Abstract
Rho termination factor is an essential hexameric helicase responsible for terminating 20–50% of all mRNA synthesis in E. coli. We used single- molecule force spectroscopy to investigate Rho-RNA binding interactions at the Rho- utilization (rut) site of the ? tR1 terminator. Our results are consistent with Rho complexes adopting two states, one that binds 57 ±2 nucleotides of RNA across all six of the Rho primary binding sites, and another that binds 85 ±2 nucleotides at the six primary sites plus a single secondary site situated at the center of the hexamer. The single-molecule data serve to establish that Rho translocates 5′-to-3′ towards RNA polymerase (RNAP) by a tethered-tracking mechanism, looping out the intervening RNA between the rut site and RNAP. These findings lead to a general model for Rho binding and translocation, and establish a novel experimental approach that should facilitate additional single- molecule studies of RNA-binding proteins.
Keywords: RNA polymerase, transcription, optical trap, optical tweezers, single- molecule biophysics, force spectroscopy
INTRODUCTION
To ensure proper gene expression, transcription by RNA polymerase (RNAP) must be terminated as the transcription elongation complex (EC) reaches the end of a coding region. Intrinsic termination, one of two bacterial termination mechanisms, relies upon the destabilization of the EC at specific DNA terminator sequences and does not require any accessory proteins.1,2 The other mechanism, Rho-dependent termination, relies upon the action of RhoATPase, which translocates 5′? 3′ along the nascent RNA and displaces RNAP from the DNA template.3,4 Because the efficiency of termination is modulated by regulatory cofactors and anti-termination signals,2,5 termination constitutes a fundamental mechanism for controlling the level of gene expression.
Rho, a highly-conserved termination factor, is an essential protein in Escherichia coli6,7 that is present in nearly all prokaryotes.8 In addition to its role in terminating the transcription of native mRNA, Rho may modulate antisense transcription,9 influence the synthesis of tRNAs and small regulatory RNAs,9 silence foreign DNA,10 and prevent the formation of R-loops during transcription.11 In E. coli, each 47-kDa Rho protomer is comprised of 419 amino acids that fold into two major domains. The 130-aminoacid N-terminal domain constitutes the primary RNA-binding region. The C-terminal domain is homologous to F1-ATPase,12 and hydrolyzes ATP via residues found at the interface between adjacent protomers.13 The six C-terminal domains collectively form a secondary RNA-binding site in the central channel of the hexamer that interacts with RNA via a “spiral staircase” of loops.13,14 Thomsen and Berger have proposed a model for translocation in which RNA is pulled through the center of the hexamer by conformational changes in these loops which are, in turn, driven by a rotary mechanism of ATP hydrolysis.13
During Rho-dependent termination, Rho loads onto the transcript at a Rho- utilization (rut) site, acytidine-rich 5′-element of approximately 70–80 nucleotides (nt).15,16 Once Rho binds, RNA must pass through the center of the hexamer ring for ATP hydrolysis to occur,17 a topological constraint that is accommodated by Rho initially adopting an open, “lock-washer” state.18,19 However, the precise manner in which Rho initially associates with RNA is presently unknown. Kim and Patel20 have proposed that Rho first interacts with RNA via its six primary binding sites, then binds via its central secondary site, and finally transitions to a closed, hexameric conformation. Their kinetic analysis is consistent with the fact that RNA binds to Rho secondary sites with an affinity proportional to the ATP concentration, whereas binding to the primary sites occurs in an ATP- independent fashion.21
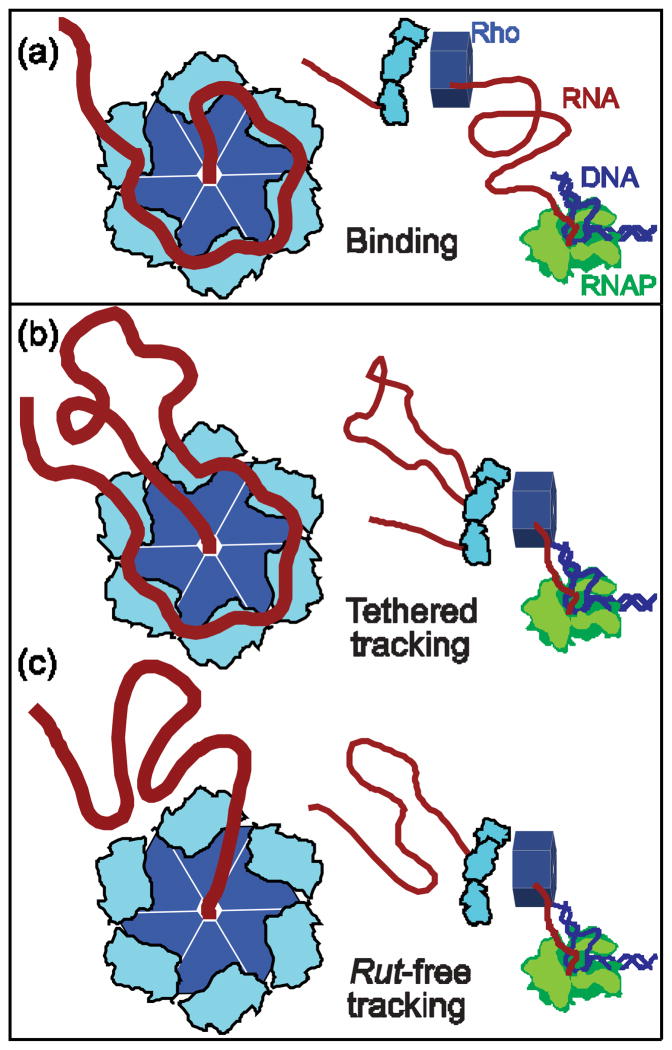
After binding to a rut site, Rho actively translocates 5′? 3′ downstream towards RNAP. Although the precise mechanism by which it tracks along mRNA is not well established, Rho motion is likely to influence how other transcription factors interact in vivo, how ribosomes behave at intragenic terminators, and whether additional Rho complexes are able to bind in succession to a single rut site. Over the last two decades, several models have been advanced to describe Rho interactions with RNA during active translocation. One model, called “pure tracking,” proposes that each N-terminal site alternates between a low- and high-affinity binding state as a result of ATP hydrolysis, producing a periodic separation from, and reattachment to, the transcript.22,23 Such a mechanism could, in principle, allow Rho to advance via a biased random walk. However, the original pure tracking model is excluded by the requirement that RNA must bind to the Rho secondary binding site (Fig. 1a) during active translocation,17 and by structural evidence suggesting that ATP hydrolysis is closely coupled to conformational changes in the Q and R loops of the secondary site.13 A second model (Fig. 1b) proposes that Rho binds continuously to the rut site via its primary binding domains while simultaneously pulling the downstream RNA through the center of the hexamer.24 In this model, called “tethered tracking,” Rho advances towards RNAP by looping out the intervening mRNA. However, the results supporting tethered tracking are controversial, and the data have alternatively been interpreted to be the consequence of multiple Rho comp lexes binding to an individual RNA transcript.23 Additionally, tethered tracking has proved to be difficult to distinguish experimentally from a third possibility, one we refer to as “rut-free tracking,” where Rho factor detaches from the rut site at the start of translocation, but continues to associate with mRNA via its secondary binding site (Fig. 1c). The rut-free tracking model finds support in the observation that Rhomay adopt a closed configuration around RNA in the absence of nucleotides bound to its N-terminal domains.13
Figure 1. Proposed Models of Rho Translocation.
(a) A Rho hexamer (left; light and dark blue) binds RNA (red) at a rut site. It subsequently translocates towards a downstream molecule of RNAP (right; green). Rho binds via six N-terminal primary domains (light blue) and a single secondary binding site (center).
(b) The tethered-tracking mechanism. Rho maintains binding interactions with the rut site via its primary binding domains while moving 5′ to 3′, threading the downstream nucleotides through the secondary site, and consequently forming a growing loop of RNA, until it reaches the position of RNAP.
(c) The rut-free tracking mechanism. The primary binding domains release the rut site once translocation begins, and Rho interacts with RNA exclusively via its secondary site. No loop is formed in the RNA as Rho moves towards RNAP.
Single-molecule optical trapping assays have previously been shown to be capable of inducing the mechanical release of DNA-binding proteins via the application of longitudinal loads that stretch the DNA substrate. Proteins that have been studied in this fashion include nucleosomes,25,26 histone- like nucleoid structuring proteins (H-NS),27 and MutSfamily proteins.28 In particular, Hall et al.26 mapped histone-DNA interactions to reveal regions of strong binding with near-base-pair accuracy. Here, we extend this general approach to study a protein bound to RNA, examining Rho binding and translocation, and determining the binding footprint of Rho with near-nucleotide resolution. A longitudinal load, applied by two optical traps to the opposite ends of a nascent transcript, was used to displace individual complexes of Rho bound to anRNA substrate transcribed by a single molecule of RNAP. The data for substrates of different length indicate that Rho moves via a tethered-tracking mechanism. We present evidence for the existence of two RNA-binding states, with one of these states binding via the secondary site. Additionally, we identify two transient intermediate states in which RNA is bound to Rho via four, or five, of its six N-terminal domains. Our work was carried out in the context of the? tR1 terminator,29,30 which allowed us to address a proposed role for the recruitment of Rho to the rut site by the box B hair pin of the terminator.31 By relating single-molecule unbinding data to the crystal structures for Rho, we arrive at a general model for Rho binding and translocation.
RESULTS
Single-molecule Dumbbell Assay
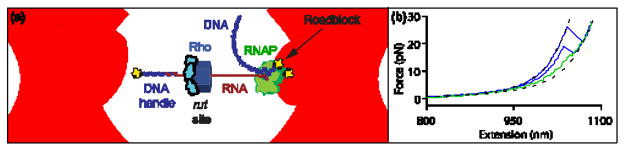
We employed a “dumbbell” assay to apply tension between the 5′ end of a nascent mRNA molecule transcribed in situ and its 3′ end, which remains associated with a molecule of RNAP that is transcriptionally stalled at a biotin-streptavidin roadblock32(Fig. 2a). The dumbbell was comprised of two beads, held in separate optical traps, with one bead attached to a 3,057-bp double-stranded DNA (dsDNA) “handle” hybridized to the 5′ end of the nascent RNA via a 25-nt complementary over hang33. The opposite bead was bound directly to RNAP via abiotin-avidin linkage.
Figure 2. Single-Molecule Dumbbell Assay.
(a) Experimental geometry of the dumbbell assay (not to scale). Tension was applied by moving apart the two trapped beads. The beads (light blue) were maintained in separate optical traps (pink), with a dsDNA handle (dark blue) attached to one and an RNAP molecule (green) to the other by biotin-avidin linkages (yellow). The handle was hybridized to the 5′ end of the RNA transcript (red) via a 25- nt overhang. The RNA was transcribed in situ from a template that carried the rut site of the? tR1 terminator followed by a downstream sequence of variable length (30 bp, 75 bp or 150 bp) ending in a transcriptional roadblock (yellow) that stalls RNAP. The assay permits Rho to bind to the nascent RNA and, in the presence of ATP, translocate towards RNAP.
(b) Representative force-extension curves (FECs). Most FECs (green) resemble those obtained in the absence of Rho and exhibited a small rip due to unfolding of the boxB hairpin in the rut site. FECs displaying larger, high- force rips were observed only in the presence of Rho(blue), and correspond to Rho release from the RNA.
The RNA substrates employed in these experiments were encoded by DNA templates comprised of three segments. The first segment of each template consisted of a short (11 bp) spacer placed adjacent to the 25-bp hybridization region used to attach the DNA handle. The second segment consisted of a 68-bp element derived from a sequence immediately downstream of the cro gene of bacteriophage?, which contains the rut site of the ? tR1 terminator, but is missing the remainder of the terminator region.29,30 The third segment was derived from sequences of the pALB3 plasmid32 and had a length that varied with the experiment, consisting of 30, 75, or 150 bp, with a biotinylated nucleotide at the 3′ end, used to bind streptavidin: these templates were named RB30, RB75, and RB150. The final~25 bp segment of each template was not transcribed, or generated RNA that was inaccessible to Rho binding due to the footprint of RNAP.34,35 Transcriptional run-off assays using polyacrylamide gel electrophoresis were used to confirm that Rho-dependent termination did not take place on a longer template that carried only the rut element, but could be restored to a frequency of 45% on a variant of this same template carrying the full- length ?tR1 terminator (data not shown). A termination efficiency of 60% was observed in a single-molecule assay of this same template (see Supplementary Data).
Dumbbells were assembled and transcription was initiated in vitro by the introduction of NTPs, whereupon elongation continued until RNAP reached the roadblock position. We thereafter collected force-extension curves(FECs)for the RNA transcripts in the presence of 20 nM Rho protein. Records were acquired by moving the optically-trapped beads apart at constant velocity until a preset (maximal) force was attained, and such measurements were repeated once every ~30 s, to allow sufficient time for the RNA to relax and any translocation events to occur. FECs provide information about the displacements and energetics associated with the underlying transitions. Similar force spectroscopy has been used to probestructural changes in a variety of nucleic acids, where “rips”—abrupt, discontinuous changes in the extension—correspond to structural features, such as unfolding or unbinding.32,36,37 We calculated the associated change in extension for rips by fitting the pre-rip and post-rip segments of FECs to a worm- like chain (WLC) model (see Materials and Methods). In the absence of Rho, we consistently observed a small rip in FECs(Fig. 2b), corresponding to a release in the tether length of 10 ±1 nm (N = 207; 14 molecules). We attributed this rip to the unfolding of the 15-nt box B hairpin located within the rut site, because the measured size is consistent with a release of 16 ± 1 nt of RNA, assuming a rise perbase of 0.59 nm.38 In the presence of Rho, the majority of FECs (~95%)were identical to those observed in its absence, however, we also obtained a number of FECs(observed in 26% of transcripts) displaying more prominent rips, characterized by larger changes in tether extension (>18 nm)at high force (blue traces; Fig. 2b). Based on their characteristics, we concluded that these larger, Rho-dependent rips corresponded to the displacement of Rho from the RNA under mechanical load.
Two Initial Rho Binding States
To achieve termination, Rho must first bind to RNA. This loading process, which does not require ATP hydrolysis,39,40 is proposed to occur in a minimum of two steps.20,21 In the first step, Rho binds RNA with high affinity (Ka ~1010 M−1)at the six primary sites. In the second step, Rho bind sRNA at the secondary site with a weaker affinity(Ka ~ 4 × 106 M−1 at saturating ATP) and undergoes a conformational change. This change is thought to correspond to a transition from an open-ring, lock-washer configuration state to a closed state, forming a catalytically active ATPase poised to translocate.18
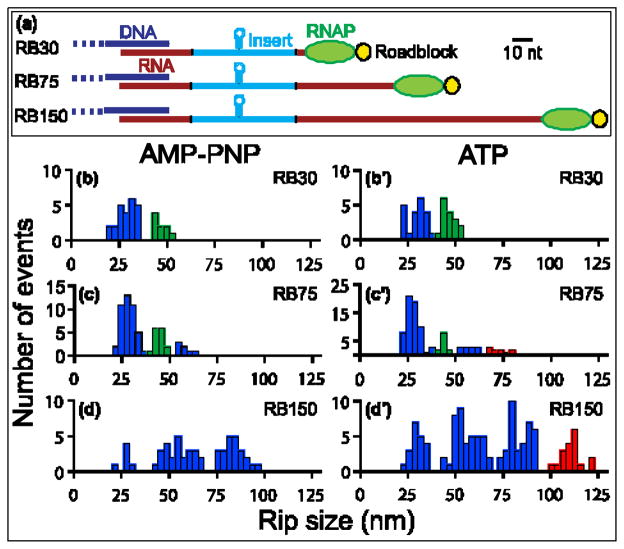
To investigate the initial binding states, we collected FECs in the presence of the non-hydrolyzable ATP-analog AMP-PNP[adenosine 5′-(β,?- imido)triphosphate]. We began by examining Rho interaction with RB30, a short template carrying a rut site plus a few nucleotides of exposed upstream and downstream sequence(Fig. 3a). We observed two well-separated populations of Rho-dependent rips (Fig. 3b) centered at 29 ±1 nm and 46 ±1 nm. Based on their characteristics (discussed below), we refer to these populations as “primary rips” or “secondary rips,” where the first set corresponds to the release of RNA from Rho primary sites, and the second set corresponds to the release of RNA from both primary and secondary sites. Additional populations of rips were observed on longer templates, however, as we explain below, all rips scored in the presence of AMP-PNP could be interpreted in the context of these two binding states.
Figure 3. Distributions of Rho Unbinding Rips.
(a) Diagram of the RB30, RB75, and RB150 transcripts, drawn to approximate scale along the pulling coordinate (scale bar, 10 nt). Two segments of RNA (red) flank an insert from ?phage(light blue) carrying the rut site of ?tR1 (including the boxB hairpin). The transcripts were transcribed in situ from templates carrying 30, 75, or 150 bp downstream of the insert prior to a 3′ roadblock (yellow). Transcripts were hybridized to DNA handles (dark blue) via their 5′ ends (Fig. 2); ~25 nt at the 3′ end of each transcript lies under the footprint of RNAP (green) and is inaccessible to Rho. (B–D′) Histograms of the rip sizes for RNA release obtained for the three templates in the presence of AMP-PNP(left) or ATP (right). Population peaks are color-coded as follows: primary rips, with sizes of ~28 nm or multiples thereof (blue bars); secondary rips, with sizes of ~45 nm (green bars); translocation rips, with sizes that depend upon transcript length (red bars).
(b) RB30 with AMP-PNP. Two populations, at 29 ±1 nm and 46 ±1 nm(mean ± SEM). Data from 8 molecules (N =33). (b′) RB30 with ATP. Two populations, at 30 ±1 nm and 46 ±1 nm. Data from 8 molecules (N =37). One population, at 27 ± 1 nm, was obtained for the RB30 boxB deletion construct (Fig. S1)
(c) RB75 with AMP-PNP. Three populations, at 28 ±1 nm, 45 ±1 nm and 57 ±1 nm. Data from 22 molecules (N =65). (c′) RB75 with ATP. Four populations, three of which were observed in the presence of AMP-PNP (at 28 ±1 nm, 45 ±1 nm, and 57 ±1 nm), as well as a new peak at 73 ±1 nm. Data from 33 molecules (N =96).
(d) RB150 with AMP-PNP. Three evenly spaced populations at 27 ±1 nm, 55 ±1 nm and 84 ±1 nm. Data from 17 molecules (N =54). (d′) RB150 with ATP. Four populations, three of which were observed in the presence of AMP-PNP, (30 ±1 nm, 56 ±1 nm and 84 ±1 nm) as well as a new peak at 111 ±1 nm. Data from 37 molecules (N =114).
Multiple Rho Bind Long Templates
We also examined Rho binding to a series of longer transcripts in the presence of AMP-PNP. FECs for RB75 exhibited both primary and secondary rips (as with RB30), as well as a new rip population centered at 57 ±1 nm (Fig. 3c). On the RB150 template, we observed primary rips plus two additional rip populations centered at 55 ±1 nm and 84 ±1 nm (Fig. 3d). About 20% of the two new populations observed with the longer transcripts also exhibited sub-rips of roughly the same size as primary rips (28 nm). Because the sizes of the two new populations were integral multiples of 28 nm, we concluded that these likely corresponded to the release of multiple complexes of Rho from RNA. Although our templates carried a single rut site, the observation of multiple Rho release events was not entirely unexpected, given that Rho is known to bind to RNA cooperatively.23,41,42 Based on the size of the RNA footprint of Rho, up to three complexes of Rho could bind to RB150 at a time, two could bind to RB75, but just one could bind to RB30(see Discussion)
Rho Translocates Via Tethered Tracking
The two mechanisms for translocation considered here—rut-free tracking and tethered tracking—lead to different predictions for the sizes of rips exhibited after Rho moves from the rut site towards RNAP. If Rho moves via rut-free tracking, contacting RNA exclusively via its secondary binding site (Fig. 1c), one would expect to observe only small rips (~2 nm, estimated from Thomsen & Berger13), with sizes independent of the length of the underlying template. However, if Rho moves via tethered tracking, one would expect to observe ever-larger rips, proportional to the lengths of the templates and only taking place in the presence of ATP, corresponding to the release of the tether length of RNA looped out during translocation (Fig. 1b).
We collected FECs on RB30 in the presence of ATP. Here again, we observed both primary and secondary rips(30 ±1 nm and 46 ±1 nm; Fig. 3b′)that matched those previously seen with AMP-PNP, and did not resolve a distinct translocation rip (i.e., a rip corresponding to the release of Rho complexes that translocated). Rho can potentially translocate only a short distance (<5 nt) on this transcript before encountering RNAP, and therefore some of the 46-nm rips observed could, in principle, correspond to the release of Rho complexes that translocated via tethered tracking. However, we could not distinguish these from secondary rips based on size alone. This ambiguity was resolved by examining a series of longer templates (see below).
When the RB150 template was examined, four populations of Rho-dependent rips were observed (Fig. 3d′). Three of these populations were observed both in the presence of ATP or AMP-PNP, centered at 30 ± 1 nm, 56 ± 1 nm and 84 ± 1 nm, and were interpreted as corresponding to the release of one, two or three Rho complexes from the template, respectively. The fourth population, observed only in the presence of ATP, was centered at 111 ±1 nm. This rip distance is close to that expected for tethered tracking, i.e., for a Rho-unbinding event(45 nm) accompanied by the release of nearly all the downstream RNA, for an additional ~120 nt (~71 nm). However, the size of the fourth rip population was also consistent, within error, with the release of a fourth Rho molecule, bound to RNA in an ATP-dependent manner. To disambiguate this result, we studied a different- length template, RB75. Based on the average rip values obtained for RB30 and RB150, the tethered-tracking mechanism yields an estimated RNA release of 70 ± 2 nm for this template.[We note that this template was created so that the distance corresponded to a non- integral multiple (~2.5) of the primary rip size and was therefore unlikely to be mistaken for Rho release.] We again obtained four populations of rips(Fig. 3c′). Three of these populations were observed in the presence of either AMP-PNP or ATP, and centered at 28 ±1 nm, 44 ±1 nm and 57 ±1 nm, respectively. The fourth population (a “translocation rip”), observed only with ATP, measured 73 ±1 nm, and matched, within experimental error, the value predicted for tethered tracking. (The existence of this population in the presence of ATP is statistically significant at the 97% level, based on a t-test over the subset of rips >50 nm). Thus, the data support tethered tracking, and disfavor rut-free and pure tracking, because the latter mechanisms would not be expected to produce template length-dependent loops.
A Complementary Oligomer Inhibits Multiple Rho Binding While Permitting Translocation
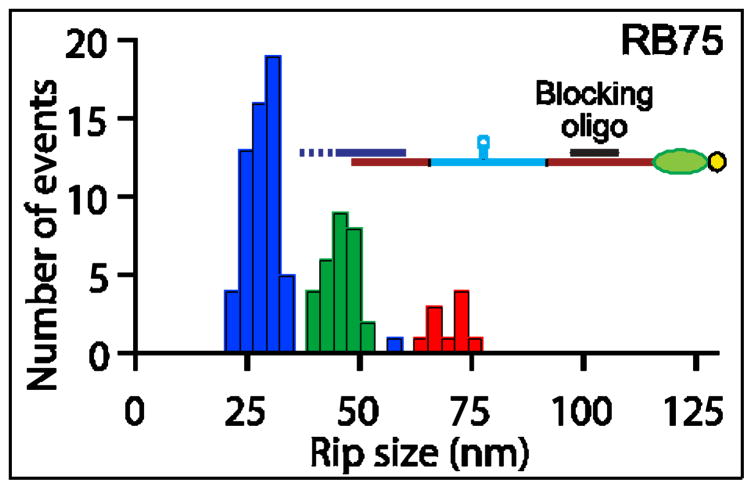
In light of observing multiple Rho-release events, we also explored experimental conditions designed to allow only a single Rho complex to bind the transcript. Rho does not bind to RNA-DNA hybrids,23,29 so we examined template RB75 in the presence of a20-nt “blocking” DNA oligomer complementary to a portion of the RNA segment located transcriptionally downstream of the rut site (Fig. 4). Because Rho is known to act in vitro as an RNA-DNA helicase and separate short (<120 nt)DNA oligomers from complementary RNA,23,43,44 we anticipated that the use of a blocking oligomer would not significantly affectany translocation.
Figure 4. Blocking Oligomers Inhibit Multiple Rho Binding but Permit Translocation.
Histogram of unbinding rip sizes in the presence of blocking oligomers and ATP on RB75 (inset: scale diagram of RB75 showing the 20 nt DNA oligomer (black) hybridized). Three populations were observed, at 28 ±1 nm, 45 ±1 nm and 71 ±2 nm (color-coding as in Fig. 3). The frequency of multiple Rho unbinding events (blue bars) was greatly diminished and the number of secondary rips (green bars) enhanced compared to conditions without blocking oligomers (see Fig. 3c′), whereas translocation rips (red bars) occurred with the same frequency (11 ±3 % with blocking oligomers vs. 10 ±3 % without). Data from 43 molecules (N =98).
In the presence of excess DNA oligomer(100 nM), we observed three populations of rips on RB75 (Fig. 4), corresponding to primary and secondary rips of 28 ±1 nm and 45 ±1 nm respectively, as well as to a third population, previously assigned to a translocation rip, of 71 ±2 nm. In these experiments, the 56 nm- long rips observed in the absence of blocking oligomer were nearly non-existent. Because the ATP-dependent rips (71 nm) persisted in the presence of complementary oligomers, these results serve as additional confirmation that such rips arise from tethered tracking, rather than multiple Rho unbinding events. We note that the fraction of Rho complexes that displayed translocation rips in the presence of blocking oligomers (11% ±3%) was similar to that observed in their absence (10% ±3%), suggesting that the presence of a DNA-RNA hybrid did not adversely affect Rho translocation.
Role of the boxB Hairpin in Binding Rho to the rut Site
The rut site of? tR1 contains a 15- nt box B hairpin, a motif known to suppress Rho-dependent and intrinsic termination in phage ? by recruiting N protein to the transcript.2,45 The deletion of this hairpin is reported to have essentially no effect on the termination efficiency in vitro or in vivo.30 Although the role of the box B hairpin is not well understood, point mutations that structurally destabilize it can dramatically reduce termination efficiency.31 Based on their results, Vieu and Rahmouni31 proposed that boxB plays a role in clamping together the high-affinity regions of the? tR1 rut site for optimal binding by Rho. A structure for boxB complexed with Rho has not yet been solved, but this model implicitly requires the hairpin to remain folded when Rho is bound to the full rut site. However, our force spectroscopy data imply that boxB gets unfolded upon Rho binding, because we never observed a corresponding ~15-nt rip, together with a Rho-release rip, within the same FEC. Furthermore, all rips corresponding to the release of multiple Rho complexes had sizes that were integral multiples of the primary rips, even though the RNAs to which these were bound carried only a single copy of boxB.
Assuming that the boxB hairpin unfolds when Rho binds the rut site, one would expect the removal of this hairpin to have no effect on the size or occurrence of primary rips, provided Rho can form alternate contacts with the RNA. To test this prediction, we collected FECs for a modified RB30 template in which boxB was deleted from the rut site. The sizes of the primary rips were unchanged(27 ±1 nm; Fig. S1). Although boxB may act to promote termination in vivo in some fashion, our results suggest that the explanation for this behavior may be more complicated than the hairpin acting as a simple clamp. Finally, because Rho may associate with RNA up to 11–18 nt upstream from the canonical? tR1 rut site (see Discussion), and our templates coded for 18 nt of exposed upstream RNA, we considered the possibility that these templates might preclude Rho from binding the rut site in the same fashion as it does in vivo. To address that possibility, we added an additional 39 nt to RB75, bringing the total of exposed upstream nucleotides to 57. We observed no significant change in the sizes or frequencies of any unbinding rips collected on this template (Table S1).
DISCUSSION
Rho Adopts at least Two Stable RNA Binding States
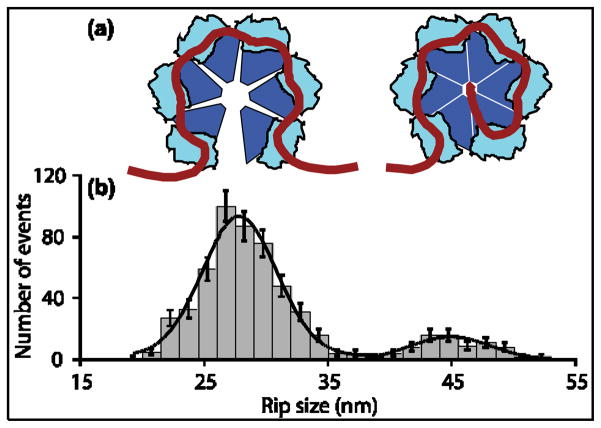
We observed two populations of Rho-dependent rips, with extensions independent of template length. Because these rips occurred under external loads in the presence of either ATP or AMP-PNP, we conclude that they correspond to the mechanical displacement of Rho from the RNA without accompanying hydrolysis or translocation. Averaging our data over all templates and conditions(Fig. 5), we obtained rip sizes of 28.0 ±0.6 nm and 45.1 ±0.9 nm (mean ±std. err.) [Cited values include a 2% error from uncertainties in the applied force and persistence length of RNA; Gaussian fits to the se distributions returned rip sizes of 27.9 ± 0.6 nm and 44.9 ± 1.0 nm (mean ± std. err.)].
Figure 5. Two RNA Binding States of Rho.
(a) Diagram depicting the two Rho binding states consistent with the observed rips. Left: RNA bound to six Rho primary sites in its “open” configuration. Right: RNA bound to six primary sites plus the central secondary site in the “closed” configuration.
(b) Global histogram (grey bars) of the sizes of RNA-release rips observed in the presence of ATP or AMP-PNP for all templates studied (N =570). Solid line: Fit to sum of two Gaussians, with peaks at 28 ±1 nm and 45 ±1 nm (mean ± S.E.M.). Data from populations centered at 56 nm and 84 nm(Fig. 3) were included in the global dataset by dividing the measured values by 2 or 3, respectively (see text).
The larger rip population (45 nm)often exhibited sub-rips: an initial release of 17 nm followed by the remaining 28 nm (29 ± 5% of all data), a characteristic indicating that any contacts formed in the 28 nm binding state are a subset of those in the 45 nm binding state. We therefore associate the 28 nm “primary rips” to RNA release from all of the six Rho primary binding sites, while the 45 nm “secondary rips ”are attributable to RNA release from both the single secondary site and six primary sites (Fig. 5). These assignments are consistent with states predicted by Kim and Patel20 and Richardson,21 and lead us to a model for Rho binding in which Rho binds RNA at the primary sites before associating with the secondary site (see below). This particular ordering of states was previously proposed by Kim and Patel20 using an independent approach that relied upon kinetic measurements. We note that the transition to a complex with contacts only at its primary sites is accompanied by the rotation of the entire Rho hexamer and the reorientation of the RNA released, which results in a comparatively large change in tether extension (here, 17 nm).
The relative frequencies of various rip populations varied from template to template. The secondary and translocation rips, which together correspond to events where RNA is released from the Rho secondary site, accounted for 36% ±6% of all rips (ATP and AMP-PNP data combined) collected on RB30, but just 22% ±3% of rips collected on RB75. One possible interpretation of this variability is that the Rho secondary site may be somewhat obstructed or inhibited with the longer transcript. However, when the blocking oligomer was added to RB75, the frequency of rips was restored to the level found in RB30 (41% ±5%). We would speculate that once Rho binds RNA at its primary sites, it may either bind to RNA at its secondary site or facilitate the cooperative binding of a second Rho molecule to the RNA, but not both. This would explain the findings, because the RB30 template is too short to bind more than a single Rho molecule. The high frequency of multiple Rho-release events on the long RB150 template may likewise explain why a distinct 45- nm peak was not observed on this template.
In the presence of ATP, Rho complexes bound to RNA produced a single, low-variance population of translocation rips that depended only upon the template length. Because we did not observe translocation rips of intermediate sizes or wider variance, we conclude that the tether looping-out process must occur rapidly under our experimental conditions (seconds or less). The observation of isolated secondary rips in the presence of ATP indicates that binding to the Rho secondary site is not sufficient, in and of itself, for translocation to occur. This is an indication that Rho complexes likely transition from a secondary-rip state to a distinct, translocation-competent state that can hydrolyze ATP (see Supplementary Data).
Rho Binding Footprints of 57 nt and 85 nt
Although Rho is generally thought to have an RNA footprint of 70–80 nt,15,46 estimates have been reported as low as 55 nt40 and as high as 84 nt.41 Typically, such footprints have been based on RNAse protection assays40,41 or the sizes of minimum templates required for termination.16,47 The number of RNA nucleotides bound to Rho, N, may be estimated from the rip size, r, and the width of the Rho enzyme projected onto the pulling coordinate, d, according to N = (r +d)/0.59, where the nucleotide pitch is taken to be 0.59 nm/nt.38 For rips involving the release of RNA from the primary sites alone, we obtained a value for d from the open crystal structure18 (PDB ID: 1PVO), where the distance between dinucleotide fragments bound to subunits A and F, the two protomers separated by a gap, was taken to be 5.4 ±0.7 nm (the uncertainty arises from the choice of atoms assumed to make contact). Based on this value, we estimate a footprint of 57 ±2 nt for the six sites, or 9–10 nt per site. For rips involving the release of RNA from both primary and secondary sites, we modeled the dinucleotide fragments into their equivalent locations in the closed crystal structure (PDB ID: 3ICE), which has a 6 nt RNA helix at the secondary site,13 and obtained an estimate for the distance between these dinucleotides and the 3′ end of the helix, d = 5.0 ±0.5 nm. Based on this value, we estimate that 85 ±2 nt of RNA associate with both primary and secondary sites. This footprint size is consistent with a minimal template for termination of 97 nt,16 assuming that an additional 12–15 nt are sequestered inside the transcription bubble and inaccessible to Rho.48
Transient Rho Unbinding Intermediates
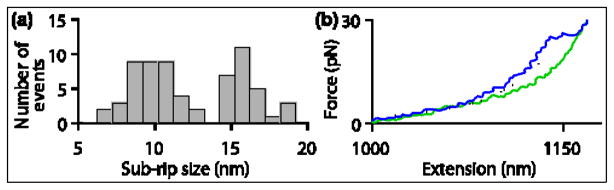
Two populations of intermediate states were observed in ~13% of all primary rips (28 nm). One population was characterized by an initial sub-rip averaging 9.6 ±0.3 nm(N =38), and the second by a sub-rip averaging 16.0 ±0.4 nm (N =27)(Fig. 6). These intermediates are likely to represent the release of RNA from a subset of the six primary sites. We therefore modeled structural pathways in which RNA unbound from 1–4 of the primary sites prior to complete release (Fig. S2). The size of the smaller sup-rip population is consistent with RNA release from a single primary site (7.9 ±0.8 nm expected) followed by release from the remaining five sites (20.1 ±0.8 nm). The larger sub-rip population is consistent with release from two primary sites (14.9 ±0.6 nm expected) followed by release from the remaining four sites (13.1 ±0.6 nm).
Figure 6. Transient Intermediate States in Rho Unbinding Rips.
(a) Histogram showing the first of two sub-rips occasionally observed within the 28 nm primary rip. The data indicate two transient intermediates, one characterized by an initial sub-rip of 9.6 ±0.3 nm (N =38) and the other by an initial sub-rip of 16.0 ±0.4 nm (N =27). The first intermediate likely corresponds to the release of RNA by one of the six primary domains; the second corresponds to release from two (see text; also Fig. S2). Rip sizes were obtained by measuring the fractional size of each sub-rip and multiplying this value by the population-average rip size (27.7 ±0.4 nm).
(b) A representative FEC that exhibits a transient intermediate rip (blue trace). The extension for the intermediate was fit to a double-WLC model (dotted line). The FEC for a molecule with no Rho bound is shown for comparison (green trace).
Rho Translocates 5′? 3′ via a Tethered-tracking Mechanism
A tethered-tracking mechanism has important implications for Rho-dependent termination. Because the rut site, once bound, remains occupied during subsequent Rho translocation, additional Rho complexes are prevented from binding sequentially to this same site. If a translocating ribosome were to encounter Rho at an intragenic rut site, it must either displace it or pause until Rho dissociates. Active translation, therefore, may act to suppress termination in transcripts bound by Rho.
Rho-dependent termination might also become coupled to translation, because the Rho footprint on the rut site may extend into the gene-coding region, as seems to be the case for? tR1. Adding the sizes of the Rho-binding footprint (85 ± 2 nt), the footprint of the RNAP transcription bubble (12–15 nt),48 and the RNAP offset from the biotin-streptavidin roadblock (10 nt)34 to the measured translocation distance for each transcript, we can estimate the location of the 5′ boundary of the Rho-binding element. Given the sequences of the associated DNA templates, this estimate positions that boundary 11–18 nt upstream of the nominal 5′ border of the canonical? tR1 rut site. In the? tR1 terminator, the start of the rut site lies just 7 nt downstream of the end of the cro gene.29,30 Based on these data, therefore, it is possible that Rho binding to this rut site may occlude the cro stop codon, and consequently affect translation (and vice versa).
Populations of intermediate states (sub-rips) were observed in 56% of records displaying translocation rips (Fig. S3). A fraction (23%) of the translocation rips themselves occurred in two steps, with a release of all but 28 nm, followed by a final, 28 nm-sub-rip (Fig. S3b). These data were interpreted as Rho unbinding RNA from the secondary site, releasing the RNA loop formed by tethered tracking, followed by RNA dissociation from the primary sites. The remainder of records (33%)exhibited sub-rips of variable sizes that could not be assigned to any particular unbinding intermediate (Fig. S3c): these may reflect the formation of secondary structure formed within the looped-out RNA (Fig. S3c′). We speculate that loops formed in the process of tethered tracking may play a role in the folding of structured, non-coding RNAs that are terminated by Rho.9
The frequency with which Rho binds via its secondary site, estimated from the ratio of secondary and translocation rips to primary rips, provides further evidence that tethered tracking is the dominant translocation mechanism. We note that the decrease in the number of secondary rips observed on RB75 in the presence of ATP (as compared to AMP-PNP) was compensated by a corresponding increase in the number of translocation rips, such that the total rip frequency remained unchanged. If instead, Rho were to engage in rut-free tracking, then the decrease in secondary rips would not be compensated in such a fashion, because the loss of this sub-population would produce no further detectable rip s. Nevertheless, on the basis of our data, one cannot formally exclude a more complicated mechanism whereby Rho first translocates via rut-free tracking, but then rebinds to the upstream rut site once it has stopped or stalled. That said, we note that experiments conducted with a blocking oligomer induce the formation of a comparatively inflexible DNA-RNA hybrid region (~6 nm), which is expected to reduce the ability of any Rho complexes engaged in rut-free tracking to rebind. However, the observed frequency of translocation rips was similar in the presence and absence of the blocking oligomer, disfavoring this modified version of rut-free tracking.
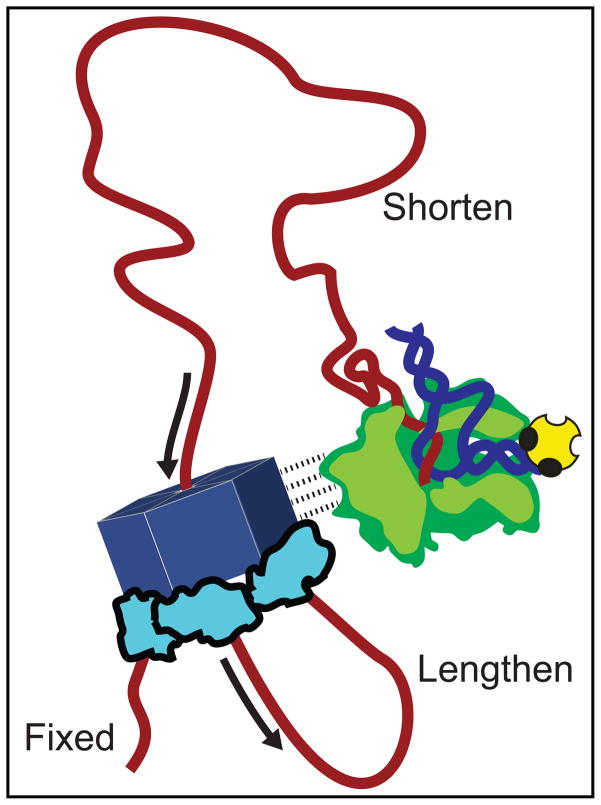
In one recent study carried out in vitro, it was proposed by E. Nudler and coworkers that Rho associates directly with RNAP throughout elongation, and that it can do so even in the absence of RNA.49 However, this model was challenged by Kalyani et al.50 who argued that Rho does not bind to an EC in the absence of a rut site, and found no evidence for a direct interaction between Rho and RNAP prior to termination. We note that if Rho and RNAP were initially conjoined as proposed by Nudler et al., the subsequent association of Rho with a rut site on the RNA would form a loop that shortened— not lengthened—during Rho translocation (Fig. 7). The mechanical disruption of such loops by tension would result in rips with sizes proportional to the number of nucleotides separating Rho from the elongation complex. In contrast to the loops formed by tethered tracking, these loops are expected to form even in the absence of ATP. However, we did not observe any rips with the required characteristics in the presence of AMP-PNP, suggesting that Rho does not associate with RNAP in our assays. It remains possible that Rho binds only very weakly to RNAP, and is consistently released during the acquisition of FECs at even the lowest tensions (<3 pN). If we assume that the binding interaction distance between Rho and RNAP is no greater than 1 nm, our force measurements place an upper limit of just 3 pN·nm, or ~0.8 kBT (0.4 kcal/mole), on the equilibrium binding energy between Rho and RNAP. We note that Rho is likely to be indirectly coupled to RNAP in vivo, because accessory factors such as NusA and NusGare known to interact with both enzymes.51,52
Figure 7. Proposed Model for Direct Interaction of Rho and RNAP.
Epshtein et al.49 proposed that Rho directly associates with RNAP throughout transcription (dashed lines). In this model, such an association would generate a loop of RNA, composed of sequences downstream of the rut site and upstream of RNAP, that shortens continuously during translocation. Furthermore, in the tethered-tracking mechanism, a second loop is expected to form, composed of sequences between the primary and secondary Rho binding sites, that lengthens continuously at the identical rate. Our results disfavor any strong, direct interaction between Rho and RNAP.
Model for Rho Binding and Translocation
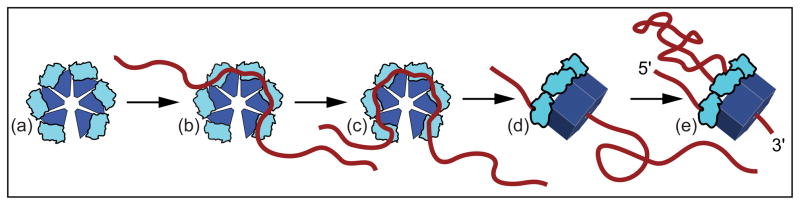
Based on the observed sizes, characteristics, and ATP-dependence of the FECs for Rho bound to templates of different lengths, a general model for Rho binding and translocation can be formulated that is quantitatively consistent with our data (Fig. 8). Initially, Rho binds to RNA at a rut site via one or more primary domains, transiently associating as the hexamer gets loaded. After 57 ±2 nt of RNA are bound by its primary sites, Rho binds via its secondary site and (presumably) undergoes a conformational transition into a closed ring form, which all together subtends a footprint of 85 ±2 nt. In the final step, Rho helicase is activated and it advances downstream, towards RNAP, while remaining bound via the rut site, looping out the intervening RNA sequence via a tethered-tracking mechanism. We anticipate that variations of this single-molecule assay will be particularly useful in studies of Rho helicase, being well-suited for characterizing basic motor properties, such as the velocity, processivity, stall force, and load-dependence. In particular, a high-resolution assay for Rho translocation may be able to resolve whether its step size is 1 nt,13 7 nt,53 or some other value. Finally, we expect single- molecule assays to shed additional light on the means by which Rho terminates transcription, and on the modulatory roles of accessory factors, such as NusA and NusG.
Figure 8. General Model of Rho Binding and Translocation.
In its open (lock-washer) configuration, Rho(a) binds RNA via the rut site, transiently occupying one or more binding states (b) before associating with 57 ±2 nt of RNA via all six primary domains (c). RNA is then passed into the center of the hexamer and Rho adopts the closed (ring) configuration, binding the transcript via its secondary site as well, with an overall RNA footprint of 85 ±2 nt (d). Finally, Rho translocates via a tethered-tracking mechanism (e), hydrolyzing ATP and proceeding downstream towards RNAP, looping out the intervening RNA sequence (see also Fig. S3).
MATERIALS AND METHODS
Assay
To produce a dumbbell assay (Fig. 2a), biotinylated RNAP molecules were initiated at a T7A1 promoter and stalled after transcribing 29 nt.54 Purified elongation complexes (ECs) were then incubated at roughly 1:1 stoichiometry with 730-nm diameter avidin- or streptavidin-coated beads at room temperature (RT; 21.5 ±0.5 °C). DNA handles(3,057 bp)produced by auto sticky PCR of the M13mp18 plasmid33 were coupled to 600- nm diameter avidin-coated beads via biotin linkages (30 min incubation; washed twice). The handles carried 5′-single-stranded overhangs complementary to the first 25 nt of RNA transcribed, leaving 11 nt of RNA between the handle and rut site insert. Handles and ECs were mixed and incubated for 1 hr at RT to form dumbbell tethers.
Transcription in dumbbells was restarted inside a flow chamber (~5 μl)by introducing 1 mM of ATP, GTP, CTP and UTP in the presence or absence of Rho protein (20 nM). Where appropriate, 1 mMAMP-PNP replaced ATP. Experiments were carried out in elongation buffer (EB; 50 mM HEPES [pH 8.0], 50 mM KCl, 5 mM MgCl 2, 0.1 mM EDTA and 0.1 mM DTT; 26 ± 1 °C) plus an oxygen-scavenging system (8.3 mg/ml glucose (Sigma), 46 U/ml glucose oxidase (Calbiochem), 94 U/ml catalase (Sigma)). Catalase and glucose oxidase were FPLC-purified (GE Healthcare) by a Superdex 200 10/300 GL column and verified to be RNAse- free (AmbionRNaseAlert); 0.2 U/μlSUPERase·In(Ambion) was added to EB to inhibit RNAse activity.
E. coli Rho protein was expressed from plasmid pCB111 by inducing a culture with an M13 derivative encoding T7 RNA polymerase under lac control55 with IPTG and purified as described.56
Data Collection
The optical trapping instrument was described previously.57 Uncertainties in force, due to variations in bead size and systematic calibration errors, were estimated at 15%. Position data were acquired at 2 kHz using custom software (Labview), filtered at 1 kHz using an 8-pole low-pass Bessel filter, and analyzed in Igor Pro (Wavemetrics). Tether lengths and additional sources of error were estimated as described.32,33 FECs were collected by slewing the movable trap with an acousto-optic deflector (IntraAction Inc.) at ~190 nm/s as the position of the bead in the stationary trap was recorded.32 Data points in FECs represent 1.25 ms integration time. For experiments involving blocking oligomers (introduced along with NTPs), tethers were maintained at an initial extension of 1.10 μm for ~35 s (at F = 15 pN) to facilitate annealing and minimize the formation of secondary structure; this procedure was repeated every time a potential translocation rip was observed.
Force-Extension Curves
To compute rip size, we subtracted the extensions returned by fits to worm- like chain (WLC) models before and after the associated rip. The pre-rip portion of each FEC was fit to a WLC model using the modified Marko-Siggia relationship ; because the pre-rip segment corresponds almost entirely to dsDNA, the elastic modulus was set to 1200 pN/nm.58 To insure single- molecule behavior, any tethers exhibiting too short a persistence length (<18 nm) or an incorrect contour length were rejected from further analysis. The post-rip portion of each FEC was fit to a double-WLC model, with the parameters of the first WLC set to those returned by the pre-rip fit phase. For the additional WLC, we used a persistence length of 1.0 nm for ssRNA59,60 and an elastic modulus of 1600 pN/nm.60 To estimate the size of the boxB rip, we assumed an A-form dsRNAhelix width of 2.2 nm38 which we subtracted from the extension of the pre-rip portion when fitting FECs.32
Our analysis excluded any small rips observed at low forces which were seen in both the presence and absence of Rho: we imposed a minimum force requirement, such that only events above the cutoff (from 9.0 pN to 12.0 pN, depending upon the length of the template) were considered. Similarly, we excluded rips smaller than 25 nt that were Rho- independent and due to the unfolding of RNA secondary structures, such as the 15 nt boxB hairpin.
Supplementary Material
First single-molecule study of Rho termination factor
Rho adopts two RNA-binding states with footprints of 57 nt and 85 nt
Rho translocates via tethered tracking
No evidence for direct interaction between Rho and RNAP prior to termination
Develop general model for Rho binding and translocation
Acknowledgments
We thank J. Andreasson and E. Koslover for reading the manuscript, J. Andreasson and P. Anthony for advice on experimental procedures, and J. Gelles for helpful discussions. FMF was supported by an NSF Graduate Research Fellowship. This work was supported by a grant from the NIGMS.
Abbreviations used
- AMP-PNP
adenosine 5′-(β,?-imido)triphosphate
- EC
Elongation Complex
- FEC
Force-extension curve
- nt
nucleotides
- RNAP
RNA Polymerase
- rut
Rho-utilization
- WLC
Worm-like chain
Footnotes
Supplementary data associated with this article can be found, in the online version, at
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Larson MH, Greenleaf WJ, Landick R, Block SM. Applied force reveals mechanistic and energetic details of transcription termination. Cell. 2008;132:971–82. doi: 10.1016/j.cell.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nudler E, Gottesman ME. Transcription termination and anti- termination in E. coli. Genes Cells. 2002;7:755–68. doi: 10.1046/j.1365-2443.2002.00563.x. [DOI] [PubMed] [Google Scholar]
- 3.Richardson JP. Rho-dependent termination and ATPases in transcript termination. Biochim Biophys Acta. 2002;1577:251–260. doi: 10.1016/s0167-4781(02)00456-6. [DOI] [PubMed] [Google Scholar]
- 4.Peters JM, Vangeloff AD, Landick R. Bacterial Transcription Terminators: The RNA 3′-end Chronicles. J Mol Biol. 2011 doi: 10.1016/j.jmb.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciampi MS. Rho-dependent terminators and transcription termination. Microbiology. 2006;152:2515–28. doi: 10.1099/mic.0.28982-0. [DOI] [PubMed] [Google Scholar]
- 6.Roberts JW. Termination factor for RNA synthesis. Nature. 1969;224:1168–74. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- 7.Das A, Court D, Adhya S. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc Natl Acad Sci U S A. 1976;73:1959–63. doi: 10.1073/pnas.73.6.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Opperman T, Richardson JP. Phylogenetic analysis of sequences from diverse bacteria with homology to the Escherichia coli rho gene. J Bacteriol. 1994;176:5033–43. doi: 10.1128/jb.176.16.5033-5043.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters JM, Mooney RA, Kuan PF, Rowland JL, Keles S, Landick R. Rho directs widespread termination of intragenic and stable RNA transcription. Proc Natl Acad Sci U S A. 2009;106:15406–11. doi: 10.1073/pnas.0903846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cardinale CJ, Washburn RS, Tadigotla VR, Brown LM, Gottesman ME, Nudler E. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320:935–8. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gowrishankar J, Harinarayanan R. Why is transcription coupled to translation in bacteria? Mol Microbiol. 2004;54:598–603. doi: 10.1111/j.1365-2958.2004.04289.x. [DOI] [PubMed] [Google Scholar]
- 12.Dombroski AJ, Platt T. Structure of rho factor: an RNA-binding domain and a separate region with strong similarity to proven ATP-binding domains. Proc Natl Acad Sci U S A. 1988;85:2538–42. doi: 10.1073/pnas.85.8.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–34. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miwa Y, Horiguchi T, Shigesada K. Structural and functional dissections of transcription termination factor rho by random mutagenesis. J Mol Biol. 1995;254:815–37. doi: 10.1006/jmbi.1995.0658. [DOI] [PubMed] [Google Scholar]
- 15.Morgan WD, Bear DG, Litchman BL, von Hippel PH. RNA sequence and secondary structure requirements for rho-dependent transcription termination. Nucleic Acids Res. 1985;13:3739–54. doi: 10.1093/nar/13.10.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu AQ, von Hippel PH. Rho-dependent termination within the trp t′ terminator. I. Effects of rho loading and template sequence. Biochemistry. 1998;37:11202–14. doi: 10.1021/bi9729110. [DOI] [PubMed] [Google Scholar]
- 17.Wei RR, Richardson JP. Identification of an RNA-binding Site in the ATP binding domain of Escherichia coli Rho by H2O2/Fe-EDTA cleavage protection studies. J Biol Chem. 2001;276:28380–7. doi: 10.1074/jbc.M102444200. [DOI] [PubMed] [Google Scholar]
- 18.Skordalakes E, Berger JM. Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell. 2003;114:135–46. doi: 10.1016/s0092-8674(03)00512-9. [DOI] [PubMed] [Google Scholar]
- 19.Canals A, Uson I, Coll M. The structure of RNA- free Rho termination factor indicates a dynamic mechanism of transcript capture. J Mol Biol. 2010;400:16–23. doi: 10.1016/j.jmb.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 20.Kim DE, Patel SS. The kinetic pathway of RNA binding to the Escherichia coli transcription termination factor Rho. J Biol Chem. 2001;276:13902–10. doi: 10.1074/jbc.M011043200. [DOI] [PubMed] [Google Scholar]
- 21.Richardson JP. Activation of rho protein ATPase requires simultaneous interaction at two kinds of nucleic acid-binding sites. J Biol Chem. 1982;257:5760–6. [PubMed] [Google Scholar]
- 22.Geiselmann J, Wang Y, Seifried SE, von Hippel PH. A physical model for the translocation and helicase activities of Escherichia coli transcription termination protein Rho. Proc Natl Acad Sci U S A. 1993;90:7754–8. doi: 10.1073/pnas.90.16.7754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walstrom KM, Dozono JM, von Hippel PH. Kinetics of the RNA-DNA helicase activity of Escherichia coli transcription termination factor rho. 2. Processivity, ATP consumption, and RNA binding. Biochemistry. 1997;36:7993–8004. doi: 10.1021/bi963180r. [DOI] [PubMed] [Google Scholar]
- 24.Steinmetz EJ, Platt T. Evidence supporting a tethered tracking model for helicase activity of Escherichia coli Rho factor. Proc Natl Acad Sci U S A. 1994;91:1401–5. doi: 10.1073/pnas.91.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennink ML, Leuba SH, Leno GH, Zlatanova J, de Grooth BG, Greve J. Unfolding individual nucleosomes by stretching single chromatin fibers with optical tweezers. Nat Struct Biol. 2001;8:606–10. doi: 10.1038/89646. [DOI] [PubMed] [Google Scholar]
- 26.Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, Wang MD. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat Struct Mol Biol. 2009;16:124–9. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dame RT, Noom MC, Wuite GJ. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444:387–90. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- 28.Jiang J, Bai L, Surtees JA, Gemici Z, Wang MD, Alani E. Detection of high-affinity and sliding clamp modes for MSH2–MSH6 by single- molecule unzipping force analysis. Mol Cell. 2005;20:771–81. doi: 10.1016/j.molcel.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 29.Chen CY, Galluppi GR, Richardson JP. Transcription termination at lambda tR1 is mediated by interaction of rho with specific single-stranded domains near the 3′ end of cro mRNA. Cell. 1986;46:1023–8. doi: 10.1016/0092-8674(86)90701-4. [DOI] [PubMed] [Google Scholar]
- 30.Graham JE, Richardson JP. rut Sites in the nascent transcript mediate Rho-dependent transcription termination in vivo. J Biol Chem. 1998;273:20764–9. doi: 10.1074/jbc.273.33.20764. [DOI] [PubMed] [Google Scholar]
- 31.Vieu E, Rahmouni AR. Dual role of boxB RNA motif in the mechanisms of termination/antitermination at the lambda tR1 terminator revealed in vivo. J Mol Biol. 2004;339:1077–87. doi: 10.1016/j.jmb.2004.04.022. [DOI] [PubMed] [Google Scholar]
- 32.Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Direct observation of hierarchical folding in single ribo switch aptamers. Science. 2008;319:630–3. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dalal RV, Larson MH, Neuman KC, Gelles J, Landick R, Block SM. Pulling on the nascent RNA during transcription does not alter kinetics of elongation or ubiquitous pausing. Mol Cell. 2006;23:231–9. doi: 10.1016/j.molcel.2006.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–62. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 35.Darst SA. Bacterial RNA polymerase. Curr Opin Struct Biol. 2001;11:155–62. doi: 10.1016/s0959-440x(00)00185-8. [DOI] [PubMed] [Google Scholar]
- 36.Liphardt J, Onoa B, Smith SB, Tinoco I, Jr, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–7. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 37.Woodside MT, Behnke-Parks WM, Larizadeh K, Travers K, Herschlag D, Block SM. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc Natl Acad Sci U S A. 2006;103:6190–5. doi: 10.1073/pnas.0511048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saenger W. Principles of Nucleic Acid Structure. Springer-Verlag; New York: 1984. [Google Scholar]
- 39.Sharp JA, Galloway JL, Platt T. A kinetic mechanism for the poly(C)-dependent ATPase of the Escherichia coli transcription termination protein, rho. J Biol Chem. 1983;258:3482–6. [PubMed] [Google Scholar]
- 40.Galluppi GR, Richardson JP. ATP-induced changes in the binding of RNA synthesis termination protein Rho to RNA. J Mol Biol. 1980;138:513–39. doi: 10.1016/s0022-2836(80)80016-7. [DOI] [PubMed] [Google Scholar]
- 41.Bear DG, Hicks PS, Escudero KW, Andrews CL, McSwiggen JA, von Hippel PH. Interactions of Escherichia coli transcription termination factor rho with RNA. II. Electron microscopy and nuclease protection experiments. J Mol Biol. 1988;199:623–35. doi: 10.1016/0022-2836(88)90306-3. [DOI] [PubMed] [Google Scholar]
- 42.McSwiggen JA, Bear DG, von Hippel PH. Interactions of Escherichia coli transcription termination factor rho with RNA. I. Binding stoichiometries and free energies. J Mol Biol. 1988;199:609–22. doi: 10.1016/0022-2836(88)90305-1. [DOI] [PubMed] [Google Scholar]
- 43.Brennan CA, Dombroski AJ, Platt T. Transcription termination factor rho is an RNA-DNA helicase. Cell. 1987;48:945–52. doi: 10.1016/0092-8674(87)90703-3. [DOI] [PubMed] [Google Scholar]
- 44.Steinmetz EJ, Brennan CA, Platt T. A short intervening structure can block rho factor helicase action at a distance. J Biol Chem. 1990;265:18408–13. [PubMed] [Google Scholar]
- 45.DeVito J, Das A. Control of transcription processivity in phage lambda: Nus factors strengthen the termination-resistant state of RNA polymerase induced by N antiterminator. Proc Natl Acad Sci U S A. 1994;91:8660–4. doi: 10.1073/pnas.91.18.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu AQ, von Hippel PH. Rho-dependent termination within the trp t′ terminator. II. Effects of kinetic competition and rho processivity. Biochemistry. 1998;37:11215–22. doi: 10.1021/bi972912s. [DOI] [PubMed] [Google Scholar]
- 47.Hart CM, Roberts JW. Deletion analysis of the lambda tR1 termination region. Effect of sequences near the transcript release sites, and the minimum length of rho-dependent transcripts. J Mol Biol. 1994;237:255–65. doi: 10.1006/jmbi.1994.1229. [DOI] [PubMed] [Google Scholar]
- 48.Monforte JA, Kahn JD, Hearst JE. RNA folding during transcription by Escherichia coli RNA polymerase analyzed by RNA self-cleavage. Biochemistry. 1990;29:7882–90. doi: 10.1021/bi00486a015. [DOI] [PubMed] [Google Scholar]
- 49.Epshtein V, Dutta D, Wade J, Nudler E. An allosteric mechanism of Rho-dependent transcription termination. Nature. 2010;463:245–9. doi: 10.1038/nature08669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalyani BS, Muteeb G, Qayyum MZ, Sen R. Interaction with the nascent RNA is a prerequisite for the recruitment of Rho to the transcription elongation complex in vitro. J Mol Biol. 2011;413:548–60. doi: 10.1016/j.jmb.2011.08.053. [DOI] [PubMed] [Google Scholar]
- 51.Nehrke KW, Zalatan F, Platt T. NusG alters rho-dependent termination of transcription in vitro independent of kinetic coupling. Gene expression. 1993;3:119–33. [PMC free article] [PubMed] [Google Scholar]
- 52.Schmidt MC, Chamberlin MJ. Binding of rho factor to Escherichia coli RNA polymerase mediated by nusA protein. J Biol Chem. 1984;259:15000–2. [PubMed] [Google Scholar]
- 53.Schwartz A, Rabhi M, Jacquinot F, Margeat E, Rahmouni AR, Boudvillain M. A stepwise 2′-hydroxyl activation mechanism for the bacterial transcription termination factor Rho helicase. Nat Struct Mol Biol. 2009;16:1309–16. doi: 10.1038/nsmb.1711. [DOI] [PubMed] [Google Scholar]
- 54.Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–47. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- 55.Burgess BR, Richardson JP. RNA passes through the hole of the protein hexamer in the complex with the Escherichia coli Rho factor. J Biol Chem. 2001;276:4182–9. doi: 10.1074/jbc.M007066200. [DOI] [PubMed] [Google Scholar]
- 56.Nowatzke W, Richardson L, Richardson JP. Purification of transcription termination factor Rho from Escherichia coli and Micrococcus luteus. Methods Enzymol. 1996;274:353–63. doi: 10.1016/s0076-6879(96)74030-2. [DOI] [PubMed] [Google Scholar]
- 57.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–5. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang MD, Yin H, Landick R, Gelles J, Block SM. Stretching DNA with optical tweezers. Biophys J. 1997;72:1335–46. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kienberger F, Costa LT, Zhu R, Kada G, Reithmayer M, Chtcheglova L, Rankl C, Pacheco AB, Thalhammer S, Pastushenko V, Heckl WM, Blaas D, Hinterdorfer P. Dynamic force microscopy imaging of plasmid DNA and viral RNA. Biomaterials. 2007;28:2403–11. doi: 10.1016/j.biomaterials.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 60.Seol Y, Skinner GM, Visscher K. Elastic properties of a single-stranded charged homopolymeric ribonucleotide. Phys Rev Lett. 2004;93:118102. doi: 10.1103/PhysRevLett.93.118102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.