Abstract
Ample evidence has shown key roles of inflammation in tumor promotion and carcinogenesis, and tumor-associated macrophages are known to promote tumor growth and dissemination. Programmed cell death 4 (Pdcd4) is a novel tumor suppressor, and although various studies have revealed that the functions and expression mechanisms of Pdcd4 in tumor promotion, those in regard to inflammation remain unclear. In the present study, we examined whether inflammatory stimuli regulate Pdcd4 expression. 12-O-tetradecanoylphorbol 13-acetate (TPA) suppressed expression of pdcd4 mRNA in human monocytic cell lines (U937, THP-1). Similarly, the bacterial endotoxin lipopolysaccharide (LPS) downregulated pdcd4 level in mouse RAW264.7 and peritoneal macrophages. Furthermore, conditioned medium from LPS-stimulated RAW264.7 macrophages suppressed pdcd4 mRNA in RAW264.7 macrophages, and findings obtained with recombinant tumor necrosis factor-α (TNF-α) and TNF-α-specific siRNA suggested that TNF-α partly mediates LPS-triggered Pdcd4 downregulation via an autocrine mechanism. Specific inhibitors of phosphoinositide-3-kinase (PI3K) and c-jun N-terminus kinase (JNK) restored LPS-abolished pdcd4 mRNA. Consistently, in MCF7 mammary carcinoma cells, conditioned medium from TPA-differentiated/activated U937 cells suppressed pdcd4 mRNA. Additionally, knockdown of pdcd4 in RAW264.7 macrophages using siRNA significantly enhanced LPS-induced TNF-α protein production, and interferon-γ, CC chemokine ligand (Ccl) 1, Ccl20, and interleukin-10 mRNA expression. These results suggest that Pdcd4 suppresses the induction of these inflammatory mediators. Taken together, loss of Pdcd4 in macrophages may be a critical step in establishing the inflammatory environment while that in tumor cells contributes to tumor progression.
Keywords: Pdcd4, TNF-α, inflammation
INTRODUCTION
Inflammation plays many key roles in the host defense system, the processes of which are strictly regulated in physiological conditions. In contrast, its loss of control, for example, excessive and chronic inflammation, leads to the onset of a number of diseases, including inflammatory and autoimmune disorders and cancer [1]. The mechanisms of inflammation consist of various cell signaling pathways, cell–cell interactions, and cell maturation and differentiation. For example, monocytes migrate into the inflamed area, followed by differentiation into macrophages, which are one of the predominant cell types that respond to microbial infection and are particularly important in innate immunity [2]. Macrophages recognize pathogenic substances through various sensor proteins and subsequently initiate inflammatory responses. Lipopolysaccharide (LPS), an endotoxin, is one of the most well-characterized inflammatory stimuli that interacts with macrophages and monocytes. In response to LPS, immune cells are activated and display various inflammatory phenotypes, including expression of inflammatory genes and production of various inflammatory cytokines, chemokines, and their mediators. These biological responses are activated through mitogen-activated protein kinases (MAPKs) and phosphoinositide-3-kinase (PI3K) signaling pathways, both of which lead to transactivation of nuclear factor-κB (NFκB) and activator protein-1 (AP-1) [3]. In addition, the tumor promoter 12-O-tetradecanoylphorbol 13-acetate (TPA) has been shown to stimulate similar signaling pathways and transcription factors contributing to monocyte differentiation and activation as well as to inflammatory responses in mouse skin [4].
Programmed cell death 4 (Pdcd4) was recently identified as a novel tumor suppressor and is considered to be a promising target for antineo-plastic therapy. It is expressed ubiquitously in normal tissues, whereas it is downregulated in a variety of tumors, exemplified by carcinomas of the skin [5] and colon [6]. Pdcd4 has been shown to suppress and prevent tumor promotion and progression by regulation of various proteins at transcriptional, translational, and posttranslational levels, as noted in the review presented by Lankat-Buttgereit and Göke [7]. A number of reports have shown that Pdcd4 negatively regulates the trans-activation of AP-1 [7,8], while suppression of Pdcd4 activates the PI3K/Akt pathway [9], which is constitutively activated in various diseases. Meanwhile, there is ample evidence indicating mechanistic links between cancer and inflammation, in which both AP-1 and PI3K play key roles [10,11]. Therefore, we speculate that Pdcd4 is a negative regulator in inflammatory processes, though few reports have presented this possibility [8,12].
In the present study, we examined whether inflammatory stimuli downregulate Pdcd4 in several cellular and animal models. In addition, we attempted to elucidate the molecular mechanisms underlying Pdcd4 repression and found that LPS downregulated Pdcd4 via the c-jun N-terminal kinase (JNK) and PI3K pathways in mouse macrophages and that downregulation of Pdcd4 significantly increased the production of TNF-α protein, a cytokine that has versatile roles in inflammation. Importantly, we provide evidence that mediators released by inflammatory activated macrophages attenuate Pdcd4 in tumor cells increasing transformation of the latter.
MATERIALS AND METHODS
Animals and Cells
Female ICR mice were purchased from Japan SLC (Shizuoka, Japan) at 5–6 wk of age and maintained according to the Guidelines for the Regulation of Animals, provided by the Animal Experimentation Committee of Kyoto University. All animals were housed under controlled conditions of humidity (60 ± 5%), lighting (12-h light cycle), and temperature (24 ± 2°C). RAW264.7 mouse macrophages, U937 human monocytic lymphoma cells, THP-1 human acute monocytic leukemia cells, and MCF7 human mammary carcinoma cells were purchased from American Type Culture Collection (Manassas, VA). RAW264.7 macrophages and MCF7 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% and 10% heat-inactivated fetal bovine serum (FBS), respectively, as well as penicillin (100 U/mL) and streptomycin (100 mg/mL). U937 and THP-1 cells were cultured in Roswell Park Memorial Institute (RPMI) medium 1640 containing 10% heat-inactivated FBS, penicillin (100 U/mL), and streptomycin (100 mg/mL). Each cell line was incubated at 37°C under a humidified atmosphere of 95% air and 5% CO2.
Chemicals
DMEM, RPMI 1640, and FBS were purchased from Invitrogen (Carlsbad, CA). LPS used in vitro was from Escherichia coli 0127:B8, while that used in vivo was from Salmonella typhimurium ATCC 7823, and both were obtained from Sigma–Aldrich (St. Louis, MO). Antibodies were obtained from the following sources: rabbit anti-phospho-PI3K, anti-phospho-extracellular signal-regulated kinase 1/2 (ERK1/2), anti-phospho-p38, anti-JNK 1/2, anti-phospho-c-jun, and horseradish peroxidase (HRP)-conjugated anti-rabbit IgG were from Cell Signaling Technology (Beverly, MA); goat anti-β-actin was from Santa Cruz Biotechnology (Santa Cruz, CA); mouse anti-α-tubulin was from Oncogene (San Diego, CA); and HRP-conjugated anti-goat IgG and anti-mouse IgG were from Dako (Glostrup, Denmark). Anti-Pdcd4 antibody was described previously [13]. All other chemicals were purchased from Wako Pure Chemical Industries (Osaka, Japan), unless otherwise specified.
Mouse Tissues
The backs of 7-wk-old female ICR mouse were shaved with a surgical clipper, then TPA (8 nmol) dissolved in 200 μL of acetone was applied onto the shaved portion of each. Six hours later, the mice were killed by cervical dislocation, then the back skin was isolated and scraped with a razor to obtain epidermal protein samples [14]. Each collected epidermal layer was lysed for Western blot analysis, which is described below.
Mouse Peritoneal Macrophages
Six-week-old ICR mice were killed, and ice-cold DMEM containing 10% FBS and 1% heparin was injected into the peritoneal cavity of each. Cells were collected twice by washing with the medium and cultured in 12-well plates. After 24 h of incubation at 37°C, nonadherent cells were removed by rinsing with phosphate-buffered saline (PBS), while adherent cells were treated with LPS (500 ng/mL) in DMEM without FBS. After 18 h, total RNA was isolated, as described below.
Western Blot Analysis
RAW264.7 macrophages were seeded 12 h before treatment. Prior to the experiments cells were rinsed with PBS and medium was exchanged to DMEM without FBS. After being treated with LPS (100 ng/mL) for various time periods, the cells were lysed with lysis buffer [protease inhibitor, phosphatase inhibitor (Sigma–Aldrich), 10 mM Tris (pH 7.4), 1% sodium dodecyl sulfate (SDS), 1 mM sodium vanadate]. The lysates from cells and mouse skin were then sonicated and centrifuged at 15 000g for 5 min. The protein concentration in the supernatant was quantified using a Bio Rad Protein Assay (Bio-Rad Laboratories, Hercules, CA) and standardized using γ-globulin as the reference. The lysates were denatured in a sample buffer containing SDS and 2-mercaptoethanol. An equal amount of protein (20–30 μg) was separated using SDS gel electrophoresis, subsequently proteins were electrotransferred to Immobilon-P membranes (Millipore, Billerica, MA). Proteins were detected with specific primary and secondary antibodies, and visualized using ECL reagents (GE Healthcare, Little Chalfont, UK). Densitometric analysis was performed using Scion Image 0.4.0.3 (Scion Corporation, Frederick, MD).
Reverse Transcription-(Quantitative) Polymerase Chain Reaction (RT-qPCR)
U937 cells were stimulated with TPA (10 nM) for various time periods. RAW264.7 macrophages were treated with LPS (100 ng/mL), conditioned medium from RAW264.7 macrophages (RAW-CM), or tumor necrosis factor-α (TNF-α; 2500 pg/mL) for various time periods. MCF7 cells were exposed to CM from U937 cells (U937-CM) or TNF-α (20 ng/mL). For pathway analysis, cells were pretreated with various inhibitors (PD98059, MEK1/2 inhibitor, 50 μM; SP600125, JNK1/2 inhibitor, 10 or 50 μM; SB203580, p38 MAPK inhibitor, 10 μM; LY294002, PI3K inhibitor, 30 μM; 0.5% DMSO as control) for 30 min before exposure to LPS (100 ng/mL), RAW-CM, or TNF-α (2500 pg/mL) for 24 h. Cells were preincubated with vehicle (0.5% DMSO) or each inhibitor for 30 min, then treated with or without LPS (100 ng/mL) or TNF-α (2500 pg/mL) for 24 h, followed by incubation with Cell Counting Kit-8 (Dojindo, Kumamoto, Japan). From the result of this assay, the indicated concentration of each inhibitor did not have any effect on cell viability.
Total RNA was extracted using TRIzol Reagent (Invitrogen), according to the manufacturer’s protocol. mRNA was reverse transcribed using the iScript cDNA synthesis kit (BioRad, München, Germany) according to the manufacturer’s instructions. Expression was analyzed using PCR (10× PCR buffer, dNTP mixture, MgCl2, Taq DNA polymerase, 5 U/μL; Takara, Otsu, Japan). Amplified DNA was separated by agarose gel electrophoresis and stained with SYBR Gold (Invitrogen). Image analysis was performed using Scion Image 0.4.0.3. Lack of PCR saturation was confirmed by titrating each cDNA amount (data not shown). Alternatively, mRNA expression was analyzed by quantitative PCR using the Absolute Blue SYBR Green fluorescein kit assay (ThermoScientific, Hamburg, Germany) according to the manufacturer’s protocol. Primer sequences and PCR conditions were designed based on specificity and suitability for qPCR analysis (GC-content, length) or selected from previous reports [15–19]. They were as follows: pdcd4 (mouse), 300 bp (5′-TAATCAgTg-CAAgCgAAATTAAggAA-3′ and 5′-CCTTTCCCA-gATCTggACCgCCTATC-3′), at 94°C for 15 s, 55°C for 30 s, and 72°C for 45 s; TNF-α (mouse), 402 bp (5′-CCTgTAgCCCACgTCgTAgC-3′ and 5′-TTgACCT-CAgCgCTgAgTTg-3′), at 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s; interferon (IFN)-γ (mouse), 237 bp (5′-AACgCTACACACTgCATCTTgg-3′ and 5′-gACTT-CAAAgAgTCTgAgg-3′), at 90°C for 40 s, 62°C for 40 s, and 72°C for 60 s; CC chemokine ligand (Ccl)-1 (mouse), 125 bp (5′-CgTgTggATACAggATgTTgA-CAg-3′ and 5′-AggAggAgCCCATCTTTCTgTAAC-3′), at 95°C for 10 s, 56°C for 5 s, and 72°C for 30 s; Ccl-20 (mouse), 86 bp (5′-CAgAAgCAgCAAgCAACTACgA-3′ and 5′-CTgTCTTgTgAAACCCACAATAgC-3′), at 94°C for 20 s, 60°C for 20 s, and 72°C for 20 s; interleukin (IL)-10 (mouse), 195 bp (5′-CgggAAgA-CAATAACTg-3′ and 5′-CATTTCCgATAAggCTTgg-3′), at 94°C for 30 s, 58°C for 30 s, and 72°C for 60 s; cyclophilin (mouse), 240 bp (5′-gCCAggACCTgTAT-gCTTCA-3′ and 5′-TTgggTCgCgTCTCgTTCgA-3′), at 95°C for 30 s, 50°C for 30 s, and 72°C for 60 s; hypoxanthine guanine phosphoribosyl transferase (HPRT) (mouse), 196 bp (5′-gTAATgATCAgTCAAC-ggggAC-3′ and 5′-CCAgCAAgCTTgCAACCTTAACC-A-3′), at 95°C for 30 s, 55°C for 25 s, and 72°C for 45 s; pdcd4 (human), 134 bp (5′-ACAgTTggTgggCCAgTT-TATTgC-3′ and 5′-TCAgAAgCACggTAgCCTTATC-CA-3′), at 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s; TNF-α (human), 215 bp (5′-TCTCgAACCCC-gAgTgACA-3′ and 5′-gAggAgCACATgggTggAg-3′), at 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s; 18S (human), 151 bp (5′-gTAACCCgTTgAACCCCATT-3′ and 5′-CCATCCAATCggTAgTAgCg-3′), at 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s.
Preparation of Macrophage Conditioned Media
Following preincubation of RAW264.7 macrophages with regular growth medium, the cells were exposed to LPS (100 ng/mL) in DMEM without FBS. After 12 h of incubation, the medium was collected for the preparation of RAW-CM. U937 cells were exposed to TPA (10 nM) for 48 h. Adherent, that is, differentiated and activated, U937 cells were trypsinized, washed with PBS, and reseeded. After 24 h the medium was collected as U937-CM.
ELISA for TNF-α
RAW264.7 macrophages were seeded 12 h before experiments. After pretreatment with various inhibitors (PD98059, MEK1/2 inhibitor, 50 μM; SP600125, JNK1/2 inhibitor, 50 μM; SB203580, p38 MAPK inhibitor, 10 μM; LY294002, PI3K inhibitor, 30 μM; 0.5% DMSO as a control) for 30 min, cells were exposed to LPS (100 ng/mL) for 1 h. The concentration of TNF-α in the supernatants from culture media was determined using an ELISA kit (Endogen, Cambridge, MA), according to the manufacturer’s instructions.
Luciferase Assay for AP-1-Dependent Transactivation
Following 12 h of preincubation, RAW264.7 macrophages were transiently cotransfected with an AP-1-driven luciferase reporter (Clontech, Palo Alto, CA) and a pRL-TK plasmids (Promega, Madison, WI) using Lipofectamine (Invitrogen) in OPTI-MEM (Invitrogen). After 6 h, medium was changed to DMEM (10% FBS) for an additional 6 h. Then the cells were stimulated with LPS in DMEM without FBS. MCF7 cells were transiently transfected with the same constructs using Rotifect (Roth) according to the manufacturer’s protocol. After 16 h, medium was changed and cells were stimulated by U937-CM or direct co-culture with differentiated U937 cells for 24 h. Following stimulation, cells were lysed, and firefly and renilla luciferase activity assays were performed using a Dual Luciferase kit assay (Promega) and luminometer (PerkinElmer, Boston, MA), according to the manufacturers’ instructions.
Knockdown of TNF-α and pdcd4 With siRNA
RAW264.7 macrophages were seeded 24 h before siRNA transfection in regular growth medium. Control siRNA (Santa Cruz Biotechnology), TNF-α siRNA (Invitrogen), or Pdcd4 siRNA (Invitrogen) were mixed with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol, and added to the cells at a final concentration of 75 nM. Six hours after transfection, the cells were recovered in DMEM containing 10% FBS for 24 h, then stimulated with LPS (100 ng/mL) for 6 or 12 h. The cell lysates and media were collected for RT-PCR, Western blotting, and ELISA, as described above.
PCR Array Analysis
RT2 Profiler™ PCR Array PAMM-011 (SA Biosciences, Frederick, MD) was used to assess the expression profiles of inflammatory cytokines and receptors in LPS-stimulated, Pdcd4-silenced RAW264.7 macrophages. Control or Pdcd4 siRNA-transfected RAW264.7 macrophages were exposed to LPS (100 ng/mL) for 6 h, which was followed by cDNA synthesis from 1 μg of total RNA. Thermal cycling was performed on 7300 Real Time PCR System using SYBR green PCR mix (Applied Biosystems, Foster, CA) according to the manufacturer’s protocol.
Statistical Analysis
Data are shown as the mean ± standard deviation from more than three independent experiments and were evaluated using Student’s t-test, unless otherwise specified. Differences were considered statistically significant at the P <0.05 level.
RESULTS
Inflammatory Stimuli Decrease Pdcd4 Expression In Vitro and In Vivo
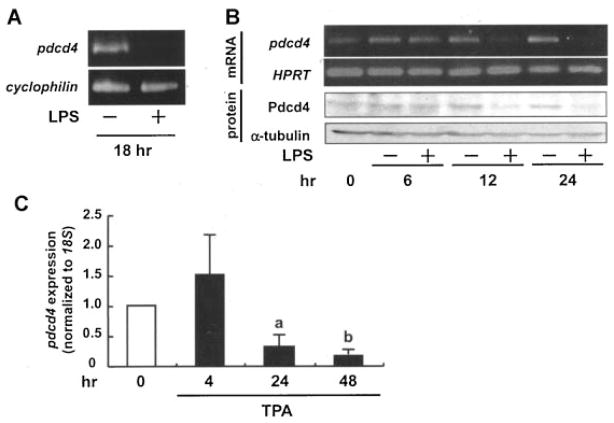
To assess if inflammatory stimuli affect Pdcd4 expression, mouse peritoneal macrophages were treated with LPS. pdcd4 mRNA expression was strongly diminished after 18 h LPS treatment (Figure 1A). Consistently, LPS suppressed the expression of mRNA and protein of Pdcd4 in RAW264.7 macrophages at 12–24 h (Figure 1B). To evaluate if this effect is a general effect, human monocytic cell lines were treated with TPA, which was previously shown to induce differentiation and maturation into activated macrophages [20]. TPA suppressed pdcd4 mRNA expression in U937 cells (Figure 1C) and THP-1 cells (data not shown) in a time-dependent manner. Initial effects on pdcd4 mRNA levels appeared at 12 and 24 h of TPA in THP-1 and U937, respectively. These results imply that Pdcd4 is lost in response to inflammatory conditions both in monocytes and macrophages.
Figure 1.
Inflammatory stimuli downregulate Pdcd4 expression in macrophages. (A) Mouse peritoneal macrophages were isolated and harvested as described in the Materials and Methods Section, then exposed to LPS (500 ng/mL) for 18 h before being subjected to RT-PCR (cyclophilin used as the control). (B) RAW264.7 mouse macrophages were stimulated with LPS (100 ng/mL) for 0, 6, 12, and 24 h. Total RNA was isolated and subjected to RT-PCR (hypoxanthine guanine phosphoribosyl transferase (HPRT) was used as control) (upper panels). Alternatively, proteins were isolated and analyzed via Western blot analysis (α-tubulin served as the loading control). Images presented are representative of three independent experiments. (C) U937 human monocytic lymphoma cells were stimulated with TPA (10 nM) for 4, 24, and 48 h, total RNA was isolated and subjected to RT-qPCR (pdcd4 expression was normalized to 18S rRNA expression and given relative to control treatment). Student’s t-test was used to determine significant differences. aP <0.05 and bP <0.005 versus control.
JNK1/2 and PI3K Pathways Participate in pdcd4 Repression
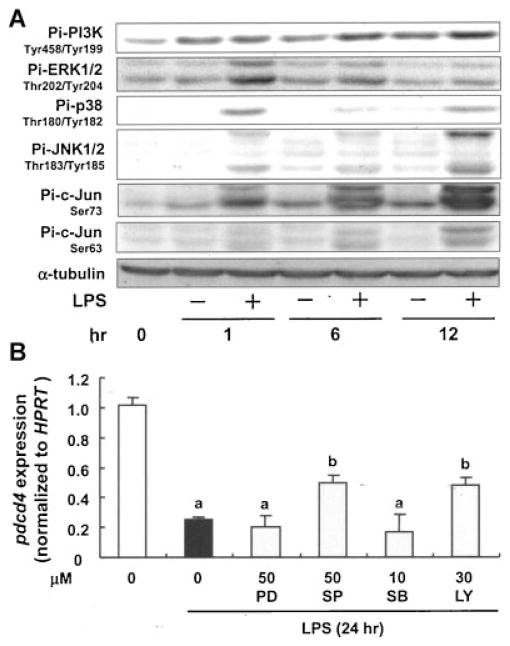
Inducers of inflammatory response generally function through activation of multiple signaling pathways and transcription factors as well as through production of inflammatory mediators [21]. Therefore, we next aimed at analyzing the molecular mechanisms underlying LPS-induced pdcd4 mRNA repression in RAW264.7 macrophages. As shown in Figure 2A, PI3K, MAPKs, and c-jun were activated in RAW264.7 cells when stimulated with LPS. To investigate the involvement of these signaling molecules in LPS-triggered pdcd4 downregulation, we employed a pharmacological approach using specific protein kinase inhibitors, that is, PD98059 (MEK1/2 inhibitor, 50 μM), SP600125 (JNK1/2 inhibitor, 50 μM), SB203580 (p38 MAPK inhibitor, 10 μM), and LY294002 (PI3K inhibitor, 30 μM). None of these inhibitors had a significant effect on cell viability after 24 h of incubation (data not shown). Pretreatment of RAW264.7 macrophages with SP600125 and LY294002, but not PD98059 and SB203580, 30 min prior to LPS exposure for 24 h significantly alleviated LPS-induced attenuation of pdcd4 mRNA (P <0.05; Figure 2B).
Figure 2.
JNK and PI3K signaling cascades contribute to LPS-induced Pdcd4 regulation. (A) RAW264.7 macrophages were stimulated with LPS (100 ng/mL) for 0–12 h, then subjected to Western blotting for the detection of phosphorylation of MAPKs (ERK1/2, JNK1/2, and p38), c-jun, and PI3K using appropriate antibodies. The expression levels of the nonphosphorylated forms did not differ significantly among the treatments (data not shown). Results shown are representative of three independent experiments. (B) RAW264.7 macrophages were preincubated with vehicle (0.5% DMSO), PD98059 (MEK inhibitor), SP600125 (JNK inhibitor), SB203580 (p38 inhibitor), or LY294002 (PI3K inhibitor) for 30 min, then treated with or without LPS (100 ng/mL). After 24 h of incubation, total RNA was isolated for RT-PCR. Each value represents the mean ± SD of at least three independent experiments. Student’s t-test was used to determine significant differences. aP <0.005 versus control, bP <0.05 versus LPS.
LPS-Induced TNF-α Secretion Partly Mediates pdcd4 Repression
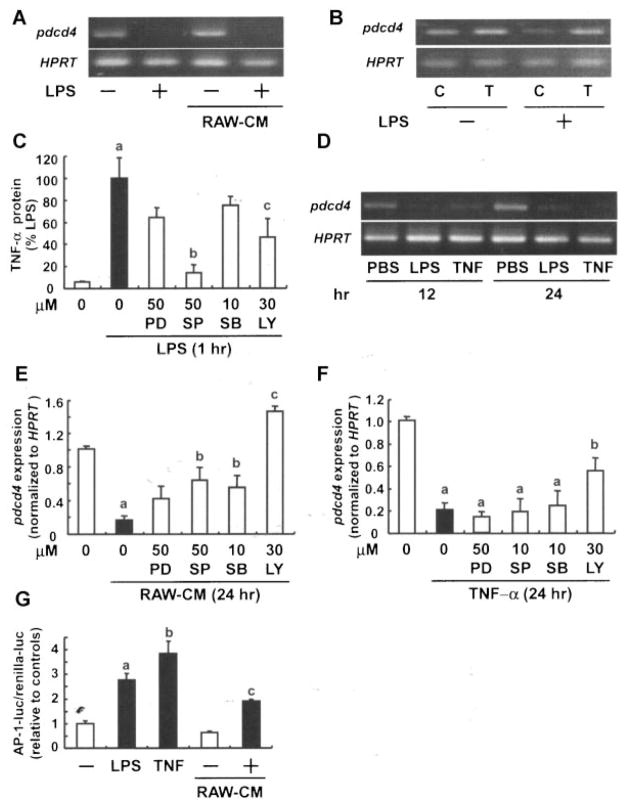
To examine whether LPS-dependent signaling directly affects pdcd4 mRNA levels or if factor(s) are released from LPS-stimulated RAW264.7 macrophages to mediate pdcd4 downregulation, we treated RAW264.7 macrophages with LPS for 12 h, changed the medium to non-LPS containing medium, and harvested the resulting medium after additional 24 h as conditioned medium (RAW-CM). Subsequent incubation of fresh RAW264.7 macrophages with RAW-CM abolished pdcd4 mRNA expression to a similar degree as LPS, whereas CM from nonstimulated cells did not have any effects (Figure 3A). Since TNF-α represents an important constituent of the proinflammatory response of macrophages to LPS, we speculated that TNF-α could function as suppressor of pdcd4 via an autocrine mechanism. To this end, we depleted TNF-α in RAW macrophages using an siRNA approach. Attenuation of TNF-α expression rescued pdcd4 mRNA expression from LPS-mediated downregulation (Figure 3B). Consistently, treatment with recombinant TNF-α abolished pdcd4 mRNA to a similar extent as LPS (Figure 3D). To address the signaling cascades involved in LPS-induced TNF-α production various chemical inhibitors were employed. Both inhibition of JNK (by SP600125) and PI3K (by LY294002) significantly reduced the LPS-triggered TNF-α protein increase (P <0.01 and <0.05, respectively), while the other tested inhibitors were less active (Figure 3C). In addition, CM-abolished pdcd4 mRNA was inhibited by SP600125 and SB203580 (P <0.05) and markedly by LY294002 (P <0.005) (Figure 3E), while TNF-α-abolished pdcd4 mRNA was significantly restored by LY294002 alone (P <0.05) (Figure 3F). Thus, we conclude that JNK and PI3K activation is critical for LPS-induced production of TNF-α and TNF-α is an essential autocrine mediator contributing to the LPS-dependent loss of pdcd4 mRNA. Interestingly, though, the latter mechanism only requires intact PI3K signaling. Transformation, measured as AP-1 transactivation, was previously reported to be inhibited by Pdcd4 [7,8]. Consistently, TNF-α and RAW-CM induced AP-1 transactivation in RAW264.7 macrophages to a extent similar to LPS (Figure 3G).
Figure 3.
LPS-released TNF-α contributes to Pdcd4 regulation in macrophages. (A) RAW264.7 macrophages were exposed to LPS (100 ng/mL) for 1 h, after which the medium was refreshed. After 11 h, conditioned medium (RAW-CM) was collected and added to RAW264.7 macrophages, which were incubated for another 24 h, followed by RT-PCR. (B) RAW264.7 macrophages were transfected with control (C) or TNF-α (T) siRNA using Lipofectamine 2000 in OPTI-MEM, and recovered in DMEM containing 10% FBS for 24 h, followed by exposure to LPS (100 ng/mL) for 12 h. (C) RAW264.7 macrophages were pretreated with each inhibitor for 30 min, and then exposed to LPS for 1 h. Next, supernatants from the cell cultures were collected for quantification of TNF-α by ELISA. Each value represents the mean ± SD of at least three independent experiments. Student’s t-test was used to determine significant differences. aP <0.05 versus control, bP <0.01, cP <0.05 versus LPS. (D) RAW264.7 macrophages were treated with PBS, LPS, or recombinant TNF-α for 12 or 24 h, then subjected to RT-PCR analysis (HPRT used as the control). For (E) and (F) RAW264.7 macrophages were preincubated with vehicle (0.5% DMSO), PD98059 (MEK inhibitor), SP600125 (JNK inhibitor), SB 203580 (p38 inhibitor), or LY294002 (PI3K inhibitor) for 30 min. After the preincubation, cells were treated (E) with RAW-CM (as described above) or (F) TNF-α 2500 pg/mL) for 24 h. Total RNA was isolated for RT-PCR. Each value represents the mean ± SD of at least three independent experiments. Student’s t-test was used to determine significant differences. aP <0.005 versus control, bP <0.05 versus conditioned medium and TNF-α, respectively. (G) RAW264.7 macrophages were cotransfected with AP-l luciferase construct and phRL-TK vector using Lipofectamine. Transiently transfected cells were stimulated with LPS, TNF-α, or RAW-CM (generated with or without LPS-activation) for 24 h. Luciferase activity was measured as described in the Materials and Methods Section. AP-1-luc activity was expressed relative to the respective controls. Each value represents the mean ± SD of at least three independent experiments. Student’s t-test was used to determine significant differences. aP <0.005 and bP <0.05 versus control, cP <0.05 versus CM control.
Inflammatory Macrophages Limit pdcd4 mRNA Expression in Tumor Cells
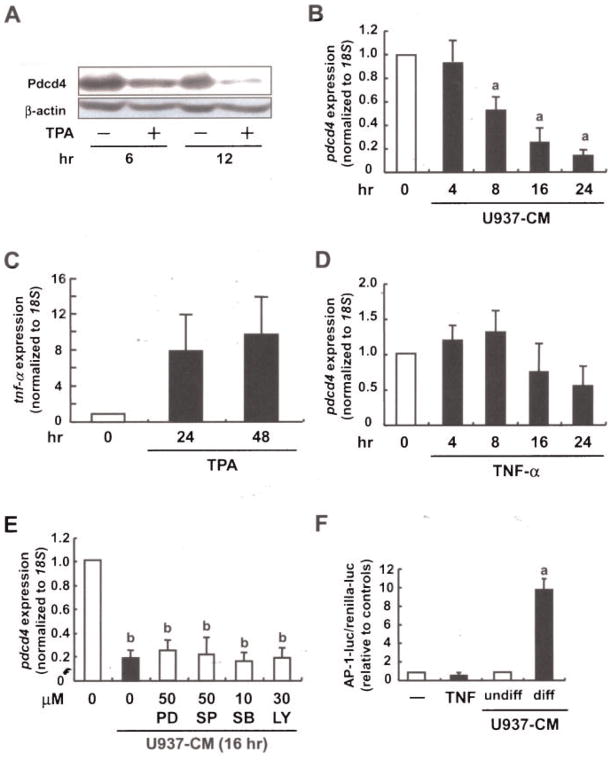
As macrophages play a pivotal role in the regulation of the inflammatory tumor microenvironment, we went on to characterize the effect of macrophages on Pdcd4 expression in tumor cells. First indications that inflammatory conditions affect Pdcd4 expression came from in vivo experiments. Specifically, Pdcd4 protein levels were strongly attenuated in mouse epidermis in response to TPA treatment (Figure 4A), which was consistent with our previous reports [15,22]. To further elucidate the impact of macrophage-primed conditions on tumor cells, we induced differentiation and activation of human monocytic cells U937 by TPA treatment for 48 h. Differentiated/activated U937 cells were washed and reseeded in fresh medium. After 24 h the resulting supernatant was used as conditioned medium of U937 cells (U937-CM). Pdcd4 mRNA expression in human MCF7 mammary carcinoma cells was strongly diminished in response to U937-CM treatment for >16 h (Figure 4B). Based on the observation that TNF-α played an important role in the autocrine signaling to deplete Pdcd4, we next analyzed expression of TNF-α in U937 cell in response to TPA. TPA treatment resulted in a rapid, strong increase of TNF-α RNA at 4 h. Longer treatments as used for differentiation (24 and 48 h) caused a slight attenuation of the initial peak expression but stayed elevated for >48 h (Figure 4C). To verify that TNF-α also acts in a paracrine manner to eliminate Pdcd4, we exposed MCF7 cells to recombinant TNF-α. Surprisingly, TNF-α had only a limited effect on pdcd4 mRNA expression in MCF7 tumor cells under prolonged incubations (Figure 4D). Interestingly, Pdcd4 mRNA expression was not rescued from U937-CM-induced downregulation by inhibitors of PI3K, MEK/ERK, p38-MAPK, or JNK signaling (Figure 4E). Taken together, these data imply that Pdcd4 is lost in both macrophages and tumor cells under inflammatory conditions. TNF-α appears to be an important factor mediating autocrine effects to macrophages, while other mediators seem to be involved in mediating Pdcd4 regulating effects to tumor cells. Meanwhile, AP-1-activity in MCF7 cells was strongly induced by U937-CM from differentiated U937 as compared to U937-CM from undifferentiated U937, while TNF-α did not increase AP-1 activity (Figure 4F) different from the results in RAW264.7 macrophages (Figure 3G). Thus, AP-1-dependent transcription, which is like pdcd4 mRNA expression, responds differently to TNF-α in macrophages and tumor cells, while CM from macrophages appears active in both cell lines. Therefore, we conclude that while TNF-α is a critical autocrine factor sustaining the inflammatory response in macrophages, Pdcd4 expression in and transformation of tumor cells appears to be affected by other inflammatory mediators.
Figure 4.
Macrophage-released factors contribute to Pdcd4 regulation in tumor cells. (A) Shaved dorsal skin areas of 6-wk-old ICR mice were treated with the vehicle (acetone) or TPA (8 nmol). After 6 or 12 h, the mice were killed and the epidermis from each was collected, after which lysates from the epidermis samples were subjected to Western blot analysis (β-actin used as the control). (B) U937 cells were differentiated and activated with TPA (10 nM) for 48 h. Differentiated U937 cells were reseeded and allowed to condition medium for 24 h (U937-CM). Human MCF7 breast tumor cells were incubated with U937-CM for 4–24 h. (C) U937 cells were stimulated with TPA (10 nM) for 4, 24, or 48 h. (D) MCF7 cells were incubated with recombinant TNF-α (20 ng/mL) for 4–24 h. (E) MCF7 cells were preincubated with the indicated inhibitors for 30 min. After the preincubation, cells were treated with (C) U937-CM (as described above) or (D) TNF-α (20 ng/mL) for 24 h. For (B)–(E) total RNA was isolated and subjected to RT-qPCR (expression was normalized to 18S rRNA expression and given relative to control treatment). Student’s t-test was used to determine significant differences. aP <0.005 and bP <0.001 versus control. (F) MCF7 cells were cotransfected with AP-l luciferase construct and phRL-TK vector using Rotifect reagent. Transiently transfected cells were stimulated with TNF-α, or U937-CM (undifferentiated vs. differentiated) for 24 h. Luciferase activity was measured as described in the Materials and Methods Section. AP-1-luc activity was expressed relative to the respective controls. Student’s t-test was used to determine significant differences. aP <0.005 versus control.
Pdcd4 Limits LPS-Dependent Induction of Inflammatory Mediators
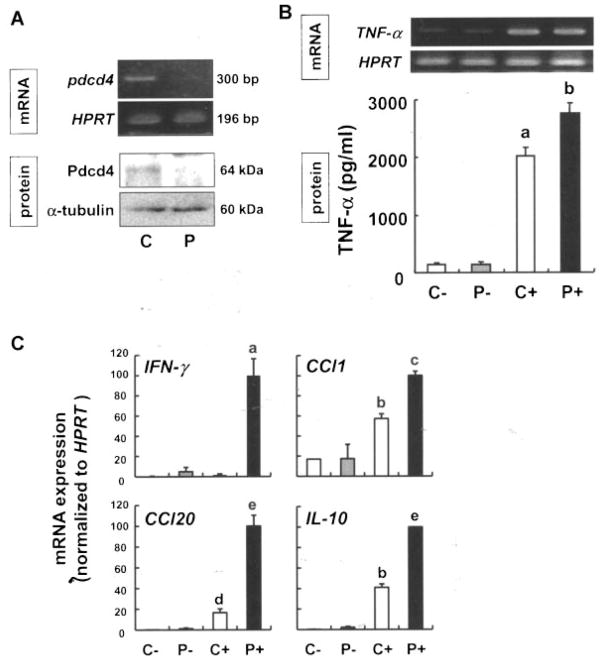
As Pdcd4 has previously been proposed to influence expression of inflammatory cytokines [23], we next aimed at elucidating the influence of Pdcd4 on the inflammatory setting described above. Therefore, RAW264.7 macrophages were transfected with control or Pdcd4-specific siRNAs (Figure 5A) to determine the effects on their proinflammatory responses. Although the expression levels of TNF-α mRNA did not differ between control and Pdcd4-silenced cells, Pdcd4 protein concentration in the media was significantly higher in the Pdcd4-silenced cells (P <0.05; Figure 5B).
Figure 5.
Knockdown of Pdcd4 increases expression of proinflammatory genes. (A) RAW264.7 macrophages were transfected with the control (C) or Pdcd4 (P) siRNA using Lipofectamine 2000 in OPTI-MEM, and recovered in DMEM containing 10% FBS for 24 h. Total RNA and protein were extracted for RT-PCR and Western blot analysis, respectively. (B) After recovery, cells were exposed to vehicle or LPS (100 ng/mL) for 6 h then the total RNA and supernatants from the cell cultures were subjected to RT-PCR and ELISA, respectively. C−, control siRNA-transfected cells; P−, Pdcd4 siRNA-transfected cells; C+, control siRNA-transfected cells; P+, Pdcd4 siRNA-transfected cells incubated with LPS. Each value represents the mean ± SD of three separate experiments. Student’s t-test was used to determine significant differences. aP <0.001 versus control siRNA without LPS, bP <0.05 versus control siRNA with LPS. (C) To verify the results obtained from the PCR array analysis (Supplementary Table 1), expression levels of INF-γ, Ccl-1, Ccl-20, and IL-10 were semi-quantified by real-time RT-PCR, and HPRT served as the internal standard. Each value represents the mean ± SD of three separate experiments. Student’s t-test was used to determine significant differences. aP <0.01 versus control siRNA with LPS, bP <0.005 versus control siRNA without LPS, cP <0.001 versus control siRNA with LPS, dP <0.05 versus control siRNA without LPS, eP <0.005 versus control siRNA with LPS.
To identify additional inflammatory mediators that are LPS-inducible, yet suppressed by Pdcd4, we analyzed the expression levels of 83 key genes involved in the inflammatory responses including chemokines, cytokines, interleukins, and their receptors with PCR array. We found that 30 genes were induced by >8.0-fold by LPS as compared with vehicle control (Supplementary Table 1). Pdcd4 downregulation markedly increased expression levels of IFN-γ, CC chemokine receptor (Ccr)-4, Ccl-1, Ccl-20, and IL-10 by >2.0-fold (Supplementary Table 1). Expression levels of four mRNAs were validated by real-time RT-qPCR analysis. As shown in Figure 5C, depletion of Pdcd4 led to a dramatic increase of IFN-γ (16- and 60-fold), Ccl-20 (4.3- and 6.0-fold), IL-10 (10- and 2.4-fold), and Ccl-1 (1.0- and 1.7-fold) mRNA expression in nonstimulated and LPS-stimulated cells, respectively, as compared with Pdcd4-normal cells.
Thus, because inflammation-induced secretion of TNF-α appears to be partly suppressed by Pdcd4 and because TNF-α attenuates pdcd4 expression, we propose a self-enhancing mechanism for inflammation-associated TNF-α production. Consistently, Pdcd4 appears to limit the expression of additional constituents of the inflammation-associated immune response such as IFN-γ, Ccl-20, IL-10, and Ccl-1.
DISCUSSION
An increasing body of evidence indicates that Pdcd4 is a novel tumor suppressor found to be deficient in various types of mammalian tumors. Suppression of AP-1 transactivation and the PI3K pathway has been put forward as molecular mechanisms underlying the tumor suppressive function of Pdcd4. Elevated AP-1 and PI3K activation are characteristic features of tumorigenesis associated with chronic inflammation, yet the regulation of Pdcd4 and its effects within the inflammatory microenvironment remain to be fully elucidated. As shown in Figure 1, the inflammatory stimuli TPA and LPS downregulated Pdcd4 expression in several monocytic cells and macrophages. Furthermore, CM from LPS-stimulated RAW264.7 cells and CM from TPA-activated U937 cells suppressed Pdcd4 mRNA expression (Figures 3A and 4B). Furthermore, our data suggest that TNF-α is a key player in the suppression of Pdcd4 mRNA by LPS through an autocrine mechanism (Figure 3B and D). Nonetheless, since inhibition of JNK or p38 MAPK was effective in blocking the downregulation of Pdcd4 by CM (Figure 3E) but not by TNF-α (Figure 3F), other factors are likely to be involved as well. Previously, Zhang and DuBois [24] reported that a cyclooxygenase (COX)-2 selective inhibitor, NS-398, induced pdcd4 mRNA. Along a similar line, Nieves-Alicea et al. [25] showed that overexpression of COX-2 led to decreased pdcd4 mRNA levels and that prostaglandin (PG) E2 downregulated Pdcd4 in breast cancer cells. COX-2 has been extensively linked to inflammation-associated carcinogenesis [26]. In spite of those previous findings, COX-2 selective inhibitors (NS-398 and nimesulide) had no effects on LPS-suppressed pdcd4 mRNA expression, and PGE2 did not downregulate pdcd4 mRNA expression in the present study (data not shown). This is attributable to differences in cell types and stimuli used. Although IL-6 was detected in the medium, anti-IL-6 neutralizing antibody showed scant effects and also the IL-1β production was not remarkable in the present conditions (data not shown). Therefore, so far, these prominent proinflammatory mediators were ruled out. Importantly, macrophage-induced Pdcd4 repression in tumor cells was not (or only minimally) attributable to TNF-α, which supports the notion that there are other factors released by macrophages under inflammatory conditions that also contribute to the loss of Pdcd4. Elucidation of responsible factor(s) for the downregulation of Pdcd4 both in macrophages and in tumor cells under inflammatory conditions is a matter of ongoing investigations.
Mechanistically, we demonstrate that LPS-activated JNK and PI3K signaling pathways are involved in suppressing Pdcd4 expression and resultant TNF-α production in macrophages (Figures 2B and 3C). Interestingly, while PI3K signaling appeared to be essential for both the induction of TNF-α as well as the Pdcd4 repression by TNF-α, the increased production of TNF-α seemed to require intact JNK signaling only (Figure 3C and F). Thus, while JNK appears to be upstream of the production of TNF-α, PI3K seems critical for both the LPS-induced TNF-α expression as well as the TNF-α signaling contributing to the suppression of Pdcd4. This is in accordance with reports linking LPS-induced TNF-α production to JNK signaling [27], or connecting the PI3K pathway with TNF-α expression and TNF-α-dependent signaling [28,29]. Interestingly, in tumor cells where the Pdcd4-regulating influence of TNF-α was limited, neither JNK nor PI3K signaling proved to be essential for U937-CM-mediated Pdcd4 down-regulation. Thus, activated JNK and PI3K signaling seems to be specific for TNF-α-related effects on and by Pdcd4. Recently, work on Pdcd4 regulation focused largely on microRNA (miR)-21-mediated translational repression [30] and phosphorylation-dependent degradation of Pdcd4 protein [22,31]. Furthermore, the expression of miR-21 in response to TPA or LPS was demonstrated to be AP-1-dependent [32,33] and induction of AP-1 activity was specifically shown to be required for miR-21-mediated Pdcd4 regulation [34]. In our study, AP-1 trans-activation was induced in macrophages by CM from LPS-activated RAW264.7 macrophages or by TNF-α alone (Figure 3G), and in tumor cells by CM from TPA-activated U937 cells (Figure 4F). Thus, while our present findings suggest TNF-α and other constituents of the inflammatory tumor microenvironment as regulators of Pdcd4 mRNA expression, we cannot exclude other mechanisms by which Pdcd4 expression might be limited under inflammatory conditions. As a side note, while CM contained comparable levels of TNF-α (Figure 5B) to those exogenously supplied for TNF-α only experiments (Figure 3F), CM activated AP-1 not as strong as TNF-α alone (Figure 3G). This could be taken as an indicator for additional, counter-acting factors involved that remain to be identified and underlines the need for further in-depth analysis of components of the CM. As indicated above, TNF-α was neither effective in attenuating Pdcd4 levels nor did it induce AP-1 activity in MCF7 cells. Therefore, the mechanism of Pdcd4 downregulation appears to be highly cell type specific.
Functionally, knockdown of Pdcd4 increased TNF-α protein production at the posttranscriptional level in LPS-stimulated RAW264.7 macrophages (Figure 5B). While the exact mechanism remains elusive, a molecular link between Pdcd4 and p38 MAPK might be possible, since TNF-α converting enzyme was shown to be activated by p38 MAPK [35]. Recently, Sheedy et al. [23] put Pdcd4 forward as an proinflammatory regulator, since they found the expression of the immune modulatory factor IL-10 to be increased in Pdcd4-knockdown cells. This is consistent with our finding of increased IL-10 expression in Pdcd4-depleted macrophages in response to LPS. Nevertheless, while Sheedy et al. describe the opposite effect for IL-6, that is, loss of Pdcd4 resulted in a decrease of IL-6 production, we observed an increase in the LPS inducibility of various proinflammatory mediators such as IFN-γ, Ccl-20, and Ccl-1 in macrophages depleted of Pdcd4. These factors were previously described to be induced by proinflammatory stimuli such as LPS, TNF-α, IL-1β, CD40 ligand, and IFN-γ [36,37] and contribute to shaping the inflammatory microenvironment by inducing the synthesis of proinflammatory mediators and recruiting further inflammatory cells [36,38,39]. Taken together, Pdcd4 downregulation appears to be critical for the expression of numerous inflammatory mediators. The seemingly contradictory observation regarding IL-10 indicates that Pdcd4 might actually exert a tuning function in the tight regulation of pro- versus antiinflammatory events.
In conclusion, we put forward downregulation of Pdcd4 expression in inflammatory environments as both inflammation-sustaining and tumor-promoting event. Our results indicate that LPS activates JNK and PI3K pathways in macrophages to produce and release TNF-α protein, which down-regulates Pdcd4 in an autocrine manner. The observation that the production of TNF-α protein was increased in Pdcd4-silenced cells provides a positive feed-forward loop to maintain inflammatory conditions. Pdcd4 is similarly lost in tumor cells in response to inflammatory conditions resulting in increased AP-1 transactivation, indicative of transformation potential. In the case of tumor cells TNF-α was excluded as a major factor. Thus, an in-depth analysis of Pdcd4 regulatory mechanisms in various cellular components of the inflammatory tumor microenvironment might assign Pdcd4 a key role both in TNF-α-associated inflammatory diseases as well as in inflammation-associated tumorigenesis. Therapeutically, Pdcd4 can be envisioned as target for novel antiinflammatory drugs as well as for tumor therapeutic agents.
Supplementary Material
Acknowledgments
This study was supported in part by a Grant-in-Aid for Cancer Research from the Ministry of Health, Labor and Welfare of Japan (A.M.) and the LOEWE Schwerpunkt OSF (III L 4-518/55.004 (2009)) funded by the Hessian Ministry of Higher Education, Research and the Arts (T.S.).
Abbreviations
- LPS
lipopolysaccharide
- MAPK
mitogen-activated protein kinase
- PI3K
phosphoinositide-3-kinase
- AP-1
activator protein-1
- TPA
12-O-tetradecanoylphorbol 13-acetate
- Pdcd4
programmed cell death 4
- JNK
c-jun N-terminal kinase
- DMEM
Dulbecco’s modified Eagle’s medium
- FBS
fetal bovine serum
- ERK
extracellular signal-regulated kinase
- PBS
phosphate-buffered saline
- SDS
sodium dodecyl sulfate
- RT-PCR
reverse transcription-polymerase chain reaction
- CM
conditioned medium
- TNF-α
tumor necrosis factor-α
- IFN
interferon
- Ccl
CC chemokine ligand
- IL
interleukin
- COX
cyclooxygenase
- miR
microRNA
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.Schwartsburd P. Chronic inflammation as inductor of pro-cancer microenvironment: Pathogenesis of dysregulated feedback control. Cancer Metastasis Rev. 2003;22:95–102. doi: 10.1023/a:1022220219975. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R, Janeway CJ. Innate immunity: The virtues of a nonclonal system of recognition. Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 3.Guha M, Mackman N. LPS induction of gene expression in human monocytes. Cell Signal. 2001;13:85–94. doi: 10.1016/s0898-6568(00)00149-2. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein L, Colburn N. AP1/jun function is differentially induced in promotion-sensitive and resistant JB6 cells. Science. 1989;244:566–569. doi: 10.1126/science.2541502. [DOI] [PubMed] [Google Scholar]
- 5.Matsuhashi S, Narisawa Y, Ozaki I, Mizuta T. Expression patterns of programmed cell death 4 protein in normal human skin and some representative skin lesions. Exp Dermatol. 2007;16:179–184. doi: 10.1111/j.1600-0625.2006.00531.x. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q, Sun Z, Yang H. Downregulation of tumor suppressor Pdcd4 promotes invasion and activates both beta-catenin/Tcf and AP-1-dependent transcription in colon carcinoma cells. Oncogene. 2008;27:1527–1535. doi: 10.1038/sj.onc.1210793. [DOI] [PubMed] [Google Scholar]
- 7.Lankat-Buttgereit B, Göke R. The tumour suppressor Pdcd4: Recent advances in the elucidation of function and regulation. Biol Cell. 2009;101:309–317. doi: 10.1042/BC20080191. [DOI] [PubMed] [Google Scholar]
- 8.Hilliard A, Hilliard B, Zheng S, et al. Translational regulation of autoimmune inflammation and lymphoma genesis by programmed cell death 4. J Immunol. 2006;177:8095–8102. doi: 10.4049/jimmunol.177.11.8095. [DOI] [PubMed] [Google Scholar]
- 9.Lankat-Buttgereit B, Müller S, Schmidt H, Parhofer K, Gress T, Göke R. Knockdown of Pdcd4 results in induction of proprotein convertase 1/3 and potent secretion of chromogranin A and secretogranin II in a neuroendocrine cell line. Biol Cell. 2008;100:703–715. doi: 10.1042/BC20080052. [DOI] [PubMed] [Google Scholar]
- 10.Hirsch E, Ciraolo E, Ghigo A, Costa C. Taming the PI3K team to hold inflammation and cancer at bay. Pharmacol Ther. 2008;118:192–205. doi: 10.1016/j.pharmthera.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Ono M. Molecular links between tumor angiogenesis and inflammation: Inflammatory stimuli of macrophages and cancer cells as targets for therapeutic strategy. Cancer Sci. 2008;99:1501–1506. doi: 10.1111/j.1349-7006.2008.00853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azzoni L, Zatsepina O, Abebe B, Bennett I, Kanakaraj P, Perussia B. Differential transcriptional regulation of CD161 and a novel gene, 197/15a, by IL-2, IL-15, and IL-12 in NK and T cells. J Immunol. 1998;161:3493–3500. [PubMed] [Google Scholar]
- 13.Yang H, Jansen A, Nair R, et al. A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB or ODC transactivation. Oncogene. 2001;20:669–676. doi: 10.1038/sj.onc.1204137. [DOI] [PubMed] [Google Scholar]
- 14.Rho O, Bol DK, You J, Beltran L, Rupp T, DiGiovanni J. Altered expression of insulin-like growth factor I and its receptor during multistage carcinogenesis in mouse skin. Mol Carcinog. 1996;17:62–69. doi: 10.1002/(SICI)1098-2744(199610)17:2<62::AID-MC2>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 15.Yasuda M, Nishizawa T, Ohigashi H, et al. Linoleic acid metabolite suppresses skin inflammation and tumor promotion in mice: Possible roles of programmed cell death 4 induction. Carcinogenesis. 2009;30:1209–1216. doi: 10.1093/carcin/bgp106. [DOI] [PubMed] [Google Scholar]
- 16.Liu L, Zhou X, Liu H, Xiang L, Yuan Z. CpG motif acts as a ’danger signal’ and provides a T helper type 1-biased microenvironment for DNA vaccination. Immunology. 2005;115:223–230. doi: 10.1111/j.1365-2567.2005.02150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams AS, Nath P, Leung SY, et al. Modulation of ozone-induced airway hyperresponsiveness and inflammation by interleukin-13. Eur Respir J. 2008;32:571–578. doi: 10.1183/09031936.00121607. [DOI] [PubMed] [Google Scholar]
- 18.Murakami A, Shigemori T, Ohigashi H. Zingiberaceous and citrus constituents, 1′-acetoxychavicol acetate, zerumbone, auraptene, and nobiletin, suppress lipopolysaccharide-induced cyclooxygenase-2 expression in RAW264. 7 murine macrophages through different modes of action. J Nutr. 2005;135:2987S–2992S. doi: 10.1093/jn/135.12.2987S. [DOI] [PubMed] [Google Scholar]
- 19.Jun HS, Yoon CS, Zbytnuik L, van Rooijen N, Yoon JW. The role of macrophages in T cell-mediated autoimmune diabetes in nonobese diabetic mice. J Exp Med. 1999;189:347–358. doi: 10.1084/jem.189.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cochran F, Finch-Arietta M. Regulation of interleukin-1 beta and tumor necrosis factor secretion by the human monocytic leukemia cell line, THP-1. Agents Actions. 1989;27:271–273. doi: 10.1007/BF01972794. [DOI] [PubMed] [Google Scholar]
- 21.Kyriakis J, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 22.Schmid T, Jansen A, Baker A, Hegamyer G, Hagan J, Colburn N. Translation inhibitor Pdcd4 is targeted for degradation during tumor promotion. Cancer Res. 2008;68:1254–1260. doi: 10.1158/0008-5472.CAN-07-1719. [DOI] [PubMed] [Google Scholar]
- 23.Sheedy F, Palsson-McDermott E, Hennessy E, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2009;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Z, DuBois R. Detection of differentially expressed genes in human colon carcinoma cells treated with a selective COX-2 inhibitor. Oncogene. 2001;20:4450–4456. doi: 10.1038/sj.onc.1204588. [DOI] [PubMed] [Google Scholar]
- 25.Nieves-Alicea R, Colburn N, Simeone A, Tari A. Programmed cell death 4 inhibits breast cancer cell invasion by increasing tissue inhibitor of metalloproteinases-2 expression. Breast Cancer Res Treat. 2009;114:203–209. doi: 10.1007/s10549-008-9993-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang D, Dubois RN. The role of COX-2 in intestinal inflammation and colorectal cancer. Oncogene. 2010;29:781–788. doi: 10.1038/onc.2009.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu G, Chen T, Ueng Y, Chen R. Ketamine inhibits tumor necrosis factor-alpha and interleukin-6 gene expressions in lipopolysaccharide-stimulated macrophages through suppression of toll-like receptor 4-mediated c-Jun N-terminal kinase phosphorylation and activator protein-1 activation. Toxicol Appl Pharmacol. 2008;228:105–113. doi: 10.1016/j.taap.2007.11.027. [DOI] [PubMed] [Google Scholar]
- 28.Kim B, Cho J. Anti-inflammatory effect of honokiol is mediated by PI3K/Akt pathway suppression. Acta Pharmacol Sin. 2008;29:113–122. doi: 10.1111/j.1745-7254.2008.00725.x. [DOI] [PubMed] [Google Scholar]
- 29.Shima E, Katsube M, Kato T, et al. Calcium channel blockers suppress cytokine-induced activation of human neutrophils. Am J Hypertens. 2008;21:78–84. doi: 10.1038/ajh.2007.13. [DOI] [PubMed] [Google Scholar]
- 30.Allgayer H. Pdcd4, a colon cancer prognostic that is regulated by a microRNA. Crit Rev Oncol Hematol. 2010;73:185–191. doi: 10.1016/j.critrevonc.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Dorrello N, Peschiaroli A, Guardavaccaro D, Colburn N, Sherman N, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 32.Chen A, Luo M, Yuan G, et al. Complementary analysis of microRNA and mRNA expression during phorbol 12-myristate 13-acetate (TPA)-induced differentiation of HL-60 cells. Biotechnol Lett. 2008;30:2045–2052. doi: 10.1007/s10529-008-9800-8. [DOI] [PubMed] [Google Scholar]
- 33.Moschos S, Williams A, Perry M, Birrell M, Belvisi M, Lindsay M. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genomics. 2007;8:240. doi: 10.1186/1471-2164-8-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Talotta F, Cimmino A, Matarazzo M, et al. An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation. Oncogene. 2009;28:73–84. doi: 10.1038/onc.2008.370. [DOI] [PubMed] [Google Scholar]
- 35.Kotlyarov A, Neininger A, Schubert C, et al. MAPKAP kinase 2 is essential for LPS-induced TNF-alpha biosynthesis. Nat Cell Biol. 1999;1:94–97. doi: 10.1038/10061. [DOI] [PubMed] [Google Scholar]
- 36.Hu X, Ivashkiv LB. Cross-regulation of signaling pathways by interferon-gamma: Implications for immune responses and autoimmune diseases. Immunity. 2009;31:539–550. doi: 10.1016/j.immuni.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Homey B, Dieu-Nosjean M, Wiesenborn A, et al. Up-regulation of macrophage inflammatory protein-3 alpha/CCL20 and CC chemokine receptor 6 in psoriasis. J Immunol. 2000;164:6621–6632. doi: 10.4049/jimmunol.164.12.6621. [DOI] [PubMed] [Google Scholar]
- 38.Nishimura M, Kuboi Y, Muramoto K, Kawano T, Imai T. Chemokines as novel therapeutic targets for inflammatory bowel disease. Ann N Y Acad Sci. 2009;1173:350–356. doi: 10.1111/j.1749-6632.2009.04738.x. [DOI] [PubMed] [Google Scholar]
- 39.Bishop B, Lloyd C. CC chemokine ligand 1 promotes recruitment of eosinophils but not Th2 cells during the development of allergic airways disease. J Immunol. 2003;170:4810–4817. doi: 10.4049/jimmunol.170.9.4810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.