Abstract
Outer-membrane TonB-dependent transporters, such as the Escherichia coli ferric citrate transporter FecA, interact with the inner-membrane protein TonB through an energy-coupling segment termed the Ton box. In FecA, which regulates its own transcription, the Ton box is preceded by an N-terminal extension that interacts with the inner membrane protein FecR. Here, site-directed spin labeling was used to examine the structural basis for transcriptional signaling and Ton box regulation in FecA. EPR spectroscopy indicates that regions of the N-terminal domain are in conformational exchange, consistent with its role as a protein binding element; however, the local fold and dynamics of the domain are not altered by substrate or TonB. Distance restraints derived from pulse EPR were used to generate models for the position of the extension in the apo, substrate- and TonB-bound states. In the apo state, this domain is positioned at the periplasmic surface of FecA, where it interacts with the Ton box and blocks access of the Ton box to the periplasm. Substrate addition rotates the transcriptional domain and exposes the Ton box, leading to a disorder transition in the Ton box that may facilitate interactions with TonB. When a soluble fragment of TonB is bound to FecA, the transcriptional domain is displaced to one edge of the barrel, consistent with a proposed β-strand exchange mechanism. However, neither substrate nor TonB displace the N-terminus further into the periplasm. This result suggests that the intact TonB system mediates both signaling and transport by unfolding portions of the transporter.
Keywords: EPR spectroscopy, site-directed spin labeling, membrane protein, β-barrel, protein-protein interactions
In Gram-negative bacteria, the acquisition of iron and other trace nutrients is facilitated by a family of outer-membrane TonB-dependent transport proteins. These TonB-dependent transporters are believed to derive energy by coupling to TonB, an inner membrane protein that acts in a complex with ExbB and ExbD.1; 2; 3; 4; 5; 6; 7; 8 Once transported across the outer membrane, substrate binds to a periplasmic binding protein,9 and is then delivered to a specific ABC import protein on the inner membrane.
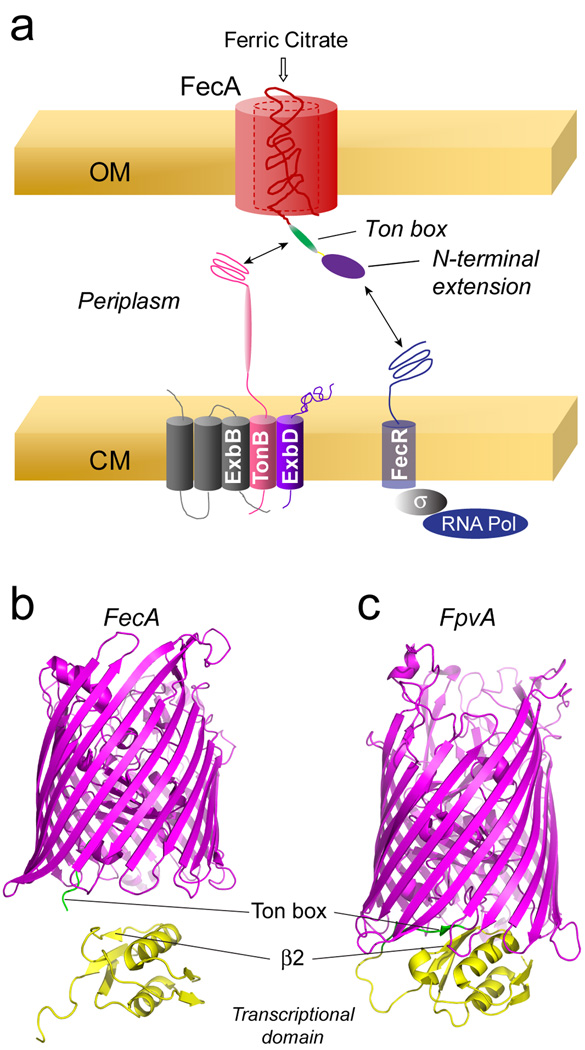
TonB-dependent transporters are structurally homologous. Each is based upon a 22 stranded β-barrel where the N-terminal region of the protein forms a conserved fold within the lumen of the barrel sometimes termed a core or plug. As illustrated in Fig. 1a, the interaction between the transporter in the outer membrane and TonB on the inner membrane is mediated by the Ton box10; 11, a highly conserved energy-coupling segment. In most TonB-dependent transporters, the Ton box is located near the N-terminus. For example in the Escherichia coli vitamin B12 transporter, BtuB, the Ton box includes residues 6–12. However, in a subset of TonB-dependent transporters that regulate their own transcription, the Ton box is preceded by an N-terminal extension or transcriptional signaling domain.6; 12; 13; 14
Figure 1.
a) Model for transcriptional regulation of the ferric citrate import operon by FecA, showing the inner membrane TonB, ExbB, ExbD complex that drives transport. Crystal structures of two TonB-dependent iron transporters: b) the Escherichia coli ferric citrate receptor, FecA (PDB ID: 1KMO), and c) the pyoverdine receptor, FpvA, from Pseudomonas aeruginosa (PDB ID: 2O5P). For FpvA, one structure in the unit cell is shown where the Ton box (shown in green) is resolved. In this structure the N-terminal transcriptional signaling motif (yellow) interacts with the Ton box through an edge interaction with β2. In FecA, the N-terminal transcriptional signaling motif is not resolved. Shown below the FecA structure is the solution NMR structure of the N-terminal signaling motif (PDB ID: 1ZZV).
The Escherichia coli ferric citrate transporter, FecA, regulates its own transcription, and the Ton box in FecA is composed of residues 80 to 86, where the first 75 residues at the N-terminus form a transcriptional signaling domain. As shown in Fig. 1a, transcription is mediated by an interaction of the N-terminal extension of FecA with FecR on the inner membrane, which in turn activates FecI, a sigma factor that directs the RNA polymerase to the promoter region of the ferric citrate import operon.15; 16; 17 Exactly how this transcriptional regulation is mediated is not understood. As shown in Fig. 1a, FecA and FecR lie in the outer and cytoplasmic membranes, respectively, and the interaction between the two must bridge a 200 Angstrom gap between these two membranes.18 It is not known whether substrate binding or TonB binding alters the conformation and position of the domain or whether other elements of the TonB system facilitate the FecA-FecR interaction.
A high-resolution structural model for FecA is shown in Fig. 1b.19 In this and other FecA crystal structures, the N-terminal transcriptional domain is not resolved, although a structure has been determined by solution NMR for the domain expressed as globular protein (see Fig. 1b).20 The transcriptional domain is resolved in the apo state of a related transporter, FpvA, from Pseudomonas aeruginosa.21; 22; 23 FpvA is a transporter for pyoverdine, a siderophore that binds iron with high affinity and competes with tranferrin for iron in mammals.24 Like FecA, FpvA also up-regulates its own expression upon the binding of its substrate. In one structure within the unit cell of the FpvA crystal, β-strand 2 in the transcriptional domain interacts with the Ton box in an edge-to-edge manner (see Fig. 1c). Based upon this model, it is proposed that substrate binding promotes an interaction between TonB and the Ton box of FpvA, which subsequently displaces the transcriptional motif from the transporter.21 In FecA, the binding of substrate is observed to disorder the Ton box,25 and this order-to-disorder transition might trigger the interaction between FecA and TonB.
In the present work, we use site-directed spin labeling (SDSL) along with continuous wave and pulse EPR to determine the position and dynamics of the N-terminal transcriptional domain in FecA. EPR spectroscopy will reveal local dynamics, structure and structural changes,26 and it is particularly well suited to verify membrane protein structure. Moreover, EPR spectroscopy is one of the best methods to examine equilibria between conformational substates in proteins.27;28;29 In FecA, EPR spectra from the N-terminal transcriptional domain show that the structure of the domain is not altered by substrate; however, regions of the domain are in conformational exchange between substates. In lipid bilayers, the domain is positioned so that the second β-strand from the N-terminus (β2) may interact with the Ton box. The binding of substrate to FecA rotates the domain and exposes the Ton box. When TonB interacts with FecA, this interaction is also disrupted and there is a change in the position of the domain, consistent with a proposed β-strand exchange mechanism. However, relative to the periplasmic surface of the outer membrane, the transcriptional domain is not displaced further into the periplasmic space. The results provide a plausible mechanism for the regulation of the FecA-TonB interaction, but the results also suggest that the TonB-energy transduction system must act to unfold portions of the transporter, thereby acting to both drive transport and bring the N-terminal transcriptional domain into proximity of the inner membrane receptor FecR.
Results
Substrate addition does not alter the fold of the N-terminal signaling domain
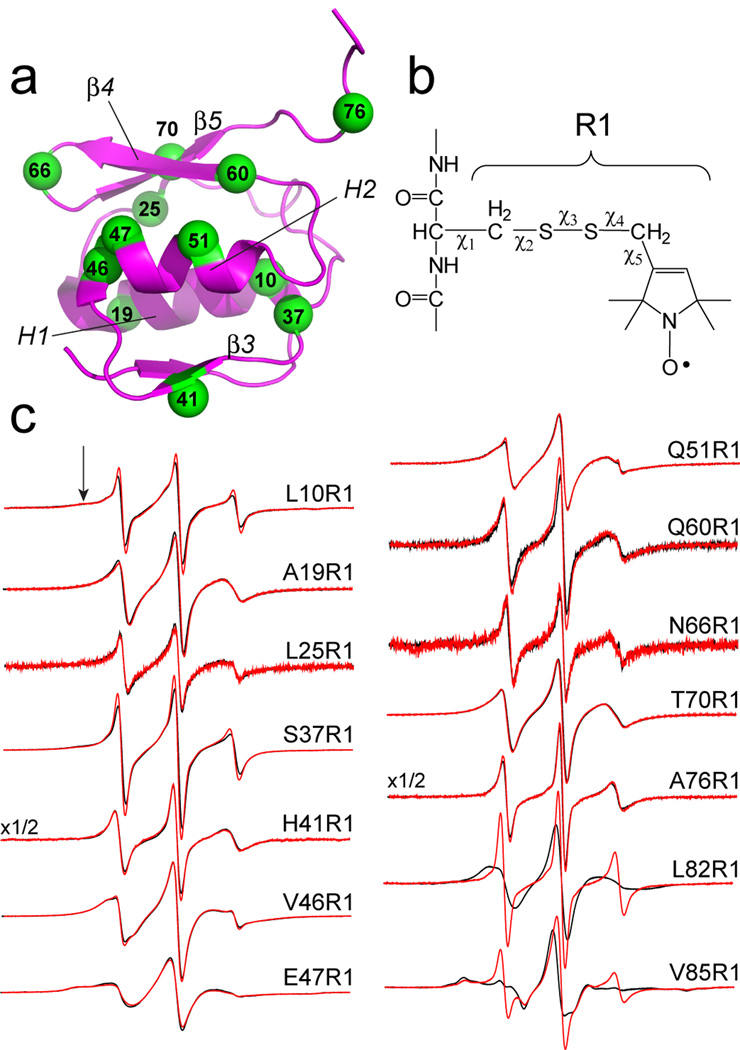
Upon the addition of substrate, the Ton box of FecA was previously shown to undergo an order-to-disorder transition.25 To determine whether the fold of the N-terminal transcriptional motif is altered by substrate binding, EPR spectra were recorded for single labeled sites in the N-terminus to which the nitroxide side-chain, R1, was incorporated (see Fig. 2a, b). The spectra shown in Fig. 2c are consistent with the structure of the N-terminal domain. As expected labels in the more ordered regions of the domain, 46R1 and 47R1 on H2, and 70R1 on β5, yield the broadest spectra, while labels in loops, 37R1 (β2–β3 connector), 66R1 (β4–β5 connector) and 76R1 (on the segment N-terminal to the Ton box), produce narrower spectra, reflecting isotropic label motion with correlation times on the order of 1 ns. EPR spectra are shown for twelve sites in the N-terminal domain in Fig. 2c, and addition substrate produces no significant change in any of the EPR spectra. These results indicate the N-terminal extension is correctly folded, but that the fold and dynamics of the extension is not altered by substrate binding. For comparison, EPR spectra are also shown from sites 82 and 85 in the Ton box in the presence and absence of substrate, which reflect the EPR lineshape changes associated with the substrate-induced disorder transition in the Ton box.
Figure 2.
EPR spectroscopy of the N-terminal transcriptional domain. a) The N-terminal domain of FecA indicating the α-carbons (in green) of sites to which the spin-labeled side chain, R1, has been attached. b) Engineered cysteine residues are reacted with the MTSL spin label to produce spin-labeled side chain R1. On exposed helical sites, the EPR spectra are largely affected by backbone motion and by rotation about χ4 and χ5.31 c) EPR spectra obtained from 12 sites in the N-terminal domain, and two sites in the FecA Ton box (L82R1 and V85R1). The two spectra from the Ton box were published previously25 and are shown for comparison.
All but one of the spectra shown in Fig. 2c are obtained from outward facing sites, and are therefore unlikely to significantly perturb the fold of the domain.30 The one exception is L10R1, which faces the interior of the domain. The spectrum for a label facing the protein interior should reflect a highly immobilized nitroxide, but instead the spectrum of L10R1 is composed of two motional components, one which is typical of R1 attached to a loop or unfolded protein segment and a second characteristic of R1 that is restricted by tertiary contact (arrow in Fig 2c). EPR spectra characteristic of flexible or unstructured protein segments were obtained from other inward facing sites on this domain (see Fig. 1 Supplementary Material), suggesting that R1 destabilizes the N-terminal domain when incorporated into its interior. Since well-folded proteins tolerate the incorporation of R1,30 this observation suggests that the fold of the N-terminal domain is only marginally stable.
The EPR spectra shown in Fig. 2c indicate that the domain may have significant backbone motion on the ns time-scale. EPR spectra for the R1 side chain at surface exposed helical sites have been well-characterized,31 and two such sites in the N-terminal domain are 19R1 and 51R1. These spectra may be simulated using the MOMD model developed by Freed and co-workers32 with parameters used previously for aqueous exposed helical sites.31 In the case of 51R1, the spectrum is fit well with two motional components, where the major component (approximately 90% of the spins) has a correlation time of about 1.7 ns and order parameter that is approximately half that at a well-ordered helical site. The spectrum for 19R1 is also well modeled with two motional components, where the major component (approximately 90%) is simulated by motion that is also about 1.7 ns and nearly isotropic (see Supplementary Material, Fig. 2). The motional models that fit these spectra indicate these helices have a high-degree of motion on the ns time-scale,33 and that the domain is not tightly folded. These are properties that would be expected for a domain designed for protein-protein interactions.
Regions of the N-terminal signaling motif are in conformation exchange
Many of the spectra in Fig. 2c appear to arise from nitroxide side chains executing more than one mode of motion. Multiple modes of R1 motion may result from different label rotamers or from different protein structural substates. One approach to distinguish between these two possibilities is to test the spectra for sensitivity to stabilizing osmolytes, such as sucrose or a polyethylene glycol (PEG).25;27; 28 Osmolytes appear to be preferentially excluded from the protein surface, thereby raising the energy of the protein.34; 35; 36; 37 In the case where two conformers are in equilibrium, osmolytes will lower the energy of the least hydrated, or more compact, protein form.38 In contrast to protein equilibria, the conversion between label rotamers is not sensitive to the addition of osmolytes.28
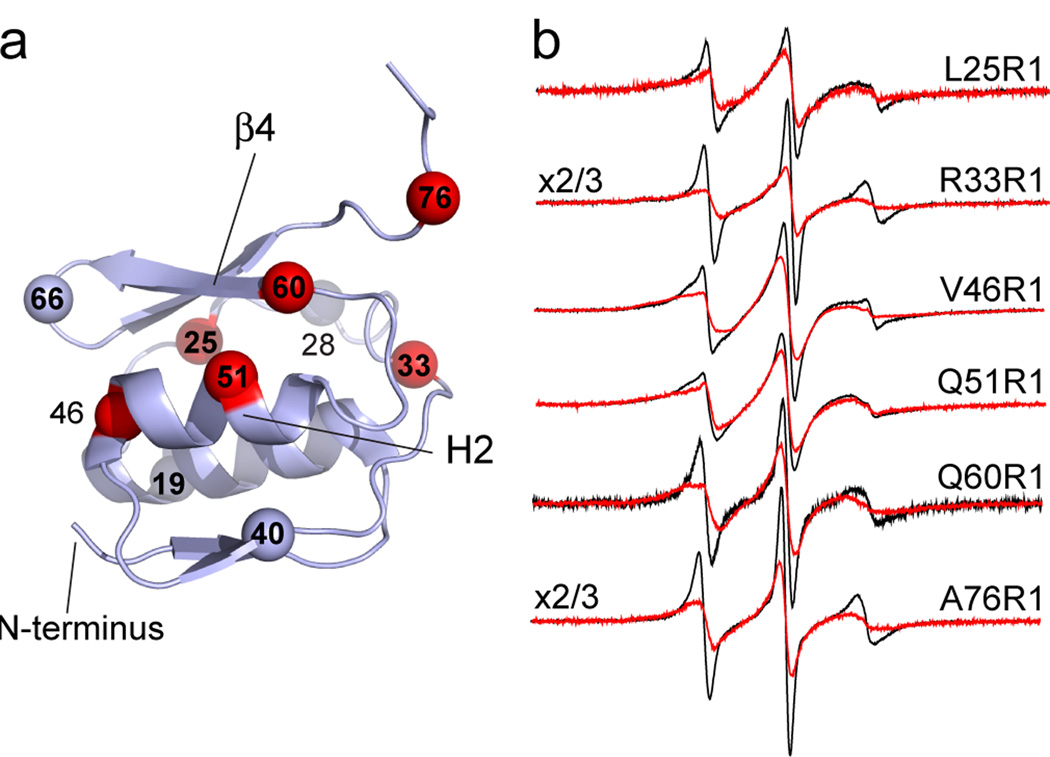
Shown in Fig. 3 are EPR spectra from the N-terminal transcriptional domain in the absence and presence of PEG 3350. Of the ten sites examined, six are sensitive to PEG addition while 4 are not. At these 6 sites, PEG addition shifts the populations of spins to the less mobile state. These changes indicate a shift to a more compact protein state and the existence of a conformational equilibrium at these sites. For sites 33 and 76, which lie in flexible loop regions, conformational exchange is expected. For the N-terminal domain, evidence for exchange in H2 and in β4 suggests that this face of the protein may be converting between two or more conformers.
Figure 3.
Effect of osmolytes on EPR spectra from the N-terminal domain. a) The N-terminal transcriptional domain showing the α-carbons of sites where evidence for conformational exchange is observed in red. Sites where conformational exchange is not observed are shown in gray. b) EPR spectra from 6 sites in the absence (black traces) and presence (red traces) of 25% w/v PEG 3350.
Distance measurements with pulse EPR define a position for the FecA transcriptional signaling motif in bilayers
The position of the N-terminal transcriptional domain is not resolved in high-resolution crystal structures of FecA. To determine the position and orientation of this domain in FecA, double electron-electron resonance, DEER, was used to estimate distances between pairs of R1 labels, where one label is incorporated into the N-terminal signaling domain and a second is incorporated into the Ton box or the FecA barrel. The DEER experiment produces a time-dependent spin-echo, where the strength of the dipolar interaction is encoded as a frequency in the time-dependent spin-echo.39 This signal yields both a distance and a distance distribution between the dipolar coupled spin pairs (see Experimental). The DEER experiment produces a signal from excited spin pairs, and unlabeled or single-labeled protein do not contributed to the DEER signal.
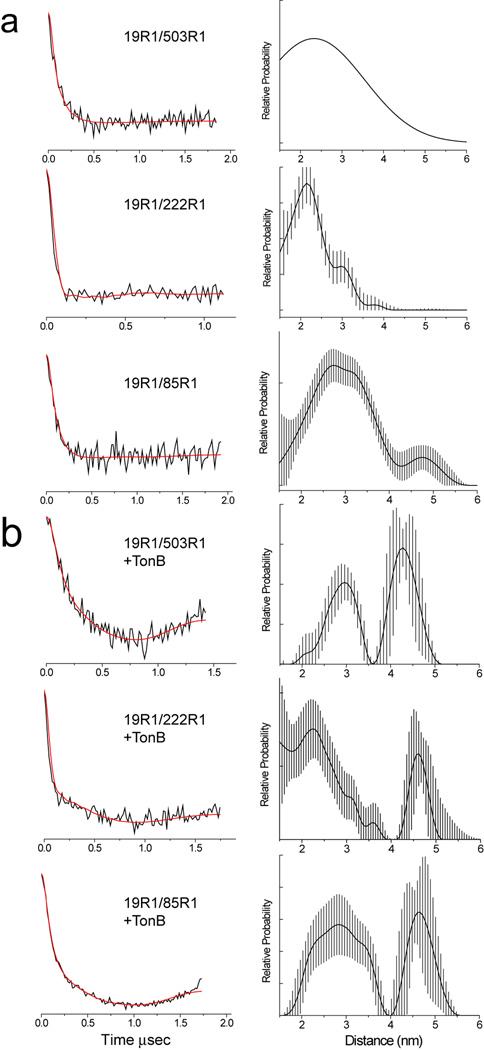
Shown in Fig. 4 are examples of DEER data obtained for pairs of labels where one label is at position 19R1 in the N-terminus and a second is at 85R1 in the Ton box, or at 222R1 and 503R1 in the barrel. The fits to the data are shown as red traces, and the distances and distributions corresponding to these fits are shown on the right side of Fig. 4. These data were obtained for FecA which was reconstituted into DLPC and diluted 1:3 with wild-type protein (see Methods). Both the incorporation of wild-type protein and reconstitution into DLPC extended the R1 phase memory times and improved signal-to-noise. These DEER data are typical of those obtained when distances to the transcriptional motif are measured. The signals are highly dampened and show little oscillation, indicating that there is a broad distribution of distances. For the three pairs shown, there is a major distance in the 2 to 3 nm range, but minor longer distance components are also present. These data indicate that in bilayers, the transcriptional domain does not assume a highly defined position, but samples multiple positions relative to the barrel of FecA.
Figure 4.
Dipolar evolution data and corresponding distance distributions for several spin pairs in FecA in a) the apo state and b) when bound to TonBΔTM. The red traces represent the best fits to the dipolar evolution data for the corresponding distributions. The error bars on the distribution are estimates using DeerAnalysis 2011 and are a result of uncertainty in the background correction. Two spin pairs are shown where one label is in the N-terminal signaling domain and a second in the FecA barrel (19R1/503R1, 19R1/222R1) and where one label is in the signaling domain and a second in the FecA Ton box (19R1/85R1). The fit shown for the spin pair 19R1/503R1 in the apo form was fit with a single Gaussian. It should be noted that substrate binding is not necessary to mediate binding between FecA and TonB under the conditions of this experiment (Freed and Cafiso, unpublished). The most populated distances obtained from these measurements are shown in Table 1.
Shown in Fig. 4b are data obtained from the same three spin pairs shown in Fig. 4a following the addition of the TonB fragment (33–239) (or TonBΔTMD). In each case, the DEER signals change and the fits indicate the emergence of a longer distance component, which is significantly populated. This TonB-induced component is 1 to 2 nm longer than the major component obtained in the absence of TonB. A summary of the most populated distances obtained and width of the distributions in the apo, TonB and substrate bound states is shown in Table 1.
Table 1.
The most probable distances and distance distributions (in Angstroms) obtained using DEER for double labeled mutants of FecA reconstituted into DLPC.†
| FecA Mutant | Apo | + Ferric citrate | + TonB (33–239) |
|---|---|---|---|
| A19R1/V85R1 | 24 ± 12 | 22.8 ± 3.6 | 28 ± 5.3 |
| 41.6 ± 3.3 | 48.8 ± 4 | ||
| A85R1/N506R1 | 20 ± 5 | ||
| A19R1/G259R1 | 30 ± 15 | ||
| A19R1/K353R1 | 37 ± 13 | ||
| A19R1/N506R1 | 28 ± 12 | ||
| R33R1/G259R1 | 23 ± 6 | 22 ± 4.2 | 22 ± 4.2 |
| 43.7 ± 2.5 | 43.7 ± 2.5 | ||
| N66R1/K353R1 | 21 ± 14 | 23.9 ± 3.9 | 24.9 ± 5.7 |
| 37.1 ± 3.9 | 42.1 ± 2.3 | ||
| A19R1/D222R1 | 22 ± 5 | 21.8 ± 4.8 | 23.6 ± 5.8 |
| 38.5 ± 4.3 | 46.9 ± 3.9 | ||
| A19R1/D503R1 | 23 ± 14 | 26.8 ± 4.3 | 29.3 ± 3.6 |
| 41.4 ± 2.6 | 44.8 ± 4.2 |
Distances were determined using the program DEER Analysis (see Methods) and are given as r ± σ(r), where σ(r) is the standard deviation in the distance distribution. For distance less than 30 Å, the uncertainty in the most probable distance is less than 2 A with an uncertainty in the distribution of about 20%. The longer distances near 40 Å have a larger uncertainty of 3 to 5 Å and an uncertainty in the distribution of about 40%. Each measurement was repeated at least twice and gave similar results.
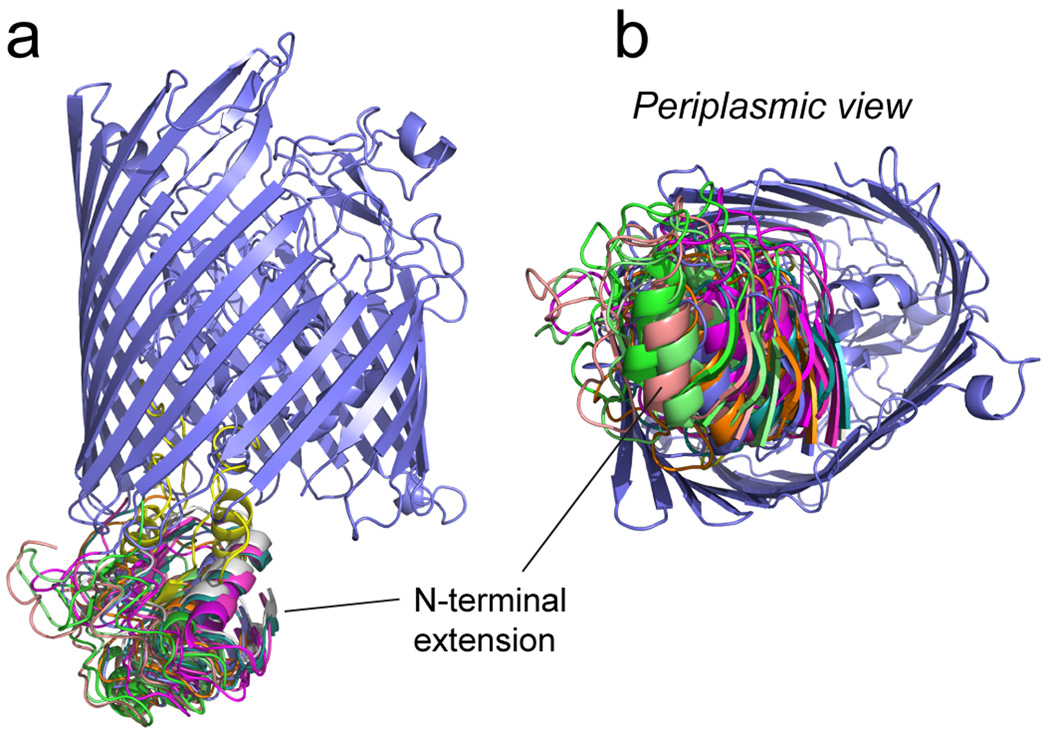
The distances and distributions shown in Table 1 were used in combination with simulated annealing to obtain a position for the N-terminal transcriptional domain in the apo, TonB and substrate bound states. These models were generated using the crystal structure of FecA, the solution NMR structure of the N-terminal extension and Xplor-NIH40; 41 to incorporate the EPR restraints (see Methods). A family of structures obtained using the EPR restraints in the apo state is shown in Fig. 5. This figure displays the 10 lowest energy structures aligned by the FecA barrel (which was invariant in the simulation). Although this group of structures displays a similar orientation for the N-terminal extension, there is considerable variability among the structures, consistent with the broad distribution in distances. The RMSD for the backbone atoms of the N-terminus when the structures are aligned to the FecA barrel is approximately 7 Angstroms.
Figure 5.
Structure for the apo state of FecA obtained by incorporating EPR-derived distance restraints into XPLOR-NIH. As indicated in Methods, the docking of the N-terminal extension obtained by NMR (PDB ID: 1ZZV) was performed to the crystal structure of FecA, (PDB ID: 1KMO). The ten lowest energy structures are shown in various colors, where each structure is aligned against the FecA barrel (purple).
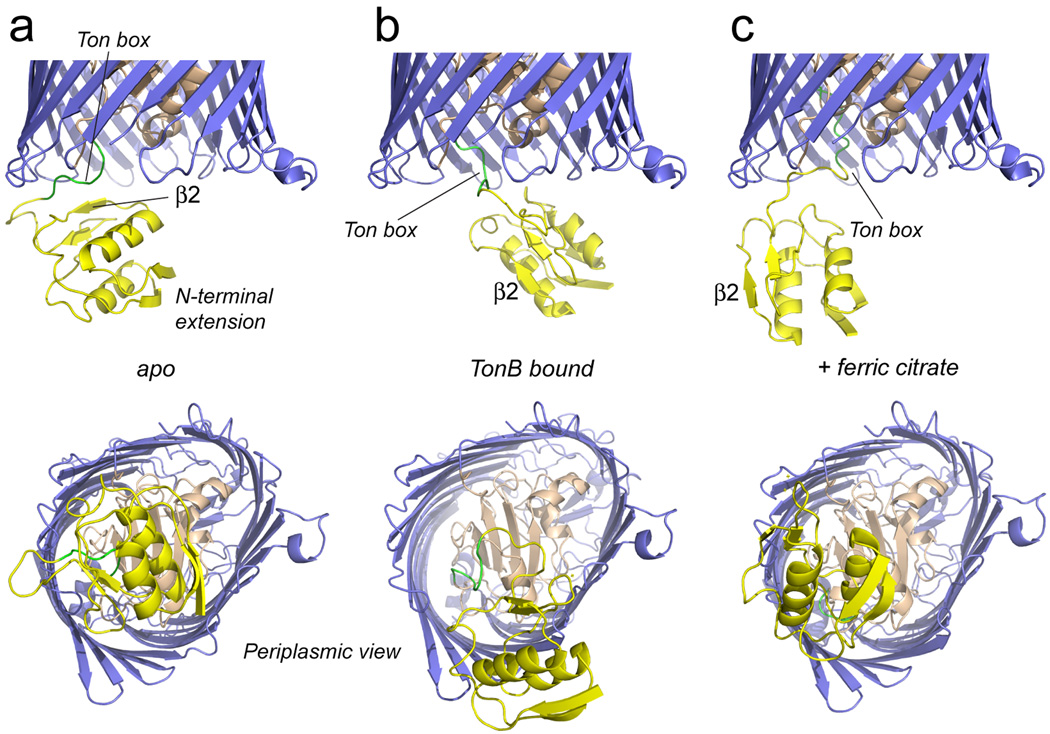
Shown in Fig. 6a is the lowest energy structure for FecA in the apo state. In this structure, the N-terminal domain has an orientation similar to that assumed in the FpvA structure (Fig. 1b), and strand 2 is in a position to interact closely in an edge-to-edge manner with the Ton box as seen for FpvA.21 A double FecA mutant, where one label is at position 26 in strand 2 and second is at position 80 in the Ton box, yields an EPR spectrum showing evidence for strong dipolar coupling (see Fig. 3 Supplementary Material), consistent with this structure. It should be noted that the structure of the region near the Ton box is underdetermined, since there are few restraints to define its position.
Figure 6.
Average (lowest energy) structures obtained for FecA, where the N-terminal extension (yellow) has been docked to the FecA barrel and hatch region using EPR derived distance restraints and XPLOR-NIH. Shown in a) is the complete apo structure of FecA, in b) is shown the structure when complexed to TonB (33–259) and is c) is shown the structure in the presence of the substrate, ferric citrate (appended to 1KMN). Substrate produces a rotation of the domain, so that β2 is disengaged from the Ton box. TonB binding displaces the N-terminal extension from the central region on the periplasmic surface of the transporter. In each case, the signaling domain is not displaced further into the periplasm.
The N-terminal domain is not as closely associated with the FecA barrel as is the domain in the crystal structure of FpvA, and this less intimate association is consistent with the EPR spectra from FecA. For example, the EPR spectra from 19R1 reflect a side chain that is exposed and not in tertiary contact with other regions of the protein. In the FpvA structure, the homologous site (R64) is buried and in contact with sites in the barrel. Previous spectra obtained from the FecA Ton box indicate that the Ton box has a considerable degree of backbone motion, and is not rigidly fixed by an interaction with β2 in the N-terminal domain.25 The broad distance distributions seen in Table 1 are consistent with these earlier data. Thus, although there are remarkable similarities between the model obtained here for FecA and the FpvA structure, in bilayers the N-terminal domain in FecA exhibits significant heterogeneity in its position.
The binding of TonBΔTMD or substrate alters the position of the signaling motif and is consistent with a strand-exchange mechanism
When TonBΔTMD is added to the spin labeled FecA samples, distances measured between the N-terminus and spin-labeled sites in the barrel increase substantially. Longer distance components are also seen upon the addition of the ferric citrate substrate (see Table 1). Shown in Fig. 6b is the structure obtained by simulated annealing using the longer distance constraints that are induced by the binding of TonB. Comparison to the model obtained without TonB indicates that the N-terminal domain has been displaced from the center towards the side of the barrel, so that the loop connecting H2 and β4 is located near the turn connecting barrel strands 11 and 12. In this structure, the N-terminal domain is no longer closely associated with the Ton box, and is consistent with previous models obtained for BtuB and FhuA, which indicate that TonB interacts in an edge-to-edge fashion with the Ton box.10; 11
Figure 6c shows a model obtained using the longer distance constraints obtained in the presence of substrate, ferric citrate. In this model, the N-terminal transcriptional domain has rotated so that β-strand 2 of the domain has disengaged from the Ton box. This rotation will free the Ton box and expose it to the periplasmic space, although it does not displace the Ton box into the periplasm. The model shown in Fig. 6c also accounts for the increase in backbone motion observed previously in the FecA Ton box upon substrate addition.25 It should be noted that neither substrate nor TonB displace the N-terminal signaling domain further into the periplasmic space.
Discussion
Substrate binding to FecA is believed to have two consequences. First, the inner membrane protein TonB engages the transporter and drives the movement of ferric citrate across the outer membrane. Second, substrate binding induces the transcription of genes on the ferric citrate uptake operon, a process that is mediated by an interaction between the N-terminal extension of FecA and the inner membrane protein FecR. To determine how substrate or TonB initiate these protein-protein interactions, the position of the N-terminal extension relative to the FecA barrel was examined in the apo state, and in the presence of either substrate or a TonB fragment lacking its transmembrane segment (TonBΔTMD).
EPR-derived distance restraints and simulated annealing yield a model for the apo state of FecA where the N-terminal transcriptional domain is in a position to interact with the Ton box through the second β-strand in the domain (Fig. 6a). While this structure resembles one of the crystal structures obtained for FpvA,21 pulse EPR data indicate that there is considerable conformational heterogeneity in the position of the N-terminal extension relative to the FecA barrel. This finding is consistent with EPR lineshapes obtained from the FecA Ton box,25 and it may explain why the N-terminal extension is not resolved in any crystal structures of FecA. As shown previously, addition of substrate enhances disorder in the Ton box,25 and the measurements made here indicate that this disorder transition is accompanied by a change in orientation of the N-terminal extension so that strand 2 is no longer in a position to interact with the Ton box (Fig. 6c). Thus, the loss of tertiary contact between the Ton box and strand 2 apparently accounts for changes seen previously in EPR lineshapes from the Ton box.
The affinity of TonB for TonB-dependent transporters is enhanced in when substrate binds the transporter,42; 43; 44 and this interaction appears to be regulated in different ways in FecA and BtuB (the Escherichia coli vitamin B12 transporter). In BtuB, the Ton box is folded within the interior of the barrel where it is part of the plug or core region of the transporter.45 Substrate binding shifts the equilibrium of the Ton box so that the unfolded state is a dominant population,46 and this unfolded population projects as much as 30 Angstroms into the periplasm.47 This induced unfolding initiates a “fly-casting” mechanism that will promote BtuB-TonB interactions.48 In FecA, the Ton box is sterically blocked from interacting with TonB by the N-terminal extension, and as described above, substrate may promote the interaction with TonB by displacing the extension from the Ton box. Unlike the BtuB Ton box, the FecA Ton box is not displaced into the periplasmic space. But, a dramatic displacement may not be necessary to initiate FecA-TonB interactions since the Ton box in the substrate bound state is disordered and dynamic.
The addition of a soluble form of TonB (33–259) produces a significant shift in the position of the N-terminal extension (Fig. 6b); however, neither substrate nor TonB displace a significant population of the N-terminus further into the periplasmic space. How then is transperiplasmic signaling in FecA accomplished? The results obtained here indicate that the N-terminal extension is in conformational equilibrium between two or more structural forms. Dynamics is often characteristic of regions of proteins that mediate interactions,49 and this observation indicates that the N-terminal extension of FecA may be primed to mediate a protein-protein interaction. Changes in orientation of the N-terminal domain, such as those seen here in the presence of substrate (Fig. 6c) or TonB (Fig. 6b), might place the domain in a geometry that will increase the likelihood of collisions with FecR and promote the FecA/FecR interaction. However, an obvious obstacle to this interaction is the 200 Angstrom gap between the inner and outer membrane that must be spanned in Escherichia coli.18 It is conceivable that this interaction might be mediated in regions where the inner and outer membranes come into close proximity and form adhesion sites (termed Bayer Bridges); however, the existence of these structural features is controversial, and there is evidence indicating that these features are an artifact of the fixation techniques used in microscopy.50
In FecA, substrate transport can occur independently of transcriptional regulation of the ferric citrate import genes, however, transcriptional regulation requires FecA and the TonB, ExbB, ExbD proteins.15; 16 One proposed action of TonB in transport is an energy-dependent unfolding of the interior hatch or core region of TonB-dependent transporters,51 and recent work has shown that in BtuB a portion of this interior domain, perhaps involving 60 or 65 residues, may be reversibly unfolded while maintaining the structure of other regions of the transporter.52 This type of unfolding would be sufficient to extend the N-terminal transcriptional domain and promote interactions with FecR. Although we have no direct evidence for this mechanism, the activity of the intact TonB/ExbB/ExbD system could facilitate both transport and transcriptional regulation by unfolding a portion of the core or plug domain on the interior of the transporter.
In summary, using site-directed spin labeling and continuous wave and pulse EPR spectroscopy, we determined a position for the N-terminal extension of FecA, which is not resolved in any available crystal structures. EPR spectroscopy is particularly well-suited to examine dynamic regions of proteins that are difficult to study but play important roles in mediating protein-protein interactions. In FecA, the addition of substrate releases the N-terminal extension from the Ton box exposing the Ton box to the periplasmic space where it may interact with TonB. The addition of a TonB fragment displaces the N-terminal domain to the edge of the barrel, consistent with a previously proposed strand-exchange mechanism based upon the structure of FpvA. However, neither substrate nor TonB significantly displace the transcriptional domain further into the periplasmic space, suggesting that additional structural changes, such as a partial unfolding of the core or hatch region of FecA (driven by the TonB, ExbB, ExbD complex), facilitate transcriptional regulation of the ferric citrate import genes.
Experimental Procedures
Materials
The sulfhydryl-reactive spin-label methanethiosulfonate, S-(1-oxy-2,2,5,6-tetramethylpyrroline-3-methyl)methanethiosulfonate (MTSL), was purchased from Toronto Research Chemicals (Ontario, Canada). DTT (dl-dithiothreitol) was obtained from Sigma (St. Louis, MO) and sarkosyl was from Fisher Chemical Co. (Pittsburgh, PA). 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine was purchased from Avanti Polar Lipids (Alabaster, AL).
Mutagenesis, expression and purification for FecA
The plasmid harboring wild-type FecA, pIS711,16 was generously provided by Volkmar Braun (University of Tübingen, Tübingen, Germany). The mutations were introduced into FecA using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), and was subsequently verified by nucleotide sequencing. Expression, purification and reconstitution of FecA into vesicle bilayers followed a procedure described previously.25 In the present case, all FecA mutants were spin-labeled prior to purification by ion-exchange chromatography. For labeling, 30 mL of supernatant were concentrated to 10 mL and 200 µL of 16mM MTSL stock solution of MTSL was added to the mixture. The reaction was allowed to proceed overnight at room temperature before purification by HPLC. Each labeled mutant was pure as judged by SDS PAGE, and each appeared to be correctly folded. The labeled mutants exhibited a behavior similar to that of wild-type protein upon purification and reconstitution into POPC bilayers, and the EPR spectra of labels used for distance measurements were consistent with the local structure at the labeled site. From the amplitudes or modulation depths in the DEER signals (see below), the labeling efficiency was 70% or greater.
Cloning, expression and purification of TonB
The TonB fragment lacking the transmembrane segment (33–239 or TonBΔTMD) was obtained from Robert Nakamoto (Molecular Physiology, Univeristy of Virginia) and coded into a pHIS-parallel1 vector.53 The plasmid encoding TonBΔTMD was then transformed into T7 Express lysY/Iq competent cells (New England Biolabs, Ipswich, MA). The cells were grown in 2xYT media at 37°C until the O.D. at 600 nm was approximately 0.6. The cells were then induced with 0.8mM IPTG, allowed to grow for an additional 6 hours at 25°C and then centrifuged at 3000 g for 20 min. at 4°C.
To limit proteolysis, the purification of TonBΔTMD was performed on ice. The pellet was resuspended in 20mL of resuspension buffer (25mM Tris pH 7.0 with 10% glycerol) to which protease inhibitor AEBSF (20mg/400µL, 200µL per 1L) was added. The sample was homogenized by hand and cells were disrupted by passing the sample three times through a French Press. Following 15 min centrifugation at 10,500g, the supernatant was retained and loaded to a His-Trap column equilibrated in equilibration buffer (25mM Tris, 300mM NaCl and 20mM Imidazole). The bound protein was eluted with 20–30 ml elution buffer (25mM Tris, 300mM NaCl and 250mM Imidazole), the fractions checked using SDS-PAGE, and the pure fractions of TonB pooled. The sample was then concentrated by ultrafiltration using a 3 kDa molecular-weight cut-off membrane to yield approximately 1mM of a 150 µL solution.
Electron Paramagnetic Resonance measurements
EPR spectroscopy was performed on a continuous wave X-band EMX spectrometer (Bruker Biospin, Billerica, MA) equipped with a ER 4123D dielectric resonator. All EPR spectra were recorded with a 100 G magnetic field sweep, 1 G modulation, 2.0 mW incident microwave power at a temperature of 298 K. The measurements were performed on 6 µL samples in glass capillary tubes 0.60 ID × 0.84 OD round capillary (VitroCom, Mountain Lakes, NJ). The phasing, normalization, and subtraction of EPR spectra was performed using LabVIEW software provided by Dr. Christian Altenbach (University of California, Los Angeles, California).
Pulse EPR experiments were preformed on a Bruker Elexsys E580 EPR spectrometer running either at X or Q band. Two to three measurements were carried out on each sample, and at least one measurement at Q-band was carried out on each sample. Measurements at X-band used either an ER4118X-MS3 or -MS2 split-ring resonator. Measurements at Q-band used an EN5107D2 dielectric resonator. Pulse-EPR measurements were performed on 20–25 µL of sample loaded into quartz capillaries with 2 ID × 2.4 OD (Fiber Optic Center, Inc. New Bedford, MA). Spin-labeled samples of double-labeled FecA mutants had protein concentrations that varied from 150 to 250 µM.
DEER samples were flash frozen in liquid nitrogen, and the data were recorded at 80 K. Data in pulse mode were acquired using a four-pulse DEER sequence54 with a 16-ns π/2 and two 32-ns π observe pulses separated by a 32-ns π pump pulse. The dipolar evolution times were typically 1.2 to 2.0 µs. The pump frequency was set to the center maximum of the nitroxide spectrum and the observer frequency was set to the low-field maximum, typically 15–25 MHz higher.
The phase-corrected dipolar evolution data were processed and distance distributions determined using either Gaussian fitting or Tikhonov regularization incorporated into the DeerAnalysis2011 software package.55 This program also contains an error analysis routine that was used to assess the effect of the background subtraction upon the distance distribution. The phase memory time of FecA reconstituted in different lipids such as POPC, DMPC and DLPC was studied between 5K and 100K. FecA reconstituted in DLPC produced the longest phase memory times; and as a result, DLPC was chosen for the reconstitution. The spin-labeled protein was also diluted with wild-type protein in a 1:3 ratio (protein:wild type). This approach was used previously for pulse EPR experiments on BtuB,47 and was found to improve the phase memory times for FecA by reducing undesired intermolecular dipolar interactions.
The FecA substrate, diferric dicitrate, is paramagnetic, and pulse EPR experiments in the presence of substrate lead to shortened relaxation times. This limited the echo time that could be used in these experiments, and hence limited the accuracy and distance range of our measurements. The effect of the diferric dicitrate could be minimized by maintaining FecA and substrate near stoichiometrically equivalent levels.
Generation of FecA models from distance restraints and simulated annealing
Models for the position of the N-terminal extension relative to the FecA barrel were generated using Xplor-NIH40; 41 in a similar manner to that described previously.56 Briefly, the spin-label side chain R1 was appended to the structure at appropriate locations and the first two dihedral angles for the side chain, χ1 and χ2, were set to −60°. A linker was manually constructed between the residue 79 of the NMR structure of the N-terminal extension and residue 80 from the Ton box of the crystal structure of FecA. A simulated annealing routine was performed without any experimental restraints to allow other side-chain atoms to find energetically reasonable conformations. The result of this procedure was the starting structure for the simulation which fixed spin-label side-chain conformations, backbone atoms of the N-terminal domain (residues 1 to 71) and residues in FecA from (88 to 741). The major distances obtained from analysis of the DEER data were chosen as restraints and included a distribution that represented 2/3 of a standard deviation in the distribution. Simulated annealing was performed using the program Xplor-NIH with restraints based on DEER distances between the barrel and N-terminus, between the Ton box and the barrel and between the Ton box and the N-terminus. Structures were visualized and analyzed with the program PyMOL (DeLano Scientific LLC, Palo Alto, CA).
Supplementary Material
Highlights.
FecA controls its own transcription by regulating FecA-FecR interactions
Models for the FecA N-terminal domain were obtained in apo and ligand-bound states
Substrate and TonB binding dissociate the domain from the FecA Ton box
Changes in the domain position indicate how FecA-protein interactions are regulated
Acknowledgements
LabVIEW programs that were used to process and simulate EPR spectra were generously provided by Dr. Christian Altenbach (UCLA). This work was supported by a grant from the National Institutes of Health, NIGMS, GM 035215.
Abbreviations used
- DEER
double electron-electron resonance
- DLPC
dilauroylphosphatidylcholine
- DMPC
dimyristoylphosphatidylcholine
- EPR
electron paramagnetic resonance
- MTSL
methanethiosulfonate spin label
- C8E4
n-octyl tetraoxyethylene
- PC
phosphatidylcholine
- POPC
palmitoyloleoylphosphatidylcholine
- R1
spin-labeled side chain produced by derivatization of a cysteine with MTSL
- SDSL
site-directed spin labeling
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moeck GS, Coulton JW. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol Microbiol. 1998;28:675–681. doi: 10.1046/j.1365-2958.1998.00817.x. [DOI] [PubMed] [Google Scholar]
- 2.Klebba PE, Newton SM. Mechanisms of solute transport through outer membrane porins: burning down the house. Curr Opin Microbiol. 1998;1:238–247. doi: 10.1016/s1369-5274(98)80017-9. [DOI] [PubMed] [Google Scholar]
- 3.Faraldo-Gomez JD, Sansom MS. Acquisition of siderophores in gram-negative bacteria. Nat Rev Mol Cell Biol. 2003;4:105–116. doi: 10.1038/nrm1015. [DOI] [PubMed] [Google Scholar]
- 4.Postle K, Kadner R. Touch and go: tying TonB to transport. Mol. Microbiol. 2003;49:869–882. doi: 10.1046/j.1365-2958.2003.03629.x. [DOI] [PubMed] [Google Scholar]
- 5.Wiener MC. TonB-dependent outer membrane transport: going for Baroque? Curr Opin Struct Biol. 2005;15:394–400. doi: 10.1016/j.sbi.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schauer K, Rodionov DA, de Reuse H. New substrates for TonB-dependent transport: do we only see the 'tip of the iceberg'? Trends Biochem Sci. 2008;33:330–338. doi: 10.1016/j.tibs.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Krewulak KD, Vogel HJ. TonB or not TonB: is that the question? Biochem Cell Biol. 2011;89:87–97. doi: 10.1139/o10-141. [DOI] [PubMed] [Google Scholar]
- 9.Quiocho FA, Ledvina PS. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol Microbiol. 1996;20:17–25. doi: 10.1111/j.1365-2958.1996.tb02484.x. [DOI] [PubMed] [Google Scholar]
- 10.Pawelek PD, Croteau N, Ng-Thow-Hing C, Khursigara CM, Moiseeva N, Allaire M, Coulton JW. Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science. 2006;312:1399–1402. doi: 10.1126/science.1128057. [DOI] [PubMed] [Google Scholar]
- 11.Shultis DD, Purdy MD, Banchs CN, Wiener MC. Outer membrane active transport: structure of the BtuB:TonB complex. Science. 2006;312:1396–1399. doi: 10.1126/science.1127694. [DOI] [PubMed] [Google Scholar]
- 12.Schalk IJ, Yue WW, Buchanan SK. Recognition of iron-free siderophores by TonB-dependent iron transporters. Mol Microbiol. 2004;54:14–22. doi: 10.1111/j.1365-2958.2004.04241.x. [DOI] [PubMed] [Google Scholar]
- 13.Koebnik R. TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol. 2005;13:343–347. doi: 10.1016/j.tim.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Braun V, Endriss F. Energy-coupled outer membrane transport proteins and regulatory proteins. Biometals. 2007;20:219–231. doi: 10.1007/s10534-006-9072-5. [DOI] [PubMed] [Google Scholar]
- 15.Harle C, Kim I, Angerer A, Braun V. Signal transfer through three compartments: transcription initiation of the Escherichia coli ferric citrate transport system from the cell surface. EMBO J. 1995;14:1430–1438. doi: 10.1002/j.1460-2075.1995.tb07129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim I, Stiefel A, Plantor S, Angerer A, Braun V. Transcription induction of the ferric citrate transport genes via the N-terminus of the FecA outer membrane protein, the Ton system and the electrochemical potential of the cytoplasmic membrane. Mol Microbiol. 1997;23:333–344. doi: 10.1046/j.1365-2958.1997.2401593.x. [DOI] [PubMed] [Google Scholar]
- 17.Enz S, Mahren S, Stroeher UH, Braun V. Surface signaling in ferric citrate transport gene induction: interaction of the FecA, FecR, and FecI regulatory proteins. J Bacteriol. 2000;182:637–646. doi: 10.1128/jb.182.3.637-646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matias VR, Al-Amoudi A, Dubochet J, Beveridge TJ. Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J Bacteriol. 2003;185:6112–6118. doi: 10.1128/JB.185.20.6112-6118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferguson AD, Chakraborty R, Smith BS, Esser L, van der Helm D, Deisenhofer J. Structural basis of gating by the outer membrane transporter FecA. Science. 2002;295:1715–1719. doi: 10.1126/science.1067313. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Herrero A, Vogel HJ. Nuclear magnetic resonance solution structure of the periplasmic signalling domain of the TonB-dependent outer membrane transporter FecA from Escherichia coli. Mol Microbiol. 2005;58:1226–1237. doi: 10.1111/j.1365-2958.2005.04889.x. [DOI] [PubMed] [Google Scholar]
- 21.Brillet K, Journet L, Celia H, Paulus L, Stahl A, Pattus F, Cobessi D. A beta strand lock exchange for signal transduction in TonB-dependent transducers on the basis of a common structural motif. Structure. 2007;15:1383–1391. doi: 10.1016/j.str.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 22.Wirth C, Meyer-Klaucke W, Pattus F, Cobessi D. From the periplasmic signaling domain to the extracellular face of an outer membrane signal transducer of Pseudomonas aeruginosa: crystal structure of the ferric pyoverdine outer membrane receptor. J Mol Biol. 2007;368:398–406. doi: 10.1016/j.jmb.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 23.Greenwald J, Nader M, Celia H, Gruffaz C, Geoffroy V, Meyer JM, Schalk IJ, Pattus F. FpvA bound to non-cognate pyoverdines: molecular basis of siderophore recognition by an iron transporter. Mol Microbiol. 2009;72:1246–1259. doi: 10.1111/j.1365-2958.2009.06721.x. [DOI] [PubMed] [Google Scholar]
- 24.Poole K, McKay GA. Iron acquisition and its control in Pseudomonas aeruginosa: many roads lead to Rome. Front Biosci. 2003;8:d661–d686. doi: 10.2741/1051. [DOI] [PubMed] [Google Scholar]
- 25.Kim M, Fanucci GE, Cafiso DS. Substrate-dependent transmembrane signaling in TonB-dependent transporters is not conserved. Proc Natl Acad Sci U S A. 2007;104:11975–11980. doi: 10.1073/pnas.0702172104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fanucci GE, Cafiso DS. Recent advances and applications of site-directed spin labeling. Curr Opin Struct Biol. 2006;16:644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Flores Jimenez RH, Do Cao MA, Kim M, Cafiso DS. Osmolytes modulate conformational exchange in solvent-exposed regions of membrane proteins. Protein Sci. 2010;19:269–278. doi: 10.1002/pro.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez CJ, Fleissner MR, Guo Z, Kusnetzow AK, Hubbell WL. Osmolyte perturbation reveals conformational equilibria in spin-labeled proteins. Protein science : a publication of the Protein Society. 2009;18:1637–1652. doi: 10.1002/pro.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCoy J, Hubbell WL. High-pressure EPR reveals conformational equilibria and volumetric properties of spin-labeled proteins. Proc Natl Acad Sci U S A. 2011;108:1331–1336. doi: 10.1073/pnas.1017877108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McHaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 31.Columbus L, Kalai T, Jeko J, Hideg K, Hubbell WL. Molecular motion of spin labeled side chains in alpha-helices: analysis by variation of side chain structure. Biochemistry. 2001;40:3828–3846. doi: 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]
- 32.Budil DE, Lee S, Saxena S, Freed JH. Nonlinear-least-squares analysis of slow-motion EPR spectra in one and two dimensions using a modified Levenberg-Marquardt algorithm. Journal of Magnetic Resonance Series A. 1996;120:155–189. [Google Scholar]
- 33.Columbus L, Hubbell WL. Mapping backbone dynamics in solution with site-directed spin labeling: GCN4-58 bZip free and bound to DNA. Biochemistry. 2004;43:7273–7287. doi: 10.1021/bi0497906. [DOI] [PubMed] [Google Scholar]
- 34.Timasheff SN. Control of protein stability and reactions by weakly interacting cosolvents: the simplicity of the complicated. Adv Protein Chem. 1998;51:355–432. doi: 10.1016/s0065-3233(08)60656-7. [DOI] [PubMed] [Google Scholar]
- 35.Parsegian VA, Rand RP, Rau DC. Osmotic stress, crowding, preferential hydration, and binding: A comparison of perspectives. Proc Natl Acad Sci U S A. 2000;97:3987–3992. doi: 10.1073/pnas.97.8.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Timasheff SN. Protein hydration, thermodynamic binding, and preferential hydration. Biochemistry. 2002;41:13473–13482. doi: 10.1021/bi020316e. [DOI] [PubMed] [Google Scholar]
- 37.Rosgen J, Pettitt BM, Bolen DW. An analysis of the molecular origin of osmolyte-dependent protein stability. Protein Sci. 2007;16:733–743. doi: 10.1110/ps.062671607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim M, Xu Q, Murray D, Cafiso DS. Solutes alter the conformation of the ligand binding loops in outer membrane transporters. Biochemistry. 2008;47:670–679. doi: 10.1021/bi7016415. [DOI] [PubMed] [Google Scholar]
- 39.Larsen RG, Singel DJ. Double Electron-Electron Resonance Spin-Echo Modulation - Spectroscopic Measurement of Electron-Spin Pair Separations in Orientationally Disordered Solids. Journal of Chemical Physics. 1993;98:5134–5146. [Google Scholar]
- 40.Schwieters CD, Kuszewski JJ, Clore GM. Using Xplor-NIH for NMR Molecular Structure Determination. Progress in Nuclear Magnetic Resonance Spectroscopy. 2006;48:47–62. [Google Scholar]
- 41.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH Molecular Structure Determination Package. Journal of Magnetic Resonance. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 42.Moeck GS, Coulton JW, Postle K. Cell envelope signaling in Escherichia coli. Ligand binding to the ferrichrome-iron receptor fhua promotes interaction with the energy-transducing protein TonB. J Biol Chem. 1997;272:28391–28397. doi: 10.1074/jbc.272.45.28391. [DOI] [PubMed] [Google Scholar]
- 43.Cadieux N, Kadner RJ. Site-directed disulfide bonding reveals an interaction site between energy coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc. Natl. Acad. Sci. USA. 1999;96:10673–10678. doi: 10.1073/pnas.96.19.10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogierman M, Braun V. Interactions between the outer membrane ferric citrate transporter FecA and TonB: studies of the FecA TonB box. J Bacteriol. 2003;185:1870–1885. doi: 10.1128/JB.185.6.1870-1885.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chimento DP, Mohanty AK, Kadner RJ, Wiener MC. Crystallization and initial X-ray diffraction of BtuB, the integral membrane cobalamin transporter of Escherichia coli. Acta crystallographica. Section D, Biological crystallography. 2003;59:509–511. doi: 10.1107/s0907444903000052. [DOI] [PubMed] [Google Scholar]
- 46.Freed DM, Horanyi PS, Wiener MC, Cafiso DS. Conformational Exchange in a Membrane Transport Protein Is Altered in Protein Crystals. Biophysical journal. 2010;99:1604–1610. doi: 10.1016/j.bpj.2010.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu Q, Ellena JF, Kim M, Cafiso DS. Substrate-dependent unfolding of the energy coupling motif of a membrane transport protein determined by double electron-electron resonance. Biochemistry. 2006;45:10847–10854. doi: 10.1021/bi061051x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trizac E, Levy Y, Wolynes PG. Capillarity theory for the fly-casting mechanism. Proc Natl Acad Sci U S A. 2010;107:2746–2750. doi: 10.1073/pnas.0914727107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mittag T, Kay LE, Forman-Kay JD. Protein dynamics and conformational disorder in molecular recognition. J Mol Recognit. 2010;23:105–116. doi: 10.1002/jmr.961. [DOI] [PubMed] [Google Scholar]
- 50.Kellenberger E. The 'Bayer bridges' confronted with results from improved electron microscopy methods. Mol Microbiol. 1990;4:697–705. doi: 10.1111/j.1365-2958.1990.tb00640.x. [DOI] [PubMed] [Google Scholar]
- 51.Buchanan SK, Smith BS, Venkatramani L, Xia D, Esser L, Palnitkar M, Chakraborty R, van der Helm D, Deisenhofer J. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat Struct Biol. 1999;6:56–63. doi: 10.1038/4931. [DOI] [PubMed] [Google Scholar]
- 52.Flores Jimenez RH, Cafiso DS. The N-Terminal Domain of a TonB-Dependent Transporter Undergoes a Reversible Stepwise Denaturation. Biochemistry. 2012;51:3642–3650. doi: 10.1021/bi300118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of "parallel" expression vectors. Protein Expression and Purification. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 54.Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-time free measurement of dipole-dipole interactions between electron spins. J Magn Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 55.Jeschke G, Chechik V, Ionita P, Godt A, Zimmermann H, Banham J, Timmel CR, Hilger D, Jung H. DeerAnalysis2006 - a comprehensive software package for analyzing pulsed ELDOR data. Applied Magnetic Resonance. 2006;30:473–498. [Google Scholar]
- 56.Herrick DZ, Kuo W, Huang H, Schwieters CD, Ellena JF, Cafiso DS. Solution and membrane-bound conformations of the tandem C2A and C2B domains of synaptotagmin 1: Evidence for bilayer bridging. J Mol Biol. 2009;390:913–923. doi: 10.1016/j.jmb.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.