Abstract
In mice, microRNAs (miRNAs) are required for embryonic viability, and previous reports implicate miRNA participation in brain cortical neurogenesis. Here, we provide a more comprehensive analysis of miRNA involvement in cortical brain development. To accomplish this we used mice in which Dicer, the RNase III enzyme necessary for canonical miRNA biogenesis, is depleted from Nestin expressing progenitors and progeny cells. We systematically assessed how Dicer depletion impacts proliferation, cell death, migration and differentiation in the developing brain. Using markers for proliferation and in vivo labeling with thymidine analogs, we found reduced numbers of proliferating cells, and altered cell cycle kinetics from embryonic day 15.5 (E15.5). Progenitor cells were distributed aberrantly throughout the cortex rather than restricted to the ventricular and subventricular zones. Activated Caspase3 was elevated, reflecting increased cortical cell death as early as E15.5. Cajal-Retzius positive cells were more numerous at E15.5 and were dysmorphic relative to control cortices. Consistent with this, Reelin levels were enhanced. Doublecortin and Rnd2 were also increased and showed altered distribution, supporting a strong regulatory role for miRNAs in both early and late neuronal migration. In addition, GFAP staining at E15.5 was more intense and disorganized throughout the cortex with Dicer depletion. These results significantly extend earlier works, and emphasize the impact of miRNAs on neural progenitor cell proliferation, apoptosis, migration, and differentiation in the developing mammalian brain.
Keywords: Corticogenesis, microRNA, Dicer, post-transcriptional regulation
1. Introduction
Cortical neurogenesis is a highly regulated process broadly consisting of proliferation, selective cell death, migration, and differentiation. Disruptions to these controlled processes cause neurodevelopmental disorders. For instance, a decrease in proliferation or an increase in cell death at early stages of corticoneurogenesis can cause a microcephalic disorder (Chae and Walsh, 2007; Kuijpers and Hoogenraad, 2011), whereas disrupted differentiation or migration during cortical development can induce disorganized cortical laminae, a phenotype of lissencephalic patients (Pramparo et al., 2010; Valiente and Marin, 2010). Genetic dissection of loci important in the control of cortical neurogenesis has improved our understanding of neurodevelopment. These loci include not only protein coding genes (Chae and Walsh, 2007; Liu, 2011), but also noncoding RNAs (Bian and Sun, 2011; Christensen and Schratt, 2009; Fineberg et al., 2009).
miRNAs are one class of noncoding RNAs, and they provide a mechanism for post-transcriptional gene silencing upon base pairing with target mRNAs to induce translational repression, often through de-adenylation of mRNAs (Bartel, 2009; Filipowicz et al., 2008; Standart and Jackson, 2007). miRNA control of gene expression has been implicated in developmental regulation and mature cell maintenance (Cheng et al., 2009; Christensen and Schratt, 2009; Fineberg et al., 2009; Harfe, 2005; Lau and Hudson, 2010; Manakov et al., 2009). In the nervous system, gross evaluation of total miRNA expression has been profiled, revealing strong miRNA enrichment in developing and mature neuronal tissue (Krichevsky et al., 2003; Krichevsky et al., 2006; Miska et al., 2004). And in CNS disorders, their expression can be impaired, which may contribute to underlying disease phenotypes (Choi et al., 2008; Cuellar et al., 2008; Damiani et al., 2008; Davis et al., 2008; Kim et al., 2007; Packer et al., 2008; Perkins et al., 2007; Schaefer et al., 2007; Song et al., 2011). Many reports have dissected relationships between miRNAs and disease phenotypes, and deficiencies in miRNAs that regulate embryonic neurogenesis might contribute to CNS defects and other neuropathies. We sought herein to define the impact of miRNA loss in the development of the CNS, by assessing multiple aspects of cortical neurogenesis upon blockade of canonical miRNA generation. An important goal of this survey was to define primary disturbances, for future delineation of specific miRNAs and their potential disease-related roles.
Dicer is a RNase III family ribonuclease that is important in miRNA maturation, and is required for embryonic development; Dicer depleted embryos lack primitive streak markers and arrest at embryonic day 7.5 (E7.5) (Bernstein et al., 2003). To analyze the gross impact of miRNAs in the developing CNS, conditional Dicer depleted neural tissue has been studied in several settings using mouse lines expressing various Cre drivers crossed to mice harboring lox-P flanked Dicer loci. The FoxG1 promoter, active in telencephalic progenitor cells from around E8, was used to drive Cre expression, and resulted in disorganized cortical laminae, pronounced microcephaly and lethality in utero (Choi et al., 2008; Makeyev et al., 2007). Two different groups used the Emx-1 promoter to drive expression of Cre in the cortex starting at E9.5 and concluded that miRNAs were required for neuronal maturation but differed in their interpretation of how Dicer depletion affects progenitor pools (De Pietri Tonelli et al., 2008; Kawase-Koga et al., 2009).
Here, we performed extensive additional studies in mice with Dicer depletion using the Nestin-Cre driver. Using BrdU/IdU double labeling, we found reduced numbers of proliferating cells and altered cell cycle kinetics. We also identified early cell death in all zones of the developing cerebral cortex. We found dysregulation of important markers of differentiation and migration. Finally, we show an increased expression level of a mature astrocyte marker at E15.5, well before normal induction of large scale gliogenesis. Our data considerably supplement earlier surveys and clarify the extent of impairments induced by depletion of Dicer in the developing mammalian forebrain.
2. Experimental Procedures
2.1 Animal Care and Use
Floxed Dicer (fd) mice were kindly provided by Michael McManus at UCSF and re-derived at the University of Iowa Animal Care Facility. Animals were bred to homozygosity, and genotyped using published protocols (Harfe et al., 2005). Dicer excision was validated by PCR amplification of DNA harvested from cortices using methods described earlier (Harfe et al., 2005). Nestin-Cre (nCre) mice were purchased from Jackson Labs (Strain name B6.Cg-Tg(Nes-Cre)1Kln/J; Stock number 003771) and genotyped using the following primers: forward, AGCGATCGCTGCCAGGAT; reverse, ACCAGCGTTTTCGTTCTGCC. Animals were housed and handled according to protocols approved by the University of Iowa Institutional Animal Use and Care Committee.
2.2 Embryo collection
For timed matings, male (fd/+;nCre/+) and female (fd/fd; +/+) mice were co-housed overnight. On the day of harvest, pregnant dams were over anaesthetized with inhaled isoflurane and euthanasized. Embryos were harvested and anesthetized for five minutes in weigh boats on wet ice. Appendages were removed for genotyping. For histology sections, decapitated embryo heads were stored in 4% paraformaldehyde (PFA) solution for two or 24 hrs for in situ hybridization (ISH) or immunohistochemistry (IHC) analysis respectively. IHC tissue was incubated in 30% sucrose/0.05% sodium azide at 4° C for 48 hours. Tissues were embedded in OCT matrix, sectioned using a micron cryostat at 10 μm thickness onto Superfrost plus slides (Sigma). Tissues were stored at −80 °C until used.
2.3 Immunohistochemistry
Sections were post fixed in 4% PFA for 15 minutes and for antigen retrieval, immersed in sodium citrate buffer pH 6.0 in microwave at 95° C for 3 × 5 min. Sections were blocked for one hr in 10% serum, 0.03% Triton-100 in 1 × PBS at room temperature, then incubated in primary antibody in 2% serum, 0.03% Triton-100 in 1 × PBS overnight at 4° C. Primary antibodies used were Nestin (mouse anti-Rat401, Developmental Studies Hybridoma Bank, 1:5), MAP2 (mouse anti-MAP2, Sigma, 1:200), PH3 (rabbit anti-phosphohistone 3 (ser10), Upstate division of Millipore, 1:200), activated Caspase3 (rabbit anti-cleaved Caspase3 (Asp75), Cell Signaling Technologies, 1:100), DCX (goat anti-Doublecortin (N-19), Santa Cruz, 1:200), Reelin (mouse anti-Reelin, Millipore, 1:500), Calretinin (rabbit anti-Calretinin, Swant, 1:200), Cre (mouse anti-Cre, Sigma, 1:200), BrdU/IdU (mouse anti-5′-Bromo-2′-deoxyuridine/5′-Iodo-2′-deoxyuridine, Becton Dickinson, 1:50), and BrdU (rat anti-BrdU, Serotec, 1:200). For fluorescent IHC, sections were incubated with fluorescent secondary antibodies at 1:1000 in 2% serum, 0.03% Triton-100 in PBS for one hour, then stained with 1:1000 Hoechst 33258 (Molecular Probes) in 1 × PBS for one minute for nuclear visualization. Coverslips were mounded using Vectashield (Vector Labs). For DAB IHC, sections were incubated in biotin-labeled secondary antibodies (Jackson Immunoresearch) at 1:200 in 2% serum, 0.03% Triton-100 in PBS for one hour at room temperature. Tissues were developed via the Vectastain ABC Elite Kit (Vector Labs) according to the manufacturer’s instructions. Coverslips were mounted on dehydrated sections using Fluoro Gel (Electron Microscopy Science).
2.4 RNA in situ hybridization
Sections were fixed at room temperature in 4% PFA in 1 × TBS for 20 minutes. Prior to hybridization, sections at room temperature were dehydrated sequentially in 70%, 90%, 100% ethanol for five min each, treated with 1.32% triethanolamine solution for three min, and incubated in acetic anhydride solution for ten min. Sections were then pretreated with pre-hybridization solution (100 μL/slide; 50% formamide, 5 × SSC, 0.5% Roche blocking reagent, 0.1% Tween-20, 0.1% CHAPS, 50 μg/mL yeast tRNA, 5 mM EDTA, 50 μg/mL heparin in ddH20) for one hour in a humid chamber at 55° C. Hybridization solution is composed of pre-hybridization solution plus Rnd2 probe (5′-CAGAAGATCGGGAGGAACATTC-3′) was designed to the reverse complement of the targeted Rnd2 mRNA using 2′-O-Methyl RNA (2′OMe) bases with phosphodiester linkages, ZEN non-nucleotide chemical modifier between the last and next to last base on both the 5′- and 3′-ends, and DIG group added to both ends to allow for detection (Lennox and Behlke, 2011). Sections were incubated in 1.25 pmol Rnd2 probe or scrambled negative control probe (Integrated DNA Technologies, Coralville, IA, USA) with pre-hybridization solution (100 μL/slide) for 48 hrs at 55° C under parafilm coverslips. Sections were washed three times for 30 min each in preheated 2 × SSC/0.1% CHAPS and 0.2 × SSC/0.01%CHAPS at 55° C prior to blocking sections in 20% sheep serum in KTBT for one hr at room temperature. Sections were then incubated in 1:500 sheep anti-DIG AP Fab fragment in blocking buffer at 4° C overnight. After washing 3 × 5 min each in 100 mM Tris-HCl, 50 mM MgCl2, 100 mM NaCl, 0.1% Tween-20 in ddH20 at room temperature, sections were developed using Pierce NBT/BCIP one-touch solution (34042) in the dark for approximately three hrs. Once developed, sections were washed briefly in 1 × TBS prior to dehydration and mounting of coverslips using Fluoro Gel (Electron Microscopy Science).
2.5 Microscopy and statistics
Histology sections were imaged either with a Leica Leitz DM R fluorescent microscope connected to a Olympus DP72 camera using the Olympus DP2-BSW software or on the confocal 710 microscope using Zen software. For cell count quantification, counts were obtained from the motor cortex and surrounding neocortex as defined by the prenatal brain atlas (gestational day 16 sagittal image 8/9, Schambra 2008). Sections were analyzed from both control and Nestin-Cre Dicer depleted tissue using mice from three independent breedings. To determine IdU positive cells, we subtracted the total number of BrdU only positive cells (Serotec) from BrdU/IdU positive cells (BD Biosciences). All cell count results were normalized to total cells. Statistical significance was tested using the two-tailed Student’s t-test for unpaired differences with GraphPad (San Diego, CA) Prism software. For signal intensity quantification, medial-lateral plane matched sections were compared using ImageJ software (Rasband, W.S., ImageJ, U.S. National Institute of Health, Bethesta, Maryland, USA, http://imagej.nih.gov/ij, 1997–2011). DCX signal intensity was calculated from a 100 pixels2 area within the cortical plate (CP) and normalized to a 100 pixel2 background area within the ventricular zone (VZ).
2.6 RNA isolation and qRT-PCR analysis
Total RNA was isolated from the cortex of E15.5 Dicer depleted and control embryos using Trizol reagent according to manufacturer’s protocol (Invitrogen, CA). RNA quantity and quality was measured using a ND-1000 (Nanodrop, Wilmington, DE). Reverse transcription was performed on 1 μg of total RNA using the Superscript III Reverse Transcriptase kit according to manufacturer’s instructions (Invitrogen, CA). The cDNA was diluted 1:15 in ddH20. Taqman relative quantification PCR was performed on the diluted cDNA of total cortical RNA following the manufacturer’s protocol (Applied Biosystems, Foster City, CA) and results normalized to total RNA. Analysis was performed using average adjusted relative quantification on the following probes: Nestin (Applied Biosystems, assay ID: Mm00450205_m1), DCX (Applied Biosystems, assay ID: Mm00438401_m1*), Reelin (Applied Biosystems, assay ID: Mm00465200_m1*), and Rnd2 (Applied Biosystems, assay ID: Mm00501561_m1*).
3. Results
Female mice with loxP sites flanking the essential RNase III domain of Dicer (fd/fd mice) were bred to male mice harboring Cre recombinase driven by the Nestin promoter (nCre mice). Embryos without Nestin-Cre expression (fd/fd;+/+) were used as controls for fd/fd:nCre/+ mice in all studies. The Nestin promoter is active in embryonic neural progenitor cells (NPCs) as early as E9.5 in mouse cerebral cortex (Dubois et al., 2006; Kawaguchi et al., 2001). Like Kawase-Koga and colleagues, we noted reduced numbers of fd/fd:nCre embryos; only 13.5% of all E15.5 embryos and 9% of all E16.5 embryos were Dicer depleted (data not shown). No Dicer depleted mice were recovered at birth.
We focused our study between E15.5 and E16.5 to capture the effects of Dicer depletion at the height of neuronal proliferation and the onset of gliogenesis (Sauvageot and Stiles, 2002). At this time there was RNase III excision of Dicer, as assessed by PCR of genomic DNA (data not shown). This is congruent with others work showing Dicer expression by western blot (Kawase-Koga et al., 2009). Additionally, a recent study calculated the average half-life of miRNAs to be ~119hrs (Gantier et al., 2011). Thus to allow for Cre expression, Dicer excision, and miRNA depletion, we choose E15.5 as the earliest time point of interest. Notably, while a small number of Dicer depleted embryos exhibited massive cell loss and extremely small cortices at E15.5, similar to that reported by De Pietri Tonelli et al, 2008, we found these embryos to be outliers in our colony. Therefore, in this study we analyzed sections from more representative embryos.
3.1 Gross cortical abnormalities of Dicer depleted mice
To evaluate the effects of Dicer ablation on the developing CNS, we prepared E16.5 whole embryo sections in the sagittal plane and stained for Nissl to examine brain size. Specific analysis of the cerebral cortex showed microcephalic characteristics, including gross enlargement of the lateral ventricles and marked cortical thinning in the Dicer depleted embryos compared to control embryos (data not sown). These findings are analogous to previously published results where embryos were analyzed at a later time point, E18.5 (Kawase-Koga et al., 2009), wherein they ascribed the cortical reduction to abnormal developmental of the late-born neurons. However, effects on late-born neurons cannot explain the decrease in cortical thickness and ventricular enlargement we observed in E15.5 embryos. We thus investigated other consequences of Dicer depletion.
Simply stated, developmental malformations of the cerebral cortex can be categorized into defects in NPC proliferation, cell death, migration and/or differentiation (Chae and Walsh, 2007). To assess which of these defects in early corticoneurogenesis are disrupted in the Dicer depleted cortex, we first surveyed Nestin, a marker present in all NPCs, to determine if proliferating cell populations differed. We quantified transcript levels via RT-qPCR using cortical RNA harvested from E15.5 embryos and found Nestin mRNA increased by ~8-fold in Dicer depleted cortical tissue relative to control brains (Fig. 1A). In normal mice embryos, Nestin expressing cells are abundant within the VZ and sparse throughout the SVZ. However, in Dicer depleted cortical tissue, Nestin expression was observed aberrantly distributed throughout the cortical parenchyma at E15.5 (Fig. 1B). The novel finding of an altered pattern of Nestin expressing cells prompted us to examine mature neurons using MAP2. In Dicer-depleted E15.5 cortical sections, we detected no significant decrease in MAP2 transcript levels (Fig. 1A), but the cortical layering was disorganized relative to control tissues (Fig. 1B). These results are consistent with the poor organization reported throughout the normally non-proliferative region of the developing cortex in the Emx1-Cre Dicer knockout mice (De Pietri Tonelli et al., 2008). Kawase-Koga and colleagues examined MAP2 and NeuN at E18.5 and P0 in the Nestin-Cre Dicer depleted mice, and detected a reduction in mature neuron population (Kawase-Koga et al., 2009). Our data of MAP2 at E15.5 suggests that dysregulation precedes mature neuron decline.
Figure 1.
Nestin and MAP2 dysregulation through Nestin-Cre Dicer depleted cerebral cortices. (A) RT-qPCR quantification of Nestin and MAP2 RNA expression from total RNA isolated from E15.5 cerebral cortex of both control (fd/fd;+/+) and conditional Dicer depleted (fd/fd;nCre/+) mice, with results normalized to total RNA. Ctrl, control (fd/fd;+/+); Dd, conditional Dicer depleted (fd/fd;nCre/+). Error bars represent mean ± SEM. ns= not significant, ***p<0.0001 using a two-tailed unpaired t-test. (B) Representative photomicrographs showing IHC for Nestin and MAP2 in the cerebral cortex at E15.5. Scale bars, 100 μm. VZ/SVZ, ventricular zone/subventricular zone; CP, cortical plate
3.2 Dicer inactivation reduces cell division
Since reduced cortical thickness is commonly caused by decreased NPC proliferation and/or increased cell death, we assessed the proliferative regions of the developing cerebral cortex. Proliferating NPC density was ascertained by immunostaining for the M-phase marker, PH3 at E15.5 and E16.5. Dicer depleted cortices showed a significant decrease in PH3 positive cells in the VZ and SVZ by greater than 20% at both E15.5 and E16.5 (Fig. 2A and B). Our results appear to contrast the data from Kawase-Koga and colleagues, who reported no difference in PH3 positive cells in E15.5 Dicer depleted cerebral cortex, although no quantification was reported in that work (Kawase-Koga et al., 2009). To substantiate Dicer depletion effects on proliferation, and gain information on cell cycle kinetics, we used the thymidine analogues BrdU and IdU to quantify cells in S or G2/M phase, respectively (Sunabori et al., 2008). IdU was intraperitoneally injected into pregnant dams four hours before sacrifice, and BrdU 30 minutes before sacrifice (Takahashi et al., 1995). Sections were immunostained for BrdU and IdU; BrdU positive cells are in S phase, and IdU-only positivity represent cells that were in S phase at the time of IdU injection but subsequently progressed into G2/M. In agreement with our PH3 immunostaining, we found ~40% fewer immunopositive cells (cells that had incorporated either BrdU and/or IdU) in Dicer depleted cortices relative to controls at E15.5 (Fig. 3A and B), indicating significant loss of proliferating cells. Amongst the immunopositive cells, Dicer depletion shifted the proportions to increase the S:G2/M phase ratio, indicating that Dicer depleted cells take longer to progress through S-phase (Fig. 3C). According to calculations defined by Burns and Kuan, the interval time at which the ratio of IdU-only cells to total labeled cells is 0.5 is considered the length of S-phase. Thus, our results indicate that control cortical cell S-phase length approximates 4 hrs for control cortex, while in Dicer depleted cortical progenitors, S-phase is greater than 4 hrs (Burns and Kuan, 2005) (Fig. 3C). Together these data indicate that in Dicer depleted proliferating zones, the progenitor pool reduced and there is delayed cell cycle kinetics.
Figure 2.
Decrease in progenitor cell proliferation in Dicer depleted cortical tissue. (A) Representative photomicrograph showing IHC for PH3 in cerebral cortices at both E15.5 and E16.5. Scale bars, 100 μm. (B) Quantification of IHC staining of E15.5 and E16.5 PH3 positive cells within the medial-lateral plane in matched sections shows decreased proliferating cells within Dicer depleted cortical tissue. Ctrl, control (fd/fd;+/+); Dd, conditional Dicer depleted (fd/fd;nCre/+). Y-axis denotes number of PH3 positive cells per 1000 total cells as defined by Hoechst staining in a 20x field. Error bars represent mean ± SEM. ***p<0.0001 using a two-tailed unpaired t-test.
Figure 3.
Reduction in proliferation and cell cycle kinetics in Dicer depleted brains. (A) Representative photomicrograph showing immunofluorescence staining for BrdU and IdU in the medial-lateral plane in matched 10 μm sagittal cryosections from control (fd/fd;+/+) and conditional Dicer depleted (fd/fd;nCre/+) mice. Scale bars, 20 μm. VZ, ventricular zone; SVZ, subventricular zone. (B) Quantification of total immunofluorescence staining of E15.5 BrdU and BrdU/IdU positive cells showing reduction in overall proliferating cell number. Y-axis denotes number of labeled cells per 100 total cells as defined by Hoechst staining in a 40x field. (C) Quantification of immunofluorescence staining of E15.5 BrdU or IdU positive cells, showing a retardation of the cell cycle kinetics in Dicer depleted brains as defined by the increase in S Phase cells and decrease in G2/M Phase cells. Y-axis denotes percentage of BrdU labeled or IdU-only labeled cells within the total labeled cell count. Ctrl, control (fd/fd;+/+); Dd, conditional Dicer depleted (fd/fd;nCre/+); S Phase = percentage of labeled cells positive for BrdU; G2/M Phase = percentage of labeled cells positive only for IdU. Error bars represent mean ± SEM. *p<0.5, ***p <0.0001 using a two-tailed unpaired t-test.
3.3 Dicer inactivation induces increased apoptosis in late corticogenesis
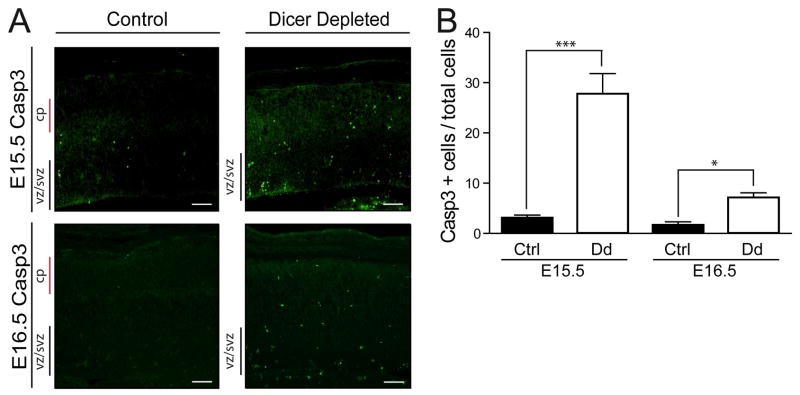
Increased cell death could also contribute significantly to the decrease in cortical thickness found in E15.5 Dicer depleted mice. Earlier work reported no differences in TUNEL staining between the cerebral cortex of Nestin-Cre Dicer depleted mice and control tissue, at least up to E18.5 (Kawase-Koga et al., 2009). However, we observe a ~9- and ~4-fold increase in cells positive for the apoptotic marker activated Caspase 3 at E15.5 and E16.5, respectively (Fig. 4A and B). Apoptotic cells were dispersed throughout the cortical parenchyma. In support of our finding, De Pietri Tonelli and colleagues also showed increased apoptotic cells throughout the cortex when examining cortices from Emx1-Cre Dicer depleted mice from E12.5 to E14.5 (De Pietri Tonelli et al., 2008).
Figure 4.
Increased apoptotic cell death in Dicer depleted cortical tissues. (A) Representative photomicrographs of immunofluorescence staining for activated Caspase3 (Casp3) in cerebral cortices at both E15.5 and E16.5. Scale bars, 100 μm. VZ/SVZ, ventricular zone/subventricular zone; CP, cortical plate. (B) Quantification of immunofluorescence staining of E15.5 and E16.5 Casp3 positive cells completed in the medial-lateral plane in matched sections, show significantly increased apoptotic cell death within Dicer depleted cortices. Y-axis denotes number of Casp3 positive cells per 1000 total cells as defined by Hoechst staining in a 20x field. Ctrl, control (fd/fd;+/+); Dd, conditional Dicer depleted (fd/fd;nCre/+). Error bars represent mean ± SEM. *p<0.01, ***p <0.0001 using a two-tailed unpaired t-test.
3.4 Dicer deficient mice have dysregulated neuronal migration and differentiation
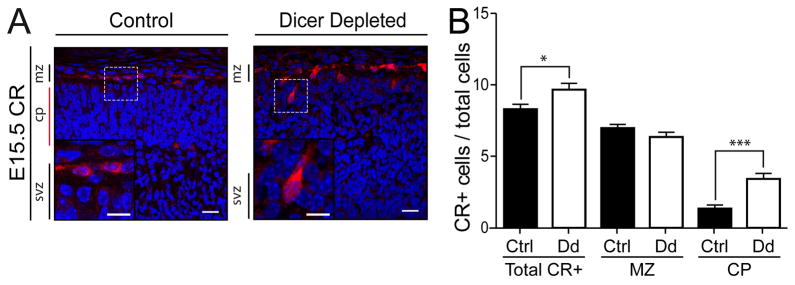
In E15.5 Dicer depleted embryonic cortices, we found ectopic Nestin expressing cells in the upper layers (Fig. 1B). Also, mature neurons within the cortical structure at E15.5 were disorganized (Fig. 1B). We therefore evaluated radial migration. The first wave of radial migration, beginning around E11.5, is largely composed of Cajal-Retzius (C-R) neurons relocating from the VZ to the marginal zone, the uppermost area of the developing cortex. The effects of Dicer depletion on C-R neuronal migration and differentiation at E15.5 was assessed using the calcium binding protein, Calretinin as a specific marker of C-R neurons. We found statistical increases in Calretinin positive cells in Dicer depleted cortex relative to controls (Fig. 5A and B). Moreover, a substantial proportion of the C-R neurons exhibited a significant defect in migration, being dispersed within the cortical plate rather than confined to the marginal zone at E15.5 (Fig. 5B). Interestingly, the C-R positive cells with migration defects were dysmorphic relative to cells that migrated properly to the marginal zone (Fig. 5A).
Figure 5.
Impaired migration of C-R neurons in the Dicer depleted cerebral cortex. (A) Immunofluorescence staining for activated Calretinin (CR; red) and Hoechst33258 (blue) at E15.5 with higher magnification inset. Scale bars, 20 μm. VZ/SVZ, ventricular zone/subventricular zone; CP, cortical plate; MZ, marginal zone. (B) Quantification of immunofluorescence staining of E15.5 CR positive cells completed in the medial-lateral plane in matched sections show significant changes in CR positive cells numbers, with the contributing increase of CR+ neurons showing impaired migration to the marginal zone in Dicer depleted cortical tissue. Y-axis denotes number of CR positive cells per 500 total cells as defined by Hoechst staining in a 40x field. Ctrl, control (fd/fd;+/+); Dd, conditional Dicer depleted (fd/fd;nCre/+). Error bars represent mean ± SEM. *p<0.05, ***p <0.0001 using a two-tailed unpaired t-test.
C-R neurons secrete Reelin, which is important for neuronal migration (Honda et al., 2011). Contrary to a recently published study, which found no change in Reelin levels within Nestin-Cre Dicer depleted cortices using histological assays at E15.5 and E18.5 (Li et al., 2011), we show a 3-fold increase in Reelin mRNA levels in Dicer depleted cortex relative to control at E15.5 (Fig. 6A). Reelin immunohistochemistry (IHC) corroborated this result; expression throughout the cortex was enhanced (Fig. 6B).
Figure 6.
Overexpression of Reelin in Dicer depleted cerebral cortices. (A) RT-qPCR quantification of Reelin RNA expression from total RNA isolated from E15.5 cerebral cortex of control (fd/fd;+/+) and conditional Dicer depleted (fd/fd;nCre/+) mice. Error bars represent mean ± SEM. **p<0.001 using a two-tailed unpaired t-test. (B) Representative photomicrograph of IHC for control (fd/fd;+/+) and Dicer depleted (fd/fd;nCre/+) mice cortices show a gradient of Reelin overexpression throughout Dicer depleted brains with the strongest signal intensity in the lateral portion of the cortical plate. Scale bars, 100 μm. VZ/SVZ, ventricular zone/subventricular zone; CP, cortical plate.
We next assessed migrating immature neurons. We found that DCX mRNA was ~7-fold higher in Dicer depleted tissue relative to controls at E15.5 (Fig. 7A), and DCX staining was regionally restricted to a significant degree (Fig. 7B and D). DCX signal intensity analysis also revealed a ~3-fold increase in E16.5 cortices relative to controls (Fig. 7C). Rnd2, a maturation marker, was evaluated by in situ hybridization (ISH) (Fig. 8B) and quantitation of mRNA levels (Fig. 8A). We found in both analyses the levels of Rnd2 mRNA to be increased in Dicer depleted brains relative to control. Additionally, Rnd2 mRNA was aberrantly localized throughout the cortical parenchyma in Dicer depleted embryos relative to control embryos (Heng et al., 2008). The overproduction of both DCX and Rnd2 in the Dicer depleted cerebral cortex may contribute to the premature maturation of neurons in inappropriate locations, which in turn causes the overall cortical lamina disorganization.
Figure 7.
Reduced immature migrating neurons in Dicer depleted cortices. (A) RT-qPCR quantification of DCX RNA expression from total RNA isolated from E15.5 cerebral cortex of control (fd/fd;+/+) or conditional Dicer depleted (fd/fd;nCre/+) mice with results normalized to total RNA. Error bars represent mean ± SEM. Values are statistically significant (*p<0.01) using a two-tailed unpaired t-test. (B) Representative photomicrograph of IHC for DCX in control (fd/fd;+/+) and conditional Dicer depleted (fd/fd;nCre/+) embryos showing reduced spread of DCX positive cells throughout the Dicer depleted cortex. Scale bars, 100 μm. (C) Quantification of average IHC DAB signal intensity per 100 pixels2 within the cortical plate relative to the VZ, showed increased DCX signal intensity within E16.5 Dicer depleted cortices. Error bars represent mean ± SEM. ***p<0.0001 using a two-tailed unpaired t-test. (D) Quantification of the DCX spread across the cortex is presented as a ratio of IHC staining of DCX compared to total cortex thickness, with decreased DCX spread within E15.5 Dicer depleted cortices. Error bars represent mean ± SEM. *p<0.01 using a two-tailed unpaired t-test. VZ/SVZ, ventricular zone/subventricular zone; CP, cortical plate.
Figure 8.
Overexpression of Rnd2, a multipolar to bipolar transition regulator in Dicer depleted cortical tissue. (A) RT-qPCR quantification of Rnd2 RNA expression from total RNA isolated from E15.5 cerebral cortex of both control (fd/fd;+/+) and conditional Dicer depleted (fd/fd;nCre/+) mice with results normalized to total RNA. Error bars represent mean ± SEM. *p<0.01 using a two-tailed unpaired t-test. (B) In situ hybridization staining for Rnd2. Shown are representative photomicrographs from 10 μm sagittal cryosections through the dorsal telencephalon of control (fd/fd;+/+) and conditional Dicer depleted (fd/fd;nCre/+) mice, with overexpression evident of Rnd2 mRNA throughout the Dicer depleted cortical tissue. Scale bars, 100 μm. VZ/SVZ, ventricular zone/subventricular zone; CP, cortical plate.
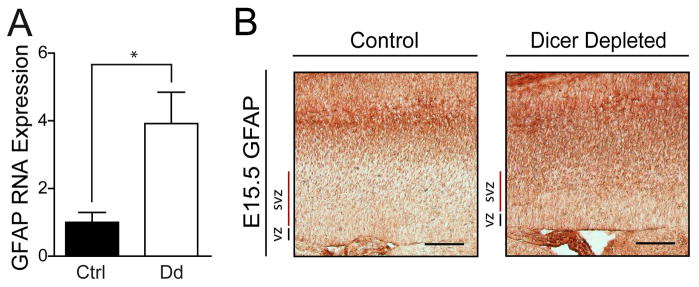
To test if Dicer depletion induces precocious or enhanced astrocyte differentiation, we assessed GFAP levels in cortical sections. We found ~4-fold elevated GFAP mRNA levels in Dicer depleted cortex relative to control (Fig. 9A). IHC for GFAP confirmed increased expression, with GFAP-positive cells distributed throughout Dicer depleted cortical parenchyma (Fig. 9B).
Figure 9.
The astrocytic marker GFAP is overexpressed in Dicer depleted cortices. (A) RT-qPCR quantification of GFAP RNA expression from total RNA isolated from E15.5 cerebral cortex of both control (fd/fd;+/+) and conditional Dicer depleted (fd/fd;nCre/+) with results normalized to total RNA. Error bars represent mean ± SEM. *p<0.05 using a two-tailed unpaired t-test. (B) Representative photomicrographs of IHC staining in control (fd/fd;+/+) and conditional Dicer depleted (fd/fd;nCre/+) mice cortices show disorganized and broad expression of GFAP in the Dicer depleted mice cortex. Scale bars, 100 μm. VZ/SVZ, ventricular zone/subventricular zone; CP, cortical plate.
4. Discussion
We validate several key aspects of prior published reports in which Dicer was depleted in the telencephalon of developing mice NPCs. More importantly, we extend these results significantly by examining the effect of Dicer depletion on proliferating progenitor cells, cell cycle kinetics, and cell death. In addition, we provide evidence for a strong regulatory role of miRNAs in both early and late neuronal migration and differentiation that may provide candidates for future exploration of miRNA regulation in neuronal development. Finally, we implicate a role for miRNA regulation of astrocyte differentiation during cortical development.
4.1 Dicer depletion impacts proliferating cells and induces cell death
We found significant decreases in proliferating cells in vivo and delayed cell cycle kinetics. Previous work using PH3 staining did not detect significant differences in dividing cells until E18.5 in this model (Kawase-Koga et al., 2009). However, our quantitative data demonstrates reduced PH3 positive cells in the VZ and SVZ at both E15.5 and E16.5. Our findings are the first to report cell cycle kinetic defects within the cerebral cortex of Dicer depleted mice. Specifically, we showed an increase in S-phase length in Dicer depleted proliferating cells compared to the 4 hrs S-phase length in control proliferating cells. These results are congruent with reports indicating the importance of specific miRNAs in proliferating cell regulation (Gaughwin et al., 2011; Shi et al., 2010). The most well described miRNA associated with neural cell proliferation is miR-9 (Delaloy et al., 2010; Shibata et al., 2011), which we found reduced in Dicer depleted NPCs (data not shown).
Specific miRNAs have been identified as cell cycle regulators in many model systems including those studied in cancer (Chivukula and Mendell, 2008; Gillies and Lorimer, 2007; Lal et al., 2008; Linsley et al., 2007; Medina et al., 2008; Xia et al., 2009; Xia et al., 2010), and several have been specifically implicated in regulating cell cycle kinetics within normal cortical neurogenesis (Andersson et al., 2010; Bonev et al., 2011; Sun et al., 2011). Our thymidine analog data provides solid evidence that miRNAs are required for appropriate cell cycle kinetics within the developing cerebral cortex. Our data also suggests that the increase in Nestin positive cells is not due to increased cell cycle kinetics or increased progenitor pool populations as determined via PH3 and BrdU/IdU analysis. Instead, cells retaining Nestin expression are likely unable to progress past the progenitor state, similar to the effects of miR-9 loss described by Delaloy and colleagues (Delaloy et al., 2010).
We also found increased apoptotic cell death beginning as early as E15.5 throughout the cortex, which may explain why the abundant immature cells do not result in an increased mature population over time. Analysis of the Emx1-Cre Dicer depleted model also revealed increased cell death, beginning largely in the VZ, by E12.5 (De Pietri Tonelli et al., 2008), however TUNEL analysis of the Nestin-Cre Dicer depleted cortex by others did not reveal differences until at least E18.5 (Kawase-Koga et al., 2009). One possible explanation for the discrepancy between our work and the earlier Nestin-Cre report may result from the better retention of activated Caspase 3 within fixed tissue than the antigens for the TUNEL assay, thus providing a more sensitive test for cell death (Bressenot et al., 2009; Duan et al., 2003; Singh et al., 2009).
4.2 Effect of microRNA depletion in migration and differentiation in developing cortex
Upon evaluation of the first wave of migration, we found significant increases in C-R neurons within the Dicer depleted cortex. A recent report using the FoxG1 promoter to drive Cre beginning at ~E8 also found increased C-R neurons but no elevation of the secretory molecule of C-R neurons, Reelin, upon Dicer depletion at E11.5 (Nowakowski et al., 2011). Using the Nestin-Cre model with Dicer excision beginning ~E9, we also showed a significant increase in C-R neurons, but with a significant increase in Reelin at E15.5. Likely, the discrepancy between our study and the Nowakowski et al., report is due to activation of Cre expression and the timing in which analysis was completed. It is possible that the overproduction of Reelin is a consequence of elevated C-R neurons that is not discernible until after E11.5. Whether this in vivo increase in Reelin levels is due to increased proliferation of C-R neurons and/or if Reelin levels are directly regulated by miRNAs is an avenue for future exploration. Interestingly, we also report an abnormal morphology of the migrationally delayed C-R neurons in the Dicer depleted cortex. Our finding that some C-R neurons were well-formed may reflect the fact that some miRNAs persisted for several days beyond the onset of Dicer expression (expected at ~E9) and C-R differentiation around E11. This is consistent with the finding that some miRNAs persist weeks after Dicer depletion in other models (Gantier et al., 2011; Kawase-Koga et al., 2009). One explanation of the dysmorphic C-R neurons may be due specifically to the greater overall reduction of miR-9 in the migrationally stunted C-R neurons. A previous report established that miR-9 modulated C-R cell differentiation by suppressing FoxG1 expression (Shibata et al., 2008).
The importance of miRNAs in migration and differentiation is implicated primarily in tumor invasion, and is less well described with regard to neuronal differentiation/migration in the developing cortex. Of particular interest are the potential for miRNAs in cortical organization that, when disrupted or ablated, may predispose a brain to aberrant wiring as found in disorders such as epilepsy and schizophrenia (Frotscher, 2010; Liu, 2011). Specifically, we showed i) increased Reelin expression, possibly due to increased C-R neuron numbers, ii) increased DCX mRNA with a reduction in the normally constrained layers of DCX positive cells, and iii) increased numbers of Rnd2 positive cells and Rnd2 mRNA levels. These findings indicate that miRNA-mediated regulation is likely required at multiple stages of neuronal differentiation and migration. Specifically, Rnd2 is vital for neurite branching and retraction, important in the transition from multipolar to bipolar neuronal orientation, and is directly regulated by Neurogenin2 in the developing cortex (Chardin, 2006; Heng et al., 2008). Whether miRNA repression of Rnd2 is direct or indirect via an upstream regulator such as Neurogenin2 is yet to be defined. In neuroblastoma cells, miR-128 upregulation inhibits Reelin and DCX expression and reduces neuroblastoma cell motility and invasiveness (Evangelisti et al., 2009). Interestingly, one recently published study showed miR-134 directly targets DCX in vivo, repressing differentiation and maintaining the proliferation state of neural progenitors (Gaughwin et al., 2011).
4.3 Dicer depletion leads to a precocious induction of astrocyte differentiation
At the end of neurogenesis, Nestin positive radial glia differentiate into GFAP positive mature astrocytes (Mori et al., 2005). The onset of this process normally occurs at ~E16 in the developing cerebral cortex, but in our Dicer depleted cortices we find an advanced increase in both GFAP mRNA levels and protein expression throughout the cortical parenchyma relative to control brains at E15.5. Although previous Dicer depletion models investigated the role of miRNA on gliogenesis in the developing spinal cord (Zheng et al., 2010) and interneurons in the telencephalon (Kawase-Koga et al., 2009), we are the first to identify the gross effect of Dicer depletion on astrocyte differentiation with the developing cerebral cortex. Our finding suggests an important regulatory role for miRNAs in the binary switch from neurogenesis to gliogenesis within the developing cerebral cortex. An interesting candidate for this transitional regulation may be miR-125b, which is reported to play a direct role in the development of glioma stem cells (Pogue et al., 2010; Wan et al., 2012).
In summary, we significantly extend the growing body of evidence indicating a strong regulatory role of miRNAs in proliferation and cell cycle kinetics, as well as in neuronal and astrocyte migration and differentiation in the developing mammalian brain. It will be interesting to test in future studies the extent to which miRNAs modulate the expression of RNA-binding proteins important in neural development (Bolognani and Perrone-Bizzozero, 2008), transcription factors or repressors (Cheng et al., 2009), or splicing factors (Makeyev et al., 2007). Dissecting the complex effects of reducing miRNAs that modulate master regulators vs. the direct impact on proteins with more specialized activities will provide a wealth of information about their overall importance in the developing brain.
Highlights.
Dicer loss causes mis-localized progenitor cells and disorganized cortical lamina
Dicer ablation reduces cell cycle kinetics in the developing cortex
Without Dicer there is early cell death in the developing cerebral cortex
Dicer loss causes dysregulated differentiation and migration markers
Dicer depletion causes a precocious induction of astrocyte differentiation
Acknowledgments
The authors would like to acknowledge the members of the Davidson Lab. This work was supported by the March of Dimes, the Roy J. Carver Trust and DA025132.
Abbreviations
- miRNA
miR, microRNA
- E15.5
embryonic day 15.5
- fd
floxed Dicer
- nCre
Nestin-Cre
- PFA
paraformaldehyde
- ISH
in situ hybridization
- IHC
immunohistochemistry
- NPC
neural progenitor cell
- PH3
phosphohistone 3
- BrdU
5′-bromo-2′-deoxyuridine
- IdU
5′-iodo-2′-deoxyuridine
- C-R
Cajal-Retzius
- DCX
Doublecortin
- VZ
ventricular zone
- SVZ
subventricular zone
- CP
cortical plate
- MZ
marginal zone
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersson T, Rahman S, Sansom SN, Alsio JM, Kaneda M, Smith J, O’Carroll D, Tarakhovsky A, Livesey FJ. Reversible block of mouse neural stem cell differentiation in the absence of dicer and microRNAs. PLoS One. 2010;5:e13453. doi: 10.1371/journal.pone.0013453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–7. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Bian S, Sun T. Functions of Noncoding RNAs in Neural Development and Neurological Diseases. Mol Neurobiol. 2011 doi: 10.1007/s12035-011-8211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognani F, Perrone-Bizzozero NI. RNA-protein interactions and control of mRNA stability in neurons. J Neurosci Res. 2008;86:481–9. doi: 10.1002/jnr.21473. [DOI] [PubMed] [Google Scholar]
- Bonev B, Pisco A, Papalopulu N. MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev Cell. 2011;20:19–32. doi: 10.1016/j.devcel.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressenot A, Zimmer O, Marchal S, Gauchotte G, Montagne K, Plenat F. [Detection of apoptosis in vivo: comparison of different methods in histological sections of subcutaneous xenografts of HT29 human colon adenocarcinoma] Ann Pathol. 2009;29:370–5. doi: 10.1016/j.annpat.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Burns KA, Kuan CY. Low doses of bromo- and iododeoxyuridine produce near-saturation labeling of adult proliferative populations in the dentate gyrus. Eur J Neurosci. 2005;21:803–7. doi: 10.1111/j.1460-9568.2005.03907.x. [DOI] [PubMed] [Google Scholar]
- Chae TH, Walsh CA. Genes that control the size of the cerebral cortex. Novartis Found Symp. 2007;288:79–90. doi: 10.1002/9780470994030.ch6. discussion 91–8. [DOI] [PubMed] [Google Scholar]
- Chardin P. Function and regulation of Rnd proteins. Nat Rev Mol Cell Biol. 2006;7:54–62. doi: 10.1038/nrm1788. [DOI] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivukula RR, Mendell JT. Circular reasoning: microRNAs and cell-cycle control. Trends Biochem Sci. 2008;33:474–81. doi: 10.1016/j.tibs.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi PS, Zakhary L, Choi WY, Caron S, Alvarez-Saavedra E, Miska EA, McManus M, Harfe B, Giraldez AJ, Horvitz RH, Schier AF, Dulac C. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron. 2008;57:41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen M, Schratt GM. microRNA involvement in developmental and functional aspects of the nervous system and in neurological diseases. Neurosci Lett. 2009;466:55–62. doi: 10.1016/j.neulet.2009.04.043. [DOI] [PubMed] [Google Scholar]
- Cuellar TL, Davis TH, Nelson PT, Loeb GB, Harfe BD, Ullian E, McManus MT. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc Natl Acad Sci U S A. 2008;105:5614–9. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiani D, Alexander JJ, O’Rourke JR, McManus M, Jadhav AP, Cepko CL, Hauswirth WW, Harfe BD, Strettoi E. Dicer inactivation leads to progressive functional and structural degeneration of the mouse retina. J Neurosci. 2008;28:4878–87. doi: 10.1523/JNEUROSCI.0828-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, Ullian EM. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–30. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pietri Tonelli D, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–21. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, Young WL, Ivey KN, Gao FB. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell. 2010;6:323–35. doi: 10.1016/j.stem.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan WR, Garner DS, Williams SD, Funckes-Shippy CL, Spath IS, Blomme EA. Comparison of immunohistochemistry for activated caspase-3 and cleaved cytokeratin 18 with the TUNEL method for quantification of apoptosis in histological sections of PC-3 subcutaneous xenografts. J Pathol. 2003;199:221–8. doi: 10.1002/path.1289. [DOI] [PubMed] [Google Scholar]
- Dubois NC, Hofmann D, Kaloulis K, Bishop JM, Trumpp A. Nestin-Cre transgenic mouse line Nes-Cre1 mediates highly efficient Cre/loxP mediated recombination in the nervous system, kidney, and somite-derived tissues. Genesis. 2006;44:355–60. doi: 10.1002/dvg.20226. [DOI] [PubMed] [Google Scholar]
- Evangelisti C, Florian MC, Massimi I, Dominici C, Giannini G, Galardi S, Bue MC, Massalini S, McDowell HP, Messi E, Gulino A, Farace MG, Ciafre SA. MiR-128 up-regulation inhibits Reelin and DCX expression and reduces neuroblastoma cell motility and invasiveness. FASEB J. 2009;23:4276–87. doi: 10.1096/fj.09-134965. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64:303–9. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Frotscher M. Role for Reelin in stabilizing cortical architecture. Trends Neurosci. 2010;33:407–14. doi: 10.1016/j.tins.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Gantier MP, McCoy CE, Rusinova I, Saulep D, Wang D, Xu D, Irving AT, Behlke MA, Hertzog PJ, Mackay F, Williams BR. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 2011;39:5692–703. doi: 10.1093/nar/gkr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaughwin P, Ciesla M, Yang H, Lim B, Brundin P. Stage-Specific Modulation of Cortical Neuronal Development by Mmu-miR-134. Cereb Cortex. 2011 doi: 10.1093/cercor/bhq262. [DOI] [PubMed] [Google Scholar]
- Gillies JK, Lorimer IA. Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle. 2007;6:2005–9. doi: 10.4161/cc.6.16.4526. [DOI] [PubMed] [Google Scholar]
- Harfe BD. MicroRNAs in vertebrate development. Curr Opin Genet Dev. 2005;15:410–5. doi: 10.1016/j.gde.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng JI, Nguyen L, Castro DS, Zimmer C, Wildner H, Armant O, Skowronska-Krawczyk D, Bedogni F, Matter JM, Hevner R, Guillemot F. Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature. 2008;455:114–8. doi: 10.1038/nature07198. [DOI] [PubMed] [Google Scholar]
- Honda T, Kobayashi K, Mikoshiba K, Nakajima K. Regulation of cortical neuron migration by the Reelin signaling pathway. Neurochem Res. 2011;36:1270–9. doi: 10.1007/s11064-011-0407-4. [DOI] [PubMed] [Google Scholar]
- Kawaguchi A, Miyata T, Sawamoto K, Takashita N, Murayama A, Akamatsu W, Ogawa M, Okabe M, Tano Y, Goldman SA, Okano H. Nestin-EGFP transgenic mice: visualization of the self-renewal and multipotency of CNS stem cells. Mol Cell Neurosci. 2001;17:259–73. doi: 10.1006/mcne.2000.0925. [DOI] [PubMed] [Google Scholar]
- Kawase-Koga Y, Otaegi G, Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev Dyn. 2009;238:2800–12. doi: 10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–4. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–81. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–64. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijpers M, Hoogenraad CC. Centrosomes, microtubules and neuronal development. Mol Cell Neurosci. 2011 doi: 10.1016/j.mcn.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Lal A, Kim HH, Abdelmohsen K, Kuwano Y, Pullmann R, Jr, Srikantan S, Subrahmanyam R, Martindale JL, Yang X, Ahmed F, Navarro F, Dykxhoorn D, Lieberman J, Gorospe M. p16(INK4a) translation suppressed by miR-24. PLoS One. 2008;3:e1864. doi: 10.1371/journal.pone.0001864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P, Hudson LD. MicroRNAs in neural cell differentiation. Brain Res. 2010;1338:14–9. doi: 10.1016/j.brainres.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennox KA, Behlke MA. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. 2011 doi: 10.1038/gt.2011.100. [DOI] [PubMed] [Google Scholar]
- Li Q, Bian S, Hong J, Kawase-Koga Y, Zhu E, Zheng Y, Yang L, Sun T. Timing specific requirement of microRNA function is essential for embryonic and postnatal hippocampal development. PLoS One. 2011;6:e26000. doi: 10.1371/journal.pone.0026000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, Chau N, Cleary M, Jackson AL, Carleton M, Lim L. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–52. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JS. Molecular genetics of neuronal migration disorders. Curr Neurol Neurosci Rep. 2011;11:171–8. doi: 10.1007/s11910-010-0176-5. [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–48. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manakov SA, Grant SG, Enright AJ. Reciprocal regulation of microRNA and mRNA profiles in neuronal development and synapse formation. BMC Genomics. 2009;10:419. doi: 10.1186/1471-2164-10-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina R, Zaidi SK, Liu CG, Stein JL, van Wijnen AJ, Croce CM, Stein GS. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008;68:2773–80. doi: 10.1158/0008-5472.CAN-07-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Buffo A, Gotz M. The novel roles of glial cells revisited: the contribution of radial glia and astrocytes to neurogenesis. Curr Top Dev Biol. 2005;69:67–99. doi: 10.1016/S0070-2153(05)69004-7. [DOI] [PubMed] [Google Scholar]
- Nowakowski TJ, Mysiak KS, Pratt T, Price DJ. Functional dicer is necessary for appropriate specification of radial glia during early development of mouse telencephalon. PLoS One. 2011;6:e23013. doi: 10.1371/journal.pone.0023013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci. 2008;28:14341–6. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J, Hammond SM. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue AI, Cui JG, Li YY, Zhao Y, Culicchia F, Lukiw WJ. Micro RNA-125b (miRNA-125b) function in astrogliosis and glial cell proliferation. Neurosci Lett. 2010;476:18–22. doi: 10.1016/j.neulet.2010.03.054. [DOI] [PubMed] [Google Scholar]
- Pramparo T, Youn YH, Yingling J, Hirotsune S, Wynshaw-Boris A. Novel embryonic neuronal migration and proliferation defects in Dcx mutant mice are exacerbated by Lis1 reduction. J Neurosci. 2010;30:3002–12. doi: 10.1523/JNEUROSCI.4851-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageot CM, Stiles CD. Molecular mechanisms controlling cortical gliogenesis. Curr Opin Neurobiol. 2002;12:244–9. doi: 10.1016/s0959-4388(02)00322-7. [DOI] [PubMed] [Google Scholar]
- Schaefer A, O’Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, Greengard P. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204:1553–8. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Zhao X, Hsieh J, Wichterle H, Impey S, Banerjee S, Neveu P, Kosik KS. MicroRNA regulation of neural stem cells and neurogenesis. J Neurosci. 2010;30:14931–6. doi: 10.1523/JNEUROSCI.4280-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Kurokawa D, Nakao H, Ohmura T, Aizawa S. MicroRNA-9 modulates Cajal-Retzius cell differentiation by suppressing Foxg1 expression in mouse medial pallium. J Neurosci. 2008;28:10415–21. doi: 10.1523/JNEUROSCI.3219-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 Regulates Neurogenesis in Mouse Telencephalon by Targeting Multiple Transcription Factors. J Neurosci. 2011;31:3407–22. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SS, Mehedint DC, Ford OH, 3rd, Jeyaraj DA, Pop EA, Maygarden SJ, Ivanova A, Chandrasekhar R, Wilding GE, Mohler JL. Comparison of ACINUS, caspase-3, and TUNEL as apoptotic markers in determination of tumor growth rates of clinically localized prostate cancer using image analysis. Prostate. 2009;69:1603–10. doi: 10.1002/pros.21019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YJ, Tian XB, Zhang S, Zhang YX, Li X, Li D, Cheng Y, Zhang JN, Kang CS, Zhao W. Temporal lobe epilepsy induces differential expression of hippocampal miRNAs including let-7e and miR-23a/b. Brain Res. 2011;1387:134–40. doi: 10.1016/j.brainres.2011.02.073. [DOI] [PubMed] [Google Scholar]
- Standart N, Jackson RJ. MicroRNAs repress translation of m7Gppp-capped target mRNAs in vitro by inhibiting initiation and promoting deadenylation. Genes Dev. 2007;21:1975–82. doi: 10.1101/gad.1591507. [DOI] [PubMed] [Google Scholar]
- Sun G, Ye P, Murai K, Lang MF, Li S, Zhang H, Li W, Fu C, Yin J, Wang A, Ma X, Shi Y. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat Commun. 2011;2:529. doi: 10.1038/ncomms1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunabori T, Tokunaga A, Nagai T, Sawamoto K, Okabe M, Miyawaki A, Matsuzaki Y, Miyata T, Okano H. Cell-cycle-specific nestin expression coordinates with morphological changes in embryonic cortical neural progenitors. J Cell Sci. 2008;121:1204–12. doi: 10.1242/jcs.025064. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Nowakowski RS, Caviness VS., Jr The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J Neurosci. 1995;15:6046–57. doi: 10.1523/JNEUROSCI.15-09-06046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiente M, Marin O. Neuronal migration mechanisms in development and disease. Curr Opin Neurobiol. 2010;20:68–78. doi: 10.1016/j.conb.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Wan Y, Fei XF, Wang ZM, Jiang DY, Chen HC, Yang J, Shi L, Huang Q. Expression of miRNA-125b in the new, highly invasive glioma stem cell and progenitor cell line SU3. Chin J Cancer. 2012 doi: 10.5732/cjc.011.10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, Qi Y, Ng SS, Chen X, Chen S, Fang M, Li D, Zhao Y, Ge R, Li G, Chen Y, He ML, Kung HF, Lai L, Lin MC. MicroRNA-15b regulates cell cycle progression by targeting cyclins in glioma cells. Biochem Biophys Res Commun. 2009;380:205–10. doi: 10.1016/j.bbrc.2008.12.169. [DOI] [PubMed] [Google Scholar]
- Xia W, Li J, Chen L, Huang B, Li S, Yang G, Ding H, Wang F, Liu N, Zhao Q, Fang T, Song T, Wang T, Shao N. MicroRNA-200b regulates cyclin D1 expression and promotes S-phase entry by targeting RND3 in HeLa cells. Mol Cell Biochem. 2010;344:261–6. doi: 10.1007/s11010-010-0550-2. [DOI] [PubMed] [Google Scholar]
- Zheng K, Li H, Zhu Y, Zhu Q, Qiu M. MicroRNAs Are Essential for the Developmental Switch from Neurogenesis to Gliogenesis in the Developing Spinal Cord. J Neurosci. 2010;30:8245–50. doi: 10.1523/JNEUROSCI.1169-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]