Abstract
Background
Recently, genome-wide association studies identified a pleiotropic gene locus, ABO, as being significantly associated with hematological traits. To confirm the effects of ABO on hematological traits, we examined the link between the ABO locus and hematological traits in Korean population-based cohorts.
Results
Six tagging SNPs for ABO were analyzed with regard to their effects on hematological traits [white blood cell count (WBC), red blood cell count (RBC), platelet (Plat), mean corpuscular volume (MCV), and mean corpuscular haemoglobin concentration (MCHC)]. Linear regression analyses were performed, controlling for recruitment center, sex, and age as covariates. Of the 6 tagging SNPs, 3 (rs2073823, rs8176720, and rs495828) and 3 (rs2073823, rs8176717, and rs687289) were significantly associated with RBC and MCV, respectively (Bonferroni correction p-value criteria < 0.05/6 = 0.008). rs2073823 and a reported SNP (rs8176746), as well as rs495828 and a reported SNP (rs651007), showed perfect linkage disequilibrium status (r2s = 0.99). Of the remaining 3 SNPs (rs8176720, rs8176717 and rs687289), rs8176717 generated an independent signal with moderate p-value (= 0.045) when it was adjusted for by rs2073823 (the most significant SNP). We also identified a copy number variation (CNV) that was tagged by the SNP rs8176717, the minor allele of which correlated with the deletion allele of CNV. Our haplotype analysis indicated that the haplotype that contained the CNV deletion was significantly associated with MCV (β ± se = 0.363 ± 0.118, p =2.09 × 10-3).
Conclusions
Our findings confirm that ABO is one of the genetic factors that are associated with hematological traits in the Korean population. This result is notable, because GWASs fail to evaluate the link between a CNV and phenotype traits.
Keywords: ABO, GWAS, CNV, Hematological trait, Korean
Background
The ABO gene encodes isoforms for terminal glycosyltransferases, which transfer N-acetylgalactosamine and galactose to a common precursor (H substance), and lies on chromosome 9q34.2, containing 7 exons [1]. Exon 7 contains a domain that distinguishes between the A and B activities of the glycosyltransferase [2]. Several genomewide association studies (GWASs) have identified ABO as a candidate marker of the risk for coronary artery disease (CAD) [3], in addition to established CAD markers (sE-selectin, sP-selectin, and s-ICAM1) [4-6].
Hematological traits, such as red blood cell count (RBC), white blood cell count (WBC), platelet number (Plat), hemoglobin level (Hb), and hematocrit (Hct), are measured routinely to diagnose and monitor hematologic diseases and ascertain overall patient health. Recent GWASs on hematological traits have been reported for Caucasian [7], Japanese [8], and African-American [9] cohorts. These studies have identified more than 30 loci that carry common DNA polymorphisms that are linked to hematological traits.
The pleiotropic gene ABO correlated significantly with hematological traits in a Japanese [8] and African-American study [9], 3 SNPs of which (rs8176746, rs651007, rs495828) were reported in previous GWASs. rs8176746 is a nonsynonymous SNP and a deterministic variant of the B-type blood group [10]. rs651007 and rs495828 lie in the promoter region and are associated with CAD [4]. To confirm the effects of ABO on hematological traits, we examined the link between the ABO locus and hematological traits in Korean population-based cohorts.
Results
Hematological traits
The population characteristics and mean hematological traits are described in Table 1. Six hematological traits [WBC, RBC, Hb, Hct, Plat, and mean corpuscular volume (MCV)] were measured experimentally, and 2 other traits [mean corpuscular haemoglobin (MCH) and mean corpuscular hemoglobin concentration (MCHC)] were calculated using the RBC, Hb, and Hct values. Among hematological traits, RBC correlated with Hb and Hct, with Pearson’s r = 0.86 and 0.84, respectively. Also, MCV was linked to MCH, with Pearson’s r = 0.81. WHR, Plat, and MCHC correlated moderately (r < 0.7). Thus, we conducted a genetic association study of the ABO gene region with the 5 unrelated hematological traits.
Table 1.
Summary of participant characteristics and hematological traits
| Variables | Mean ± standard deviation |
|---|---|
| Sample size (n) |
6675 |
| Age (years) |
50.1 ± 8.8 |
| Male (%) |
47.7 |
| Ansung (%) |
50.5 |
| WBC count (103/μl) |
6.4 ± 1.9 |
| RBC count (106/μl) |
4.4 ± 0.4 |
| Hb (g/dl) |
13.8 ± 1.6 |
| Hct (%) |
41.2 ± 4.4 |
| Platelet count (103/μl) |
244.3 ± 60.1 |
| MCV (fl) |
93.9 ± 5.3 |
| MCH (pg) |
31.2 ± 2.0 |
| MCHC (g/dl) | 33.2 ± 1.3 |
Note. WBC: white blood cell, RBC: red blood cell, Hb: hemoglobin, Hct: hematocrit, MCV: mean corpuscular volume, MCH: mean corpuscular hemoglobin, MCHC: mean corpuscular hemoglobin concentration.
SNP selection
SNPs in Affymetrix 5.0 SNP array and imputation SNP data were obtained from the Korean Genome Epidemiology Study (KoGES) of the National Institute of Health, Korea, and the genotype data were Korea Association Resource consortium (KARE) data. The genomewide SNPs have been examined in genomewide association studies for anthropometric [11] and biochemical traits [12]. In this study, we focused on the ABO region that was reported by a Japanese study. Population stratification of the genotyped samples was also tested in an earlier report [11]; there was no population stratification that was demonstrated by Multidimensional Scaling (MDS) Analysis and Principal Component Analysis (PCA) (Additional file 1: Figure S1). Genomic inflation factors were low ranging from 1.01 (WBC) to 1.03 (Hct), suggesting that population stratification was well controlled (Additional file 2: Table S1)
We initially used 76 SNPs around ABO on chromosome 9 from 135,070 kbp to 135,152 kbp. The ABO gene boundaries were established by linkage disequilibrium (LD) analysis (Additional file 3: Figure S2). Three LD blocks encompassed ABO and its promoter region. The 3 LD blocks included 58 SNPs, 10 of which were genotyped by Affymetrix 5.0 SNP array; the remaining 48 SNPs were imputed by IMPUTE, based on the HAPMAP database. The characteristics of the 58 SNPs are described in Additional file 2: Table S1. The SNPs were classified as 8 nonsynonymous SNPs, 1 synonymous SNP, 8 upstream SNPs, and 41 intron SNPs.
ABO gene SNP association study
For the association analysis, we isolated 6 tagging SNPs for ABO. In Additional file 4: Table S2, we describe the 6 SNP groups with high LD (r2 > 0.9) and underlined the tagging SNPs. The association results are described in Table 2. In this study, we used Bonferroni correction p-value criteria (< 8.3 × 10-3) for multiple comparisons, and the significant effect sizes and p-values are underlined in Table 2. Three SNPs (rs2073823, rs8176720, and rs495828) and 3 SNPs (rs2073823, rs8176717, and rs687289) were significantly associated with RBC and MCV, respectively.
Table 2.
Association analysis of 6 high-LD-group tagging SNPs with five hematological traits by linear regression analysis, controlling for area, age, and sex as covariates
| |
|
|
|
WBC |
RBC |
Plat |
MCV |
MCHC |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CHR | SNP | BP | A1 | BETA ± SE | P | BETA ± SE | P | BETA ± SE | P | BETA ± SE | P | BETA ± SE | P |
| 9 |
rs2073823 |
135122337 |
A |
0.056 ± 0.039 |
0.15 |
0.036 ± 0.008 |
2.13 × 10-6† |
−1.213 ± 1.253 |
0.33 |
−0.480 ± 0.118 |
5.06 × 10-5† |
0.075 ± 0.029 |
9.55 × 10-3 |
| 9 |
rs8176720 |
135122694 |
C |
0.043 ± 0.032 |
0.19 |
0.018 ± 0.006 |
5.58 × 10-3 |
−0.232 ± 1.033 |
0.82 |
−0.071 ± 0.098 |
0.47 |
0.025 ± 0.024 |
0.30 |
| 9 |
rs8176717 |
135122855 |
T |
0.001 ± 0.039 |
0.97 |
−0.012 ± 0.008 |
0.13 |
0.885 ± 1.253 |
0.48 |
0.356 ± 0.118 |
2.62 × 10-3† |
−0.035 ± 0.029 |
0.22 |
| 9 |
rs687289 |
135126927 |
A |
0.001 ± 0.032 |
0.97 |
0.001 ± 0.006 |
0.89 |
1.017 ± 1.025 |
0.32 |
−0.274 ± 0.097 |
4.59 × 10-3 |
0.020 ± 0.024 |
0.40 |
| 9 |
rs8176681 |
135129575 |
C |
0.016 ± 0.035 |
0.64 |
0.009 ± 0.007 |
0.19 |
−1.488 ± 1.109 |
0.18 |
0.063 ± 0.105 |
0.55 |
0.003 ± 0.026 |
0.92 |
| 9 | rs495828 | 135144688 | T | −0.050 ± 0.037 | 0.18 | −0.030 ± 0.007 | 2.69 × 10-5† | 2.217 ± 1.180 | 0.06 | 0.061 ± 0.112 | 0.58 | −0.042 ± 0.027 | 0.12 |
Note. Chr: chromosome, BP: base pair, A1: minor allele, WBC: white blood cell count, RBC: red blood cell count, Platelet, MCV: mean corpuscular volume, MCHC: mean corpuscular hemoglobin concentration.
Underlined p-values were passed the multiple correction criteria of Bonferroni correction (p<0.008).
†Significant result after adjusted by rs2073823.
To identify independent association signals, we performed a conditional analysis by including rs2073823 in the linear regression model of other significant SNP associations. For RBC, the association signal of rs8176720 disappeared (p-value = 0.803), but that of rs495828 was significant (p-value = 0.004) after adjusting for rs2073823. rs8176717 was moderately associated with MCV (p-value = 0.045), but the association signal with rs687289 disappeared (p-value =0.492). Thus, we identified 3 independent associations (rs2073823, rs8176717 and rs495828) between ABO and hematological traits.
Identification of copy number variation
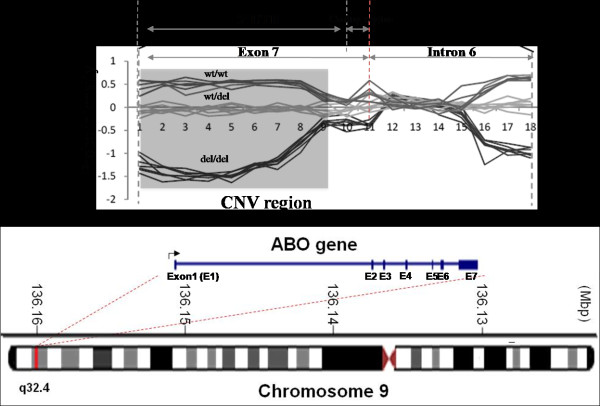
A copy number variation (CNV) region was detected on chromosome 9, 135,120,477–135,122,527 (Figure 1), which includes the 3′ untranslated region of the ABO gene. Because the array CGH experiment was conducted using a subset (n = 4694) of all KoGES samples, to maximize the sample size, we surveyed a tagging SNP that correlated well with CNV region genotypes. We determined the SNP rs8176717 to correlate with the CNV region (r2 = 0.96), the minor allele of which (T allele) implied the minor allele (deletion allele) of CNV.
Figure 1.
Probe intensity of copy number variation region: Log2ratio plot of the test sample and the reference (NA10851) signal intensity.
Haplotype analysis
We estimated the haplotyes for the 6 SNPs (Table 3). A total of 6 haplotypes were predicted, comprising 4 common haplotypes and 2 rare haplotypes (frequencies < 0.05). A haplotype (Hap 3) included the minor allele of rs8176717, which tagged the CNV and was significantly associated with MCV (beta ± se = 0.363 ± 0.118, p-value = 2.09 × 10-3). The other significant haplotype was Hap 4, which was linked to RBC (beta ± se = 0.036 ± 0.008, p-value = 4.27 × 10-6) and MCV (beta ± se = −0.512 ± 0.119, p-value = 1.81 × 10-5).
Table 3.
Haplotype frequencies and association results of six SNPs with red blood cell count (RBC) and mean corpuscular volume (MCV)
|
Haplotypes |
Tagging SNPs for ABO gene region |
Haplotype frequency |
RBC |
MCV |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2073823 | rs8176720 | rs8176717 | rs687289 | rs8176681 | rs495828 | beta ± se | p | beta ± se | p | ||
| Hap 1 |
G |
T |
G |
G |
C |
G |
0.297 |
0.008 ± 0.007 |
0.249 |
0.064 ± 0.106 |
0.544 |
| Hap 2 |
G |
T |
G |
A |
T |
T |
0.261 |
−0.030 ± 0.007 |
2.63 × 10-5 |
0.064 ± 0.111 |
0.563 |
| Hap 3 |
G |
C |
T |
G |
T |
G |
0.214 |
−0.013 ± 0.008 |
0.103 |
0.363 ± 0.118 |
2.09 × 10-3 |
| Hap 4 |
A |
C |
G |
A |
T |
G |
0.208 |
0.036 ± 0.008 |
4.27 × 10-6 |
−0.512 ± 0.119 |
1.81 × 10-5 |
| Hap 5 |
G |
T |
G |
G |
T |
G |
0.012 |
NA |
NA |
NA |
NA |
| Hap 6 | A | C | G | A | C | G | 0.007 | NA | NA | NA | NA |
Discussion
In this study, we confirmed the association between ABO and hematological traits in a large Korean population. Also, we found a copy number variation that influenced hematological traits.
Of the 6 tagging SNPs in the ABO gene, rs2073823 was the most significant, in perfect LD (r2 = 0.995) with rs8176746, an SNP from the Japanese GWAS on hematological traits [8]. The minor allele of rs8176746 is the variant that encodes the B-type blood group. [10]. However, this SNP was not reported in a GWAS of hematological traits in Caucasians [7,9], possibly due to ethnic differences in the minor allele frequency in Caucasian (0.08), Chinese (0.23), and Japanese (0.17) individuals. The allele frequencies correspond well to the frequency of blood type B in Caucasian (~8%) and East Asian (~22%) individuals, as inferred from the BLOODBOOK website (http://www.bloodbook.com/world-abo.html). Using the minor allele frequency (0.008) and the mean RBC (± sd) = 4.82 ± 0.50 of Caucasians, we estimated the number of individuals required for the 80% power at the alpha = 5 × 10-8 (genome-wide significant levels) [7,9]. To be replicated the rs2073823 (LD with rs8176746) association, 51,876 individuals would be necessary. However, the previous European study [7] used 33,623 individuals which it was smaller than the estimated individual number at the genome-wide significant level. In our study, individuals with a minor allele of rs2073823 had elevated RBC counts but decreased MCV. Thus, individuals with the blood type B might have higher RBC counts and lower MCV than those with other blood types, at least among Asians.
The second highest signal was generated from an upstream SNP, rs495828, which was also was reported in the Japanese GWAS [8]; this SNP was in perfect LD with rs651007, which was reported in an African-American GWAS [9]. Notably, the 3 proximal SNPs (rs651007, rs579459, and rs649129) were in complete LD (r2 = 0.99) with rs495828. Because carriers of the minor allele of these 3 SNPs have significantly lower levels of sP-selectin [5], sE-selectin [6], and risk of CAD [4], the relationship between hematological traits and coronary artery disease phenotypes should be examined.
The Japanese GWAS reported complete LD between rs8176746 and rs495828. To confirm the LD, we estimated the LD in Europeans (r2 = 0.010 and D’ = 0.150), Africans (r2 = 0.035 and D’ = 1.000), Chinese, Japanese (r2 = 0.050 and D’ = 1.000), and Koreans in this study (r2 = 0.087 and D’ = 1.000). Even though it was reported that rs8176746 and rs495828 are in complete LD in the Japanese study, the data from publically available databases suggests some inconsistencies with high D’ and low r2. This suggests that rs495828 may represent an independent association signal for RBC. A limitation of our study is that the 2 most significant SNPs—rs8176746 and rs495828—were not genotyped directly, although the minor allele frequencies of these SNPs are similar to those reported in the Japanese GWAS [2].
The CNV region that we identified has been reported by 7 other studies [13-18]. The minor allele of CNV was a deletion mutation of the 3′ untranslated region of ABO; thus, the CNV might influence its expression. In our results, the haplotype included the minor allele of the CNV-tagging SNP (rs8176717) and was significant associated with MCV. This result is notable, because most GWASs do not evaluate the link between a CNV and phenotype traits. Thus, our study is a model that can be used to correlate SNPs and CNV.
Conclusions
ABO is one of the genetic factors that are associated with hematological traits in East Asian populations. Also, we identified a novel association with a SNP that tags a common CNV with MCV. This result is notable, because GWASs fail to evaluate the link between a CNV and phenotype traits.
Methods
Study participants
This study was conducted as part of an ongoing population-based cohort of the Korean Genome and Epidemiology Study (KoGES). All participants were recruited from the cities of Ansung and Ansan in Gyeonggi-do Province, Korea. This study was approved by the Institutional Review Board of the Korea National Institute of Health, and all participants provided written informed consent for study participation.
Hematological trait measures
A total of 6675 samples were available for hematological trait analysis, as described in Table 1, Venous blood samples were drawn from all participants into 4.5-ml tubes that contained K3-EDTA as an anticoagulant and were analyzed within 30 min to 4 h of collection. Hematological traits were measured by Seoul Clinical Laboratories Company Ltd. The ADIVA 120 hematology system (Bayer Diagnostics, USA) was calibrated per the manufacturer’s guidelines. WBC count, RBC count, platelet count, Hb level, Hct, mean corpuscular volume (MCV), mean corpuscular hemoglobin (MCH) level, and mean corpuscular hemoglobin concentration (MCHC) were determined automatically for all samples.
SNP determination
The ABO gene is located on chromosome 9 from 135,120,384–135,140,451 bp. SNP genotypes were determined using the Affymetrix 5.0 SNP array, the experimental procedures of which are detailed elsewhere [11]. Further, to increase the number of genotype markers, we imputed additional SNPs using the Affymetrix 5.0 SNP array and the HapMap database (HAPMAP 3, http://www.hapmap.org); the imputation methods have been described [19]. The final SNPs were selected using the following criteria: minor allele frequency > 0.1; missing rate < 10%; and Hardy-Weinberg equilibrium test p-value > 0.05 for experimentally determined SNPs and imputation SNPs. Information on the SNPs was obtained from the dbSNP database (http://www.ncbi.nlm.gov/snp), and the genetic distance between the Korean and other populations was calculated using F-statistic [20]. LD blocks and pairwise LD (D’ and r2) of SNPs were estimated and determined for the tagging SNPs in the ABO gene region using Haploview [21].
CNV determination
To identify regions of CNV, samples from 4694 participants were genotyped using the NimbleGen HD2 2x720K array comparative genomic hybridization (aCGH) assay with DNA from peripheral blood. All samples passed experimental quality control metrics, such as the chromosome X shift and mad.1dr, as determined using NimbleScan version 2.5 per the manufacturer’s guidelines. After quality control procedures, the signal intensity ratio between the test and reference sample (NA10851 from the HapMap cell line DNA) of each probe was log2-transformed.
Regions of CNV were identified using the Genome Alteration Detection Analysis algorithm [22], which was used for samples from 4694 participants, with T = 10, alpha = 0.2, and MinSegLen = 10. The threshold for defining regions of CNV was set to an average log2 ratio of ± 0.25 Additional file 5: Figure S3.
CNV-tagging SNP
We tagged SNPs to maximize the sample size. To find SNPs that tagged the identified CNVs well, we performed a correlation analysis that was similar to that in the Wellcome Trust Case Control Consortium CNV study [23] using calls that were identified in a GWAS with the Affymetrix 5.0 array [11]. For each CNV, we calculated the squared Pearson's r value between CNV regions and SNPs. We considered all SNPs within 1 Mb of the estimated 2 breakpoints (i.e., start and end points) of each CNV region. We selected the SNP with the highest r2 value for each CNV region.
Association tests
Linear regression analysis was used to analyze the association between ABO SNPs or haplotypes of tagging SNPs and hematological trait, controlling for gender, age, and recruitment center as covariates. The asymptotic Hardy-Weinberg equilibrium test was conducted using PLINK (version 1.07) [24], and all reported p-values were two-sided (α = 0.05). Associations between SNPs and hematological traits were significant at p < = 8.3 × 10-3 after Bonferroni correction for multiple testing of 6 SNPs. The sample size was estimated for rs2073823 association in the European with the 80% statistical power at the genome-wide significance level by the QUANTO software (version 1.2.4, http://hydra.usc.edu/gxe/).
Authors’ contributions
KWH participated in the design of the study, genetic analysis, and drafted the manuscript. SM participated in the design of the study, CNV genotype determination, and drafted the manuscript. YJK participated in the CNV-tagging SNP identification and statistical analysis. YKK participated in the CNV genotype experiments. DJK participated in the SNP and CNV genotype experiments. CK participated in the hematological trait measurements. SSK participated in writing the manuscript and discussion. BJK participated in writing the manuscript and discussion.
Acknowledgments
This work was supported by an intramural grant from the Korea National Institute of Health (2010-N73001-00) and by grants from the Korea Centers for Disease Control and Prevention (4845–301, 4851–302, and 4851–307).
Supplementary Material
Figure S1. Multidimensional scaling (MDS) analysis and principal component analysis (PCA) [Cho et al., 2009].
Table S1. Genomic inflation factor for hematological trait genome-wideassociation studies.
Figure S2. Linkage disequilibrium blocks of ABO gene region.
Table S2. SNP list of ABO gene region, minor allele frequency comparison, and genetic distance calculation between KARE and other populations. Underlined SNPs indicate the tagging SNPs for the ABO gene region used in the main paper.
Figure S3. CNV clustering results. We used CNV tools to summarize the signal intensity data and assign a [specific OR discrete] CNV genotype within the CNV region. (A) Histogram of the clustering procedure using data, transformed by the linear discriminant function (LDF). (B) Cluster plot of the CNV region predicted from the LDF signal.
Contributor Information
Kyung-Won Hong, Email: zknightm@khu.ac.kr.
Sanghoon Moon, Email: sanghoon.moon@cdc.go.kr.
Young Jin Kim, Email: in_time@korea.kr.
Yun Kyoung Kim, Email: yk_kim@korea.kr.
Dong-Joon Kim, Email: dodari11@nih.go.kr.
Cheong-sik Kim, Email: kcdc215@korea.kr.
Sung Soo Kim, Email: ksungsoo@korea.kr.
Bong-Jo Kim, Email: kbj6181@cdc.go.kr.
References
- Storry JR, Olsson ML. The ABO group system revisited: review and update. Immunohematology. 2009;25:48–59. [PubMed] [Google Scholar]
- Yamamoto F, McNeill PD, Hakomori S. Genomic organization of human histo-blood group ABO genes. Glycobiology. 1995;5:51–58. doi: 10.1093/glycob/5.1.51. [DOI] [PubMed] [Google Scholar]
- Schunkert H, König IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C. et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Cornelis MC, Kraft P, Jensen M, van Dam RM, Sun Q, Girman CJ, Laurie CC, Mirel DB, Hunter DJ. et al. Genetic variants in ABO blood group region, plasma soluble E-selectin levels and risk of type 2 diabetes. Hum Mol Genet. 2010;19:1856–1862. doi: 10.1093/hmg/ddq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AD, Lopes-Virella MF, Waggott D, Boright AP, Hosseini SM, Carter RE, Shen E, Mirea L, Bharaj B, Sun L. et al. Genome-wide association identifies the ABO blood group as a major locus associated with serum levels of soluble E-selectin. Arterioscler Thromb Vasc Biol. 2009;29:1958–1967. doi: 10.1161/ATVBAHA.109.192971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbalic M, Dupuis J, Dehghan A, Bis JC, Hoogeveen RC, Schnabel RB, Nambi V, Bretler M, Smith NL, Peters A. et al. Large-scale genomic studies reveal central role of ABO in sP-selectin and sICAM-1 levels. Hum Mol Genet. 2010;19:1863–1872. doi: 10.1093/hmg/ddq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesh SK, Zakai NA, van Rooij FJ, Soranzo N, Smith AV, Nalls MA, Chen MH, Kottgen A, Glazer NL, Dehghan A. et al. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat Genet. 2009;41:1191–1198. doi: 10.1038/ng.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamatani Y, Matsuda K, Okada Y, Kubo M, Hosono N, Daigo Y, Nakamura Y, Kamatani N. Genome-wide association study of hematological and biochemical traits in a Japanese Population. Nat Genet. 2010;42:210–215. doi: 10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- Lo KS, Wilson JG, Lange LA, Folsom AR, Galarneau G, Ganesh SK, Grant SF, Keating BJ, McCarroll SA, Mohler ER 3rd. et al. Genetic association analysis highlights new loci that modulate hematological trait variation in Caucasians and African Americans. Hum Genet. 2011;129:307–317. doi: 10.1007/s00439-010-0925-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip SP. Sequence variation at the human ABO locus. Ann Hum Genet. 2002;66:1–27. doi: 10.1017/S0003480001008995. [DOI] [PubMed] [Google Scholar]
- Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban HJ, Yoon D, Lee MH, Kim DJ, Park M. et al. A large-scale genome-wide association study of Asian population uncover genetic factors influencing eight quantitative traits. Nat Genet. 2009;41:527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Go MJ, Hu C, Hong CB, Kim YK, Lee JY, Hwang JY, Oh JH, Kim DJ, Kim S. et al. Large-scale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat Genet. 2011;43:990–995. doi: 10.1038/ng.939. [DOI] [PubMed] [Google Scholar]
- Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, Krauss RM, Myers RM, Ridker PM, Chasman DI. et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung HC, Szpiech ZA, Degnan JH, Wang K, Guerreiro R. et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P. et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PI, Maller JB, Kirby A. et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- Shaikh TH, Gai X, Perin JC, Glessner JT, Xie H, Murphy K, O’Hara R, Casalunovo T, Conlin LK, D’Arcy M. et al. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res. 2009;19:1682–1690. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong KW, Lim JE, Kim YJ, Cho NH, Shin C, Oh B. KARE Genomewide Association Study of Blood Pressure Using Imputed SNPs. Genomics & Informatics. 2010;8:103–107. doi: 10.5808/GI.2010.8.3.103. [DOI] [Google Scholar]
- Wright S. The genetical structure of populations. Nature. 1950;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;28:1323–1328. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Pique-Regi R, Monso-Varona J, Ortega A, Seeger RC, Triche TJ, Asgharzadeh S. Sparse representation and Bayesian detection of genome copy number alterations from microarray data. Bioinformatics. 2008;24:309–318. doi: 10.1093/bioinformatics/btm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, Robson S, Vukcevic D, Barnes C, Conrad DF. Wellcome Trust Case Control Consortium et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Tomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ. et al. PLINK: a tool set for whole-genome association and population based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Multidimensional scaling (MDS) analysis and principal component analysis (PCA) [Cho et al., 2009].
Table S1. Genomic inflation factor for hematological trait genome-wideassociation studies.
Figure S2. Linkage disequilibrium blocks of ABO gene region.
Table S2. SNP list of ABO gene region, minor allele frequency comparison, and genetic distance calculation between KARE and other populations. Underlined SNPs indicate the tagging SNPs for the ABO gene region used in the main paper.
Figure S3. CNV clustering results. We used CNV tools to summarize the signal intensity data and assign a [specific OR discrete] CNV genotype within the CNV region. (A) Histogram of the clustering procedure using data, transformed by the linear discriminant function (LDF). (B) Cluster plot of the CNV region predicted from the LDF signal.