Summary
Regulation of genes that initiate and amplify inflammatory programs of gene expression is achieved by signal-dependent exchange of co-regulator complexes that function to read, write and erase specific histone modifications linked to transcriptional activation or repression. Here, we provide evidence for the role of trimethylated histone H4 lysine 20 (H4K20me3) as a repression checkpoint that restricts expression of toll like receptor 4 (TLR4) target genes in macrophages. H4K20me3 is deposited at the promoters of a subset of these genes by the SMYD5 histone methyltransferase through its association with NCoR corepressor complexes. Signal-dependent erasure of H4K20me3 is required for effective gene activation and is achieved by NF-κB-dependent delivery of the histone demethylase PHF2. Liver X receptors antagonize TLR4-dependent gene activation by maintaining NCoR/SMYD5-mediated repression. These findings reveal a histone H4K20 tri-methylation/de-methylation strategy that integrates positive and negative signaling inputs that control immunity and homeostasis.
Introduction
The survival of vertebrate organisms relies on innate and adaptive immune mechanisms to detect, combat and eliminate foreign pathogens. Recognition of foreign pathogens by pattern recognition receptors (PRRs) triggers transcriptional activation of genes that amplify inflammatory responses, enable antimicrobial activities and initiate the development of acquired immunity (Kawai and Akira, 2010; Takeuchi and Akira, 2010). The conservation of key signaling pathways required for these responses, including the Toll, JAK/STAT and Immune Deficiency (Imd) pathways, throughout metazoan evolution underscores their importance in effectively defending against bacterial and viral infection (Lemaitre and Hoffmann, 2007; Martinelli and Reichhart, 2005). Macrophages represent a crucial cell type for initial recognition of pathogens based on their expression of numerous PRRs, including the toll-like receptors (TLRs). The TLR family is comprised of a highly conserved set of membrane receptors that recognize specific components of bacterial and viral pathogens including lipoproteins (TLR1/2/6), lipopolysaccharide (TLR4), flagellin (TLR5), single-stranded RNA (TLR7/8), double-stranded RNA (TLR3), and double stranded DNA (TLR9) (Kawai and Akira, 2010; Martinelli and Reichhart, 2005). Upon TLR ligation by their respective ligands, adapter proteins are recruited to the cytoplasmic domains of TLRs and initiate signaling cascades, which ultimately result in the activation of signal-dependent transcription factors such as nuclear factor-κ B (NF-κB), activator protein 1 (AP1) and interferon regulatory factor (IRFs). These factors function in a combinatorial manner to promote the induction of inflammatory cytokines, type I interferons, and numerous other regulatory and effector molecules required for host defense (Kawai and Akira, 2010). Although required for effective immunity, persistent or inappropriate activation of inflammatory gene programs can contribute to chronic diseases that include type II diabetes (Hotamisligil, 2006), atherosclerosis (Tedgui and Mallat, 2006), cancer (Karin et al., 2006) and neurodegenerative diseases (Glass et al., 2010). Precise control of innate and adaptive immune responses is therefore critical for maintaining proper tissue homeostasis. Multiple mechanisms have been identified that function to prevent signal-independent activation of inflammatory responses and to mediate their resolution upon eradication of the inciting stimulus. Several studies have suggested that many highly inducible inflammatory response genes are maintained in a repressed state under basal conditions by co-repressor complexes containing nuclear receptor co-repressor 1 (NCoR), silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) complexes, and/or the co-repressor of REST (CoREST) (Ghisletti et al., 2009; Hargreaves et al., 2009; Hoberg et al., 2006; Huang et al., 2009; Huang et al., 2011; Ogawa et al., 2004; Pascual et al., 2005; Saijo et al., 2009; Venteclef et al., 2010). These complexes are actively removed in response to pro-inflammatory signals as a prerequisite to transcriptional activation and thereby serve as transcriptional checkpoints that regulate the transition from basal to activated states (Ghisletti et al., 2009; Huang et al., 2009; Huang et al., 2011; Ogawa et al., 2004; Pascual et al., 2005; Saijo et al., 2009). Several members of the nuclear receptor family of transcription factors, including glucocorticoid receptors, peroxisome proliferator-activated receptors (PPARs) and liver X receptors (LXRs), exert potent anti-inflammatory effects as an important aspect of their biological function (Flammer and Rogatsky, 2011; Hong and Tontonoz, 2008; Saijo et al., 2009). Investigation of the molecular basis for these actions has revealed diverse points of regulation that can be selectively targeted to achieve receptor- and gene-specific control of the inflammatory response. In the cases of PPARγ and LXRs, for example, ligand-dependent SUMOylation of specific residues in their respective ligand binding domains results in their interaction with NCoR/SMRT complexes o n inflammatory promoters, where they inhibit gene activation by interfering with signal-dependent co-repressor clearance (Blaschke et al., 2006; Ghisletti et al., 2009; Pascual et al., 2005; Venteclef et al., 2010). Additional mechanisms utilized by the glucocorticoid receptor and the orphan nuclear receptor Nurr1 include inhibition of specific activator/co-activator interactions and gene-specific recruitment of co-repressors (Ogawa et al., 2005; Saijo et al., 2009). Investigation of nuclear receptor repression pathways has thus led to insights into molecular mechanisms that both positively and negatively regulate inflammatory responses at a gene-specific level. To further advance our understanding of these mechanisms, we undertook a genome-wide screen in Drosophila to identify novel proteins required for nuclear receptor trans-repression. This screen led to the identification and characterization of SMYD5, a previously unrecognized component of the NCoR complex that specifically trimethylates histone H4 at lysine 20 (H4K20me3) on a subset of pro-inflammatory promoters. This serves as a repression mark that is required for LXR repression and is removed, upon TLR4 activation, by PHF2, a member of the jumonji domain family of lysine demethylases. These findings highlight a role for active histone H4K20 methylation and demethylation as a molecular checkpoint in the regulation of a subset of pro-inflammatory gene programs.
Results
Identification of SMYD5 as a Negative Regulator of Inflammatory Response Genes
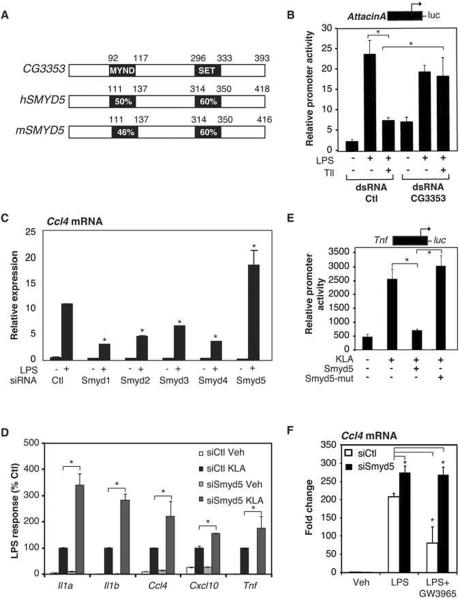
Nuclear receptors exert potent anti-inflammatory effects as important aspects of their biological functions, but these mechanisms remain poorly understood. As Drosophila melanogaster utilizes a conserved pathway involving signal-dependent activation of the NF-κB homologue Relish (Rel) to initiate inflammatory responses (Lazzaro, 2008; Lemaitre and Hoffmann, 2007) (Supp. Fig. 1A) and also express nuclear receptors (King-Jones and Thummel, 2005), we explored the use of Drosophila Schneider (S2) cells as a model to identify genes involved in nuclear receptor-mediated repression of inflammatory gene expression. A screen of Drosophila nuclear receptors identified tailless (Tll) as an orphan nuclear receptor capable of repressing LPS-dependent activation of an Attacin A promoter-luciferase reporter gene (Supp. Fig. 1B). Using this assay, we evaluated a dsRNA library targeting 21,300 genes for silencing RNAs (Boutros et al., 2004) that could reverse Tll-repression of Attacin A promoter activity. In addition to known suppressors of inflammatory responses, this screen led to the identification of CG3353 (Supp. Fig. 1C), encoding a 393 amino acid protein containing a SET domain. CG3353 is most closely related to mammalian SMYD5, a member of the SMYD family of SET and MYND domain containing proteins (Fig 1A). Using an independent dsRNA, we confirmed that knockdown of CG3353 abolished Tll transrepression, as well as resulted in an increase in basal expression of the Attacin A reporter gene (Fig. 1B).
Figure 1. SMYD5 is a Negative Regulator of Inflammatory Response Genes and is Required for LXR Transrepression.
A. Percent amino acid identity between Drosophila CG3353 and mammalian SMYD5 proteins, which share conserved MYND and SET domains.
B. dsRNA knockdown of CG3353 in Drosophila S2 cells reverses Tll-dependent repression of the Attacin A luciferase reporter. Values represent the mean ± SEM of three experiments, *p<0.05.
C. Effect of control (Ctl) or Smyd1-5 siRNAs transfected into thioglycollate elicited macrophages on KLA-induced expression of Ccl4 mRNA 4 h following treatment. Values represent the average of three experiments −/+ SEM, *p<0.05 as compared to siCtl LPS treatment.
D. Effect of control (Ctl) or Smyd5 siRNAs transfected into thioglycollate elicited macrophages on KLA-induced expression of ll1a, ll1b, Ccl4, Cxcl10 and Tnf mRNAs 4 h following treatment. Values represent the average of three experiments −/+ SEM, *p<0.05.
E. Effect of overexpression of WT SMYD5 and mutant (H315L) SMYD5 on KLA induction of the Tnf promoter. Values represent the mean ± SEM of three experiments. *p<0.05.
F. Effect of SMYD5 knockdown on LXR repression of Ccl4 mRNA in thioglycollate elicited macrophages treated with GW3965 for 1 hour followed by 4 hours of KLA treatment. Values represent the average of three experiments −/+ SEM, *p<0.05 relative to siCtl, KLA treated sample.
Primary mouse macrophages express all five members of the mammalian SMYD subfamily (Supp. Fig. 1D), providing a biologically relevant model system to evaluate their potential roles in innate immune responses. Using specific siRNAs to deplete expression of each SMYD family member (Supp. Fig. 1E), we observed that knockdown of SMYD5 resulted in exaggerated transcriptional responses of a subset of TLR4-responsive genes following stimulation with a purified LPS, Kdo2 lipid A (KLA) (Fig. 1C, 1D, Supp. Fig. 1F). In contrast, siRNAs directed against the other members of the mammalian SMYD family (Smyd1–4), resulted in suppressed responses (Fig. 1C, Supp. Fig. 1F), indicating distinct molecular functions. Overexpression of wild type SMYD5 markedly repressed KLA induction of a TNFα-luciferase reporter gene, while overexpression of SMYD5 containing a mutation predicted to abolish methyltransferase activity (H315L; SMYD5-mut) did not (Fig. 1E). Knockdown of SMYD5 expression also abolished or compromised the ability of the LXR-specific agonist (GW3965) to inhibit LPS-induced expression of Ccl4 (Fig.1F) and other LXR-sensitive genes such as IL1a (Supp. Fig.1G), suggesting a conserved function in Drosophila and mammalian cells. In contrast, knockdown of SMYD5 did not alter inhibitory effects of the glucocorticoid receptor (data not shown), suggesting that SMYD5 contributes to receptor-specific repression mechanisms.
SMYD5 trimethylates H4K20 at TLR4-responsive Promoters
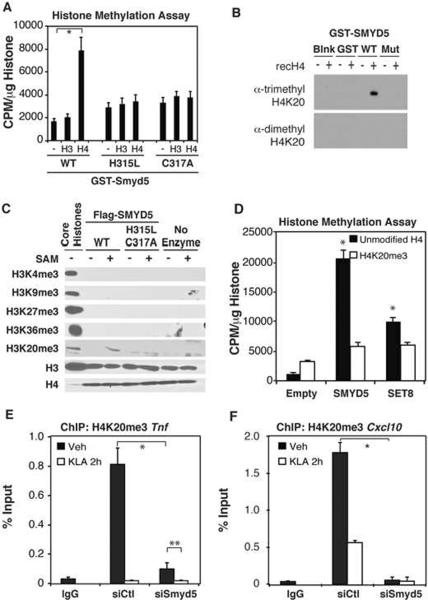
Next, we investigated the ability of bacterially expressed SMYD5 to methylate recombinant histones. Although SMYD1–3 have been shown to methylate histone H3 (Albert and Helin, 2010), a GST-SMYD5 fusion protein purified from bacteria exhibited methyltransferase activity on recombinant histone H4, but not histone H3 (Fig 2A). Mutations in the catalytic domain of SMYD5 (H315L or C317A) abrogated this activity (Fig 2A). To date there are only two identified methylation sites on histone H4: arginine at position 3 and lysine at position 20. Since SET domains typically methylate only lysine residues, our data suggested that SMYD5 most likely functions as a H4K20 methyltransferase. Consistent with this, methylation-specific antibodies detected trimethylation of lysine 20 on histone H4 incubated with wild type bacterially expressed SMYD5 but no methylation was observed with mutant SMYD5 (Fig 2B). Immunoprecipitates of Flag-tagged wild type and mutant SMYD5 expressed in mammalian cells also catalyzed trimethylation of H4K20, while no methylation was observed for H3K4me3, H3K9me3, H3K27me3, or H3K36me3 (Fig 2C). The specificity of the H4K20me3 antibody was confirmed using recombinant H4 proteins chemically mono-, di- or tri-methylated at position 20 (Supp. Fig 2A).
Figure 2. SMYD5 Trimethylates H4K20 at TLR4-responsive Promoters.
A. Methyltransferase activity of SMYD5, and two SET domain mutants (H315L and C317A), on recombinant histones H3 and H4. GST-Purified SMYD5 proteins were incubated with radiolabeled H3-SAM and recombinant histones for 4 hours. Activity was measured as CPM/μm histone. Values represent the average of three experiments −/+ SEM,*p<0.05
B. GST-SMYD5 and GST-SMYD5 (H315L,C317A) were incubated with recombinant histone H4. Reaction products were detected by immunoblotting with antibodies that recognize di- or tri- H4K20 methylation.
C. Immunoprecipitated Flag-SMYD5 and Flag-SMYD5 (H315L/C317A) were incubated with recombinant histone H3 or H4 in the presence or absence of S-adenosylmethionine (SAM). Reaction products were detected by immunoblotting with indicated antibodies.
D. Flag-SMYD5 or recombinant SET8 were incubated with recombinant histone H4, or a chemically modified histone H4 trimethylated at K20. Activity was measured as CPM/μg histone. Values represent the average of three experiments −/+ SEM, *p<0.05 relative to Empty, unmodified H4 treatments.
E. Chromatin immunoprecipitation assays assessing the H4K20me3 levels on the Tnf promoter after treatment of thioglycollate elicited macrophages with siCtl and siSmyd5 for 48 hours followed by 2 hours of Veh or KLA stimulation. Values represent the average of three experiments −/+ SEM, **,*p<0.05.
F. Chromatin immunoprecipitation assays assessing H4K20me3 levels on the Cxcl10 promoter before and after KLA stimulation for 2 hours. Values represent the average of three experiments −/+ SEM,*p<0.05.
To determine whether H4K20 is the only residue in histone H4 modified by SMYD5 methylation, we performed methyltransferase reactions for SET8, a SET domain protein that monomethylates H4K20, or SMYD5, using recombinant unmodified histone H4 or recombinant H4 chemically tri-methylated at position 20. SMYD5 and SET8 methylated unmodified histone H4 but not chemically tri-methylated H4K20 (Fig. 2D). In addition, Flag-tagged SMYD5 and SET8 methylated recombinant histone H4, but not recombinant histone H4 in which lysine 20 was mutated to alanine (H4K20A, Supp. Fig. 2B). Collectively, these results indicate that SMYD5 functions to catalyze trimethylation of H4K20.
H4K20me3 has been established as a mark for transcriptional repression that is primarily associated with heterochromatin (Gonzalo et al., 2005; Kourmouli et al., 2005; Schotta et al., 2004). However, genome-wide chromatin immunoprecipitation with parallel high-throughput sequencing (ChIP-Seq) studies have identified specific promoters that are enriched for H4K20me3 (Barski et al., 2007; Gonzalo et al., 2005; Mikkelsen et al., 2007). Therefore, we hypothesized that SMYD5 functions as a corepressor of inflammatory genes by catalyzing local trimethylation of H4K20. Consistent with this, chromatin immunoprecipitation (ChIP) experiments detected H4K20me3 near the transcriptional start sites of several pro-inflammatory promoters, exemplified by the Tnf (Fig. 2E) and Cxcl10 (Fig. 2F) promoters, under basal conditions. Notably, there was a marked reduction in this mark upon stimulation of TLR4 with KLA (Fig 2E, F). Furthermore, knockdown of SMYD5 by siRNA treatment resulted in an almost complete loss of H4K20me3 on the Tnf and Cxcl10 promoters (Fig 2E, F), while not affecting total H4 occupancy (Supp. Fig 2C, D). Smyd5 siRNA did not reduce mRNA expression of the previously identified H4K20 methyltransferases: Set8, Nsd1, Suv420H1, or Suv420H2, indicating that the observed loss of H4K20me3 on pro-inflammatory promoters is most likely dependent on reduced SMYD5 expression (Supp. Fig 2E). Furthermore, siRNA mediated knockdown of the previously identified H4K20me3 methyltransferases SUV420H1 and SUV420H2 did not alter the H4K20me3 status of the Tnf (Supp. Fig. 2F) or Cxcl10 (Supp Fig. 2G) promoters and knockdown of SUV420H1 or SUV420H2 reduced, rather than enhanced, KLA-dependent activation of Tnf (Supp. Fig. 2H) or Cxcl10 mRNA (Supp Fig. 2I).
SMYD5 is a Component of NCoR Co-repressor Complexes
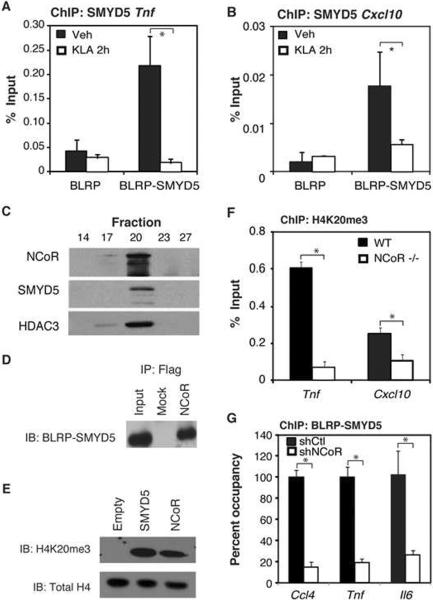
Next, we sought to determine whether SMYD5 resides at inflammatory promoters. Since available antibodies were not suitable for chromatin immunoprecipitation, we implemented a strategy to biotin-tag SMYD5 in RAW264.7 macrophages (de Boer et al., 2003). A biotin ligase recognition peptide (BLRP) was fused to the amino terminus of SMYD5 and the resulting protein was co-expressed with the bacterial biotin ligase BirA in RAW264.7 macrophages. This strategy enabled streptavidin-based ChIP assays to detect enrichment for SMYD5 on the Tnf and Cxcl10 promoters in the absence of inflammatory signals, whereas activation of TLR4 by KLA resulted in the dismissal of SMYD5 from these promoters (Fig. 3A, B).
Figure 3. The NCoR Complex Directs SMYD5 to TLR4-Responsive Promoters.
A. Chromatin immunoprecipitation assays assessing BLRP-SMYD5 occupancy of the Tnf promoter before and after KLA stimulation for 2 hours. Values represent the average of three experiments −/+ SEM,*p<0.05.
B. Chromatin immunoprecipitation assays assessing BLRP-SMYD5 occupancy of the Cxcl10 promoter before and after KLA stimulation for 2 hours. Values represent the average of three experiments −/+ SEM,*p<0.05.
C. Gel filtration experiments for HEK293 cell lysates transfected with Flag-NCoR and GFP-SMYD5. The lysates were then separated using SDS-PAGE analysis and immunoblotted with anti-NCoR, HDAC3, and GFP antibodies.
D. Co-Immunoprecipitation of BLRP-SMYD5 with Flag-NCoR in BLRP-SMYD5 RAW264.7 cells.
E. Methyltransferase reaction for SMYD5 and NCoR. HEK293 cells were transfected with vectors containing Flag-Empty, Flag-SMYD5, or Flag-NCoR and immunoprecipitated using Flag M2 beads. Lysates were subjected to a methyltransferase reaction and immunoblotted using a specific H4K20me3 antibody.
F. Chromatin immunoprecipitation assays assessing the H4K20me3 levels on the Tnf and Cxcl10 promoters in WT and NCoR−/− bone marrow derived macrophages. Values represent the average of three experiments −/+ SEM, *p<0.05.
G. Chromatin immunoprecipitation assays assessing the BLRP-SMYD5 recruitment to the Ccl4, Tnf and ll6 promoters following shCtl and shNCoR transfected Raw-BLRP-SMYD5 cells. Values represent the average of three experiments −/+ SEM, *p<0.05.
The amino-terminus of SMYD5 contains a MYND domain, which was previously demonstrated to mediate interactions with co-repressor proteins including NCoR and SMRT (Liu et al., 2007). Gel filtration analysis of nuclear extracts from HEK 293 cells transfected with GFP-SMYD5 and Flag-NCoR indicated that GFP-SMYD5 eluted with NCoR and HDAC3 in high molecular weight fractions (Fig. 3C). Co-immunoprecipitation assays in 293T cells detected interaction between full-length SMYD5 and NCoR (Fig. 3D). This interaction suggested that the NCoR complex could possess H4K20 histone methyltransferase activity. To test this hypothesis, we expressed Flag NCoR in 293T cells, purified the complex using an anti-Flag affinity matrix and subjected the precipitate to histone methyltransferase assays using recombinant histone H4 as a substrate. Interestingly, both the SMYD5 and NCoR complexes methylated H4K20me3 (Fig. 3E). Genetic deletion of NCoR in bone marrow-derived macrophages resulted in the loss of H4K20me3 on pro-inflammatory promoters, including Tnf and Cxcl10, as compared to Wt macrophages (Fig. 3F). Furthermore, SMYD5 occupancy on pro-inflammatory promoters was significantly reduced in BLRP-SMYD5 expressing RAW264.7 cells following shRNA-mediated knockdown of NCoR (Fig. 3G).
PHF2 is Required for Removal of H4K20me3 and Activation of TLR4-Responsive Promoters
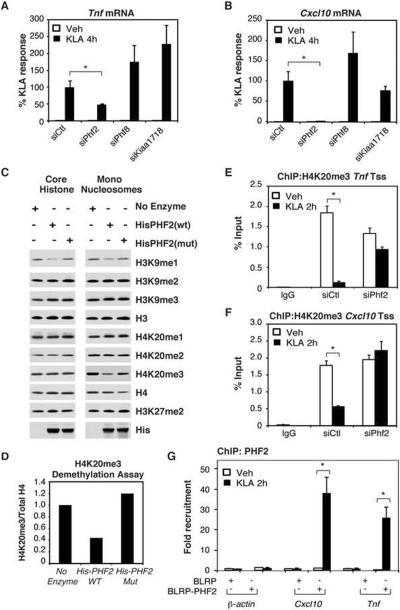
The observation that the H4K20me3 mark at inducible promoters was markedly reduced shortly after TLR4 ligation suggested the possibility of signal-dependent demethylation. Although no H4K20me3 demethylases have been described, recent studies have shown that PHF8 demethylates H4K20me1 (Liu et al., 2010; Qi et al., 2010), raising the possibility that two closely related proteins previously shown to possess demethylase activity, PHF2 (Wen et al., 2010) and KIAA1718 (Horton et al., 2010), may target H4K20me3 on inflammatory gene promoters. Knockdown of Phf2, but not Kiaa1718 or Phf8 (Supp. Fig. 3A–C), significantly diminished KLA induction of the SMYD5-sensitive Tnf, and Cxcl10 genes (Fig. 4A,B).
Figure 4. PHF2 Demethylates H4K20me3 and is Required for TLR4-dependent Gene Activation.
A. Quantitative real time PCR for Tnf mRNA isolated from thioglycollate elicited macrophages treated with siRNA for Control, Phf2, Phf8 and Kiaa1718 and subsequently treated with KLA for 4 hours. Values represent the average of three experiments −/+ SEM, *p<0.05.
B. Quantitative real time PCR for Cxcl10 mRNA isolated from thioglycollate elicited macrophages treated with siRNA for Control, Phf2, Phf8 and Kiaa1718 and subsequently treated with KLA for 4 hours. Values represent the average of three experiments −/+ SEM,*p<0.05.
C. Histone demethylase assay for PHF2 and a PHF2 mutant performed on core histone and mononucleosomes. His-PHF2(wt) or His-PHF2(mut)(H248A, D250A) were purified and incubated with core histones or mononucleosomes in histone demethylation buffer for 4 hours. Demethylation activity was evaluated by immunoblotting with specific antibodies.
D. Quantification of H4K20me3 intensities from figure 3C normalized to H4 signal.
E. Chromatin immunoprecipitation assays assessing the occupancy of H4K20me3 on the Tnf promoter in thioglycollate elicited macrophages treated with siCtl or siPhf2 for 48 hours followed by Veh or KLA stimulation for 2 hours. Values represent the average of three experiments −/+ SEM,*p<0.05.
F. Chromatin immunoprecipitation assays assessing the occupancy of H4K20me3 on the Cxcl10 promoter in thioglycollate elicited macrophages treated with siCtl or siPhf2 for 48 hours followed by Veh or KLA stimulation for 2 hours. Values represent the average of three experiments −/+ SEM,*p<0.05.
G. Chromatin immunoprecipitation assays assessing the recruitment of BLRP-PHF2 to the β-actin, Cxcl10, and Tnf promoters upon KLA stimulation for 2 hours. Values represent the average of three experiments −/+ SEM,*p<0.05.
To investigate PHF2 enzymatic function, wild type PHF2 and a mutant PHF2-A/A (H248A, D250A), which alters conserved residues required for iron binding in the active site of the Jumonji domain, were purified from bacteria cell extracts and subjected to an in vitro histone demethylation assay using purified core histones and mononucleosomes. PHF2 effectively removed H4K20me3 methylation on mononucleosomes, while the mutant PHF2-AIA was much less active (Fig. 4C, D). PHF2 also demethylated H3K9me1 on mononucleosomes as well as on core histones, consistent with recent observations (Wen et al., 2010) (Fig. 4C).
ChIP experiments were next performed to investigate whether PHF2 was involved in the removal of the H4K20me3 mark at TLR4-responsive promoters. Primary bone marrow derived macrophages were transfected with control siRNAs or siRNAs directed against Phf2 and the cells were then treated with vehicle or KLA for 4h. The H4K20me3 mark was depleted from the Tnf and Cxcl10 promoters in siCtl treated cells upon KLA treatment as previously seen, but not in PHF2 knockdown macrophages (Fig. 4E, F). Next, we sought to determine whether PHF2 is recruited to promoters exhibiting loss of H4K20me3 in response to LPS signaling. Because available antibodies were not suitable for the chromatin immunoprecipitation studies of PHF2, we implemented the above described biotin tagging strategy to perform chromatin immunoprecipitation studies of PHF2 in RAW264.7 macrophages. These studies demonstrated that PHF2 is recruited to the pro-inflammatory promoters Tnf and Cxcl10, but not β-actin, only upon activation of TLR4 signaling, correlating with the removal of H4K20me3 (Fig. 4G).
Exchange of SMYD5 and PHF2 Controls the Regulation of TLR4-Dependent Gene Expression
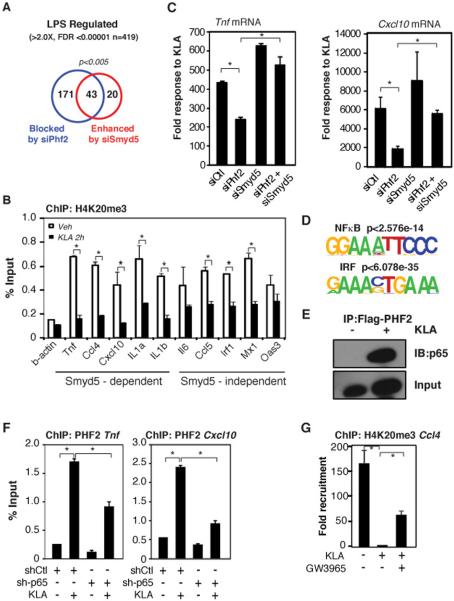
To globally evaluate the roles of PHF2 and SMYD5 in the regulation of TLR4-dependent gene activation, we performed RNA-sequencing of polyA RNA isolated from primary macrophages following transfections with Ctrl, Smyd5 or Phf2 siRNAs for 48 hours and subsequently treated with KLA for 4 hours (Supp. Table 1S). Of the 419 mRNAs that were significantly (FDR<0.00001) stimulated greater than 2 fold by KLA treatment (Fig. 5A), knockdown of PHF2 blunted the response of 51% (214 of 419) of these mRNAs. PHF2-dependent mRNAs were significantly enriched for functional annotations linked to cytokine activity, chemokine receptor binding and chemotaxis (Supp. Fig 4A). Conversely, knockdown of SMYD5 resulted in significantly exaggerated KLA responses for 63 of these mRNAs, which were significantly enriched for functional annotations linked to immune, defense and inflammatory responses. (Supp. Fig. 4B). Notably 68% (43 of 63) of mRNAs that were hyper-induced by knockdown of SMYD5 required PHF2 for activation (p<0.005). However, the observation that the PHF2-dependent subset of TLR4 responsive genes was substantially larger than the subset of genes hyper-induced following SMYD5 knockdown raised the question of whether the actual set of H4K20me3-marked promoters are more broadly distributed than that defined by hyper-induction. Consistent with this possibility, ChIP experiments indicated that H4K20me3 was present on the promoters of 10/10 PHF2-dependent KLA target genes evaluated, regardless of whether they were hyper-activated following SMYD5 depletion (Fig. 5B). These results suggest that SMYD5 and PHF2 function in a reciprocal manner to regulate the transcriptional response to KLA. Consistent with this possibility, knockdown of Smyd5 mRNA, which depletes SMYD5-dependent promoters of H4K20me3, circumvents the necessity for PHF2 in LPS-dependent gene activation of the Tnf andCxcl10 mRNAs (Fig. 5C).
Figure 5. Exchange of SMYD5 and PHF2 Controls the Regulation of TLR4-Dependent Gene Expression.
A. Venn diagram comparing the overlap of LPS genes that are blocked by siPhf2 treatment, enhanced by siSmyd5 treatment or unaffected by either treatment identified by polyA mRNA sequencing.
B. Chromatin immunoprecipitation assays for H4K20me3 on SMYD5 dependent and SMYD5 independent promoters in thioglycollate elicited macrophages. Values represent the average of three experiments −/+ SEM,*p<0.05.
C. Quantitative real time PCR for Tnf and Cxcl10 mRNAs isolated from thioglycollate elicited macrophages treated with siRNA for Control, Phf2, Smyd5, or a combination of Smyd5 and Phf2 siRNA and subsequently treated with KLA for 4 hours. Values represent the average of three experiments −/+ SEM,*p<0.05.
D. De Novo motif analysis of PHF2-dependent promoters.
E. Co-immunoprecipitation assay for transfected Flag-PHF2 with endogenous p65 in RAW 264.7 cells treated with Veh or KLA for 1 hour.
F. Chromatin immunoprecipitation assays for BLRP-PHF2 on the Tnf and Cxcl10 promoters in cells transfected with shCtl or sh-p65 for 48 hours and subsequently treated with KLA for 3 hours. Values represent the average of three experiments −/+ SEM,*p<0.05.
G. Chromatin immunoprecipitation assays for H4K20me3 on the Ccl4 promoter in thioglycollate elicited macrophages treated with Veh, KLA or a combination of GW3965 and KLA. Values represent the average of three experiments −/+ SEM,*p<0.05.
To gain insights into potential proteins involved in the signal dependent recruitment of PHF2 to these promoters, we analyzed the promoters of PHF2-dependent target genes and found NF-κB and IRF recognition motifs to be highly enriched (Fig. 5D). Therefore, we performed co-immunoprecipitation assays to determine whether PHF2 interacts with p65, a member of the NF-κB family, which is required for activation of a large subset of TLR4-dependent genes (Lim et al., 2007; Medzhitov and Horng, 2009). We detected KLA-dependent interaction between p65 and PHF2 (Fig. 5E). Notably, knockdown of p65 resulted in diminished recruitment of PHF2 to both the Cxcl10 and Tnf promoters, demonstrating that p65 is required for full recruitment of PHF2 (Fig. 5F).
LXRs exert repressive effects on TLR4 target genes by preventing signal-dependent NCoR clearance (Ghisletti et al., 2009). Since SMYD5 is delivered to inflammatory promoters by NCoR complexes and is required for LXR-mediated transrepression, we tested whether LXR agonists result in retention of H4K20me3 following TLR4 ligation. Consistent with this possibility, the LXR agonist GW3965 inhibited KLA-induced erasure of H4K20me3 on the Ccl4 promoter (Fig. 5G).
Discussion
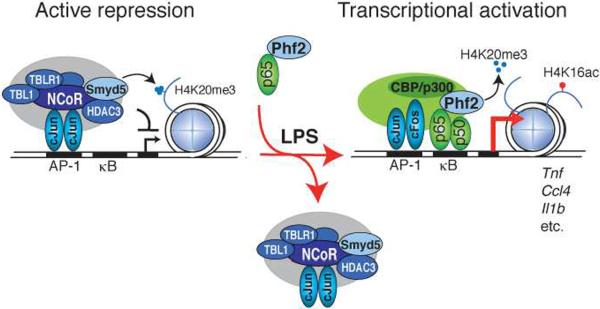
In concert, these findings reveal remarkable specificity in the growing spectrum of biological processes controlled by regulated methylation/demethylation of H4K20, previously established to play roles in cell cycle (H4K20me1) (Liu et al., 2010; Wu et al.), DNA damage repair (H4K20me2) (Pei et al., 2011; Sanders et al., 2004), and Wnt signaling (Li et al., 2011). Here, we provide evidence for an H4K20me3 methylation/demethylation mechanism, catalyzed by SMYD5 and PHF2 respectively, which is required for signal-dependent regulation of a subset of inflammatory response genes. We propose that under resting conditions, SMYD5 is recruited to a subset of TLR4-responsive promoters through its association with NCoR co-repressor complexes, where it trimethylates H4K20 (Fig. 6). Thus, in addition to mediating repression through the removal of acetylation by associated histone deacetylases, such as HDAC3, NCoR complexes also contribute to repression through the histone methyltransferase activity of associated SMYD5. The basis for the repressive function of H4K20me3 remains to be established; however recent studies have suggested that H4K20me3 inhibits acetylation of H4K16, resulting in the pausing of RNA polymerase II (Kapoor-Vazirani et al., 2011). Consistent with this, paused Pol II is a characteristic feature of many genes that are rapidly and highly induced by TLR4 signaling (Escoubet-Lozach et al.; Hargreaves et al., 2009).
Figure 6. Role of H4K20me3 in Regulation of TLR4 Responsive Genes.
Integrated model for SMYD5 and PHF2 regulation of inflammatory gene promoters.
The present studies expand on the emerging recognition of distinct biochemical activities and biological functions of members of the SMYD family of histone methytransferases. SMYD1–3 have been reported to mediate trimethylation of histone H3K4 and reduced deposition of this activation mark could potentially account for why siRNA knockdown of Smyd1, 2 and 3 resulted in reduced responses to TLR4 activation (Albert and Helin, 2010). It will thus be of interest to define the molecular mechanisms that dictate distinct substrate specificities. The preference of SMYD5 for H4K20 appears to be an intrinsic property of the enzyme based on the observed activity of the purified GST-SMYD5 expressed in bacteria. In addition to SMYD5 and SUV420H1/2, SMYD3 has also recently been reported to trimethylate H4K20 (Foreman et al., 2011). However, the observations that knockdown of each of these enzymes resulted in reduced activation of TLR4 target genes, and that the presence of the H4K20me3 mark at TLR4-responsive promoters was largely dependent on SMYD5, strongly argue that SMYD3 and SUV420H1/2 primarily act on different substrates and/or are directed to H4K20 at different genomic locations by alternative protein complexes. Of note, we find that knockdown of SUV420H1/2 results in significant reduction in the expression of Phf2, which could at least partly explain the effect of their knockdown on TLR4-dependent gene activation (J.S., unpublished).
We were initially led to SMYD5 by an RNAi screen that identified the Drosophila homologue CG3353 as being required for Tailless repression of LPS signaling. The observation that SMYD5 is required for transrepression activities of LXRs on at least a subset of its target genes suggests evolutionarily conserved molecular functions of SMYD5 that link nuclear receptor signaling to antagonism of innate immune responses. The finding that SMYD5 is required for LXR-dependent, but not glucocorticoid receptor-dependent antagonism of TLR4-induced gene expression is also consistent with prior observations that LXR, but not glucocorticoid receptor, transrepression requires NCoR (Blaschke et al., 2006; Ghisletti et al., 2009; Venteclef et al., 2010; Ogawa et al., 2005).
We further show that TLR4-induced removal of H4K20me3 is catalyzed by PHF2,which is delivered to responsive promoters by the p65 subunit of NF-κB (Fig. 6). The observation that more genes require PHF2 for optimal responses to TLR4 signaling than are hyperinduced in the setting of SMYD5 knockdown has several potential explanations. One is that while removal of H4K20me3 may be required for signal-dependent activation, lack of this mark may not be sufficient for hyperactivation, suggesting that it serves a repression `checkpoint' function. Consistent with this possibility, many PHF2-dependent promoters that were not hyper-responsive in response to SMYD5 knockdown were nevertheless marked by H4K20me3 under basal conditions. It is also possible that PHF2 targets additional methylated residues, such as H3K9me1, on pro-inflammatory promoters that are deposited by other histone methyltransferases. It is of interest that PHF2 was capable of catalyzing removal of H3K9me1, but not H4K20me3, on core histones. We speculate that the requirement for a nucleosome substrate for demethylation of H4K20me3 may be due to the ability of the Plant Homeodomain in PHF2 to recognize H3K4me3 (Wen et al., 2010). This interaction may anchor the enzyme in the correct conformation or allosterically regulate substrate specificity, as previously suggested for H4K20me1 demethylation by PHF8 (Liu et al., 2010). As our findings suggest that demethylation of H4K20me3 is a critical step in gene activation of inflammatory response genes such as Tnf, modulation of the expression or activity of PHF2, may provide novel approaches for treatment of inflammatory diseases.
Experimental Procedures
Drosophila RNAi Screen
Schneider S2 cells were plated in 6cm dishes and transfected with 1.5 μg Attacin A reporter and 1.5 μg Tailless expression vector using Effectene (Qiagen) transfection reagent according to the manufacture's guidelines. The cells were then plated in 384 well plates containing 250 ng of dsRNA and incubated for 48 hours. Cells were then treated with 100 ng/ml LPS for 8 hours, lysed and luciferase activity was monitored using a luminometer.
Gel Filtration
HEK293 cells were transfected with Flag-tagged NCoR WT and GFP-SMYD5 WT using Lipofectamine™ 2000. 48 hrs post-transfection, HEK293 cultured cells were harvested, washed twice with ice-cold PBS, and cytoplasmic extracts were prepared using a low-salt buffer (10 mM HEPES pH 7.9, 50 mM NaCl, 1mM DTT). Nuclei were spun down and extracted in 20 mM HEPES pH 7.9, 20% glycerol, 350 mM NaCl, 0.2 EDTA, 1 mM DTT and protease inhibitors. 1 mg sample was then applied onto a 250 ml column of Sephacryl S-300 (Amersham Biosciences, Inc.) previously equilibrated with gel filtration buffer (20 mM HEPES pH 7.9, 120 mM NaCl, 0.2 mM EDTA, 1 mM DTT) and 60, 1-ml fractions were collected. Fractions were analyzed by western blot.
Cell Culture and Western Blots
Raw 264.7 cells were grown in DMEM supplemented with 10% FBS and penicillin/streptomycin. Raw 264.7 cells were transfected using Superfect (Qiagen) according to the manufacture's guidelines and as previously published (Ghisletti et al., 2009). Primary Thioglycollate-elicited macrophages and bone-marrow derived macrophage cells were generated as previously described (Ogawa et al., 2005). BLRP stables for SMYD5 and PHF2 were generated as previously reported (Heinz et al., 2010). LPS (Sigma) and KLA (Avanti) were used at a concentration of 100 ng/ml unless otherwise stated. The shRNA constructs were constructed using pSilencer 3.0 H1 (Ambion). Antibodies used are as follows: Flag (Sigma, F3165), GFP (Abcam, Ab290), p65 (Santa Cruz, sc-8008), BLRP(Genscript, A00674), HDAC3 (Santa Cruz, sc-11417), H4 (Active Motif, 39269), H4K20me1 (Abcam, Ab9051), H4K20me2 (Abcam, Ab9052), H4K20me3 (Abcam, Ab9053), H3K9me1 (Abcam, Ab8896), H3K9me2 (Abcam, Ab1220), H3K9me3 (Abcam, Ab8898), H3K27me2 (Abcam, Ab24684),H3K27me3 (Millipore, 07-449) H3K4me3 (Abcam, AB8580), H3K36me3 (Abcam, Ab9050), H3 (Active Motif, 39163) and His (Santa Cruz, SC-803). Chemically methylated H4 proteins were purchased from Active Motif.
RNA Isolation, Quantification and Sequencing
RNA was purified using RNeasy Mini Kit (Qiagen) and enriched for Poly(A)-RNA with MicroPoly(A) Purist Kit (Ambion, Austin, TX, USA). For quantification, cDNA was generated using the Superscript kit (Invitrogen) and quantitative real-time PCR was performed using gene specific primers. For sequencing, RNA was treated with TURBO DNase (Ambion), fragmented using RNA Fragmentation Reagents (Ambion) and purified by a P-30 column (Bio-Rad, Hercules, CA, USA). Fragmented RNA was dephosphorylated with Antarctic phosphatase (New England Biolabs, Ipswich, MA, USA) followed by heat-inactivation and overnight precipitation. Poly(A)-tailing and cDNA synthesis was performed as previously described (Ingolia et al., 2009). However, for reverse transcription, oligos with custom barcodes (underlined) were used: 5'-Phos CA/TG/AC/GT GATCGTCGGACTGTAGAACTCT/idSp/CAAGCAGAAGACGGCATACGATTTTTTTTTTTTTTTTTTTTVN-3'. After cDNA synthesis, exonuclease was used to catalyze the removal of excess oligo. Enzyme was inactivated and RNA hydrolyzed by alkaline treatment (100 mM NaOH) and heat (25 min, 95°C). The cDNA fragments of 50–150 nucleotide s were purified on a denaturing Novex 10% polyacrylamide TBE-urea gel (Invitrogen, Carlsbad, CA, USA). The recovered cDNA was circularized, linearized, amplified for 15 cycles and gel purified as previously described (Ingolia et al., 2009). The library was sequenced on the Illumina Genome Analyzer 2 according to the manufacturer's instructions. Gene Ontology analysis for regulated genes was performed as previously described (Sullivan et al., 2010). Promoters of regulated genes were analyzed for enriched motifs as previously described (Heinz et al., 2010).
siRNA Transfections
Transfections using 5 nM siRNA were performed using lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. SMART siRNA pools for examined genes were purchased from Dharmacon.
Histone Methylation Assays
Purification of GST-SMYD5, GST-SMYD5 mutants, His-H4, and His-H4K20A was performed using standard molecular biology techniques. Flag-NCoR and Flag-SMYD5 complexes were obtained from transient transfected HEK 293 cells and immunoprecipitated with Flag M2 beads (Sigma). Purified proteins were incubated with 2 μg recombinant histone H4 (NEB), or chemically methylated histone H4 (Active Motif),or His tagged H4 or H4K20A, 1 ml [3H] adenosyl-L-methionine (PerkinElmer) or 0.3 mM SAM (NEB), in histone methylation buffer (50 mM Tris [pH 9.0], 0.5 mM DTT) overnight at 30°C. For the radiolabled experiments the reactions were then transferred to P81 paper and washed three times with 10% trichloroacetic acid for 15 minutes followed by a wash with 95% ethanol and analyzed using a scintillation counter. Otherwise samples were separated using SDS-PAGE and immunoblotted using specific antibodies.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were performed as previously described (Heinz et al., 2010). The antibodies used in these studies are as follows: IgG (Santa Cruz Biotechnology), H4K20me3 (Abcam, Ab9053). SMYD5 and PHF2 ChIPs were performed using streptavidin immunoprecipitations from Raw 264.7 cells stably expressing SMYD5 of PHF2 tagged with a biotin recognition peptide as described previously (Heinz et al., 2010).
Demethylation Assay
His-PHF2 and His-PHF2 (H248A, D250A) were purified from bacterial extracts using standard protocols and were incubated with 2 mg mononucleosomes in DeMTase buffer (20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 50 mM [NH4]2Fe[SO4]2 for 2–5 hr at 37°C. Samples were separated using SDS-PAGE and immunoblotted using specific antibodies. Quantification of western blots were performed using ImageJ software (Abramoff, 2004).
Co-Immunoprecipitations
Flag-PHF2 or Flag-NCoR and BLRP-SMYD5 were transfected into 293T cells. Cells were lysed and sonicated in lysis buffer (10 mM Tris (pH 8.0), 420 mM NaCl, 0.5% NP40, 1mM EDTA, Protease Inhibitors (Roche). Samples were then diluted with dilution buffer (10 mM Tris (pH 8.0), 0.5% NP40, 1.0 mM EDTA) and protein complexes were purified using Flag M2 beads (Sigma). Beads were washed with dilution buffer five times and eluted with 3× Flag peptide. Lysates were then separated using SDS-PAGE and interactions were detected using western blot analysis.
Supplementary Material
Highlights
Smyd5 establishes H4K20me3 as a repression checkpoint at TLR4-responsive promoters
Smyd5 is recruited to TLR4-responsive genes by NCoR co-repressor complexes
Smyd5 is required for transrepression of TLR4-responsive genes by liver X receptors
Phf2 removes H4K20me3 and is required for TLR4-dependent gene activation
Acknowledgements
We thank Dawn Zhang and David Gosselin for critical comments and Lynn Bautista for assistance with preparation of the manuscript. These studies were supported by P01-HC088093, P30 DK063491 and CA52599 to CKG. MGR is supported by the Howard Hughes Medical Institute. JS was supported by T32 CA009523. MUK was supported by Fondation Leducq Career Development award and Sigrid Jesélius fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers Sequencing data are available in the Gene Expression Omnbibus database (http://www.ncbi.nlm.nih.goc/geo) under the accession number GSE39113.
References
- Abramoff MD, Magalhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Albert M, Helin K. Histone methyltransferases in cancer. Semin Cell Dev Biol. 2010;21:209–220. doi: 10.1016/j.semcdb.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Blaschke F, Takata Y, Caglayan E, Collins A, Tontonoz P, Hsueh WA, Tangirala RK. A nuclear receptor corepressor-dependent pathway mediates suppression of cytokine-induced C-reactive protein gene expression by liver X receptor. Circ Res. 2006;99:e88–99. doi: 10.1161/01.RES.0000252878.34269.06. [DOI] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci U S A. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escoubet-Lozach L, Benner C, Kaikkonen MU, Lozach J, Heinz S, Spann NJ, Crotti A, Stender J, Ghisletti S, Reichart D, et al. Mechanisms Establishing TLR4-Responsive Activation States of Inflammatory Response Genes. PLoS Genet. 7:e1002401. doi: 10.1371/journal.pgen.1002401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flammer JR, Rogatsky I. Minireview: Glucocorticoids in autoimmunity: unexpected targets and mechanisms. Mol Endocrinol. 2011;25:1075–1086. doi: 10.1210/me.2011-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman KW, Brown M, Park F, Emtage S, Harriss J, Das C, Zhu L, Crew A, Arnold L, Shaaban S, et al. Structural and functional profiling of the human histone methyltransferase SMYD3. PLoS One. 2011;6:e22290. doi: 10.1371/journal.pone.0022290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Jepsen K, Benner C, Hardiman G, Rosenfeld MG, Glass CK. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 2009;23:681–693. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH. Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140:918–934. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S, Garcia-Cao M, Fraga MF, Schotta G, Peters AH, Cotter SE, Eguia R, Dean DC, Esteller M, Jenuwein T, et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol. 2005;7:420–428. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoberg JE, Popko AE, Ramsey CS, Mayo MW. IkappaB kinase alpha-mediated derepression of SMRT potentiates acetylation of RelA/p65 by p300. Mol Cell Biol. 2006;26:457–471. doi: 10.1128/MCB.26.2.457-471.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C, Tontonoz P. Coordination of inflammation and metabolism by PPAR and LXR nuclear receptors. Curr Opin Genet Dev. 2008;18:461–467. doi: 10.1016/j.gde.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol. 2010;17:38–43. doi: 10.1038/nsmb.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- Huang W, Ghisletti S, Perissi V, Rosenfeld MG, Glass CK. Transcriptional integration of TLR2 and TLR4 signaling at the NCoR derepression checkpoint. Mol Cell. 2009;35:48–57. doi: 10.1016/j.molcel.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Ghisletti S, Saijo K, Gandhi M, Aouadi M, Tesz GJ, Zhang DX, Yao J, Czech MP, Goode BL, et al. Coronin 2A mediates actin-dependent de-repression of inflammatory response genes. Nature. 2011;470:414–418. doi: 10.1038/nature09703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor-Vazirani P, Kagey JD, Vertino PM. SUV420H2-mediated H4K20 trimethylation enforces RNA polymerase II promoter-proximal pausing by blocking hMOF-dependent H4K16 acetylation. Mol Cell Biol. 2011;31:1594–1609. doi: 10.1128/MCB.00524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Lawrence T, Nizet V. Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell. 2006;124:823–835. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- King-Jones K, Thummel CS. Nuclear receptors--a perspective from Drosophila. Nat Rev Genet. 2005;6:311–323. doi: 10.1038/nrg1581. [DOI] [PubMed] [Google Scholar]
- Kourmouli N, Sun YM, van der Sar S, Singh PB, Brown JP. Epigenetic regulation of mammalian pericentric heterochromatin in vivo by HP1. Biochem Biophys Res Commun. 2005;337:901–907. doi: 10.1016/j.bbrc.2005.09.132. [DOI] [PubMed] [Google Scholar]
- Lazzaro BP. Natural selection on the Drosophila antimicrobial immune system. Curr Opin Microbiol. 2008;11:284–289. doi: 10.1016/j.mib.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Li Z, Nie F, Wang S, Li L. Histone H4 Lys 20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proc Natl Acad Sci U S A. 2011;108:3116–3123. doi: 10.1073/pnas.1009353108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CA, Yao F, Wong JJ, George J, Xu H, Chiu KP, Sung WK, Lipovich L, Vega VB, Chen J, et al. Genome-wide mapping of RELA(p65) binding identifies E2F1 as a transcriptional activator recruited by NF-kappaB upon TLR4 activation. Mol Cell. 2007;27:622–635. doi: 10.1016/j.molcel.2007.06.038. [DOI] [PubMed] [Google Scholar]
- Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466:508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chen W, Gaudet J, Cheney MD, Roudaia L, Cierpicki T, Klet RC, Hartman K, Laue TM, Speck NA, et al. Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO's activity. Cancer Cell. 2007;11:483–497. doi: 10.1016/j.ccr.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli C, Reichhart JM. Evolution and integration of innate immune systems from fruit flies to man: lessons and questions. J Endotoxin Res. 2005;11:243–248. doi: 10.1179/096805105X37411. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, Hoffmann A, Subramaniam S, David M, Rosenfeld MG, et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005;122:707–721. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Lozach J, Jepsen K, Sawka-Verhelle D, Perissi V, Sasik R, Rose DW, Johnson RS, Rosenfeld MG, Glass CK. A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation. Proc Natl Acad Sci U S A. 2004;101:14461–14466. doi: 10.1073/pnas.0405786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, Bergsagel PL, Wang L, You Z, Lou Z. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470:124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, et al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010;466:503–507. doi: 10.1038/nature09261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo K, Winner B, Carson CT, Collier JG, Boyer L, Rosenfeld MG, Gage FH, Glass CK. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 2009;137:47–59. doi: 10.1016/j.cell.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan AL, Benner C, Heinz S, Huang W, Xie L, Miano JM, Glass CK. Serum response factor utilizes distinct promoter- and enhancer-based mechanisms to regulate cytoskeletal gene expression in macrophages. Mol Cell Biol. 2010;31:861–875. doi: 10.1128/MCB.00836-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Tedgui A, Mallat Z. Cytokines in atherosclerosis: pathogenic and regulatory pathways. Physiol Rev. 2006;86:515–581. doi: 10.1152/physrev.00024.2005. [DOI] [PubMed] [Google Scholar]
- Venteclef N, Jakobsson T, Ehrlund A, Damdimopoulos A, Mikkonen L, Ellis E, Nilsson LM, Parini P, Janne OA, Gustafsson JA, et al. GPS2-dependent corepressor/SUMO pathways govern anti-inflammatory actions of LRH-1 and LXRbeta in the hepatic acute phase response. Genes Dev. 2010;24:381–395. doi: 10.1101/gad.545110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Li J, Song T, Lu M, Kan PY, Lee MG, Sha B, Shi X. Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation. J Biol Chem. 2010;285:9322–9326. doi: 10.1074/jbc.C109.097667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Wang W, Kong X, Congdon LM, Yokomori K, Kirschner MW, Rice JC. Dynamic regulation of the PR-Set7 histone methyltransferase is required for normal cell cycle progression. Genes Dev. 24:2531–2542. doi: 10.1101/gad.1984210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.