Abstract
Most proteins encoded by the nuclear genome are synthesized in the cytoplasm and fold into precise 3D structures. During synthesis, the nascent polypeptide begins to fold as it traverses the large subunit of the ribosome and is assisted by molecular chaperones in attaining its precise folded/highly ordered state efficiently and in a biologically relevant timescale. Proteins that are misfolded are culled, re-routed, and marked by mechanisms such as ubiquitinylation for degradation ensuring strict quality control (QC). In addition to the highly ordered “globular” proteins, emerging evidence indicates that a large fraction of the proteome also comprises the so-called “Intrinsically Disordered Proteins” (IDPs). IDPs are proteins that lack rigid 3D structures and instead, exist as dynamic ensembles. The dynamic structures in the IDPs have many similarities with “normal” globular proteins such as the native (ordered), and non-native (molten globule, pre-molten globule, and coil-like) states seen during folding of “normal” globular proteins. However, unlike the case of the nascent globular proteins, IDPs evade being detected as “misfolded” and degraded by the cell’s QC system. We refer to this paradox as the order/disorder paradox and postulate that the IDPs capitalize on their intrinsic promiscuity and ability to undergo disorder-to-order transitions upon binding to biological targets (coupled folding and binding) to escape the cell’s surveillance machinery. Understanding the mechanism by which the IDPs evade the quality check has wide implications from protein folding to disease biology since the aggregation of misfolded proteins underlies several debilitating illnesses such as many neurodegenerative diseases and cancer.
Keywords: PROTEIN FOLDING, ORDER/DISORDER PARADOX, PROTEASOME, CANCER/TESTIS ANTIGENS, CANCER
BACKGROUND
The biosynthesis of nuclear coded eukaryotic proteins, regardless of their final destination, begins in the cytoplasm. As the nascent polypeptide traverses the exit channel of the large ribosomal subunit, it begins to fold into its characteristic 3D structure. Protein folding is a rapid and complex process by which a relatively well defined 3D structure is adopted from a very large ensemble of expanded, less structured conformations driven by the protein sequence as well as thermodynamics [Anfinsen, 1973]. However, due to the highly crowded environment of the cell, the constrained environment of the exit channel of the large subunit of the ribosome, and relatively slow translational times (~15–75 s for a 300-amino-acid protein), nascent chains are exposed in partially folded, aggregation-sensitive states for prolonged periods of time increasing the risk of accumulation of mis- or unfolded proteins [Hartl and Hayer-Hartl, 2009]. To minimize their premature (mis)folding, there exist in the cell, molecular chaperones that co-translationally interact with nascent polypeptides ensuring that folding is efficient for most proteins. Thus, the chaperones, defined as proteins which interact, stabilize and/or help a non-native protein to acquire its native conformation but are not part of the final functional structure, appear to have the ability to recognize certain molecular features in the nascent polypeptide’s transition states that “allows” them to discern the proper conformation and proceed with assisting the subsequent step in the sequence of events that result in a properly folded protein. Proteins that are misfolded are culled, rerouted, and marked by ubiquitinylation for degradation ensuring strict quality control (QC) [Goldberg, 2003]. However, it should be noted that this QC-related degradation is distinct from the degradation that also occurs employing the same machinery of proteins to clear them from the system following their “life time” in the cell.
In eukaryotes, the ATP-dependent proteasome is responsible for removal of misfolded/damaged proteins as well as specific degradation of regulatory proteins [Schrader et al., 2009]. Since the proteasome degrades thousands of unrelated proteins, it is unlikely to be controlled by sequence alone. At the same time however, the proteasome must possess some specificity to avoid reckless degradation. The key to this enigma is the multiple proteolytic sites of the proteasome which alone have weak sequence preference, but in combination can degrade most proteins [Nussbaum et al., 1998]. These sites are sequestered inside the proteasome and are inaccessible to most folded proteins. However, those with specific degradation signals (“degrons”) are recognized by the proteasome allowing them to be unfolded [Lee et al., 2001] and threaded into the degradation chamber for proteolytic digestion [Nussbaum et al., 1998].
Targeting for regulated degradation is accomplished with a two-part degron: an ubiquitin tag and an unstructured/disordered region where the proteasome can engage and begin degradation [Thrower et al., 2000]. In contrast, QC-associated degradation involves recognition of structural features common to damaged or misfolded proteins rather than specific features of an individual protein by the proteasome [Gardner and Hampton, 1999]. While the degradation initiation site is a discrete region of the primary sequence in the regulatory protein, the molecular features that define a recognition signal for QC are presently unknown.
Until recently, it was generally believed that proteins are highly “ordered” molecules having unique 3D structures dictated by the protein’s sequence and the resulting intramolecular forces. Furthermore, the function of a given protein was thought to depend on its structure and that a protein would lose its function if its structure was denatured. Contrary to this sequence-structure-function paradigm, it is now evident that a significant fraction of the eukaryotic proteome is made up of “intrinsically disordered proteins” (IDPs). IDPs are proteins that lack rigid 3D structures under physiological conditions in vitro either along their entire lengths or in localized regions, but instead exist as dynamic ensembles within which atom positions and backbone Ramachandran angles exhibit extreme temporal fluctuations without specific equilibrium values [Uversky and Dunker, 2010]. Despite the lack of structure, most IDPs are adept in transitioning from disorder-to-order upon binding to their targets (coupled folding and binding) via discrete motifs termed molecular recognitions regions or MoRFs [Uversky and Dunker, 2010] and exhibit a high degree of promiscuity [Fink, 1995].
THE PROBLEM
The dynamic structures within IDPs that interconvert on different timescales have many similarities with “normal” globular proteins such as the native (ordered), and non-native (molten globule, pre-molten globule, and coil-like) states seen during folding of “normal” globular proteins [Fink, 1995]. Furthermore, it has been observed that perhaps analogous to the preferred pathways defined by the sequential assembly of elementary folding units, so-called “foldons” [Englander et al., 2007], the IDPs might contain collapsed disorder (molten globules) and extended disorder (random coil or pre-molten globule) at least under physiological conditions in vitro [Uversky, 2003]. Thus, it follows that the dynamic structures in the IDP ensemble “mimic” the folding intermediates of normal globular proteins. Then why are nascent IDPs not recognized as misfolded proteins by the QC system that “polices” them?
The abundance of several IDPs has been associated with perturbed signaling that may lead to pathological conditions [Uversky et al., 2008]. Therefore, and not surprisingly, the IDPs are tightly regulated at both the transcriptional and post-transcriptional level [Gsponer et al., 2008; Edwards et al., 2009]. However, this regulation appears to be compromised in diseased states resulting in the overexpression of the IDPs. Consistent with this hypothesis, we observed that several members of the Cancer/Testis Antigen family, an important group of proteins that are typically restricted to the testis but are aberrantly expressed in several types of cancers [Scanlan et al., 2004], are highly disordered but yet appear to have significantly long half lives (Table I). We refer to this paradox as the order/disorder paradox.
TABLE I.
Examples of Highly Intrinsically Disordered Cancer/Testis Antigens Located on the X Chromosome
| Cancer/testis antigen subfamily (number of members) | Chromosome | Disorder (%) | Length of polypeptide(s) (number of amino acids) | (PSI) |
|---|---|---|---|---|
| PAGE (5) | X | 100 | 102–146 | 4.73 (1) 5.23 (4), 5.46 (5) |
| GAGE (19) | X | 100 | 111–117 | 4.43 (1), 4.38 (5), 4.31 (8) |
| XAGE (4) | X | 100 | 108–111 | 4.59 (2) |
| SPANX (10) | X | 96.9–100 | 72–180 | 2.62 (B1), 2.02 (E) |
| SSX (2) | X | 94.7–100 | 188 | 2.56 (2), 2.64 (3), 2.47 (4) |
The following members in each subfamily are listed: PAGE (1, 2/2B, 4, 5), GAGE (1, 2A-C, 4–8, 12B-J), XAGE (2/2B, 3, 5), SPANX (A1/2, B1/2, C, E, N1-5), SSX (1, 4/4B, 7). Numbers in parenthesis next to half lives (PSI units) indicate the corresponding CTA subfamily member. Yen et al. determined that PSIs of 2 and 3.5 corresponded to approximate protein half-lives of 30 min and 2 h, respectively. In contrast, the half lives of many regulatory proteins have been observed to range from 2 to 20 min [see Goldberg, 2003] indicating that most CTAs are relatively more stable. PSI, protein stability index. Disorder was predicted using the FOLDINDEX algorithm [Prilusky et al., 2005]. Data on protein half life were obtained from Yen et al. [2008]. The value of PSI ranges from 1 to 7, with a higher PSI value meaning higher relative protein stability of enhanced green fluorescence protein-fused ORFs used by the authors to determine global protein stability. Based on PSI, proteins were classified as having short (PSI ~ 1.3), medium (~3.5), long (~5.5), and extra-long (~6.5) half life [Yen et al., 2008].
THE SOLUTION (HYPOTHESIS)
An obvious question that is often asked by researchers working in the IDP field is how do the IDPs that are extremely sensitive to proteolysis in vitro not show enhanced degradation rates in vivo? Indeed, several attempts have been made to address this question as well [Dunker et al., 2002; Hegyi and Tompa, 2008; Tsvetkov et al., 2009]. However, we would like to stress that there is an important distinction between this question that concerns the “mature” IDPs postsynthesis and the paradox raised in the present manuscript that concerns the evasion of the nascent IDP by the protein folding QC machinery.
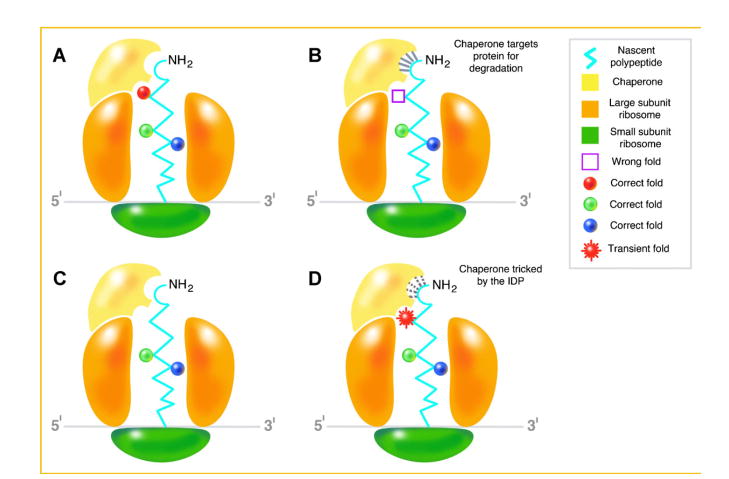
On the other hand, some intriguing explanations have also been offered to explain why the QC machinery fails to cope with the formation of potentially toxic misfolded proteins such as aging [e.g., Morley and Morimoto, 2004; Hartl and Hayer-Hartl, 2009]. However, in our view, how nascent IDPs evade being recognized and singled out for programmed destruction remains a paradox. We postulate that given their intrinsic ability to transition from disorder to order upon binding, nascent IDPs avoid the QC mechanism while engaged by the chaperones. Once disengaged, they revert to their disordered state by undergoing an order to disorder transition (Fig. 1).
Fig. 1.
Coupled folding and binding and the order/disorder paradox. A: The nascent polypeptide encoding a highly ordered globular protein begins folding as it traverses the exit channel of the large subunit of the ribosome. It is “recognized” by the chaperone as properly folded. B: The polypeptide is misfolded and therefore, is identified by the chaperone for degradation. The interaction between the misfolded polypeptide and chaperone is indicated by the solid lines near the NH2 terminal. C: The nascent polypeptide encoding an IDP begins folding as it traverses the exit channel of the large subunit of the ribosome. D: Therefore, it is “recognized” by the chaperone as misfolded/damaged and the chaperone binds to it as indicated by the dotted lines. However, by virtue of its innate ability to transition from disorder to order upon binding, the IDP assumes a “transient” fold that “tricks” the chaperone into believing that it is indeed ordered and folded to evade degradation.
While it may sound over simplistic, our hypothesis offers an experimentally tractable suggestion. However, we realize that it may not be applicable to all IDPs; for example, the so-called non-folders, proteins that are postulated to be unable to fold under any circumstances [Rauscher and Pomès, 2010]. Notwithstanding these caveats, understanding how IDPs evade the surveillance machinery may have wide implications from protein folding to disease biology as the aggregation of misfolded proteins that escape the cellular QC underlies a range of debilitating diseases, including many age-onset neurodegenerative disorders [Uversky et al., 2008]. Recent advances in NMR spectroscopy and computation techniques have enabled characterization of transient state structures that have very short lifetimes [Korzhnev et al., 2010]. We trust that in the near future, using such techniques, it may also be feasible to get a glimpse of the transient states of the disordered proteins when they interact momentarily with the chaperones to evade QC surveillance.
Acknowledgments
Grant sponsor: NCI SPORE; Grant number: 2P50CA058236-16; Grant sponsor: Patrick C Walsh Cancer Research Fund; Grant sponsor: NCI PSOC; Grant number: U54CA143803.
Supported by NCI SPORE Grant 2P50CA058236-16 and an Irene and Bernard L. Schwartz Scholar Award by the Patrick C Walsh Cancer Research Fund (to P.K.) and by a NCI PSOC grant U54CA143803 to R.H.G. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institute of Health. P.K. thanks Dr. Amita Behal for many helpful discussions and thoughtful comments on the manuscript and Mr. Timothy Phelps for the illustration.
References
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Dunker KA, Brown CJ, Lawson JD, Iakoucheva LM, Obradović Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- Edwards YJ, Lobley AE, Pentony MM, Jones DT. Insights into the regulation of intrinsically disordered proteins in the human proteome by analyzing sequence and gene expression data. Genome Biol. 2009;10:R50. doi: 10.1186/gb-2009-10-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander SW, Mayne L, Krishna MM. Protein folding and misfolding: Mechanism and principles. Q Rev Biophys. 2007;40:287–326. doi: 10.1017/S0033583508004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink AL. Compact intermediate states in protein folding. Annu Rev Biophys Biomol Struct. 1995;24:495–522. doi: 10.1146/annurev.bb.24.060195.002431. [DOI] [PubMed] [Google Scholar]
- Gardner RG, Hampton RY. A ‘distributed degron’ allows regulated entry into the ER degradation pathway. EMBO J. 1999;18:5994–6004. doi: 10.1093/emboj/18.21.5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- Gsponer J, Futschik ME, Teichmann SA, Babu MM. Tight regulation of unstructured proteins: From transcript synthesis to protein degradation. Science. 2008;322:1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol. 2009;16:574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- Hegyi H, Tompa P. Intrinsically disordered proteins display no preference for chaperone binding in vivo. PLoS Comput Biol. 2008;4(3):e1000017. doi: 10.1371/journal.pcbi.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzhnev DM, Religa TL, Banachewicz W, Fersht AR, Kay LE. A transient and low-populated protein-folding intermediate at atomic resolution. Science. 2010;329:1312–1316. doi: 10.1126/science.1191723. [DOI] [PubMed] [Google Scholar]
- Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum AK, Dick TP, Keilholz W, Schirle M, Stevanović S, Dietz K, Heinemeyer W, Groll M, Wolf DH, Huber R, Rammensee HG, Schild H. Cleavage motifs of the yeast 20S proteasome beta subunits deduced from digests of enolase 1. Proc Natl Acad Sci USA. 1998;95:12504–12509. doi: 10.1073/pnas.95.21.12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: A simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- Rauscher S, Pomès R. Molecular simulations of protein disorder. Biochem Cell Biol. 2010;88:269–290. doi: 10.1139/o09-169. [DOI] [PubMed] [Google Scholar]
- Scanlan MJ, Simpson AJ, Old LJ. The cancer/testis genes: Review, standardization, and commentary. Cancer Immun. 2004;4:1–15. [PubMed] [Google Scholar]
- Schrader EK, Harstad KG, Matouschek A. Targeting proteins for degradation. Nat Chem Biol. 2009;5:815–822. doi: 10.1038/nchembio.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov P, Reuven N, Shaul Y. The nanny model for IDPs. Nat Chem Biol. 2009;5:778–781. doi: 10.1038/nchembio.233. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Protein folding revisited. A polypeptide chain at the folding-misfolding-nonfolding cross-roads: Which way to go? Cell Mol Life Sci. 2003;60:1852–1871. doi: 10.1007/s00018-003-3096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Dunker K. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: Introducing the D2 concept. Annu Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]