Abstract
Autophagy targets pathogens, damaged organelles and protein aggregates for lysosomal degradation. These ubiquitinated cargoes are recognised by specific autophagy receptors, which recruit LC3-positive membranes to form autophagosomes. Subsequently, autophagosomes fuse with endosomes and lysosomes, thus facilitating degradation of their content, however, the machinery that targets and mediates fusion of these organelles with autophagosomes remains to be established. Here we demonstrate that myosin VI, in concert with its adaptor proteins NDP52, optineurin, T6BP and Tom1, plays a crucial role in autophagy. We identify Tom1 as a myosin VI binding partner on endosomes and demonstrate that their loss reduces autophagosomal delivery of endocytic cargo and causes a block in autophagosome-lysosome fusion. We propose that myosin VI delivers endosomal membranes containing Tom1 to autophagosomes by docking to NDP52, T6BP and optineurin thereby promoting autophagosome maturation and thus driving fusion with lysosomes.
Introduction
Autophagy is an essential lysosomal degradation pathway, in which double-membrane autophagosomes fuse and deliver their contents to lysosomes for degradation. For autophagosome expansion and maturation, membranes may be derived from the plasma membrane, ER, Golgi complex, mitochondria and endosomes1-5, however, how endosomal membranes are delivered to and incorporated into autophagosomes has yet to be established.
Autophagy receptors such as p62, nuclear dot protein 52 (NDP52) and optineurin bind protein aggregates marked with polyubiquitin to target them for autophagic degradation6 and recruit membranes for autophagosome formation through their interaction with Atg8/LC3. NDP52 and optineurin are essential for the autophagy-dependent clearance of pathogens such as Salmonella from the cytosol of eukaryotic cells7,8. We previously showed that the actin-based motor, myosin VI, directly binds to optineurin, NDP52 and its close homologue Traf6-binding protein (T6BP)9,10. Myosin VI is a unique motor protein that moves in the opposite direction along actin filaments to all the other myosins. It functions in a wide variety of intracellular processes including cargo sorting and membrane delivery in the endocytic pathway11, and like optineurin, it is linked to neurodegeneration12,13,14.
In this paper, we identify the mammalian homologue of Tom1/Tom1L2, a constituent of an alternative ESCRT-0 complex15, as a myosin VI cargo adapter and demonstrate a crucial role for myosin VI and Tom1 in autophagosome maturation and fusion with the lysosome. Tom1 mediates endosomal localisation of myosin VI by interaction with the cargo-binding WWY motif and loss of myosin VI function leads to a decrease in endocytic cargo delivery to autophagosomes. Recruitment of myosin VI to autophagosomes is dependent on its RRL motif known to interact with NDP52, optineurin, and T6BP. Therefore, myosin VI delivers Tom1-positive endosomal membranes to autophagosomes via docking to optineurin, NDP52 or T6BP, thus facilitating autophagosome maturation.
Results
Loss of myosin VI function leads to an accumulation of autophagosomes
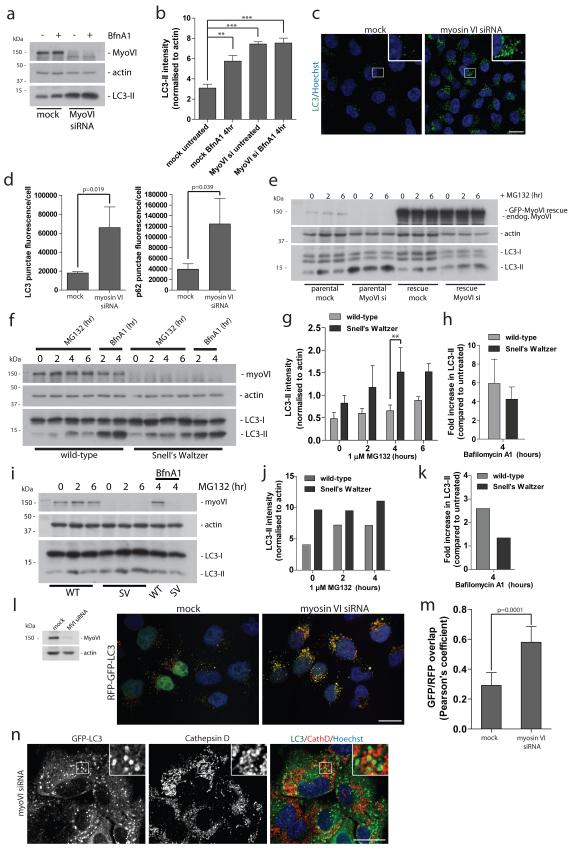
To understand the role of myosin VI and its binding partners in autophagy, we first assessed whether loss of myosin VI, in the myosin VI knockout mouse or by siRNA-mediated knockdown (KD), affects cellular autophagy levels by measuring the abundance of the lipidated autophagosome-associated LC3 (LC3-II)16. In Hela cells, myosin VI expression was suppressed with siRNA and conversion of LC3-I to LC3-II quantified in the presence or absence of the vacuolar-type H+-ATPase inhibitor BafilomycinA1, which blocks fusion of autophagosomes with lysosomes. There is a significant increase in the amount of LC3-II in myosin VI KD cells under steady state conditions as well as in response to autophagy induction following treatment with the proteasome inhibitor MG132 (Figure 1a,b; Supplementary Figure S1a). Elevated levels of LC3-II indicate an accumulation of autophagosomes, which was not due to an increase in autophagosome biogenesis, since there was no increase in LC3-II in the presence of Bafilomycin A1 (Figure 1a,b). We also observed by immunofluorescence microscopy an accumulation of LC3-positive autophagosomes and the autophagy cargo receptor p62 under basal conditions in myosin VI knockdown cells (Figure 1c,d; Supplementary Figure S1b). These results were verified using single siRNA oligonucleotides targeting myosin VI (Supplementary Figure S1c) and by rescue experiments using a Hela cell line stably expressing GFP-myosin VI containing silent mutations in the target region of a single siRNA oligonucleotide (Figure 1e). Loss of myosin VI expression in parental cells leads to an accumulation of LC3-II (Figure 1a,e), while in rescue cells expressing siRNA resistant myosin VI, loss of endogenous myosin VI has no effect on LC3-II levels (Figure 1e). Interestingly, loss of myosin VI expression also leads to enlarged, swollen GFP-LC3-positive autophagosomes (Supplementary Figure S1d,S2a), which increased from an average area of 1.39+/−0.049 (s.d.) μm2 in mock treated cells to 1.57+/− 0.055 (s.d) μm2 in myosin VI depleted cells (Supplementary Figure S1e).
Figure 1. Loss of myosin VI function leads to an accumulation of autophagosomes.
(a) Western blot analysis of myosin VI depleted Hela cells untreated or treated with 100 nM Bafilomycin A1. (b) Quantitation of Western blot LC3-II intensity (+/− s.d.) (n=3). **p<0.01, ***p<0.001 (c) Confocal immunofluorescence microscopy of LC3 punctae (green) in Hela cells following knockdown of myosin VI. Hoechst labels nuclei (blue). Scale bar, 20 μm. (d) Quantitation of LC3- and p62-positive punctae was performed and results are represented as average punctae fluorescence intensity/cell (+/− s.d.) (n=3) from >1500 cells/experiment. (e) Western blot analysis of parental Hela cells or Hela cells stably expressing siRNA resistant GFP-myosin VI transiently transfected with a single myosin VI siRNA oligonucleotide following treatment with 1 μM MG132. (f) Western blot analysis of mouse embryonic fibroblasts cultured from wild-type and Snell’s Waltzer (SV) mice treated with 1 μM MG132 or 100 nM Bafilomycin A1. (g) Quantitation of Western blot LC3-II intensity (+/− s.d) (n=3). **p<0.01. (h) To evaluate effects on autophagosome biogenesis, results are displayed as the fold increase in normalised LC3-II intensity with Bafilomycin A1 compared to untreated control. (+/− s.d.) (n=3) (i) Western blot analysis of cortical neurons from wild-type and SV mice treated with 1 μM MG132 in the absence or presence of 100 nM BafilomycinA1. (j) Quantitation of Western blot LC3-II intensity was performed. (n=2) (k) To evaluate effects on autophagosome biogenesis, results are displayed as fold increase in normalised LC3-II intensity with Bafilomycin A1 compared to untreated control. (n=2) (l) Confocal immunofluorescence microscopy of mock or myosin VI siRNA treated Hela cells stably expressing RFP-GFP-LC3 reporter. Hoechst labels nuclei (blue). (m) Quantitative data of RFP and GFP signal overlap from confocal images of mock or myosin VI siRNA treated Hela cells expressing RFP-GFP-LC3. Data is represented as the Pearson’s coefficient of RFP and GFP signal correlation from >100 cells/experiment. (+/− s.d.) (n=3) Scale bar, 20 μm. (n) Confocal immunofluorescence microscopy of myosin VI depleted RPE cells stably expressing GFP-LC3 immunostained against GFP (green) and Cathepsin D (red). Hoechst labels nuclei (blue). Insets represent magnified boxed regions. Scale bar, 20 μm.
We confirmed the myosin VI loss of function autophagy phenotype using mouse embryonic fibroblasts (MEFs) and primary cortical neurons harvested from the Snell’s Waltzer (SV) myosin VI knockout mouse17. In SV fibroblasts, treatment with the MG132 inhibitor leads to a greater accumulation of LC3-II compared to wild-type control MEFs (Figure 1f,g), while displaying no significant difference in autophagosome biogenesis (Figure 1h), thus suggesting a block in autophagy flux. Interestingly, in primary SV neurons, steady state levels of LC3-II are significantly higher compared to wild-type neurons, thus confirming the phenotype observed in myosin VI siRNA knockdown cells (Figure 1i,j,k). These results indicate that myosin VI is not required for autophagosome formation, but possibly has a role during autophagosome-lysosome fusion.
Myosin VI facilitates autophagosome maturation and fusion with the lysosome
To monitor autophagosome/lysosome fusion in myosin VI KD cells, we used Hela cells that stably expressed a pH-sensitive RFP-GFP-LC3 reporter construct18 and were treated with MG132 to induce autophagy. Loss of myosin VI expression leads to an accumulation of LC3-positive, yellow autophagosomes (Figure 1l), and quantitative analysis of these confocal images indicates an abundant correlation between the GFP and RFP signals compared to mock controls, indicating a defect in their maturation to an autolysosome, which due to the drop in pH should only appear red (Figure 1m). These yellow enlarged autophagosomes do not colocalise with lysosomal marker proteins, such as the protease Cathepsin D or Lamp1, thus supporting a defect in autophagosome/lysosome fusion in myosin VI depleted cells (Figure 1n; Supplementary Figure S2a). The lack of LC3 degradation in myosin VI depleted cells was not caused by a lysosomal defect, since we did not observe a delay or reduction in the degradation of the epidermal growth factor receptor (EGFR) (Supplementary Figure S2b). These results strongly support a role for myosin VI in autophagosome maturation and fusion with the lysosome.
Myosin VI is required for autophagosomal degradation of p62-positive protein aggregates
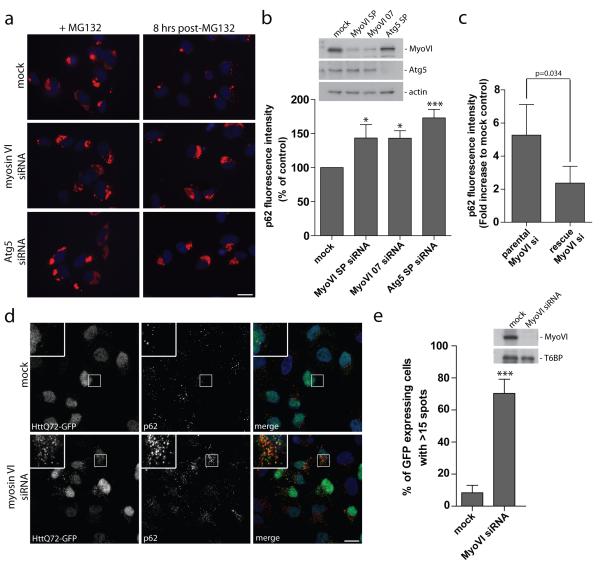
Next, we investigated whether myosin VI dysfunction has an impact on autophagy-dependent clearance of ubiquitinated cargo. RPE cells were treated with MG132 for 16 hours to allow for bulk accumulation of endogenous ubiquitinated protein aggregates19 and 8 hours after washing out this inhibitor, the remaining protein aggregates were visualised with antibodies to p62 (Figure 2a) and quantified by automated microscopy (Figure 2b). Loss of myosin VI using either a pool of four different siRNAs or a single siRNA causes a significant increase in p62-positive aggregates similar to that observed with the knockdown of Atg5, one of the essential autophagy regulators (Figure 2b). This defect in aggregate clearance was rescued by expression of a siRNA resistant myosin VI construct (Figure 2c).
Figure 2. Myosin VI is required for autophagosomal degradation of huntingtin and p62-positive protein aggregates.
(a) RPE cells transiently transfected with siRNA against myosin VI and Atg5 were treated with 1 μM MG132 for 16 hours. Cells were processed for immunofluorescence microscopy following 16 hours MG132 treatment (zero time point) and 8 hours post-washout of inhibitor. Immunolabelling for p62 was performed to visualize aggregates (red) and Hoechst was utilized to identify nuclei (blue). Scale bar, 20 μm (b) Quantitation of immunofluorescent p62 positive aggregates was evaluated using an automated Cellomics VTi microscope. Results were calculated as the average p62 fluorescence intensity at 8 hours post-MG132 washout normalized to the zero time point and represented as percent of control (+/− s.d.) (n=3). * p<0.05, ***p<0.001. (c) Parental or stable expressing siRNA resistant GFP-myosin VI Hela cells were transiently transfected with a single target myosin VI siRNA oligonucleotide and were subsequently treated with 1 μM MG132 for 16 hours. Cells were processed for immunofluorescence microscopy at 16 hours post-MG132 treatment (t=0) or allowed to recover following washout of MG132 for 8 hours (t=8). Cells were processed for quantitation with the automated Cellomics VTi microscope to evaluate p62 fluorescence intensity. Results represent the fold increase in p62 fluorescence intensity of myosin VI siRNA compared to mock control cells following recovery from MG132 washout (t=8) (+/− s.d) (n=3). (d) Hela cells with stable expression of HttQ72-GFP were transiently transfected with siRNA against myosin VI followed by saponin extraction and processing for immunofluorescence microscopy. Immunolabelling was performed for GFP (green) and p62 (red). Nuclei are labelled with Hoechst (blue). Scale bar, 20 μm. (e) Quantitation of HttQ72-GFP aggregates was performed on myosin VI siRNA transfected Hela cells. Results were calculated as the percentage of GFP expressing cells with greater than 15 GFP-positive spots/cell. Results represent the mean (+/− s.d) from n=3 independent experiments, *** p<0.001.
We tested whether the loss of myosin VI expression has an effect on clearance of huntingtin protein containing a pathological polyQ expansion, which renders this protein prone to misfolding and aggregation20. In Hela cells, we expressed a GFP-tagged huntingtin protein with a 72 amino acid polyglutamine repeat (HttQ72-GFP), which is mostly cytosolic, characterised by <10% of cells containing multiple huntingtin aggregates (>15/cell). However, after myosin VI knock down there is a dramatic increase in the number of cells (from 10% to 70%) with multiple p62- and LC3-positive HttQ72-GFP aggregates (Figure 2d,e; Supplementary Figure S3). Interestingly, myosin VI siRNA knockdown not only leads to an increase in HttQ72-GFP punctae, but also to an accumulation of p62-positive punctae (Figure 1d,2d; Supplementary Figure S1b). Collectively, these results support a role for myosin VI in the clearance of ubiquitinated p62-positive protein aggregates via autophagy.
Myosin VI localises to LC3-positive autophagosomes
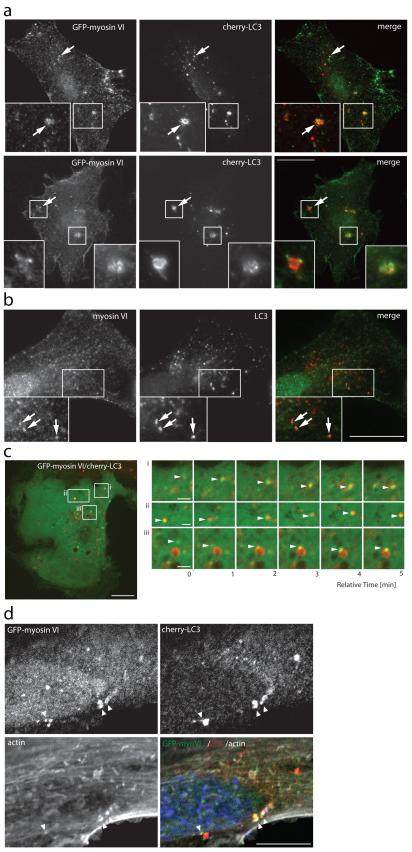
To further characterise the precise role of this motor protein in the autophagocytic pathway, we next analysed myosin VI targeting and localisation to autophagosomes. In human RPE cells, GFP-myosin VI is recruited to cherry-LC3-positive autophagosomes (Figure 3a) and endogenous myosin VI colocalises with endogenous LC3 (Figure 3b) under conditions that induce nonselective autophagy such as treatment with Torin1, a selective mTOR kinase inhibitor, or following amino acid withdrawal (Supplementary Figure S4a). Furthermore, myosin VI colocalises with LC3-positive autophagosomes containing the aggregate prone protein Ataxin3 with an 84 amino acid polyglutamine repeat, Ataxin3Q84 (Supplementary Figure S4b), highlighting a role for myosin VI in nonselective as well as in selective autophagy. A high proportion of LC3-positive autophagosomes colocalise with myosin VI (LC3/MVI), but only a distinct subpopulation of myosin VI is present on LC3-positive vesicles (MVI/LC3), reflecting the association of this multifunctional motor not only with autophagosomes but also with endocytic and secretory membrane trafficking compartments (Supplementary Figure S4c). Often, the localisation of myosin VI does not completely overlap with that of LC3 on autophagosomes, but is present in distinct areas or subdomains of the autophagosomal membrane (Figure 3a,b), reminiscent of a small membrane vesicle docking at a larger autophagocytic vacuole.
Figure 3. Myosin VI localises to LC3-positive autophagosomes.
(a) RPE cells transiently cotransfected with GFP-myosin VI and cherry-LC3 and either left untreated (top row) or treated with 250 nM Torin1 for 2 hours (bottom row) were processed for immunofluorescence microscopy. Arrows highlight areas of colocalisation and inserts provide higher magnification of boxed regions. Scale bar, 20 μm. (b) RPE cells treated with 100 nM BafilomycinA1 were processed for immunofluorescence microscopy to evaluate endogenous myosin VI and endogenous LC3 colocalisation. Arrows highlight areas of colocalisation. Scale bar, 20 μm. (c) Image stills acquired from spinning disk time-lapse video microscopy of RPE cells transiently cotransfected with cherry-LC3 and GFP-myosin VI. Boxed regions highlight areas of interest within the cell that are shown to the right (i, ii, iii) as video image stills captured at 1-minute intervals. Arrowheads highlight myosin VI positive vesicles (green) coming into contact and colocalising with LC3-positive vesicles (red). Scale bar, 10 μm (whole cell image), 2 μm (cropped images). (d) Confocal immunofluorescence microscopy of RPE cells stably expressing cherry-LC3 following transient transfection with GFP-myosin VI. Immunostaining for GFP (green) and cherry (red) was performed and actin was visualised with phalloidin (white). Nuclei were labelled with Hoechst (blue). Arrowheads indicate areas of actin rich myosin VI/LC3-positive autophagosomes. Scale bar, 20 μm
Using live cell video microscopy of RPE cells expressing GFP-myosin VI and cherry-LC3 we monitored their localisation over a period of 10 minutes. Selected still images of these movies illustrate that several myosin VI-associated vesicles (arrowheads) approach and contact LC3-positive autophagosomes (Figure 3c; Supplementary Figure S4d; Movie S1,S2,S3,S4,S5,S6), confirming a dynamic association of myosin VI with autophagosomes.
Since myosin VI is an actin-dependent motor, we probed for the presence of the actin cytoskeleton in the vicinity of autophagocytic organelles. We observed that myosin VI and actin, the motor and the track, are associated with LC3-positive autophagosomes (Figure 3d, arrowheads).
Myosin VI targets to autophagosomes via the RRL motif in the cargo binding tail domain
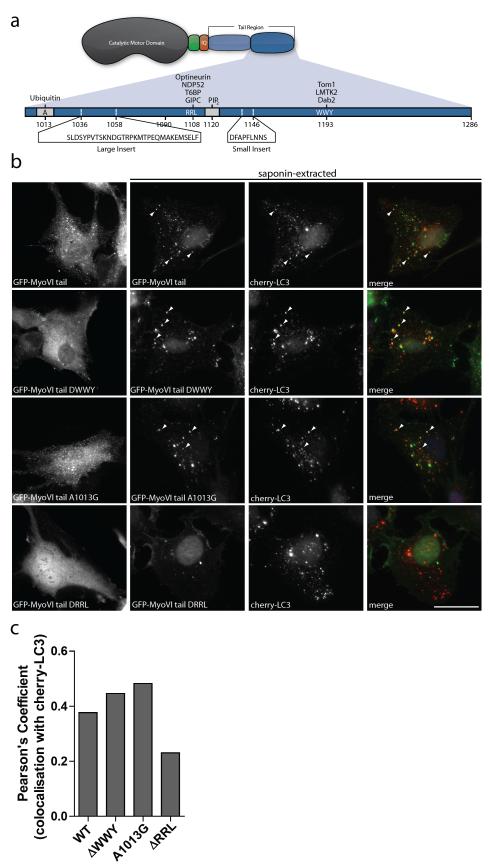
Two primary protein-protein interaction regions with either a WWY or RRL motif are present in the C-terminal cargo-binding tail region of myosin VI, which also contains a ubiquitin-binding domain (UBD)21 (Figure 4a). To identify how myosin VI is recruited to autophagosomes, GFP-myosin VI tail constructs, containing either a WWY to WLY substitution (ΔWWY), RRL to AAA substitution (ΔRRL), or A1013G point mutation in the UBD, were transiently expressed in RPE cells stably expressing cherry-LC3. To visualize the precise intracellular localisation of the GFP-myosin VI tails, the cells were briefly extracted with saponin to remove the cytosolic pool of the GFP-myosin VI tail before fixation. The wild-type myosin VI tail and the tail mutants with the ΔWWY and A1013G mutations show prominent colocalisation with LC3-positive autophagosomes (Figure 4b,c), which is significantly decreased without the RRL motif, which mediates interactions with T6BP, NDP52, and optineurin (Figure 4a,b,c).
Figure 4. Myosin VI targets to autophagosomes via the RRL motif in the cargo binding tail domain.
(a) Cartoon illustration of myosin VI domain structure. Myosin VI contains a catalytic motor domain, a unique insert (green), an IQ Calmodulin-binding motif, and a cargo-binding tail domain. The cargo-binding tail domain contains multiple protein interaction motifs (RRL and WWY) as well as regions for ubiquitin and PIP2 binding. Additionally, two alternative splicing events give rise to two insertions in the cargo-binding tail region. (b) RPE cells with stable expression of cherry-LC3 were transiently transfected with the GFP-myosin VI cargo-binding tail domain containing various mutations in the protein interaction (ΔWWY and ΔRRL) and ubiquitin-binding motifs (A1013G), followed by treatment with 250 nM Torin1 for 3 hours to induce autophagy. Immunofluorescence microscopy was performed either in the absence or presence of saponin-extraction. Arrowheads indicate areas of colocalisation. Scale bar, 20 μm. (c) A Pearson’s coefficient was calculated based on the degree of colocalisation between the different GFP-myosin VI mutant tails and cherry-LC3 from confocal immunofluorescence images. Graph represents data from more than 20 transfected cells from n=2 independent experiments.
Binding partners colocalise with myosin VI on autophagosomes
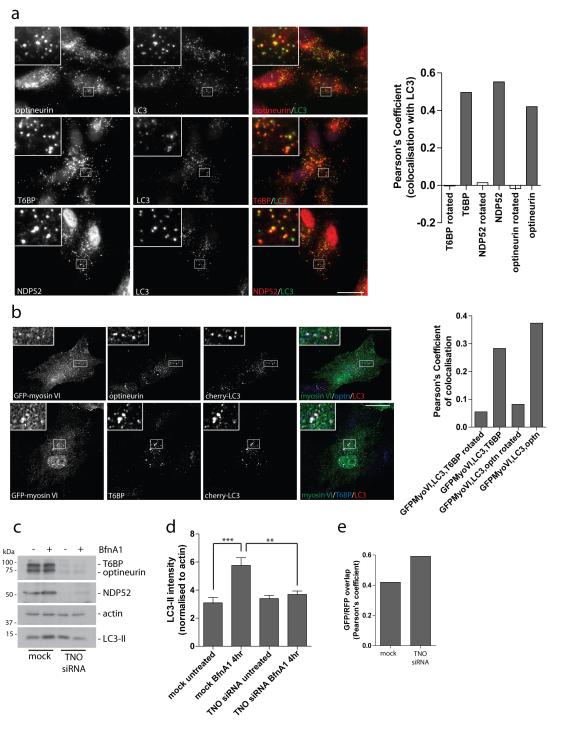
The myosin VI binding partners, optineurin, NDP52 and T6BP all contain an ubiquitin-binding domain (UBD) and a LC3 interaction region (LIR), which are typically found in autophagy receptors such as p62 and NBR122. Since previous reports demonstrated a role for NDP52 and optineurin in autophagy-dependent clearance of Salmonella from mammalian cells7,8, we tested whether the function of these autophagy receptors is restricted to pathogen clearance or whether they have a general role in autophagy. Endogenous T6BP, NDP52 and optineurin are present on LC3-positive autophagosomes under basal autophagy conditions in the presence of BafilomycinA1 (Figure 5a). Interestingly, whereas localisation of NDP52 and T6BP to autophagosomes does not require their C-terminal zinc-finger motifs that mediate ubiquitin-binding, the ubiquitin-binding motif of optineurin, within the myosin VI interaction domain, disrupts its localisation to autophagosomes (Supplementary Figure S5a).
Figure 5. The myosin VI binding partners, T6BP, NDP52, and optineurin, colocalise with myosin VI on autophagosomes and are required for autophagosome biogenesis.
(a) Immunofluorescence microscopy was performed on RPE cells following treatment with 100 nM Bafilomycin A1 for 2 hours. Immunolabelling was performed against the indicated endogenous proteins, optineurin, T6BP, and NDP52 (red), and LC3 (green). Boxed regions provide higher magnification. From confocal images, a Pearson’s coefficient was calculated to estimate the degree of colocalisation of the different autophagy adaptors with LC3. As a negative control, a Pearson’s coefficient was calculated after rotation of one color image by 90 degrees. Results represent >20 cells from n=2 independent experiments. Scale bar, 20 μm. (b) Stable cherry-LC3 expressing RPE cells were transiently transfected with GFP-myosin VI, treated with Torin1 for 3 hours to induce autophagy, and subsequently processed for confocal immunofluorescence microscopy. Areas of colocalisation appear white in the merged three-color images and boxed regions provide areas of higher magnification. Scale bar, 20 μm. A Pearson’s coefficient was calculated for the degree of colocalisation between the 3 colors. As a negative control, a Pearson’s coefficient was calculated after rotating one color image by 90 degrees. Results represent >10 cells from n=2 independent experiments. (c) Hela cells were either mock transfected or cotransfected with siRNA targeted against T6BP, NDP52, and optineurin (TNO). Cells were either left untreated or treated with100 nM Bafilomycin A1 (BfnA1) for 4 hours. Western blot analysis was performed against the indicated proteins. (d) Quantitation of LC3-II intensity (+/− s.d.) was performed from Western blot data of TNO siRNA treated Hela cells. The results represent n=3 independent experiments. **p<0.01, ***p<0.001 (e) Immunofluorescence microscopy of RFP-GFP-LC3 expressing Hela cells transfected with TNO siRNA was performed and confocal images were evaluated for the correlation in GFP and RFP punctae signal overlap using the Image JACoP plugin. Results are calculated from >100 cells from n=2 independent experiments represented as the Pearson’s coefficient of GFP/RFP signal overlap.
To test whether these binding partners/autophagy receptors recruit myosin VI to autophagosomes, RPE cells stably expressing cherry-LC3 were transiently transfected with GFP-myosin VI and subsequently treated with Torin1 before processing for immunocytochemistry with antibodies to endogenous optineurin or T6BP (Figure 5b). The triple-labelling clearly shows that myosin VI and its binding partners T6BP and optineurin are present on the same LC3-positive autophagosomes after treatment with Torin1 (Figure 5b).
Optineurin, NDP52 and T6BP are required for autophagosome biogenesis
We next tested which step along the autophagocytic pathway requires optineurin, NDP52 and T6BP. In Hela cells, a triple siRNA knockdown of T6BP, NDP52, and optineurin (TNO) does not significantly affect the steady-state level of autophagy as measured by conversion of LC3-I to LC3-II (Figure 5c,d). However, a triple knockdown for T6BP, NDP52 and optineurin causes a significant reduction in autophagosome formation under basal conditions or following induction of autophagy with the MG132 inhibitor, characterised by a decrease in LC3-I to LC3-II conversion in the presence of Bafilomycin A1 (Figure 5c,d; Figure S5b). In addition TNO siRNA in Hela cells stably expressing RFP-GFP-LC3 also leads to higher correlation of RFP/GFP signals, indicating a defect in autophagosome-lysosome fusion (Figure 5e). Thus, optineurin, T6BP and NDP52 act as autophagy receptors, linking autophagy cargo to LC3 membranes, thereby promoting autophagosome biogenesis, but also have a role during autophagosome-lysosome fusion.
Tom1 interacts with myosin VI
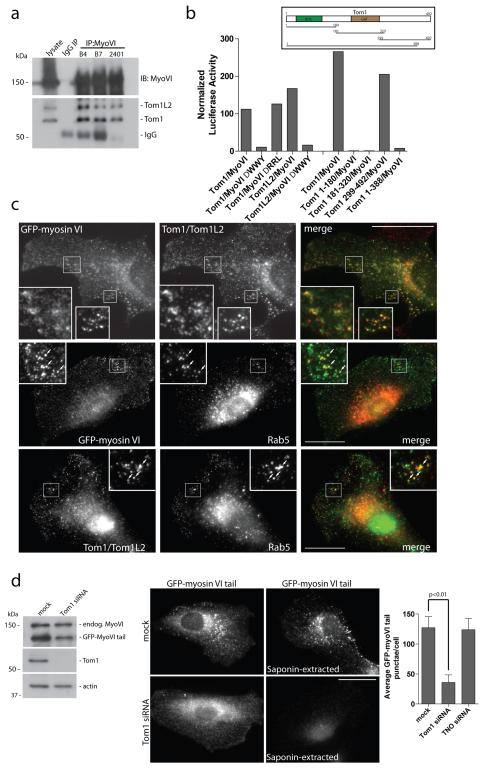
The Drosophila homologue of the Tom1 protein family, CG3529, which is an alternate endosomal sorting complex required for transport (ESCRT) class 0, was recently identified as a myosin VI binding partner23,24,25. Since the ESCRTs are required for autophagosome maturation26 , we tested whether Tom1 and myosin VI interact in mammalian cells and whether Tom1 is required for autophagy. Myosin VI and Tom1, as well as its very close homologue Tom1L2, interact in mammalian cells as they can be coimmunoprecipitated with myosin VI antibodies (Figure 6a). Using a mammalian two-hybrid assay9,10,27 we show that Tom1 binds to myosin VI via the WWY motif and not via the RRL motif, which binds optineurin, NDP52 or T6BP. The myosin VI binding site on Tom1 resides within the final 104 amino acids of the C-terminus (Figure 6b). Myosin VI and Tom1 colocalise on vesicles located along the cell periphery, that are positive for endosomal markers APPL1, GIPC, and Rab5 (Figure 6c; Supplementary Figure S6a,b). In the absence of Tom1 and Tom1L2, myosin VI is lost from endosomes (Figure 6d), and accumulates on F-actin structures throughout the cell (Supplementary Figure S7). A partial effect was observed following knockdown of the myosin VI binding partner GIPC; however, loss of T6BP, NDP52 or optineurin did not effect myosin VI localisation (Figure 6d; Supplementary Figure S6c). Thus, the Tom1 protein family is a newly identified class of myosin VI adaptor proteins that are important for targeting myosin VI to endocytic structures in mammalian cells.
Figure 6. Tom1 interacts with myosin VI and is required for myosin VI localisation to endosomes.
(a) Myosin VI and Tom1/Tom1L2 coimmunoprecipitate from LNCAP cell lysates. Immunoprecipitation of myosin VI with three separate affinity purified rabbit polyclonal antibodies (B4, B7, 2401) was performed alongside an IgG control immunoprecipitation. The immunoprecipitates were analysed by Western blot with antibodies to Tom1/Tom1L2. (b) Binding of myosin VI to Tom1 requires the WWY motif. The mammalian two-hybrid assay was used to test binding of full-length Tom1 and Tom1L2 or various truncated versions of Tom1, annotated by amino acid numbers, against wild type myosin VI tail or the myosin VI ΔWWY or ΔRRL tail. The graph shows the mean values from n=2 independent experiments. VHS = Vps-27, Hrs and STAM, GAT = GGA and Tom1. (c) RPE cells stably expressing GFP-myosin VI were processed for immunofluorescence microscopy to evaluate GFP-myosin VI (green) and endogenous Tom1/Tom1L2 (red) or Rab5 (red) colocalisation. Inserts provide higher magnification of boxed regions. Arrows indicate areas of colocalisation. Scale bar, 20 μm. (d) RPE cells with stable expression of GFP-myosin VI tail were subjected to mock or Tom1 siRNA transfection followed by Western blot analysis and immunofluorescence microscopy to evaluate GFP-myosin VI tail localisation. Cells were either fixed directly or saponin-extracted prior to fixation. Scale bar, 20 μm. Quantitation was performed on GFP-myosin VI tail stable expressing RPE cells transfected with mock, Tom1, or TNO siRNA. Cells were processed for immunocytochemistry, immunolabelled for GFP and nuclei labeled with Hoechst followed by quantitation of GFP-myosin VI tail punctae/cell using an automated Cellomics VTi microscope. More than 600 cells/group from n=3 independent experiments were analysed and represented as the mean number of GFP-myosin VI tail punctae/cell (+/− s.d.).
Loss of Tom1 inhibits the maturation of autophagosomes and their subsequent fusion with lysosomes
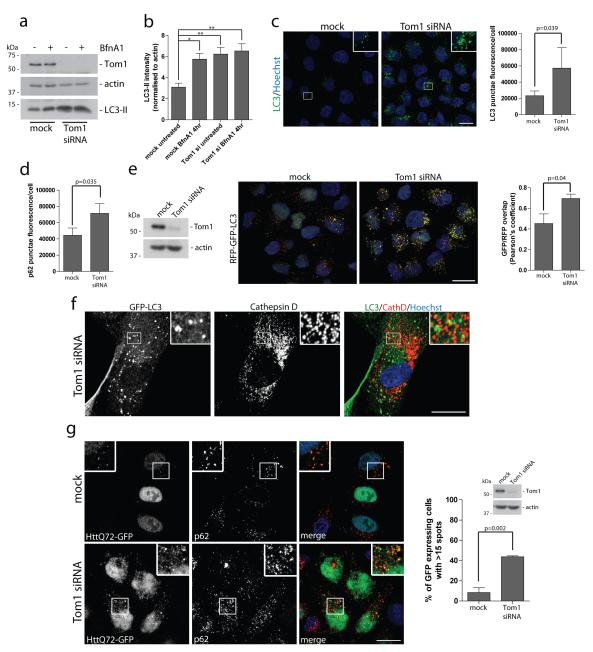
Next, we determined whether Tom1 is required for autophagy. SiRNA-mediated knockdown of Tom1 leads to an accumulation of LC3-II (Figure 7a,b; Supplementary Figure S8a), and an increase in the number of LC3-positive autophagosomes and p62-positive punctae visualised by immunofluorescence microscopy (Figure 7c,d; Supplementary Figure S8b), very similar to the phenotype observed in myosin VI knockdown cells (Figure 1a-d). In the Hela RFP-GFP-LC3 stable cell line, loss of Tom1 expression leads to an accumulation of LC3-positive, yellow autophagosomes that are Cathepsin D- and Lamp1-negative (Figure 7e,f; Supplementary Figure S2a). We also observed in Tom1 knockdown cells a defect in protein aggregate clearance and thus an increase in the number of cells with multiple p62 and LC3-positive HttQ72-GFP aggregates (Figure 7g; Supplementary Figure S3), phenocopying the knockdown of myosin VI (Figure 2d,e; Supplementary Figure S3). Given the very similar requirement for Tom1 and myosin VI during late stages of autophagy, we next tested whether Tom1 is recruited to autophagosomes and whether this localisation was myosin VI-dependent. As shown in figure 8a, endogenous Tom1 is present on LC3-positive autophagosomes. However, siRNA knockdown of myosin VI causes an increase in the number of Tom1-positive vesicles that are adjacent to, but not completely colocalising with LC3-positive autophagosomes, suggesting a defect in docking or fusion of these vesicles with autophagosomes (Figure 8a). In summary, myosin VI and Tom1 function together in the final stages of autophagy, since the loss of either protein leads to an accumulation of autophagosomes unable to mature to an autolysosome.
Figure 7. Loss of Tom1 inhibits the maturation of autophagosomes and their subsequent fusion with lysosomes.
(a) Hela cells transiently transfected with siRNA against Tom1 were left untreated or treated with 100 nM Bafilomycin A1 for 4 hours. Western blot analysis was performed on whole cell lysates against the indicated proteins. (b) Quantitation of LC3-II intensity of Western blots were performed by an infrared imaging system. (+/− s.d.) (n=3) *p<0.05, **p<0.01 (c) Confocal immunofluorescence microscopy was performed on Hela cells transfected with Tom1 siRNA to evaluate LC3 punctae formation (green). Inserts provide higher magnification of boxed regions. Nuclei are labeled with Hoechst (blue). Scale bar, 20 μm. Quantitation of LC3-positive punctae (c) and p62-positive punctae (d) was performed in 96-well format on an Arrayscan VTi HCS microscope. Cells were identified by Hoechst staining of nuclei (blue). Results are represented as the average punctae fluorescence/cell (+/− s.d.) from n=3 independent experiments each performed in triplicate wells, each with >500 cells. (e) Hela cells with stable expression of the RFP-GFP-LC3 reporter construct were subjected to mock or Tom1 siRNA mediated knockdown. Confocal immunofluorescence microscopy was performed and images were subsequently quantitated for the correlation between the GFP and RFP signals using the ImageJ JACoP plugin. Results are of at least 100 individual cells from n=3 independent experiments and are represented as the Pearson’s coefficient of GFP/RFP overlap (+/− s.d.). A higher Pearson’s coefficient of GFP/RFP signal overlap represents a greater number of autophagosomes compared to autolysosomes. Scale bar, 20 μm. (f) Confocal immunofluorescence microscopy of Tom1 depleted RPE cells stably expressing GFP-LC3 immunostained against GFP (green) and Cathepsin D (red). Hoechst labels nuclei (blue). Insets represent magnified boxed regions. Scale bar, 20 μm. (g) Hela cells with stable expression of HttQ72-GFP were transiently transfected with Tom1 siRNA, saponin-extracted, and processed for immunofluorescence microscopy to quantitate HttQ72-GFP punctae. Immunolabelling against GFP (green) and p62 (red) was performed. Nuclei (blue) were labeled with Hoechst. Results are represented as the number of GFP-expressing cells with greater than 15 GFP-positive spots/cell (+/− s.d.) (n=3). Scale bar, 20 μm.
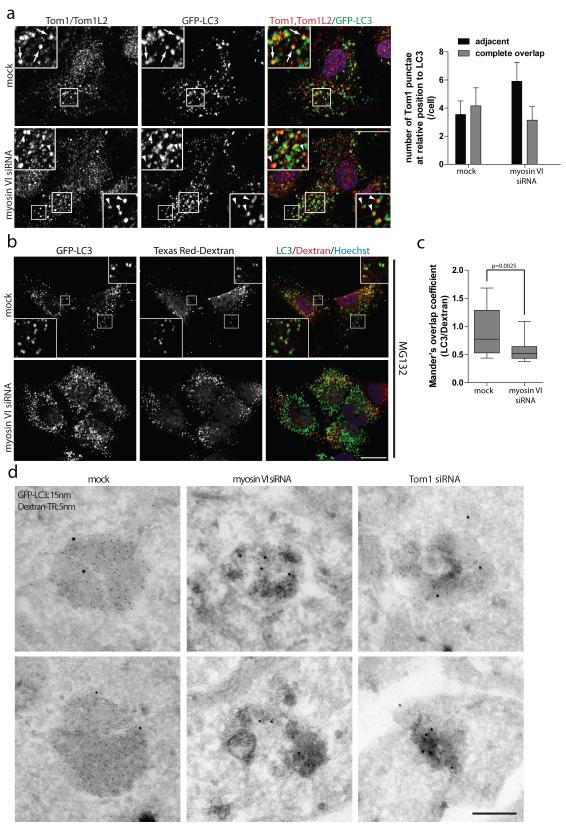
Figure 8. Myosin VI mediates delivery of endocytic cargo to autophagosomes.
(a) RPE cells with stable expression of GFP-LC3 following mock or myosin VI siRNA transfection were processed for confocal immunofluorescence microscopy and immunolabelled for Tom1/Tom1L2 (red). Nuclei (blue) were labeled with Hoechst. Inserts provide higher magnification of boxed regions and arrows highlight areas of complete overlapping colocalisation and arrowheads highlight adjacent localisation of Tom1 relative to LC3. Scale bar, 20 μm. Quantitation was performed on >75 cells for the relative position (adjacent or complete overlap) of Tom1-positive vesicles in relation to LC3-positive vesicles. Results are represented as the average number of Tom1-positive vesicles/cell categorised by their relative position to LC3 (+/− s.d.) (n=3). (b) GFP-LC3 expressing RPE cells were depleted of myosin VI by siRNA. Cells were pulse labeled with Texas Red Dextran (red) for 4 hours, prior to chase into fresh media containing 1 μM MG132 for 2 hours. Cells were processed for immunofluorescence microscopy and immunostained for GFP (green). Nuclei were labeled with Hoechst (blue). Insets represent higher magnification of boxed regions. Scale bar, 20 μm. (c) Mander’s overlap coefficients for the degree of LC3 signal colocalising with Dextran were calculated using Volocity software. Results represent >20 cells from n=3 independent experiments and are illustrated as a box and whisker plot. Box represents median, 25th and 75th percentiles and whiskers represent max and min. (d) RPE cells stably expressing GFP-LC3 were depleted of myosin VI and Tom1 by siRNA. Cells were pulse-labelled with Texas-Red Dextran for 16 hours followed by a chase period of 4 hours. Cells were processed for immunoelectron microscopy and labeled with 15 nm gold particles against GFP-LC3 and 5 nm gold particles against Texas Red-Dextran. Scale bar, 200 nm.
Myosin VI mediates delivery of endocytic cargo to autophagosomes
Since myosin VI and Tom1 are both present on the same endosomal compartment and their loss significantly effects autophagy flux, we next determined whether their requirement for autophagosome maturation is linked to their function in the endocytic pathway. To label the endocytic pathway, RPE cells stably expressing GFP-LC3 where pulse-labelled with dextran followed by treatment with the MG132 inhibitor. Confocal immunofluorescence microscopy revealed considerable colocalisation of LC3-positive autophagosomes with fluorescently-labelled dextran (Figure 8b). However, following myosin VI depletion by siRNA transfection, a significant reduction in colocalisation between LC3 and dextran was observed (Figure 8b,c). The delivery of endocytosed dextran to LC3-positive autophagosomes was confirmed at the ultrastructural level (Figure 8d). Representative immunoEM images of LC3- and dextran-positive amphisomes indicate that on average the LC3- and dextran positive organelles contain darker, denser content and more endocytosed dextran in mock treated cells compared to knockdown cells, which suggest a possible delay in maturation of LC3-positive structures in the absence of myosin VI or Tom1 (Figure 8d). We conclude that myosin VI, via docking to its autophagy adapters, delivers Tom1-positive endosomes to autophagosomes, thus facilitating autophagosome maturation and fusion with the lysosome (Supplementary Figure S8c).
Discussion
Here we demonstrate for the first time that the unique reverse-directed myosin VI plays a crucial role in autophagy. Myosin VI depletion in the knockout mouse and in siRNA knockdown cells leads to an abundant accumulation of autophagosomes due to a lysosomal fusion defect and causes a reduced rate of protein aggregate clearance, but has no significant effect on autophagosome biogenesis. These phenotypes are mirrored by Tom1, an ESCRT-0 component, which we identify as a myosin VI binding partner in mammalian cells. We demonstrate that myosin VI is required for the short-range movement and/or fusion of Tom1-positive endosomes to autophagosomes and the delivery of endocytic cargo to LC3-positive organelles. Live cell video microscopy highlights the dynamic approach, association and occasional fusion of myosin VI containing vesicles with autophagosomes. These vesicles may originate from a subpopulation of endosomes underneath the plasma membrane, which contain myosin VI, Tom1, Rab5, and APPL1 (Figure 6; Supplementary Figure S6a), suggesting that myosin VI is involved in the delivery of endocytic membranes to the autophagosome. Indeed we demonstrate that trafficking of endocytosed dextran to autophagosomes is dependent on myosin VI.
Targeting of myosin VI involves the RRL motif that binds optineurin, NDP52 and T6BP, three myosin VI adaptor proteins that contain a UBD and a LIR28. Although optineurin and NDP52 have been specifically implicated in innate immunity7,8, we demonstrate they have a more general role in selective autophagy. We also define T6BP as a newly identified autophagy receptor. In all three proteins, the ubiquitin-binding site overlaps with the myosin VI binding region, which suggests they may have a dual function. Firstly, during autophagosome biogenesis they may act like p62 as adaptors between ubiquitinated cargo and LC3-conjugated membranes28, since depletion of optineurin, T6BP or NDP52 leads to a defect in autophagosome formation. Secondly, these autophagy receptors may bind to LC3 on the outer limiting membrane of the autophagosome and to myosin VI instead of ubiquitin, thereby recruiting this motor and possibly associated membrane to autophagosomes (Supplementary Figure S8c). The failure in autophagosome/lysosome fusion, associated with myosin VI depletion may be caused by defects in autophagosome maturation due to lack of delivery, docking or fusion of endosomal membranes containing the ESCRT-0 component Tom1.
Tom1, via its WWY motif, recruits myosin VI to endosomal membranes and knockdown of Tom1 leads to an accumulation of autophagosomes attributed to a lysosomal fusion defect. This is the first time Tom1 has been implicated in autophagy, although dysfunction in other members of the ESCRT machinery have been linked to similar autophagy defects and to neurodegeneration29,26.
Autophagosomes are surrounded by an actin filament network (Figure 3d) that is required for efficient autophagosome/lysosome fusion30. Myosin VI may be involved in short-range movement and/or tethering/anchoring of Tom1-containing endosomes to actin filaments surrounding the autophagosome. The delivery of endosomes is a process shown to be required for autophagosome maturation1,31. These proposed functions of myosin VI are in line with the known motor properties of this myosin as a slow moving motor with a maximum speed of about 60–100 nm/s.
Myosin VI is expected to fulfill multiple unique and even fundamental functions in cells due to its distinct movement along actin filaments. Myosin VI functions at various steps along endocytic and secretory membrane trafficking pathways due to its interactions with a variety of adaptor proteins. The autophagy pathway is not permanently activated, but is induced upon specific stimuli, where existing proteins and pathways such as for example myosin VI and the ESCRT machinery are used to mediate high autophagic flux. Similar to myosin VI, ESCRT proteins can function in numerous different fundamental processes such as sorting in the endocytic pathway and multivesicular body formation, virus budding, cytokinesis as well as autophagy32.
These diverse cellular roles of myosin VI are reflected in the profound phenotypic abnormalities in multiple systems that are slowly emerging from the myosin VI knockout mouse and some phenotypes are suggestive of autophagy defects, including astrogliosis in the brain and decreased body weight. During autophagy, myosin VI may play a crucial, proactive role regulating the fusion of Tom1-positive endosomal membranes with autophagosomes and in addition, like its role in regulating secretory vesicle fusion events at the plasma membrane33,34, may also be involved in the fusion of the autophagosome with the lysosome and the release of its content for degradation.
Methods
Antibodies and Reagents
The antibodies used were as follows: LC3 (M152-3;1:200 IF) monoclonal antibody (MBL International, Woburn, Massachusetts, USA), Rab5 (610724;1:100 IF), EEA1 (610457;1:100 IF), EGFR (610016;1:1000 WB) and p62 (610832;1:250 IF) monoclonal antibodies (BD Biosciences, Oxford, United Kingdom), actin (A2066;1:10,000 WB), LC3 (L7543;1:2000 WB), GIPC (sc-9648;1:100 IF), APPL1 (sc-67402;1:100 IF), and Atg5 (A2859;1:1000 WB) antibodies (Sigma-Aldrich, Dorset, England), Tom1L2 (ab96320;1:100 IF, 1:1000 WB) and GFP (ab6556;1:50 EM) polyclonal antibodies and GFP (ab1218;1:1000 IF) monoclonal antibody (Abcam, Cambridge, United Kingdom), GFP (A11122;1:2000 WB, 1:1000 IF) polyclonal antibody (Invitrogen, Paisley, UK), and DsRed (632496;1:500 IF) polyclonal antibody (Clontech, Mountain View, CA, USA). Lamp1 (H4A3;1:250 IF) monoclonal antibody was purchased from Developmental Studies Hybridoma Bank (University of Iowa, USA). Affinity purified rabbit polyclonal antibodies against myosin VI, T6BP, NDP52, and optineurin were generated as previously described9,10, 35. Bafilomycin A1 (B1793) and EGF were purchased from Sigma-Aldrich. Torin1 was a kind gift from David M. Sabatini (Whitehead Institute, Boston, USA). The MG132 inhibitor was purchased from Millipore (Massachusetts, USA) and Protein A conjugated to 5 and 15 nm colloidal gold from the Department of Cell Biology, University of Utrecht. Rabbit anti-Texas Red (A-639;1:50 EM) was purchased from Life Technologies (Paisley, UK) and Texas Red lysine fixable Dextran 10,000 MW from Invitrogen.
Cell Culture and transfections
Hela cells were cultured in RPMI and Chinese hamster ovary (CHO) cells in Ham’s F-12, supplemented with 10% FBS, 2 mM Glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. RPE cells were cultured in DMEM:Ham’s F-12 (50:50), 10% FBS, 2 mM Glutamine, penicillin/streptomycin, and 30 mM sodium bicarbonate. All cDNA transfections were performed using Fugene6 according to the manufacturer’s instructions (Roche Diagnostics, Burgess Hill, UK). Stable cell lines were generated using pIRES neo2 plasmids and subsequent selection in G418 (Invitrogen). For Dextran uptake, cells were pulse labeled with Texas Red Dextran for 4 hours followed by chase into complete media with or without 1 μM MG132 for 2 hours prior to fixation. For EGFR degradation assays, cells were treated with 20 ng/ml EGF for indicated times prior to Western blot analysis.
All ON-TARGETplus SMARTpool or single target siRNA oligonucleotides were purchased from Dharmacon (Abgene House, Epsom, UK) and subsequently transfected into cells using oligofectamine according to the manufacturer’s instructions (Invitrogen). For efficient knockdown, cells were transfected twice with siRNA on Day 1 and 3 and on Day 5 cells were processed for the corresponding assay and efficiency of protein depletion was assessed by Western blot. siRNA sequences are listed in Supplementary Table 1.
Primary Cultures of embryonic fibroblasts and cortical neurons
All experiments were conducted in accordance with the United Kingdom Animals (Scientific Procedure) Act of 1986. Embryonic fibroblasts and cortical neurons were established from wild-type and Snell’s Waltzer embryos at E15-E17 of development from euthanised pregnant females. For preparation of fibroblasts, skin, limbs and muscle tissue was dissected from the embryos, the tissue was minced, and incubated in Trypsin containing 300 U/ml DNAse (Invitrogen) for 20 minutes at 37°C. Following addition of growth media (DMEM with 10% FBS, glutamine, penicillin, and streptomycin), digested tissue was triturated through a glass pasteur pipette, centrifuged, and resuspended in growth media and plated onto tissue-culture treated dishes. Cells were allowed to adhere and recover for 24 hours and cells were used at early passage. For preparation of cortical neurons, embryonic brains were dissected, meninges removed, and cortices were separated from the rest of the brain. Cortices were minced, triturated through a pasteur pipette, centrifuged, and resuspended in Neurobasal A media (Invitrogen) with B27 supplement (Invitrogen), penicillin and streptomycin. Cells were plated onto poly-L-ornithine (Sigma) coated tissue culture dishes and experiments were performed 5-7 days post-plating. Each experiment was performed from separate litters.
Plasmids
GFP-myosin VI and GFP-myosin VI tail WT, RRL to AAA and WWY to WLY constructs were generated as previously described34,36. The myosin VI ubiquitin-binding mutant A1013G was generated by Quick-change site-directed mutagenesis. The full-length GFP-myosin VI wild-type oligonucleotide 07 siRNA resistant construct was generated by the insertion of 3 silent mutations by site-directed mutagenesis. Stable cell lines were obtained by subcloning constructs into pIRESneo2 followed by transfection and selection with G418 prior to pooling positive cells and enrichment by fluorescence-activated cell sorting. GFP-tagged optineurin, T6BP, and NDP52 constructs were generated as previously described9,10. The D474N mutant of optineurin was generated by site-directed mutagenesis. Cherry-LC3 was a kind gift from Geoffrey Hesketh (University of Cambridge, UK). The GFP-LC3 construct created by Karla Kirkegaard37 (Addgene plasmid 11546) and the RFP-GFP-LC3 construct created by Tomatsu Yoshimori18 (ptfLC3; Addgene plasmid 21074) were obtained from Addgene and subcloned into pIRESneo2 for generation of stable cell lines. The HttQ72 construct created by Susan Lindquist38 (Addgene plasmid 1179) was obtained from Addgene and subsequently subcloned into pEGFPc3. To generate Lentivirus, HttQ72-GFP was cloned into the pLNCX2 vector and lentivirus was produced using the Lent-X Lentiviral expression system from Clontech. Stable expressing cell lines were generated by Lentiviral transduction in the presence of 5 μg/ml polybrene.
Mammalian two-hybrid assay
The binding site for Tom1 and Tom1L2 on myosin VI was mapped by subcloning wild-type and mutants (ΔWWY, ΔRRL) of the myosin VI tail region (aa 840-1294) into the bait vector, pM (BD Clontech), while full-length Tom1 and Tom1L2 were subcloned into the prey vector, pVP16 (BD Clontech). To map the binding site of myosin VI on Tom1, the myosin VI globular tail region (aa 1036-1294) was used as bait and various truncated Tom1 constructs (aa 1-180, 181-320, 299-492, 1-388) amplified by PCR and verified by sequencing were used as prey. The mammalian two-hybrid assay was performed in CHO cells as previously described39.
Western blot and coimmunoprecipitation
Whole cell lysates were harvested in 2X SDS-Sample buffer and boiled 5 minutes prior to being processed for Western blot analysis as previously described33. Quantitative Western blotting was performed following protein transfer to Immobilon-FL PVDF (Millipore) and immunoblotting with goat 647-conjugated secondary antibodies prior to scanning on a LI-COR Odyssey Infrared Imaging system (LI-COR Biosciences UK Ltd, Cambridge, UK). The integrated intensity of the LC3-II protein bands, following background correction, were normalised to an actin control prior to quantitation.
Coimmunoprecipitation experiments were performed from LNCAP prostate cancer cell lysates, harvested in ice-cold lysis buffer {50 mM Tris pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EGTA, 5 mM ATP, 5 mM MgCl2, and complete protease inhibitor (Roche)} and processed as previously described35. Samples were run on SDS-PAGE and immunoblotted using Clean-Blot IP detection reagent (Thermo Fisher Scientific Inc., Rockford, IL, USA).
Immunofluorescence microscopy
Cells were plated on glass coverslips, fixed in 4% formaldehyde, and permeabilised in 0.02% Triton X-100 in PBS. Alternatively, to reduce cytoplasmic background, cells were extracted in 0.02% saponin in PBS for 30 seconds prior to fixation. For Cathepsin D and Lamp1 immunostaining, fixation was performed in cold Methanol for 10 minutes. Fixed cells were blocked in 1% BSA in PBS, prior to incubation with primary antibodies. Alexa fluor 488, 568 or 647 conjugated secondary antibodies (Invitrogen) and 568-conjugated phalloidin (Invitrogen) were used for detection. Images were captured on either a Zeiss Axiovert epifluorescence microscope equipped with Hamamatsu ORCA-R2 charge-coupled device (CCD) camera driven by SIMPLE PCI Software® or on Zeiss LSM710 confocal microscope with Zeiss ZEN software®. Live cell video microscopy was acquired either on a Zeiss LSM710 confocal microscope or on a Zeiss AxioObserver Z1 inverted microscope with Plan-Apochromot 63X/1.40 oil DIC objective and an AxioCam MR3 equipped with spinning disk module using Zeiss Axiovision software®. For live cell studies, images were captured every 10 seconds for a period of 10 minutes. Images were processed with Adobe Photoshop CS4® and assembled in Adobe Illustrator CS4®.
For colocalisation analysis, Pearson’s coefficients or Mander’s overlap coefficients were calculated using either ImageJ software with Just Another Colocalisation Plugin (JACoP) (National Institutes of Health, USA), Volocity® software v.6.0.1 (PerkinElmer), or Imaris software v.7.4 (Bitplane AG, Zurich, Switzerland). For quantitation of colocalisation from cells expressing the RFP-GFP-LC3 fusion protein, ImageJ software with JACoP was used to calculate a Pearson’s coefficient based on the correlation in GFP and RFP signal overlap in greater than 100 cells from at least 2 independent experiments. All other colocalisation calculations were performed on >10 cells from at least two independent experiments.
Automated quantitation of p62-aggregates, GFP-LC3 autophagosome size, p62-punctae, LC3-punctae, and number of GFP-myosin VI tail punctae was performed on cells plated on a black PerkinElmer 96-well view plate using an ArrayScan VTi HCS Microscope (Cellomics, Pennsylvania, USA) using the Spot Detector V4 algorithm application. Immunocytochemistry was performed, nuclei were labeled with Hoechst to identify each individual cell, and the average fluorescence intensity, GFP-LC3 vesicle area, or number of GFP-myosin VI tail punctae was calculated per cell from 6 wells, with > 250 cells per well for each condition. GFP-LC3 vesicle area is represented in μm2. Results are from three independent experiments.
Immunogold Electron Microscopy
RPE cells expressing GFP-LC3, pulse-labelled with Texas Red-Dextran for 16 hours followed by a chase of 4 hours, were subsequently processed for immunogold labelling as previously described40. Sequential immunolabelling of Texas Red and GFP was performed at room temperature using the protein A-gold technique41. The sections were washed, contrasted by embedding in 1.8% methyl cellulose / 0.3% uranyl acetate, and air-dried prior to observation in a Philips CM100 transmission electron microscope at an operating voltage of 80kV.
Presentation of Data and Statistics
All graphs were produced using GraphPad Prism® software and error bars represent the standard deviation. Box and whisker plots represent the median, 25th and 75th percentile, and the minimum and maximum values. Statistics were calculated using either unpaired student t-test or one-way Anova followed by a Bonferroni multiple comparison post-hoc test.
Supplementary Material
Acknowledgements
This work was financially supported by the Wellcome Trust (F.B., D.A.T. and S.D.A.), the Medical Research Council (J.K-J. and N.A.B.) and a NIH-Oxford-Cambridge Ph.D. studentship (B.J.W.). The CIMR is in receipt of a strategic award from the Wellcome Trust (079895). We thank M. Seaman, F. Randow, L. Wartosch, D. Rubinsztein and D. Owen for critical reading of the manuscript and helpful discussions.
Footnotes
Competing financial interests The authors declare no competing financial interests.
References
- 1.Razi M, Chan EY, Tooze SA. Early endosomes and endosomal coatomer are required for autophagy. J Cell Biol. 2009;185:305–321. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hailey DW, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2257–2269. doi: 10.1091/mbc.E09-11-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011;80:125–156. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- 7.Wild P, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 9.Sahlender DA, et al. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J Cell Biol. 2005;169:285–295. doi: 10.1083/jcb.200501162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morriswood B, et al. T6BP and NDP52 are myosin VI binding partners with potential roles in cytokine signalling and cell adhesion. J Cell Sci. 2007;120:2574–2585. doi: 10.1242/jcs.007005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buss F, Kendrick-Jones J. How are the cellular functions of myosin VI regulated within the cell? Biochem Biophys Res Commun. 2008;369:165–175. doi: 10.1016/j.bbrc.2007.11.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blard O, et al. Cytoskeleton proteins are modulators of mutant tau-induced neurodegeneration in Drosophila. Hum Mol Genet. 2007;16:555–566. doi: 10.1093/hmg/ddm011. [DOI] [PubMed] [Google Scholar]
- 13.Feuillette S, et al. Filamin-A and Myosin VI colocalize with fibrillary Tau protein in Alzheimer’s disease and FTDP-17 brains. Brain Res. 2010;1345:182–189. doi: 10.1016/j.brainres.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Maruyama H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- 15.Herman EK, Walker G, van der Giezen M, Dacks JB. Multivesicular bodies in the enigmatic amoeboflagellate Breviata anathema and the evolution of ESCRT 0. J Cell Sci. 2011;124:613–621. doi: 10.1242/jcs.078436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Avraham KB, et al. The mouse Snell’s waltzer deafness gene encodes an unconventional myosin required for structural integrity of inner ear hair cells. Nat Genet. 1995;11:369–375. doi: 10.1038/ng1295-369. [DOI] [PubMed] [Google Scholar]
- 18.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 19.Janen SB, Chaachouay H, Richter-Landsberg C. Autophagy is activated by proteasomal inhibition and involved in aggresome clearance in cultured astrocytes. Glia. 2010;58:1766–1774. doi: 10.1002/glia.21047. [DOI] [PubMed] [Google Scholar]
- 20.Scherzinger E, et al. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 21.Penengo L, et al. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell. 2006;124:1183–1195. doi: 10.1016/j.cell.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 22.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 23.Finan D, Hartman MA, Spudich JA. Proteomics approach to study the functions of Drosophila myosin VI through identification of multiple cargo-binding proteins. Proc Natl Acad Sci U S A. 2011;108:5566–5571. doi: 10.1073/pnas.1101415108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang T, Liu NS, Seet LF, Hong W. The emerging role of VHS domain-containing Tom1, Tom1L1 and Tom1L2 in membrane trafficking. Traffic. 2010;11:1119–1128. doi: 10.1111/j.1600-0854.2010.01098.x. [DOI] [PubMed] [Google Scholar]
- 25.Clague MJ, Urbe S. Ubiquitin: same molecule, different degradation pathways. Cell. 2010;143:682–685. doi: 10.1016/j.cell.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 26.Rusten TE, Stenmark H. How do ESCRT proteins control autophagy? J Cell Sci. 2009;122:2179–2183. doi: 10.1242/jcs.050021. [DOI] [PubMed] [Google Scholar]
- 27.Spudich G, et al. Myosin VI targeting to clathrin-coated structures and dimerization is mediated by binding to Disabled-2 and PtdIns(4,5)P2. Nat Cell Biol. 2007;9:176–183. doi: 10.1038/ncb1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skibinski G, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 30.Lee JY, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berg TO, Fengsrud M, Stromhaug PE, Berg T, Seglen PO. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem. 1998;273:21883–21892. doi: 10.1074/jbc.273.34.21883. [DOI] [PubMed] [Google Scholar]
- 32.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 33.Bond LM, Peden AA, Kendrick-Jones J, Sellers JR, Buss F. Myosin VI and its binding partner optineurin are involved in secretory vesicle fusion at the plasma membrane. Mol Biol Cell. 2011;22:54–65. doi: 10.1091/mbc.E10-06-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chibalina MV, Seaman MN, Miller CC, Kendrick-Jones J, Buss F. Myosin VI and its interacting protein LMTK2 regulate tubule formation and transport to the endocytic recycling compartment. J Cell Sci. 2007;120:4278–4288. doi: 10.1242/jcs.014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buss F, et al. The localization of myosin VI at the golgi complex and leading edge of fibroblasts and its phosphorylation and recruitment into membrane ruffles of A431 cells after growth factor stimulation. J Cell Biol. 1998;143:1535–1545. doi: 10.1083/jcb.143.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arden SD, Puri C, Au JS, Kendrick-Jones J, Buss F. Myosin VI is required for targeted membrane transport during cytokinesis. Mol Biol Cell. 2007;18:4750–4761. doi: 10.1091/mbc.E07-02-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackson WT, et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krobitsch S, Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc Natl Acad Sci U S A. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chibalina MV, Roberts RC, Arden SD, Kendrick-Jones J, Buss F. Rab8-optineurin-myosin VI: analysis of interactions and functions in the secretory pathway. Methods Enzymol. 2008;438:11–24. doi: 10.1016/S0076-6879(07)38002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawrence SP, Bright NA, Luzio JP, Bowers K. The sodium/proton exchanger NHE8 regulates late endosomal morphology and function. Mol Biol Cell. 2010;21:3540–3551. doi: 10.1091/mbc.E09-12-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slot JW, Geuze HJ, Gigengack S, Lienhard GE, James DE. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J Cell Biol. 1991;113:123–135. doi: 10.1083/jcb.113.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.