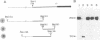Abstract
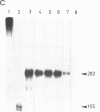
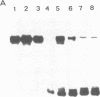
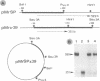
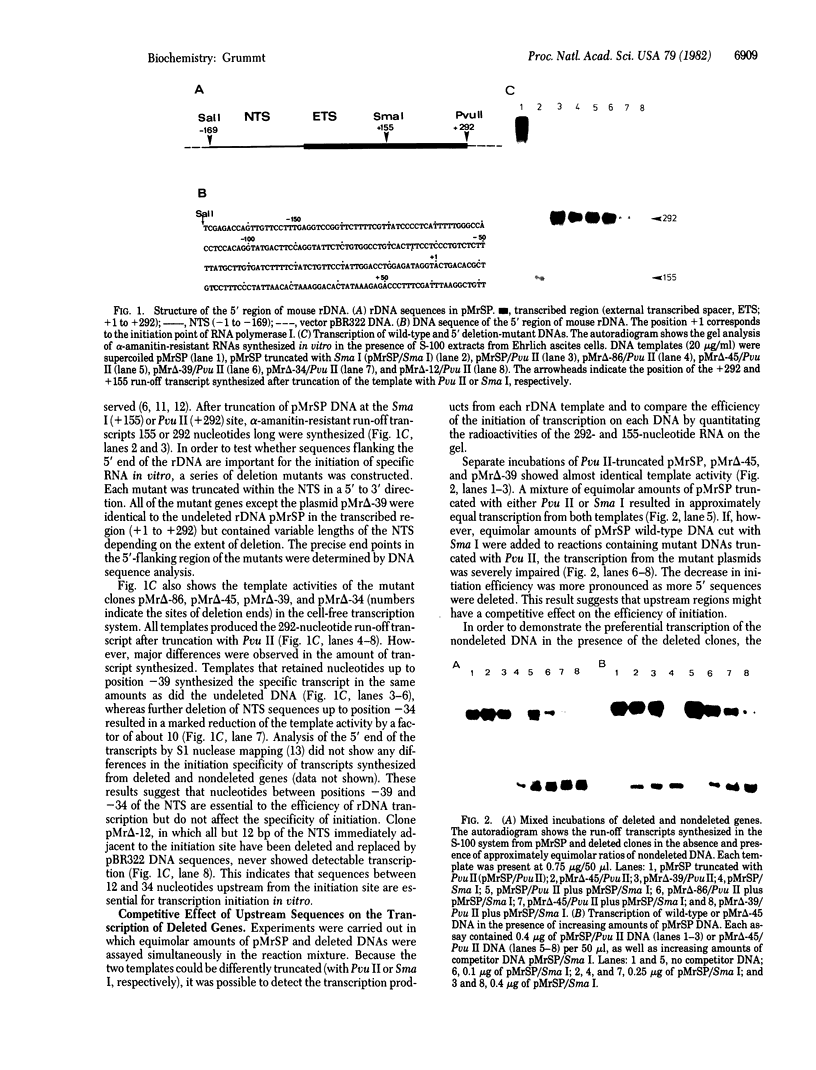
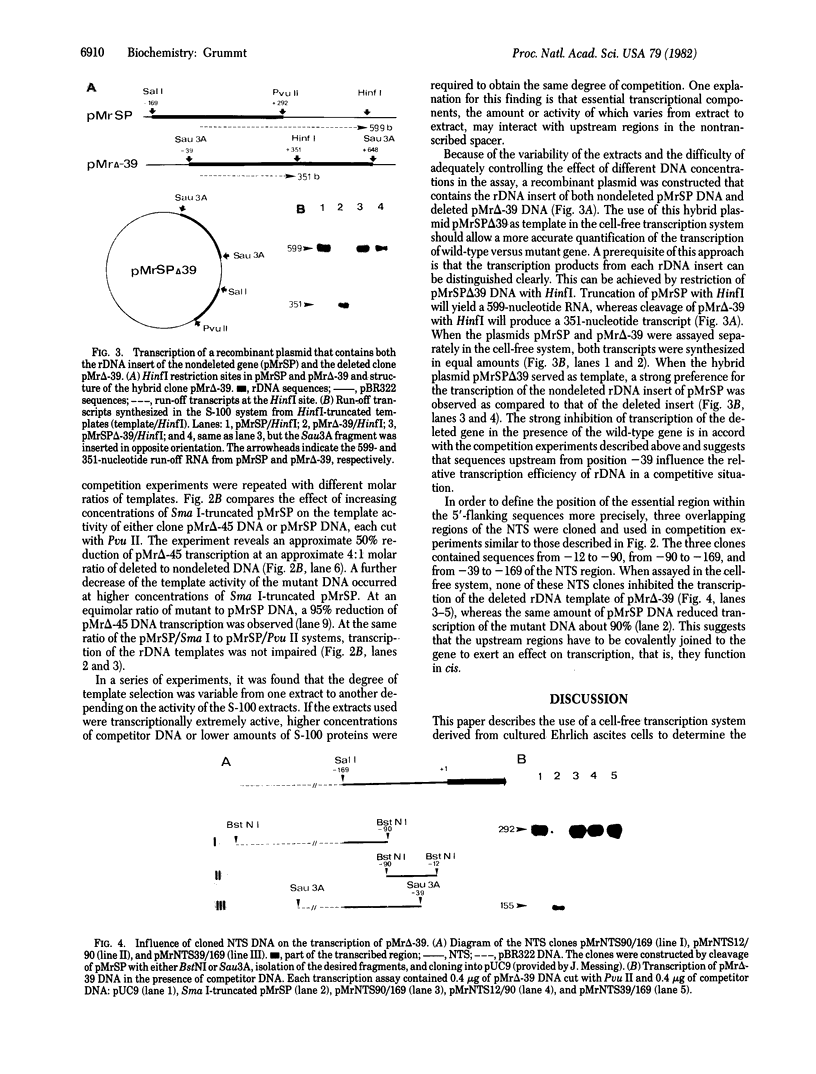
The nucleotide sequence(s) specifying RNA polymerase I initiation has been investigated by studying the transcription of deleted and nondeleted mouse ribosomal RNA gene (rDNA) templates in vitro. The deletion of 5'-flanking sequences upstream from position -- 39 did not affect transcriptional activity, but removal of sequences between positions -- 39 and -- 34 resulted in a 90% decrease of rDNA transcription. The template activity was completely eliminated by the further deletion of nucleotides -- 33 to -- 13. It is concluded that sequences between -- 34 and -- 12, upstream from the transcribed region, represent an essential control region for the initiation of transcription in vitro. Therefore, this region may be functionally analogous to the T-A-T-A box of RNA polymerase II promoters. In addition to this control region, sequences located further upstream (between positions -- 45 and -- 169) may also exert some function in efficient transcription initiation as revealed by competition experiments between wildtype and mutant rDNA templates.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bach R., Allet B., Crippa M. Sequence organization of the spacer in the ribosomal genes of Xenopus clivii and Xenopus borealis. Nucleic Acids Res. 1981 Oct 24;9(20):5311–5330. doi: 10.1093/nar/9.20.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach R., Grummt I., Allet B. The nucleotide sequence of the initiation region of the ribosomal transcription unit from mouse. Nucleic Acids Res. 1981 Apr 10;9(7):1559–1569. doi: 10.1093/nar/9.7.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayev A. A., Georgiev O. I., Hadjiolov A. A., Kermekchiev M. B., Nikolaev N., Skryabin K. G., Zakharyev V. M. The structure of the yeast ribosomal RNA genes. 2. The nucleotide sequence of the initiation site for ribosomal RNA transcription. Nucleic Acids Res. 1980 Nov 11;8(21):4919–4926. doi: 10.1093/nar/8.21.4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Gross K. J., Pogo A. O. Control of ribonucleic acid synthesis in eukaryotes. 3. The effect of cycloheximide and edeine on rna synthesis in yeast. Biochemistry. 1976 May 18;15(10):2082–2086. doi: 10.1021/bi00655a008. [DOI] [PubMed] [Google Scholar]
- Grummt I. Mapping of a mouse ribosomal DNA promoter by in vitro transcription. Nucleic Acids Res. 1981 Nov 25;9(22):6093–6102. doi: 10.1093/nar/9.22.6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Roth E., Paule M. R. Ribosomal RNA transcription in vitro is species specific. Nature. 1982 Mar 11;296(5853):173–174. doi: 10.1038/296173a0. [DOI] [PubMed] [Google Scholar]
- Grummt I., Smith V. A., Grummt F. Amino acid starvation affects the initiation frequency of nucleolar RNA polymerase. Cell. 1976 Mar;7(3):439–445. doi: 10.1016/0092-8674(76)90174-4. [DOI] [PubMed] [Google Scholar]
- Grummt I. Specific transcription of mouse ribosomal DNA in a cell-free system that mimics control in vivo. Proc Natl Acad Sci U S A. 1981 Feb;78(2):727–731. doi: 10.1073/pnas.78.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long E. O., Rebbert M. L., Dawid I. B. Nucleotide sequence of the initiation site for ribosomal RNA transcription in Drosophila melanogaster: comparison of genes with and without insertions. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1513–1517. doi: 10.1073/pnas.78.3.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. G., Sollner-Webb B. Transcription of mouse rRNA genes by RNA polymerase I: in vitro and in vivo initiation and processing sites. Cell. 1981 Nov;27(1 Pt 2):165–174. doi: 10.1016/0092-8674(81)90370-6. [DOI] [PubMed] [Google Scholar]
- Mishima Y., Yamamoto O., Kominami R., Muramatsu M. In vitro transcription of a cloned mouse ribosomal RNA gene. Nucleic Acids Res. 1981 Dec 21;9(24):6773–6785. doi: 10.1093/nar/9.24.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niles E. G., Sutiphong J., Haque S. Structure of the Tetrahymena pyriformis rRNA gene. Nucleotide sequence of the transcription initiation region. J Biol Chem. 1981 Dec 25;256(24):12849–12856. [PubMed] [Google Scholar]
- Shenk T. Transcriptional control regions: nucleotide sequence requirements for initiation by RNA polymerase II and III. Curr Top Microbiol Immunol. 1981;93:25–46. doi: 10.1007/978-3-642-68123-3_3. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Sprague K. U., Larson D., Morton D. 5' flanking sequence signals are required for activity of silkworm alanine tRNA genes in homologous in vitro transcription systems. Cell. 1980 Nov;22(1 Pt 1):171–178. doi: 10.1016/0092-8674(80)90165-8. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M., Suzuki Y. Faithful transcription initiation of fibroin gene in a homologous cell-free system reveals an enhancing effect of 5' flanking sequence far upstream. Cell. 1981 Nov;27(1 Pt 2):175–182. doi: 10.1016/0092-8674(81)90371-8. [DOI] [PubMed] [Google Scholar]
- Urano Y., Kominami R., Mishima Y., Muramatsu M. The nucleotide sequence of the putative transcription initiation site of a cloned ribosomal RNA gene of the mouse. Nucleic Acids Res. 1980 Dec 20;8(24):6043–6058. doi: 10.1093/nar/8.24.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F. L., Feigelson P. The rapid turnover of RNA polymerase of rat liver nucleolus, and of its messenger RNA. Proc Natl Acad Sci U S A. 1972 Oct;69(10):2833–2837. doi: 10.1073/pnas.69.10.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]