wu F, Mattson MP, Morris CM, Yao PJ. Evidence for CALM in directing VAMP2 trafficking. Traffic. 2008;9:417–429. doi: 10.1111/j.1600-0854.2007.00694.x. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O'Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Beaumont H, Warden D, Wilcock G, Love S, Kehoe PG, Hooper NM, Vardy ER, Hardy J, Mead S, Fox NC, Rossor M, Collinge J, Maier W, Jessen F, Ruther E, Schurmann B, Heun R, Kolsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Gallacher J, Hull M, Rujescu D, Giegling I, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, van Duijn CM, Breteler MM, Ikram MA, DeStefano AL, Fitzpatrick AL, Lopez O, Launer LJ, Seshadri S, Berr C, Campion D, Epelbaum J, Dartigues JF, Tzourio C, Alperovitch A, Lathrop M, Feulner TM, Friedrich P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefansson H, Stefansson K, Snaedal J, Bjornsson S, Jonsson PV, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido MJ, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossu P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans PA, Jonsson T, Riemenschneider M, Morgan K, Younkin SG, Owen MJ, O'Donovan M, Amouyel P, Williams J. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun G, Naj AC, Beecham GW, Wang LS, Buros J, Gallins PJ, Buxbaum JD, Ertekin-Taner N, Fallin MD, Friedland R, Inzelberg R, Kramer P, Rogaeva E, St George-Hyslop P, Arnold SE, Baldwin CT, Barber R, Beach T, Bigio EH, Bird TD, Boxer A, Burke JR, Cairns N, Carroll SL, Chui HC, Clark DG, Cotman CW, Cummings JL, DeCarli C, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Gearing M, Geschwind DH, Ghetti B, Gilman S, Giordani B, Glass J, Graff-Radford NR, Green RC, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman A, Lopez OL, Mack WJ, Markesbery W, Marson DC, Martiniuk F, Masliah E, McKee AC, Mesulam M, Miller JW, Miller BL, Miller CA, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon W, Quinn JF, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider JA, Schneider LS, Seeley W, Shelanski ML, Smith CD, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Woltjer RL, Younkin SG, Cantwell LB, Dombroski BA, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Lunetta KL, Martin ER, Montine TJ, Goate AM, Blacker D, Tsuang DW, Beekly D, Cupples LA, Hakonarson H, Kukull W, Foroud TM, Haines J, Mayeux R, Farrer LA, Pericak-Vance MA, Schellenberg GD, Arnold SE, Baldwin CT, Barber R, Beach T, Bigio EH, Bird TD, Boxer A, Burke JR, Cairns N, Carroll SL, Chui HC, Clark DG, Cotman CW, Cummings JL, DeCarli C, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Gearing M, Geschwind DH, Ghetti B, Gilman S, Giordani B, Glass J, Graff-Radford NR, Green RC, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman A, Lopez OL, Mack WJ, Markesbery W, Marson DC, Martiniuk F, Masliah E, McKee AC, Mesulam M, Miller JW, Miller BL, Miller CA, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon W, Quinn JF, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider JA, Schneider LS, Seeley W, Shelanski ML, Smith CD, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Woltjer RL, Younkin SG. Meta-analysis Confirms CR1, CLU, PICALM as Alzheimer Disease Risk Loci and Reveals Interactions With APOE Genotypes. Arch Neurol. 2010;67:1473–1484. doi: 10.1001/archneurol.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriazis GA, Wei Z, Vandermey M, Jo DG, Xin O, Mattson MP, Chan SL. Numb endocytic adapter proteins regulate the transport and processing of the amyloid precursor protein in an isoform-dependent manner: implications for Alzheimer disease pathogenesis. J Biol Chem. 2008;283:25492–25502. doi: 10.1074/jbc.M802072200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De DP, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Lee JH, Cheng R, Barral S, Reitz C, Medrano M, Lantigua R, Jimenez-Velazquez IZ, Rogaeva E, St George-Hyslop PH, Mayeux R. Identification of Novel Loci for Alzheimer Disease and Replication of CLU, PICALM, and BIN1 in Caribbean Hispanic Individuals. Arch Neurol. 2010;68:320–328. doi: 10.1001/archneurol.2010.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HL, Shi SS, Guo QH, Ni W, Dong Y, Liu Y, Sun YM, Bei W, Lu SJ, Hong Z, Wu ZY. PICALM and CR1 Variants are not Associated with Sporadic Alzheimer's Disease in Chinese Patients. J Alzheimers Dis. 2011;25:111–117. doi: 10.3233/JAD-2011-101917. [DOI] [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JS, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, George-Hyslop PS, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, Dekosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piaceri I, Bagnoli S, Lucenteforte E, Mancuso M, Tedde A, Siciliano G, Piacentini S, Bracco L, Sorbi S, Nacmias B. Implication of a Genetic Variant at PICALM in Alzheimer's Disease Patients and Centenarians. J Alzheimers Dis. 2011;24:409–413. doi: 10.3233/JAD-2011-101791. [DOI] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, Harold D, Schrijvers EM, Ramirez-Lorca R, Debette S, Longstreth WT, Jr, Janssens AC, Pankratz VS, Dartigues JF, Hollingworth P, Aspelund T, Hernandez I, Beiser A, Kuller LH, Koudstaal PJ, Dickson DW, Tzourio C, Abraham R, Antunez C, Du Y, Rotter JI, Aulchenko YS, Harris TB, Petersen RC, Berr C, Owen MJ, Lopez-Arrieta J, Varadarajan BN, Becker JT, Rivadeneira F, Nalls MA, Graff-Radford NR, Campion D, Auerbach S, Rice K, Hofman A, Jonsson PV, Schmidt H, Lathrop M, Mosley TH, Au R, Psaty BM, Uitterlinden AG, Farrer LA, Lumley T, Ruiz A, Williams J, Amouyel P, Younkin SG, Wolf PA, Launer LJ, Lopez OL, van Duijn CM, Breteler MM. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleegers K, Lambert JC, Bertram L, Cruts M, Amouyel P, van Broeckhoven C. The pursuit of susceptibility genes for Alzheimer's disease: progress and prospects. Trends Genet. 2010;26:84–93. doi: 10.1016/j.tig.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Teng EL, Chui HC. The Modified Mini-Mental State (3MS) examination. J Clin Psychiatry. 1987;48:314–318. [PubMed] [Google Scholar]
- Yu JT, Song JH, Ma T, Zhang W, Yu NN, Xuan SY, Tan L. Genetic association of PICALM polymorphisms with Alzheimer's disease in Han Chinese. J Neurol Sci. 2010;300:78–80. doi: 10.1016/j.jns.2010.09.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.