Abstract
The transcriptional coactivator CIITA regulates MHC class II genes. In the mouse, CIITA is expressed from three distinct promoters (pI, pIII, and pIV) in a developmental and cell type specific manner with pIII being responsible for B lymphocyte specific expression. Although the promoter proximal sequences that regulate CIITA in B cells have been described, nothing is known about additional distal elements that may regulate its expression in B cells. Sequence homology comparisons, DNase I hypersensitivity assays, and histone modification analysis revealed a potential regulatory element located 11 kb upstream of pIII. Deletion of this element, termed HSS1, in a bacterial artificial chromosome (BAC) encoding the entire CIITA locus and surrounding genes, resulted in a complete loss of CIITA expression from the BAC following transfection into B cells. HSS1 and pIII displayed open chromatin architecture features in B cell but not in plasma cell lines, which are silenced for CIITA expression. PU.1 was found to bind HSS1 and pIII in B cells but not in plasma cells. Depletion of PU.1 by shRNA reduced CIITA expression. Chromatin conformation capture assays showed that HSS1 interacted directly with pIII in B cells and that PU.1 was important for this interaction. These results provide evidence that HSS1 is required for B-cell specific expression of CIITA, and that HSS1 functions by interacting with pIII, forming a long distance chromatin loop that is part mediated through PU.1.
Introduction
Major histocompatibility complex class II (MHC-II) genes encode proteins that present peptide antigens to CD4+ T lymphocytes, a process that results in the initiation of adaptive immune responses (1, 2). The class II transactivator (CIITA) plays an essential role in this process by serving as the limiting transcription factor controlling MHC-II gene expression (3). Mutations in CIITA are a cause of bare lymphocyte syndrome, an inherited immunodeficiency in which MHC-II gene expression is absent (4, 5). CIITA is constitutively expressed in antigen presenting cells, such as B cells, macrophages, and dendritic cells (6). CIITA and therefore MHC-II expression can be induced by IFN-γ treatment in most cell types (7). In the mouse, three distinct promoters control CIITA expression: promoters I, III, and IV (8). Each promoter specifies the transcription of an unique exon 1, which is ultimately spliced into a downstream common exon, resulting in the production of distinct CIITA isoforms. The three CIITA promoters are activated in a cell type or cytokine dependent manner. Promoters I and III are utilized for the constitutive CIITA expression in myeloid cells and lymphoid cells, respectively (6, 8–10). Promoter IV directs CIITA expression in response to IFN-γ stimulation in non-lymphoid cells (9).
In B cells, CIITA is constitutively expressed and associates with all MHC-II promoters, recruiting histone acetyltransferases (HATs) and chromatin remodeling proteins to activate MHC-II transcription (3, 11, 12). CIITA also associates with the insulator protein CTCF to form long-range interactions with at least some human MHC-II promoter regions (13). However, when B cells terminally differentiate into plasma cells, CIITA expression is silenced, ultimately resulting in the loss of MHC-II expression (14, 15). We previously showed that during this transition histone modifications associated with active transcription were lost and replaced by at least one mark associated with gene silencing (15). Coupling these observations with those that show that the CIITA pIV DNA is methylated in cell types that are refractory to the induction of CIITA by IFN-γ (16–18), suggest that the locus has the potential for epigenetic regulation.
Five cis-regulatory elements located between −545 bp to −113 bp of the pIII transcription initiation site were identified in B cells (8, 19). These elements, termed ARE1, ARE2, site A, site B, and site C, are occupied by the transcription factors E47, PU.1/IRF1/IRF4, SP1, CREB/CBP, and Oct1 (15, 20, 21). Site C binds PU.1, and PU.1 was found to be essential for B cell specific CIITA expression (15, 21). Following differentiation to plasma cells, the occupancy of these sites by the above activators is lost and the master repressor Blimp-1 binds to this region (22, 23). The histone H3 lysine 4 demethylase LSD-1 also associates with this region and removes activation associated histone modifications (23). These latter events are likely critical to the silencing of CIITA pIII during the plasma cell differentiation process.
PU.1 (SFFV proviral integration 1) is a member of the ETS domain transcription factor family and is encoded by the PU.1-Spil-Sfpil proto-oncogene (24, 25). PU.1 expression is required for the development of common lymphoid and myeloid progenitors that give rise to B cells, T cells, natural killer cells, and macrophage (26–31). Targeted disruption of PU.1 in the mouse resulted in neonatal lethality (32). As a sequence-specific transcription factor, PU.1 binds at promoters and lymphoid specific enhancer regions either by itself or interacting with other factors, like IRF4, to regulate target genes (33–35). Although PU.1 is required for CIITA expression, it is not known how it may coordinate interactions and activate CIITA expression.
The regulation of each CIITA promoter (pI, pIII, and pIV) by their proximal promoter regulatory regions has been well studied (8, 16, 20, 21). In contrast, little is known about whether there are other distal cis-regulatory elements, which are required for CIITA regulation. Recently, Ni et al. described a series of distal elements within the CIITA gene that were responsive to IFN-γ and regulated expression through pIV (36). These results suggest the possibility that distal elements may be required for B-cell specific expression as well. To address this issue, experiments were designed to identify novel distal elements that could regulate CIITA in B cells. One element that was identified and termed HSS1 was required for B-cell specific CIITA expression through pIII. HSS1 was occupied by PU.1 in B cells but not in plasma cells. The mechanism of action of both PU.1 and HSS1 was found to involve the ability of HSS1 to interact directly with the pIII promoter region through a long-distance chromatin loop. Intriguingly, this loop was in part dependent on PU.1 expression and was significantly reduced in plasma cells, suggesting that PU.1 plays a role in loop formation and that maximum loop formation is necessary for the high levels of CIITA found in B cells. Together, these results suggest that multiple elements are required to control CIITA expression in B cells and provide these cells with the ability to present antigens.
Materials and Methods
Cells and culture
The murine BCL1 3B3 B cell line (CRL-1669, American Tissue Type Culture (ATCC), Manassas, VA) and P3X63Ag8.653 (referred to as P3X) plasma cell line (CRL-1580, ATCC) were cultured in RPMI 1640 medium (Mediatech Inc., Manassas, VA) supplemented with 10% heat inactivated FBS (Hyclone Laboratory, Logan, VT), 10 mM HEPES (HyClone Laboratory), 1 mM sodium pyruvate (HyClone Laboratory), 1X non-essential amino acid (HyClone Laboratory), and 0.05 mM β-mercaptoethanol (Sigma-Aldrich, Louis, MO). C57BL/6 mice were purchased from the Jackson Laboratory and five-week old mice were used to obtain primary B cells. Primary B cells were incubated with anti-CD43 and negatively selected using MACS paramagnetic beads and columns following the manufacturer’s protocol (Miltenyi Biotech, Inc. Auburn, CA). To induce plasma cell differentiation, purified murine primary B cells were stimulated with culture media containing IL-2 (20 ng/ml, Sigma-Aldrich), IL-5 (10 ng/ml, Sigma-Aldrich) and LPS (20 µg/ml, Sigma-Aldrich) for the indicated time as previously described (15).
DNase I hypersensitivity assay
DNase I hypersensitivity assay was performed as described by Lu et al. (37). Here, 2 × 107 cells were used to prepare nuclei. Preparations of nuclei were treated with increasing concentrations of DNase I (Roche Ltd, Indianapolis, IN). The DNA was then purified and digested to completion with EcoRV and BamHI (NEB, Inc. Beverly, MA). Nuclease sensitivity for the CIITA locus was determined by Southern blotting using radiolabeled probes and visualized by autoradiography. The probe for CIITA was derived from sequence positions 10,471,913 to 10,472,354 on chromosome 16 as described by the University California Santa Cruz Genome Browser database (http://genome.ucsc.edu/ (38)). A PCR-based DNase I hypersensitivity assay was based on that of Oestreich et al. (39). In this assay, nuclei from BCL1 and P3X cells were isolated and treated with 0, 40, 80, or 160 u/ml of DNase I for 3 minutes at 25 °C. The reaction was stopped and the DNA purified. DNA from the DNase I treated nuclei was PCR amplified using a set of primers that spanned HSS1, pI, pIII, and pIV. Detection and quantitation of these amplicons was carried out by real-time PCR. A relative hypersensitivity was compared to the results obtained for an insensitive region (Y6). The data from at least three biological replicates were normalized to Y6 for each concentration of DNase I used and then plotted as fold over the untreated sample.
PCR Primer Sequences
All PCR primer sequences are provided in Supplemental Table 1.
RNA isolation and RT-real time PCR
Total RNA was extracted from the indicated cells using RNeasy mini prep kits (Qiagen, Inc., Valencia, CA). Reverse transcriptase (Invitrogen, Inc., Carlsbad, CA) was used to produce cDNA according to the manufacturer’s directions. SYBR green dye incorporation into the PCR product using ~1/100th of the cDNA as a template was measured by an iCycler with optical assembly (Biorad, Inc., Hercules, CA). Parallel RT-PCR reactions were performed with 18s rRNA primers to normalize between samples. Following 18s rRNA normalization, the average of three biological replicates were plotted relative to the indicated control sample.
Chromatin immunoprecipitations (ChIP)
ChIP assays were conducted as previously described (11). 4 × 107 cells were crosslinked with formaldehyde (1%) for 15 min at room temperature. Cross-linked chromatin was purified and sonicated to generate an average DNA size of 600 bp. The crosslinked, sonicated chromatin (30 µg) was used for immunoprecipitations with the indicated antibody. The company and catalog number for each antibody used are listed in Supplemental Table 2. DNA was extracted by phenol-chloroform and ethanol precipitated after reversing the crosslinks. One tenth of immunoprecipitated sample was employed for quantitative real-time PCR using SYBR green incorporation for the CIITA sequence of interest. A standard curve using sonicated murine genomic DNA was used to determine the relative amount of DNA in each sample. The average of at least three experiments was normalized to an irrelevant antibody control (TCR) for each amplicon and plotted as fold over the indicated control sample.
BAC modification
The protocol for BAC modification followed that described previously (40, 41). The BAC encoding CIITA (RP23-240H17) was purchased from Children’s Hospital Oakland Research Institute (CHORI, Oakland, CA). The shuttle vector (pLD53SCA-E-B), which was used for introduction of BAC modification, was obtained from Dr. N. Heniz (Rockefeller University). The shuttle vector has an R6Kγ origin, RecA gene, SacB gene, and an ampicillin resistant gene. For deleting HSS1, the CIITA sequences between chr 16: 10,475,871 - 10,476,371 (UCSC genome Bioinformatics) and chr16: 10,477,371 - 10,477,870 were cloned into the AscI and PacI restriction sites of the shuttle vector. Similarly, for deleting pIII, chr 16: 10,487,673 - 10,488,074 and 10,488,317 - 10,488,691 were cloned into the same sites. These sequences represent the homologous targeting arms surrounding the sequences to be deleted. To generate homologous recombination events, BAC containing host bacteria were made competent and transformed with the shuttle vector by electroporation using Bio-Rad Gene Pulser. The cells are selected and grown in 1 ml of LB medium containing ampicilin (50 µg/ml) and chloramphenicol (15 µg/ml). The culture was diluted 1:1000 and incubated for 16 h at 37°C. The culture was again diluted 1:5000 and incubated for an additional 8 h at 37°C. Cells were diluted and spread onto LB plates containing the above antibiotics. Colonies were analyzed for integration of the targeting shuttle vector into the BAC by PCR. To resolve the recombinants (i.e., remove the shuttle vector DNA), co-integrated BAC containing bacteria were inoculated into Luria broth cultures supplemented with chloramphenicol (15 µg/ml) for 1 h at 37°C and plated on plates containing chloramphenicol and 5% sucrose. Colonies were screened by PCR for the resolved BAC and extensive restriction enzyme digestion was carried out to ensure that the recombination and resolution processes affected only the region mutated.
RNAi and Flow Cytometry
A series of lentivirus PU.1 shRNA constructs was purchased from Open Biosystem, Inc. (catalog number; RMM4534, Clone ID; TRCN0000009497, TRCN0000009498, TRCN0000009499, TRCN0000009500, TRCN0000009501). These lentivus constructs are based on the base vector pLKO.1.1, allowing for selection by puromycin. HEK 293FT packaging cells were used as the host for transfection of the lentivirus constructs with the pseudo envelope protein VSV-G using FuGENE 6 cationic liposomes (Roche Ltd.). After 48 h and 72 h post transfection, virus was harvested and used to infect murine B cell line BCL1 in the presence of polybrene (0.8 µg/ml). The efficiency of silencing of PU.1 was assessed by western blot as indicated.
Lentivirus infected BCL1 B cells were grown in the presence of puromycin (1 mg/ml) for 5 days. 1 × 106 cells were collected, washed with PBS containing 0.1% BSA and stained with phycoerytherin conjugated IA/E antibody for 30 min on ice. Samples were analyzed on a FACScalibur and the data analyzed by CellQuest and Flowjo 8.8.6 software.
Chromatin conformation capture (3C) assays
The 3C assays were performed essentially as described (13, 42). Briefly, 1X107 cells were crosslinked with 2% formaldehyde for 10 min. The reaction was quenched with glycine (0.125 M). Cells were lysed in 10 mM Tris pH8, 10 mM NaCl, and 0.2 % NP-40 followed by 15 strokes using a dounce homogenizer. The resulting nuclei were washed in NEB2 restriction enzyme buffer, resuspended in the same buffer containing SDS (0.3 %), and incubated for 1 h at 37°C. To sequester SDS, 2 % Triton X-100 was added, and incubated for 1 h at 37°C. 400 U HindIII was added and incubated overnight at 37°C. HindIII was inactivated with SDS (1.6 %) and incubating for 25 min at 68°C. The samples were diluted 40 fold in ligation buffer (30 mM Tris, 10 mM MgCl, 1% triton X-100, and 0.1 M ATP) and ligated with 200 U T4 DNA ligase overnight at 16°C. The crosslinks within 3C library products were reversed as per assay protocol and the DNA purified. Quantitative real-time PCR using a standard curve was conducted to measure the frequency of the 3C products within each sample. Standard curves for 3C assays were generated using a CIITA encoding BAC (RP23-240H17) that was HindIII digested and then religated to generate all possible 3C products within the locus. A 3C frequency was determined by averaging the amount of 3C product produced for a given amplicon (from three independent experiments) and dividing that value by the amount of 3C product determined for an irrelevant restriction fragment that does not interact. In this case, the pIII-H1 3C product served as the background control.
Results
A conserved sequence located −11 kb upstream of pIII is hypersensitive to DNase I
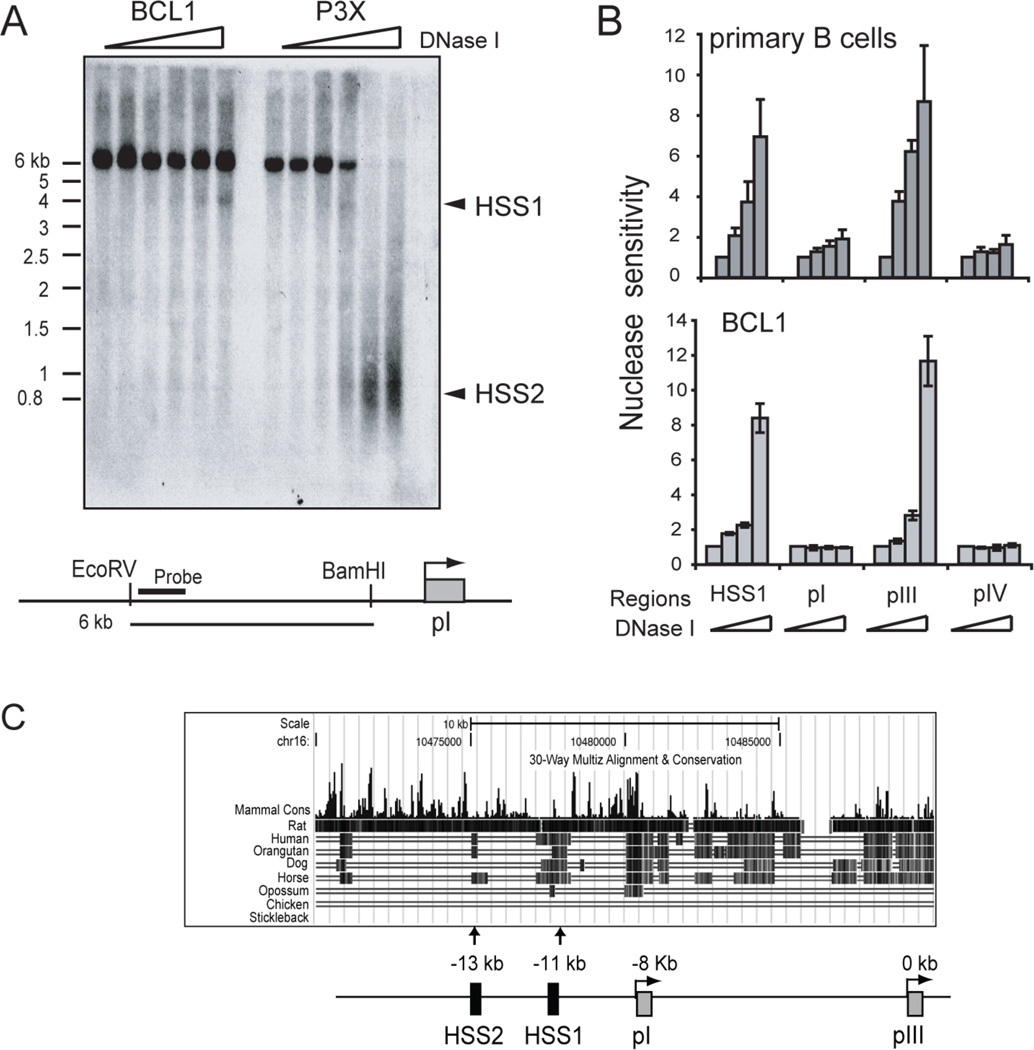
To date, only the promoter proximal sequences (within 350 bp) of CIITA pIII have been shown to regulate B-cell specific CIITA expression. To identify additional regions, a series of general assays were conducted on the sequences upstream of pIII. The sensitivity of chromatin to DNase I has been found to be associated with regulatory regions (43). Thus, as a first approach DNase I hypersensitivity assays were employed on the murine B cell line BCL1 and the plasma cell line P3X, which represent CIITA expressing and silenced cell lines, respectively. In BCL1 cells, a hypersensitive site appeared at ~ 11 kb upstream of pIII (Figure 1A). A strong DNase I hypersensitive site was also observed at −13 kb from pIII, but only in the CIITA negative P3X plasma cell line (Figure 1A). Thus, this region appears to be differentially accessible to DNase I, suggesting that it has some regulatory function. The −11 kb site specific to B cells was termed hypersensitive site 1 or HSS1 and the −13 kb site HSS2.
Figure 1. B cell and plasma cell specific DNase I hypersensitive sites are located in conserved regions upstream of CIITA promoter I.
A. Southern blot DNase I hypersensitivity assay performed on the murine B-cell line BCL1 and plasma cell line, P3X63ag8.653 (referred to as P3X). Triangles denote increasing DNase I concentration. A schematic of the region analyzed and the probe location is shown. B. PCR based DNase I hypersensitivity assay on purified murine splenic B cells and BCL1 cells. DNase I treated and control samples were subjected to real-time PCR amplification over the designated regions. The amount of PCR product for each sample was divided into the undigested sample and the average of three reactions was plotted. C. The regions corresponding to HSS1 and HSS2 were analyzed for sequence conservation using the University of California Santa Cruz Genome Browser (38). The screen shot from that analysis is shown with an annotated schematic.
A PCR-based DNase I hypersensitivity assay was used to assess HSS1 in primary murine splenic B cells (Figure 1B). Here, HSS1 was sensitive to increasing concentrations of DNase I in both primary murine B cells and BCL1 cells (Figure 1B). Promoter III also displayed strong DNase I hypersensitivy in the primary cells. Promoters I and IV were not sensitive to DNase I in these cells, thereby providing specificity controls. A region located −14 kb from pIII was used as a control and showed no changes in DNase I hypersensitivity in BCL1 or primary B cells (data not shown).
Cross species genomic DNA comparisons using the University of California Santa Cruz Genome Browser tools, found that HSS1 was highly conserved while HSS2 showed only low levels of sequence conservation (Figure 1C). No microRNA sequences or other coding/non-coding sequence features were found within these regions. In human HeLa cells, sequences within HSS1 region were recently shown to participate in IFN-γ induction through pIV (36), suggesting that this region may function as a complex regulatory component and may contribute to B-cell specific expression of CIITA. Thus, its characterization and function were investigated further.
Chromatin modifications at HSS1 reveal an accessible and active nucleosome structure
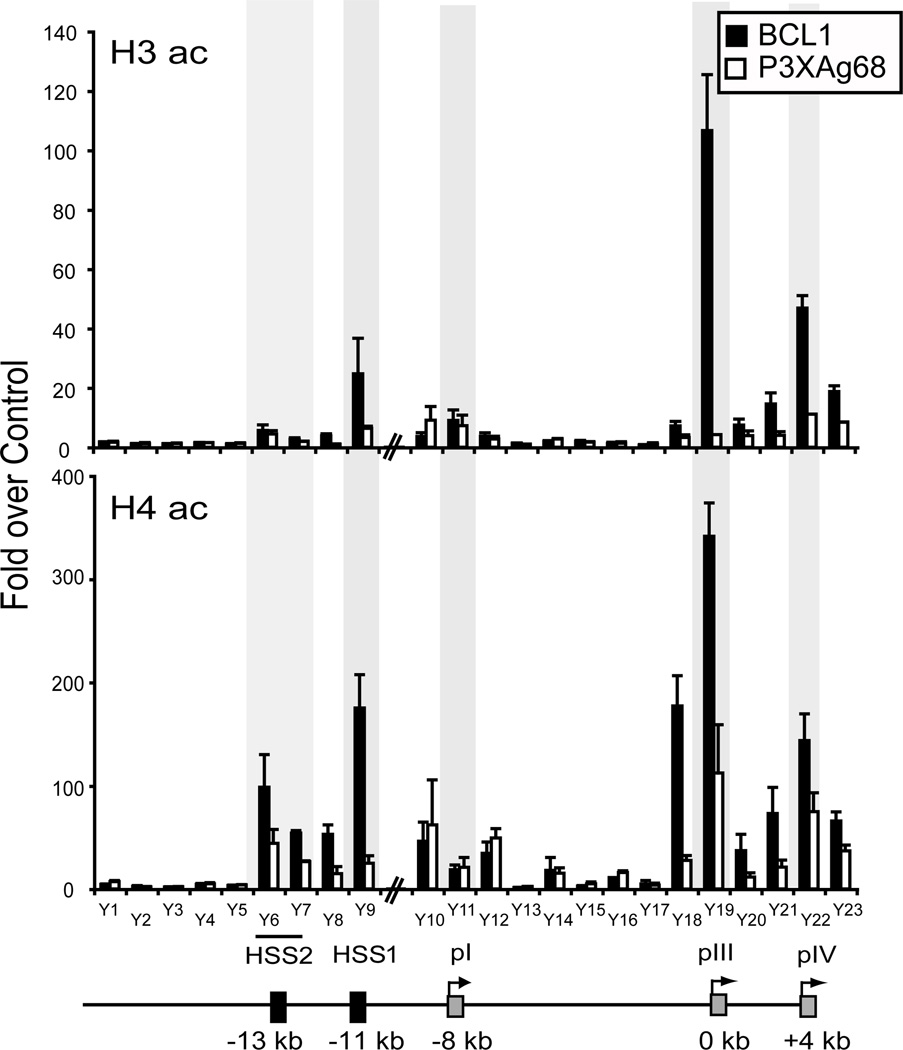
To further characterize this element and to determine if other potential regulatory regions were present within this upstream segment of the CIITA gene, a series of chromatin immunoprecipitations (ChIP) assays using antibodies to the acetylated forms of histones H3 and H4 were conducted. These modifications are typically associated with regulatory regions and expressed genes (44). Here, we sought to find regions that were differentially modified between B cells (BCL1) and plasma cells (P3X) as such regions could indicate a putative role in B-cell meditated expression. Twenty-three amplicons spaced at ~1 kb intervals, spanning the region between pIV and −18 kb upstream of pIII were used to analyze precipitated chromatin. For acetylated histone H3 ChIP, the regions corresponding to pIII and pIV showed high levels of this histone modification in BCL1 B cells but not in the P3X plasma cell line (Figure 2). This result was similar to that observed for human B and plasma cell lines (15). Promoter I displayed low H3 acetylation levels that were not differentially modulated between the two cell lines. HSS1 displayed moderate levels of acetylated histone H3 in the B cell line and low levels in the plasma cell line, suggesting that histone H3 acetylation of this region was B-cell specific. Promoter III and pIV showed high levels of histone H4 acetylation in BCL1 cells but not in P3X cells. As with histone H3 acetylation, pI showed low levels of histone H4 acetylation that did not change between the B and plasma cell lines. In agreement with the histone H3 ChIP result, HSS1 also showed high levels of H4 acetylation in B cells and low levels in plasma cells. The sequences extending upstream of HSS1 to −13 kb (HSS2), also showed high histone H4 acetylation levels; although the differences between B and plasma cells was not more than 2 fold.
Figure 2. HSS1 and CIITA promoters display reduced histone acetylation in plasma cells compared to B cells.
Chromatin prepared from the murine B-cell line BCL1 and plasma-cell line P3X were subjected to ChIP analysis using antibodies to the general acetylated (ac) forms of histone H3 and H4. A non-specific control antibody to the TCR was used to normalize the values for each amplicon. The position of the twenty-three amplicons used are schematically represented over the CIITA locus encompassing pIV through −18 kb upstream of pIII, which was designated as the origin (0 kb). All ChIP assays were performed three times from independent chromatin preparations. Real-time PCR was used with a standard curve generated for each amplicon to determine values for each of the ChIP reactions. These values were averaged for the three experiments, normalized to the amount of chromatin in each set of reactions prior to ChIP, and plotted with respect to the values obtained with the TCR antibody as fold over control. The shaded sections highlight ChIP amplicons for HSS2, HSS1, pI, pIII, and pIV.
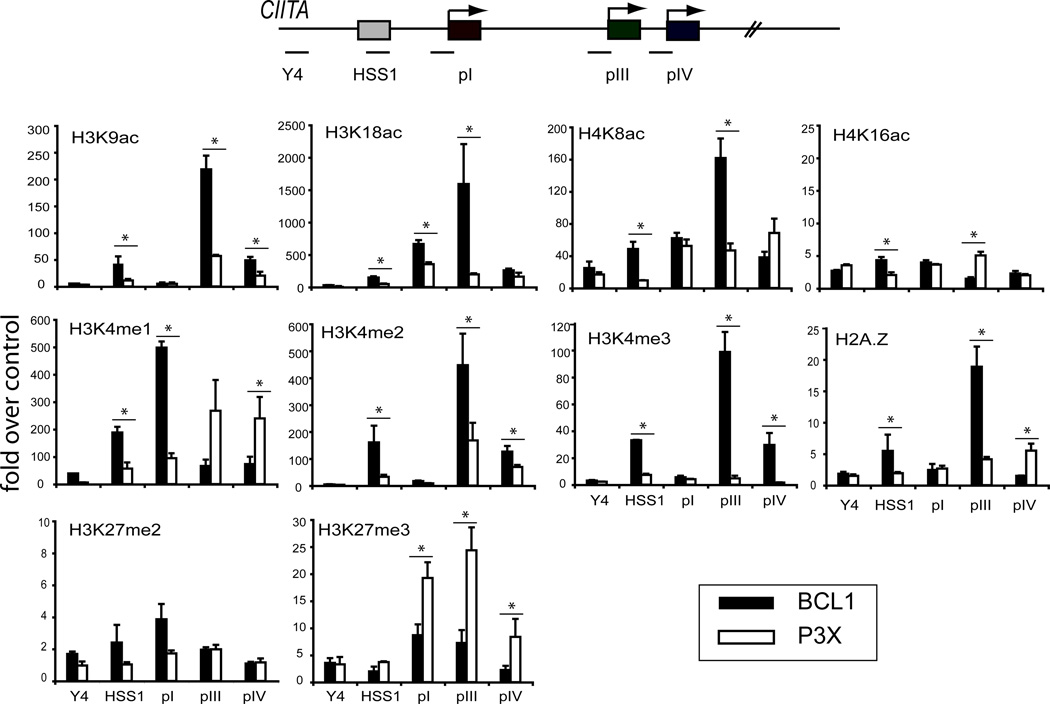
Analysis of activation/silencing associated chromatin modifications for select regions can provide a fundamental assessment of the general chromatin architecture of a region (accessibility), as well as the general gene expression activity of a region (active/inactive) (45). Additional ChIP assays were conducted on HSS1 and comparisons were made between the three promoter regions and a control region, Y4 (Figure 3). Y4 (−16 kb from pIII) represents a region with no known regulatory or promoter function. Four histone acetylation modifications were assessed: H3K9, H3K18, H4K8, and H4K16 (Figure 3). Nucleosomes at pIII were highly acetylated at histone H3K9, H3K18, and H4K8. Each of these marks was differential between BCL1 and P3X cells and therefore associated with active transcription of CIITA. Although not at the level of pIII, HSS1 showed differential levels (BCL1 vs. P3X cells) for these three modifications as well. Promoter I showed acetylation of H3K18 and H4K8; whereas pIV displayed H3K9, H3K18, and H4K8 acetylation modifications. However, pI and pIV modifications were not highly differential between the different cell types, suggesting that they may represent a general feature of the region in the lymphocyte lineage. None of the regions displayed histone H4 K16 acetylation levels that were greater than the control region Y4.
Figure 3. HSS1 and the CIITA promoters display differential histone modifications.
The modifications of histones at regulatory regions and promoters are indicative of the chromatin architecture and transcriptional state. ChIP for the indicated histone modifications were performed on BCL1 and P3X cells as above. The relative location of the five amplicons used in this series is shown in the schematic. Y4 represents an amplicon located at −16 kb from pIII and serves as a non-regulatory region control for comparison across the locus. ChIP assays were performed three times from independent chromatin preparations as above. Asterisks indicate B and plasma differences that were significant (p<0.05) as determined by student T tests. ac, acetylation; me, methylation (mono, di, tri).
The degree of methylation of H3 K4 is associated with gene transcription or poised RNA polymerase binding (47, 48). Histone H3 K4 monomethylation has been associated with enhancer activity and dimethylation with gene expression and accessibility; whereas trimethylation of this residue is associated with RNA polymerase binding and active transcription (46–48). All three configurations of histone H3 lysine methylation were observed at HSS1 in B cells and were reduced in plasma cells (Figure 3). Intriguingly, pI also displayed strong H3K4me1 at this residue in B cells; whereas this modification appeared at high levels at pIII and pIV in plasma cells. In contrast, H3K4me2 and K4me3 levels showed a direct relationship with CIITA expression. Promoter III showed the highest levels of these marks as anticipated as this promoter is predominantly used in B cells. Promoter IV also showed these two active marks, as some transcription from pIV is observed in B cells. Promoter I did not display H3K4me2 or K4me3 modifications, suggesting that this promoter region is not transcriptionally active in BCL1 cells. RT-PCR for pI in BCL1 cells showed no detectable transcripts (data not shown). The histone variant H2A.Z, which is often associated with active promoter regions (49), was observed at the highest levels in BCL1 cells at pIII and HSS1 and at lower levels at the other sites.
Repressive chromatin modifications H3K27me2 and K27me3, which are associated with the Polycomb repressor complex, were examined (Figure 3). Low levels of H3K27me2 were associated with pI and possibly with HSS1 in BCL1 cells. By contrast, P3X plasma cells showed high levels of H3K27me3 associated with all three promoter regions but not HSS1. Considering the high degree of DNase I hypersensitivity in the region in plasma cells it is likely that this region remains accessible. Thus, the three promoters display histone code patterns that are mostly associated with gene expression, and in general, the histone modification patterns of HSS1 also follow the transcriptional state of the locus in B cells.
HSS1 is required for CIITA expression
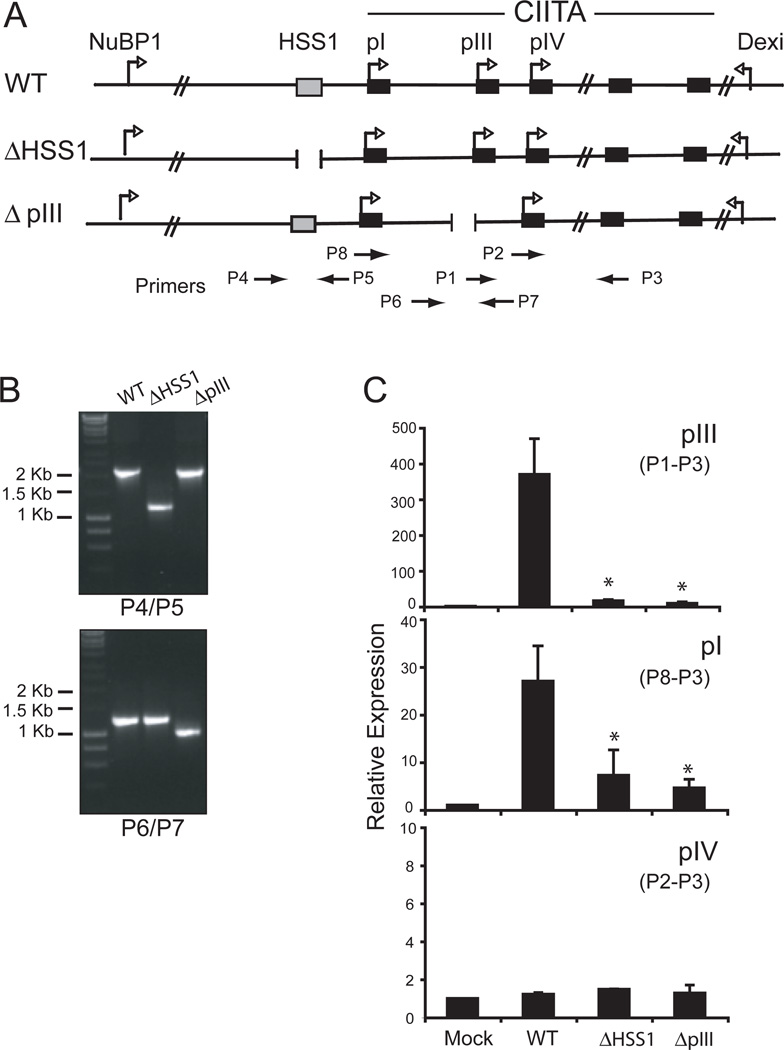
The above data suggest the possibility that HSS1 may function in the regulation of CIITA in B cells. To examine the regulatory potential of HSS1, we employed a bacterial artificial chromosome (BAC) modification system (40, 41) that would allow the introduction of a mutation into the CIITA gene and test its effect on controlling CIITA expression in the context of the locus. Thus, using the BAC system, the effect of deleting HSS1 on the expression of CIITA from each of the promoters could be assessed. The BAC (RP23-240H17) encodes the murine CIITA locus with 88,771 bp upstream of pIII and 51,386 bp downstream of CIITA’s last exon (Figure 4A). To modify the BAC, two homologous recombination-targeting shuttle vectors were created. These vectors were designed to generate a 1 kb and 500 bp deletion of HSS1 and pIII, and were termed ΔHSS1 and ΔpIII, respectively (Figure 4A). The pIII deletion includes both the transcription start site and all promoter proximal B-cell specific regulatory motifs that were characterized previously to control expression from pIII (8, 50). The targeting vectors were used to transform E. coli containing the CIITA BAC and co-integrations were screened and isolated, followed by resolution of the targeting vector sequences from the BAC DNA. The mutated BACs were assessed for integrity by restriction enzyme digestion (data not shown) and by PCR (Figure 4B). The modified BACs contained only the intended deletions.
Figure 4. HSS1 is required for B cell expression of CIITA from pIII.
A. A schematic representation of the murine CIITA encoding BAC and the surrounding loci is shown with the indicated deletion mutations that were created in HSS1 (ΔHSS1) and pIII (ΔpIII). Primers used to assess the integrity of the BAC constructs are shown. B. PCR across the regions that were deleted in each of the mutant CIITA BAC constructs using the indicated primer pairs showed specificity of the intended deletions. C. Wild-type and mutant BACs were transfected into the CIITA-deficient human B-cell line RJ2.2.5 by nucleoporation. Forty-eight h post transfection, the cells were harvested, and the RNA analyzed by real-time RT-PCR using primers specific for each of the CIITA promoter regions as shown in A. Three independent transfections were performed and the results were averaged with respect to the levels of 18s rRNA and plotted as relative expression with standard error. Student T tests showed that the differences between wt and ΔHSS1 and ΔpIII were highly significant (*) for promoters I and III (p<0.04). No differences were observed at pIV.
To assess the expression of CIITA from the BAC DNA, wild-type and modified CIITA BACs were transfected into the human B-cell line RJ2.2.5. RJ2.2.5, originally derived from Raji cells contains heterozygous deletions for CIITA and is MHC-II negative (4, 51, 52). Following transfection of wild-type and mutant (ΔHSS1 and ΔpIII) CIITA BAC DNAs into RJ2.2.5 cells, RNA was purified and CIITA promoter I, III, and IV specific mRNA levels were assessed by real-time RT-PCR. The wild-type BAC expressed CIITA from pIII and pI at levels of ~370-fold and 27-fold over the mock transfected cells, respectively (Fig 4C). The ΔHSS1 mutation resulted in nearly a complete loss of pIII expression, suggesting that HSS1 element is critical to pIII-mediated expression. The ΔHSS1 mutation had a similar influence on pI. As anticipated, the ΔpIII mutation caused a complete loss of pIII directed CIITA expression. Surprisingly, the ΔpIII mutation also affected expression from pI. This result suggests that the B-cell specific elements contained within the ΔpIII deletion were responsible for expression from pI. In contrast, no expression was detected from pIV irrespective of the CIITA BAC constructs assayed. Low levels of both pI and pIV CIITA mRNA isoforms can be detected from Raji cells but these levels are 43–64 fold less than the pIII isoform (data not shown). Thus, with the exception of pIV, the BAC mimics to a large extent the properties of the CIITA locus, and HSS1 plays a critical role in expression from pIII.
PU.1 binds to HSS1 and CIITA pIII
To predict the identity of factors that may bind HSS1 in B cells, the DNA sequence of HSS1 was analyzed using the Genomatix software program. The analysis found that HSS1 had two highly significant potential PU.1 binding sites (GGAA) located at chr16: 10,476,423 and 10,476,550 using the positions on the University of California Santa Cruz (UCSC) genome bioinformatic (1.0 core similarity score). Other putative sites, including those for CREB and PLZF were also identified but their similarity scores were lower (0.89 and 0.86, respectively). Because PU.1 is a transcription factor that binds to lymphocyte specific enhancer regions (33, 34) and is responsible for B cell and myeloid cell fate decisions (30, 53) its role was examined further. To determine the breadth of PU.1 binding in vivo, ChIP assays were conducted across the locus as above, using chromatin prepared from BCL1 and P3X cells (Figure 5A). The result showed that PU.1 bound to HSS1 in BCL1 cells (Figure 5A). Robust PU.1 binding was also detected at pIII in BCL1 cells, which was in agreement with previous studies that showed PU.1 binding to pIII (15). Low levels of PU.1 binding were detected in BCL1 cells at pI and at a region located 2 kb upstream of pIII. No binding of PU.1 at any site was detected in P3X plasma cells, suggesting that the recruitment of this factor to at least two and possibly four sites may be important for the B cell specific expression of CIITA.
Figure 5. PU.1 binds to HSS1 and CIITA pIII.
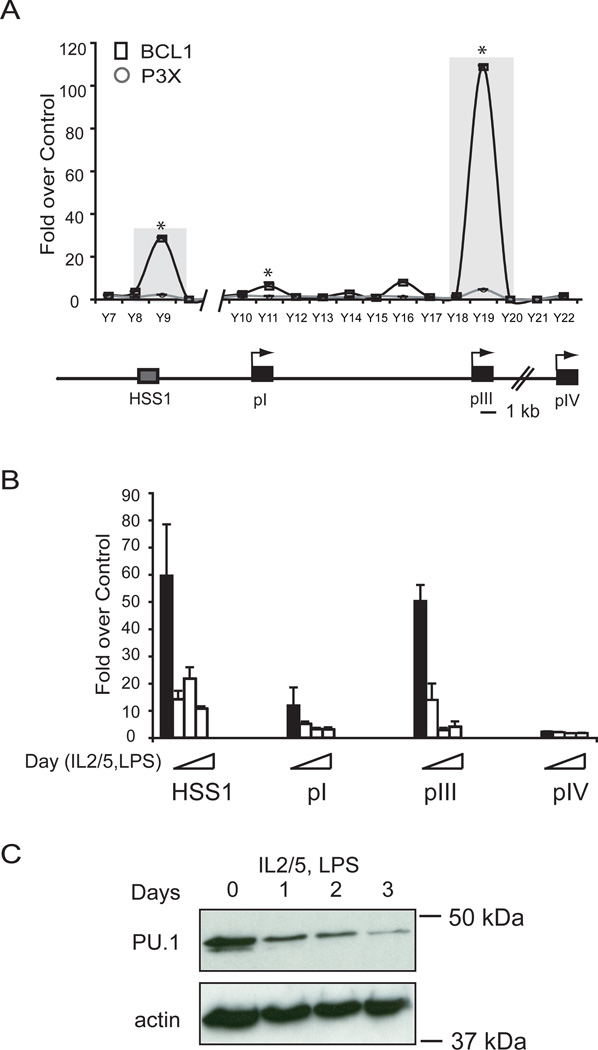
A. ChIP assays were conducted to assess the binding of PU.1 to HSS1 and other sites across the locus using chromatin prepared from BCL1 and P3X cells. The binding of PU.1 was analyzed on the CIITA gene using primers that represent sequences spaced at intervals of ~1 kb between −11 kb through +4 kb relative to CIITA promoter III. B. Purified splenic B cells were used for PU.1 ChIP assays either immediately (black bars) or ex vivo differentiated in LPS, IL-2, and IL-5 for 1, 3, or 5 days (gray bars) as represented by the slope of the triangle. The regions analyzed by ChIP are stated. In all of these assays three independent replicates were carried out, averaged and plotted vs the non-specific control with standard error. All real-time PCR values were determined as above by comparison to a standard curve for the amplicon generated from genomic DNA. Student T tests were used to determine the significance of the differences from undifferentiated cells. HSS1, pI, and pIII showed statistical significance of p<0.05 between untreated and all days post treatment. C. Western blot of murine splenic B cells (day 0) ex vivo differentiated to plasma cells (days 1, 2, and 3) that was stained for PU.1 and actin.
To examine the dynamics of PU.1 occupancy at HSS1 during the B cell to plasma cell transition, chromatin preparations from primary murine B cells ex vivo stimulated with IL-2, IL-5, and LPS to induce plasma cell differentiation (15) from day 0 through day 5 were examined. This treatment results in the silencing of CIITA expression (15). Similar, robust levels of PU.1 binding were observed at HSS1 and pIII in the untreated freshly isolated murine B cells. PU.1 occupancy was lost rapidly during the differentiation process (Figure 5B). This correlated with the loss of PU.1 protein that occurs during the differentiation process (Figure 5C).
PU.1 is required for CIITA expression in BCL1 B cells
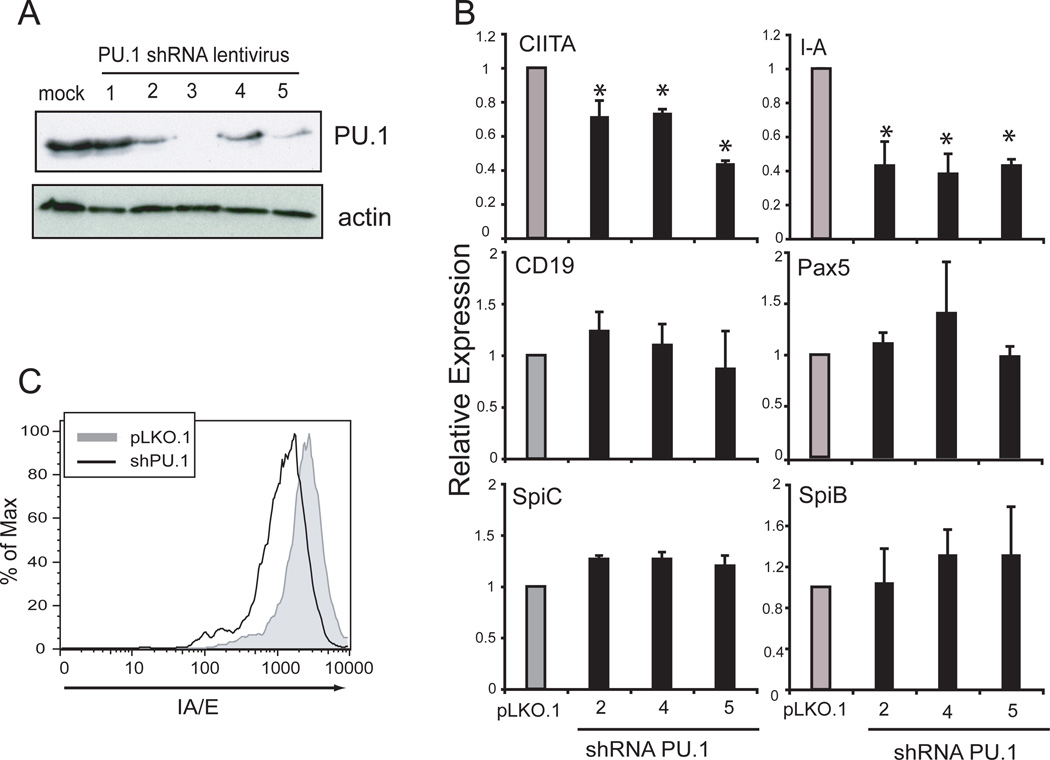
The ChIP results indicate that PU.1 binds directly to HSS1 and pIII and may regulate CIITA expression. To determine the role of PU.1 for CIITA regulation in B cells, the expression of PU.1 was depleted from BCL1 B cells using RNAi. A set of five lentivirus vectors expressing different shRNAs specific to PU.1 and the selectable marker puromycin were produced and used to infect BCL1 cells. Infected cells were selected with puromycin for 5 days. PU.1 shRNA vectors 2, 3, 4, and 5 reduced PU.1 protein to varying degrees (Figure 6A). Although vector 3 had the most pronounced effect on PU.1 expression levels, cell viability with this vector was an issue and therefore it was not used further. To assess CIITA levels after PU.1 knockdown from B cells, total RNA was purified and real time RT-PCR was carried out to determine the level of CIITA and other B cell specific gene mRNAs. PU.1 knockdown using shRNA vectors 2, 4, and 5 resulted in a significant reduction of CIITA and I-A expression (Figure 6B) despite the fact that none of the vectors were able to completely ablate PU.1 expression in the culture. Other B cell specific genes, including Pax5, Spi-C, Spi-B, and CD19 were not altered in their expression by any of the shRNAs (Figure 6B). A reduction in MHC-II surface expression was also observed following infection with the lentivirus containing shRNA5 (Figure 6C). These data suggest that PU.1 plays an essential role in CIITA expression in B cells.
Figure 6. PU.1 is required for CIITA expression in BCL1 cells.
A. Five different shRNA lentiviral expression vectors were used to knockdown the expression of PU.1 in BCL1 B cells. Infected cells were selected using puromycin for five days. Lysates prepared from the resistant cells were assayed by immunoblotting for PU.1 and actin protein. B. RNA from PU.1-shRNA-depleted BCL1 cells was purified and analyzed by real-time RT-PCR for the levels of the indicated transcripts. These experiments were performed three times from independent shRNA infections. The results of each experiment were averaged and plotted with respect to the parent lentiviral construct (pLKO.1) infected cells. Significance was determined using a student T test and comparisons between control and PU.1 shRNA with p values of <0.05 are indicated with an asterisk. C. Flow cytometry was used to assess the level of MHC-II surface expression on pLKO.1 and shPU.1 vector 5 infected BCL1 B cells.
Long distance interactions between HSS1 and CIITA form in B cells
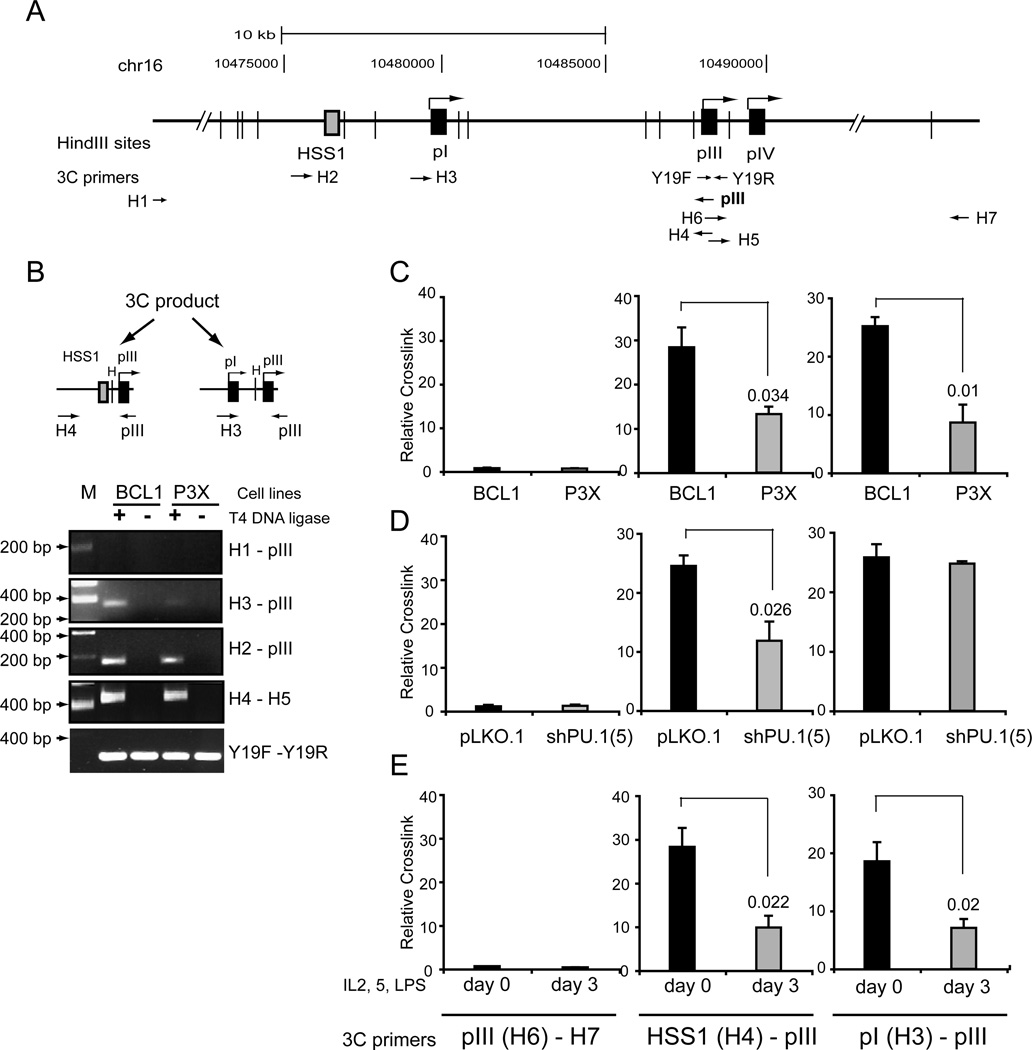
One way in which HSS1 may contribute to the regulation of CIITA in B cells is for the region to interact directly with the pIII regulatory region. To determine if HSS1 can interact with pIII, the chromatin conformation capture assay (3C) was employed (54). In 3C assays, chromatin is crosslinked with formaldehyde to fix interactions within the cells. Purified chromatin is then digested to completion with a restriction enzyme, such as HindIII. The samples are diluted greatly such that non-crosslinked DNAs are no longer spatially associated, and the crosslinked DNAs are ligated. Following reversal of the crosslinks and DNA purification, the formation of these novel “3C” junctions are evaluated by PCR. A map of the CIITA locus with primer positions and HindIII sites used is shown (Figure 7A). In these experiments, the pIII encoding restriction fragment was used as the anchor to test for interactions between it and other sequences. A 3C product was observed between HSS1 (primer H4) and pIII (Figure 7B and 7C), indicating that an interaction between these DNAs occurs. This 3C product was reduced by 53% in its frequency in P3X cells (Figure 7B and C). No interactions between pIII and Primer H1 (Figure 7B) or H7 (Figure 7C), which represent regions upstream of HSS1 and downstream of pIII, respectively were observed. In BCL1 cells, a 3C product was also observed between pI and pIII (Figure 7B and C). The pI/pIII 3C product was reduced by 65% in P3X cells, suggesting that it is B-cell specific. All 3C interactions were found to require formaldehyde crosslinking (data not shown) and the presence of DNA ligase (Figure 7B). Equal loading of samples was controlled by the internal amplification of a single restriction fragment (Figure 7B, primers H1 and H2). All restriction sites assessed were equally accessible to restriction enzymes irrespective of cell type or conditions of the assay (Supplemental Data Figure 1). Additionally, the ability of a digested fragment to self ligate and form a 3C product was also evaluated (Figure 7B, primers H4-H5) and demonstrated that these sites were equally accessible between B cells and plasma cells.
Figure 7. Long distance interaction was found between HSS1 and CIITA pIII.
A. A schematic of CIITA region analyzed by 3C assays is shown with HindIII sites, 3C primers, and relative position of HSS1 and the three CIITA promoters is shown. B. Schematic representation of 3C products for pIII interacting with HSS1 (primer H2) and pI (primer H3) are shown. These 3C products were cloned and sequenced to verify their location. Agarose gel electrophoresis of 3C reactions was carried out with the indicated primer pairs using BCL1 and P3X cells. C. Real-time PCR analysis of 3C products formed between pIII and the H7 downstream region, HSS1, and pI in BCL1 and P3X cells. A relative crosslink frequency was determined by calculating the amount of 3C product between pIII and the experimental restriction fragment and dividing it by the amount of 3C product formed for pIII and H1 (an irrelevant upstream restriction fragment (see Fig 7B)). The average of three independent experiments is shown with standard error and a student T test evaluation of the significance. No significant interactions were observed between pIII and 3C primer H7 were observed. Relative 3C crosslink frequency for pIII interactions in (D) BCL1 cells infected with pLKO.1 and PU.1 shRNA lentiviral vector 5; and (E) undifferentiated and day 3 ex vivo differentiated murine splenic B cells are presented as in C.
To determine the role of PU.1 in the ability of pIII to interact with HSS1 and pI, 3C assays were performed on BCL1 cells infected with the PU.1 shRNA lentivirus vector 5. Compared to pLKO.1 lentivirus infected cells, the shPU.1 lentivirus showed a 52% reduction in the 3C product formed between HSS1 and pIII. Intriguingly, interactions between pIII and pI were unchanged by PU.1 knockdown. No interaction was observed with the control region (primer H7-H6). Another way to deplete PU.1 from the system is to ex vivo differentiate primary B cells to plasma cells with LPS, IL-2, and IL-5 as above. In concordance with the loss of PU.1 occupancy at HSS1 and pIII, ex vivo differentiation resulted in a 65% decrease in the frequency of 3C product formation between HSS1 and pIII and a 62% loss of the interaction between pI and pIII. These results suggest that PU.1 plays a role in HSS1 interacting with pIII but that interactions between pI and pIII are dependent on another set of factors.
Discussion
In the mouse, CIITA expression is regulated from three promoter regions, with each region initiating the transcription of a unique isoform of CIITA (8). The three promoters are regulated in a cell-type dependent manner, with pIII functioning in lymphoid cells. In B cells, pIII is expressed constitutively but is silenced as B cells transition to plasma cells. Several groups have shown that the proximal region 5’ to the pIII transcription start site encodes a complex set of binding sites that is critical to B cell specific expression (20, 50). However, no other regions associated with pIII expression in B cells have been described. Using a number of tools, we identified a region located upstream of pIII and pI that is critical for B cell specific expression. This region, termed HSS1 is highly conserved, hypersensitive to DNase I treatment, and displayed chromatin marks that were associated with an active and accessible locus. A key parameter showing that HSS1 was critical to B-cell specific expression of CIITA was the deletion of this element from a BAC construct. Due to the multiple promoters that control CIITA, this experiment provided an “in context” evaluation of the role of HSS1 with respect to all the CIITA promoters. While pIII was robustly expressed in the BAC system, low levels of pI but no levels of pIV specific CIITA expression were observed. The pI transcripts were affected by the presence of HSS1 and pIII, suggesting that transcription from pI was likely controlled by factors bound at these elements. Analysis of CIITA transcript utilization from the human B cell line Raji, which is the parent of RJ2.2.5 showed that pI and pIV produced minimal levels of steady-state transcripts compared to pIII. In contrast, no pI or pIV transcripts could be detected from the murine BCL1 cells (data not shown). Thus, there is some variation between the human and murine systems or cell lines. With some expression from pI, this CIITA BAC system may more closely model the host rather than its DNA of origin.
DNase I hypersensitivity Southern blots also identified a robust hypersensitive site that was present in the plasma cell line P3X but not in B cells. Cross species conservation of HSS2 was not high, and HSS2 displayed significant levels of histone H4 acetylation but oddly near background levels of histone H3 acetylation. Intriguingly, the region just upstream of HSS2 showed no histone modifications associated with an active locus. While such demarcations can sometimes suggest that the element functions as a boundary element, no binding of the insulator factor CTCF to HSS2 was found (data not shown). Thus, at this time the function of HSS2 is not known.
In a previous study (15), we found that chromatin histone modifications that were in general associated with active gene expression were lost at the human CIITA proximal promoter regions when human B cell lines were compared to plasma cell lines. This suggested that there were global changes that controlled chromatin accessibility and the overall architecture of the locus. Here similar findings were observed for the murine B cell line BCL1 and plasma cell line P3X. Intriguingly, in the plasma cell line P3X, chromatin marks associated with Polycomb gene silencing (45, 55) were observed at all three CIITA promoters, suggesting that there is an active mechanism that results in the recruitment of Polycomb factors to silence CIITA expression. The polycomb H3K27me3 methyltransferase EZH2 was found to play a role in regulating pIV in uveal melanoma cells (18). Preliminary data showed that EZH2 binds to pIII in plasma cells (unpublished data).
Genome wide studies have helped identify the global locations to various histone marks (46, 47). Histone H3 K4 methylation marks are mostly indicative of the gene expression state of a locus. H3K4me3 is most often associated with promoter regions of actively transcribed genes (48). For CIITA in B cells histone H3K4me3 was most associated with pIII, but was also found at lower levels at pIV and HSS1. Only background levels of histone H3K4me3 were observed in plasma cells, and thus, this mark is clearly associated with CIITA expression. Histone H3K4me2 is more broadly associated with gene expression and can be found covering large segments of the promoter and upstream regulatory regions of genes (48) and perhaps is a mark of accessibility. H3K4me2 was lower in plasma cells, suggesting that the region may be less accessible; perhaps due to silencing marks associated with Polycomb. While histone H3K4me1 was reported to be associated with enhancer regions (47), the patterns for CIITA promoters and HSS1 correlated with transcription initiation in that pIII and pIV showed high levels of this modification in plasma cells; whereas the opposite was observed for HSS1 and pI in B cells. The finding of multiple modifications within a region may reflect the number of nucleosomes assessed in the ChIP assays, as well as the dynamic nature of the marks that is likely to occur.
For B cells, CIITA expression and the interaction between HSS1 and pIII was in part dependent on PU.1. Depletion of PU.1 by RNAi or through the use of plasma cells or differentiation of B cells to plasma cells, showed significant reduction in CIITA mRNA levels and in HSS1 interactions with pIII. The inability to completely deplete PU.1 levels in the RNAi and ex vivo differentiation systems are likely to underplay PU.1’s role in transcription of CIITA. Conservatively, we conclude that PU.1 is necessary for full activity of HSS1 and B-cell specific expression of CIITA, but that other factors are likely to be involved. The finding of PU.1 at both HSS1 and pIII was intriguing and may suggest a role for PU.1 to help bridge these regions together as its reduction by shRNA resulted in a decrease in the 3C product. Whether PU.1 interacts with other DNA binding proteins or whether it interacts with a set of coactivators to stabilize the interactions between pIII and HSS1 can only be speculation at this point.
Recently, Ni et al. identified a series of sequences within the CIITA locus of human HeLa cells that bound STAT1 and IRF1 in response to IFN-γ signaling. Some of these elements were required for the IFN-γ expression through pIV, the CIITA promoter responsible for IFN-γ expression in non-hematopoietic cells (36). A role for the IFN-γ elements in B cells was not determined. We found that HSS1 is the murine homolog to the human region termed −16 (36) as referenced from the pIV transcriptional start site. In IFN-γ treated HeLa cells, −16 interacted with pIV in a BRG1-dependent manner. BRG1 is the ATPase of the Swi/Snf chromatin-remodeling complex that is associated with the expression of many genes and likely facilitates nucleosome movement to allow the interactions to occur (56).
With the discovery that HSS1 functions in CIITA regulation through pIII in B cells and that there were multiple elements found to fully orchestrate pIV regulation by IFN-γ in HeLa cells (36), two questions arise. How many total elements will be required to regulate CIITA in a tissue specific manner and how will promoter usage be derived? Five elements were required for IFN-γ signaling through pIV in Hela cells (36). Our current findings imply that at least HSS1 responds to multiple signals, controlling both B cell and IFN-γ-inducible expression. It is possible that like HSS1, the other elements that were discovered in the IFN-γ system will specify expression from the different CIITA promoters. Alternatively, it is possible that only a limited set of these elements may function as master regulators of CIITA and that a set of promoter specific elements will also exist. HSS1 has the potential to be one such master regulatory element for CIITA. For such elements, the transcription factors that bind to the master element could determine which promoter is used. For IFN-γ treatment of HeLa cells STAT1/IRF1 are the key factors that likely target pIV transcription; whereas, in B cells, PU.1 is at the very least, a key component of HSS1 and pIII B cell regulatory mechanism. While little is known about even the proximal promoter elements for pI, it is possible that PU.1, which is also expressed in myeloid derived dentritic cells (30), also plays a role in this cell lineage for CIITA expression. Considering that HSS1 has the potential to bind PU.1, it is possible that it will also function in the regulation of pI. Nonetheless, the current data suggest that the HSS1/-16 element may function in multiple cell type contexts with its use dependent on the specific set of transcription factors that are expressed or induced.
Supplementary Material
Supplemental Figure 1. Restriction site accessibility
All restriction sites assessed for 3C were equally accessible to digestion by HindIII irrespective of the cell line or the condition used. A. A schematic of CIITA region analyzed by 3C assays is shown with Hind III sites, and primers that were used to assess HindIII accessibility. Nuclei from crosslinked cells prepared for 3C was digested with Hind III or left undigested. 30 ng of the purified DNA from the above reactions was used to perform PCR with indicated primers. Agarose gel electrophoresis used to assess each of the PCR products with (B) comparing BCL1 and P3X cells; and (C) undifferentiated and 3 day ex vivo differentiated primary splenic B cells.
Acknowledgements
We thank members of the lab and Dr. S. Speck for critical conversations and comments on this work. We thank Dr. N. Heinz for providing the BAC shuttle vector.
Abbreviations used in this paper
- 3C
chromatin conformation capture
- ChIP
chromatin immunoprecipitaiton
- CTCF
CCCTC transcription factor
- HSS1
hypersensitive site 1
- HSS2
hypersensitive site 2
- LSD-1
lysine-specific demethylase 1
- promoter I, III, IV
pI, pIII, pIV
Footnotes
This work was supported The National Institutes of Health AI34000.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Benacerraf B. Role of MHC gene products in immune regulation. Science. 1981;212:1229–1238. doi: 10.1126/science.6165083. [DOI] [PubMed] [Google Scholar]
- 2.Long EO. Antigen processing for presentation to CD4+ T cells. New Biol. 1992;4:274–282. [PubMed] [Google Scholar]
- 3.Boss JM, Jensen PE. Transcriptional regulation of the MHC class II antigen presentation pathway. Curr Opin Immunol. 2003;15:105–111. doi: 10.1016/s0952-7915(02)00015-8. [DOI] [PubMed] [Google Scholar]
- 4.Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 5.Dziembowska M, Fondaneche MC, Vedrenne J, Barbieri G, Wiszniewski W, Picard C, Cant AJ, Steimle V, Charron D, Alca-Loridan C, Fischer A, Lisowska-Grospierre B. Three novel mutations of the CIITA gene in MHC class II-deficient patients with a severe immunodeficiency. Immunogenetics. 2002;53:821–829. doi: 10.1007/s00251-001-0395-7. [DOI] [PubMed] [Google Scholar]
- 6.Pai RK, Askew D, Boom WH, Harding CV. Regulation of class II MHC expression in APCs: roles of types I, III, and IV class II transactivator. J Immunol. 2002;169:1326–1333. doi: 10.4049/jimmunol.169.3.1326. [DOI] [PubMed] [Google Scholar]
- 7.Steimle V, Siegrist CA, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 1994;265:106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 8.Muhlethaler-Mottet A, Otten LA, Steimle V, Mach B. Expression of MHC class II molecules in different cellular and functional compartments is controlled by differential usage of multiple promoters of the transactivator CIITA. EMBO J. 1997;16:2851–2860. doi: 10.1093/emboj/16.10.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waldburger JM, Suter T, Fontana A, Acha-Orbea H, Reith W. Selective abrogation of major histocompatibility complex class II expression on extrahematopoietic cells in mice lacking promoter IV of the class II transactivator gene. J Exp Med. 2001;194:393–406. doi: 10.1084/jem.194.4.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LeibundGut-Landmann S, Waldburger JM, Reis e Sousa C, Acha-Orbea H, Reith W. MHC class II expression is differentially regulated in plasmacytoid and conventional dendritic cells. Nat Immunol. 2004;5:899–908. doi: 10.1038/ni1109. [DOI] [PubMed] [Google Scholar]
- 11.Beresford GW, Boss JM. CIITA coordinates multiple histone acetylation modifications at the HLA-DRA promoter. Nat Immunol. 2001;2:652–657. doi: 10.1038/89810. [DOI] [PubMed] [Google Scholar]
- 12.Masternak K, Muhlethaler-Mottet A, Villard J, Zufferey M, Steimle V, Reith W. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 2000;14:1156–1166. [PMC free article] [PubMed] [Google Scholar]
- 13.Majumder P, Gomez JA, Chadwick BP, Boss JM. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J Exp Med. 2008;205:785–798. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silacci P, Mottet A, Steimle V, Reith W, Mach B. Developmental extinction of major histocompatibility complex class II gene expression in plasmocytes is mediated by silencing of the transactivator gene CIITA. J Exp Med. 1994;180:1329–1336. doi: 10.1084/jem.180.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green MR, Yoon H, Boss JM. Epigenetic regulation during B cell differentiation controls CIITA promoter accessibility. J Immunol. 2006;177:3865–3873. doi: 10.4049/jimmunol.177.6.3865. [DOI] [PubMed] [Google Scholar]
- 16.Morris AC, Beresford GW, Mooney MR, Boss JM. Kinetics of a gamma interferon response: expression and assembly of CIITA promoter IV and inhibition by methylation. Mol Cell Biol. 2002;22:4781–4791. doi: 10.1128/MCB.22.13.4781-4791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Elsen PJ, Holling TM, van der Stoep N, Boss JM. DNA methylation and expression of major histocompatibility complex class I and class II transactivator genes in human developmental tumor cells and in T cell malignancies. Clin Immunol. 2003;109:46–52. doi: 10.1016/s1521-6616(03)00200-6. [DOI] [PubMed] [Google Scholar]
- 18.Holling TM, Bergevoet MW, Wilson L, Van Eggermond MC, Schooten E, Steenbergen RD, Snijders PJ, Jager MJ, Van den Elsen PJ. A role for EZH2 in silencing of IFN-gamma inducible MHC2TA transcription in uveal melanoma. J Immunol. 2007;179:5317–5325. doi: 10.4049/jimmunol.179.8.5317. [DOI] [PubMed] [Google Scholar]
- 19.Lennon AM, Ottone C, Rigaud G, Deaven LL, Longmire J, Fellous M, Bono R, Alcaide-Loridan C. Isolation of a B-cell-specific promoter for the human class II transactivator. Immunogenetics. 1997;45:266–273. doi: 10.1007/s002510050202. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh N, Piskurich JF, Wright G, Hassani K, Ting JP, Wright KL. A novel element and a TEF-2-like element activate the major histocompatibility complex class II transactivator in B-lymphocytes. J Biol Chem. 1999;274:32342–32350. doi: 10.1074/jbc.274.45.32342. [DOI] [PubMed] [Google Scholar]
- 21.van der Stoep N, Quinten E, Marcondes Rezende M, van den Elsen PJ. E47, IRF-4, and PU.1 synergize to induce B-cell-specific activation of the class II transactivator promoter III (CIITA-PIII) Blood. 2004;104:2849–2857. doi: 10.1182/blood-2004-03-0790. [DOI] [PubMed] [Google Scholar]
- 22.Piskurich JF, Lin KI, Lin Y, Wang Y, Ting JP, Calame K. BLIMP-I mediates extinction of major histocompatibility class II transactivator expression in plasma cells. Nat Immunol. 2000;1:526–532. doi: 10.1038/82788. [DOI] [PubMed] [Google Scholar]
- 23.Su ST, Ying HY, Chiu YK, Lin FR, Chen MY, Lin KI. Involvement of histone demethylase LSD1 in Blimp-1-mediated gene repression during plasma cell differentiation. Mol Cell Biol. 2009;29:1421–1431. doi: 10.1128/MCB.01158-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreau-Gachelin F, Tavitian A, Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature. 1988;331:277–280. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- 25.Klemsz MJ, McKercher SR, Celada A, Van Beveren C, Maki RA. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 26.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 27.Henkel GW, McKercher SR, Yamamoto H, Anderson KL, Oshima RG, Maki RA. PU.1 but not ets-2 is essential for macrophage development from embryonic stem cells. Blood. 1996;88:2917–2926. [PubMed] [Google Scholar]
- 28.Scott EW, Fisher RC, Olson MC, Kehrli EW, Simon MC, Singh H. PU.1 functions in a cell-autonomous manner to control the differentiation of multipotential lymphoid-myeloid progenitors. Immunity. 1997;6:437–447. doi: 10.1016/s1074-7613(00)80287-3. [DOI] [PubMed] [Google Scholar]
- 29.Iwasaki H, Somoza C, Shigematsu H, Duprez EA, Iwasaki-Arai J, Mizuno S, Arinobu Y, Geary K, Zhang P, Dayaram T, Fenyus ML, Elf S, Chan S, Kastner P, Huettner CS, Murray R, Tenen DG, Akashi K. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005;106:1590–1600. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson KL, Perkin H, Surh CD, Venturini S, Maki RA, Torbett BE. Transcription factor PU.1 is necessary for development of thymic and myeloid progenitor-derived dendritic cells. J Immunol. 2000;164:1855–1861. doi: 10.4049/jimmunol.164.4.1855. [DOI] [PubMed] [Google Scholar]
- 31.Colucci F, Samson SI, DeKoter RP, Lantz O, Singh H, Di Santo JP. Differential requirement for the transcription factor PU.1 in the generation of natural killer cells versus B and T cells. Blood. 2001;97:2625–2632. doi: 10.1182/blood.v97.9.2625. [DOI] [PubMed] [Google Scholar]
- 32.McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, Klemsz M, Feeney AJ, Wu GE, Paige CJ, Maki RA. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 33.Pongubala JM, Nagulapalli S, Klemsz MJ, McKercher SR, Maki RA, Atchison ML. PU.1 recruits a second nuclear factor to a site important for immunoglobulin kappa 3' enhancer activity. Mol Cell Biol. 1992;12:368–378. doi: 10.1128/mcb.12.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eisenbeis CF, Singh H, Storb U. PU.1 is a component of a multiprotein complex which binds an essential site in the murine immunoglobulin lambda 2–4 enhancer. Mol Cell Biol. 1993;13:6452–6461. doi: 10.1128/mcb.13.10.6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marecki S, Fenton MJ. PU.1/Interferon Regulatory Factor interactions: mechanisms of transcriptional regulation. Cell Biochem Biophys. 2000;33:127–148. doi: 10.1385/CBB:33:2:127. [DOI] [PubMed] [Google Scholar]
- 36.Ni Z, Abou El Hassan M, Xu Z, Yu T, Bremner R. The chromatinremodeling enzyme BRG1 coordinates CIITA induction through many interdependent distal enhancers. Nat Immunol. 2008;9:785–793. doi: 10.1038/ni.1619. [DOI] [PubMed] [Google Scholar]
- 37.Lu Q, Richardson B. DNaseI hypersensitivity analysis of chromatin structure. Methods Mol Biol. 2004;287:77–86. doi: 10.1385/1-59259-828-5:077. [DOI] [PubMed] [Google Scholar]
- 38.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oestreich KJ, Yoon H, Ahmed R, Boss JM. NFATc1 regulates PD-1 expression upon T cell activation. J Immunol. 2008;181:4832–4839. doi: 10.4049/jimmunol.181.7.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeong Y, Epstein DJ. Modification and production of BAC transgenes. Chapter 23. Curr Protoc Mol Biol. 2005;(Unit 23):11. doi: 10.1002/0471142727.mb2311s71. [DOI] [PubMed] [Google Scholar]
- 41.Gong S, Yang XW, Li C, Heintz N. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kgamma origin of replication. Genome Res. 2002;12:1992–1998. doi: 10.1101/gr.476202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miele A, Gheldof N, Tabuchi TM, Dostie J, Dekker J. Mapping chromatin interactions by chromosome conformation capture. Chapter 21. Curr Protoc Mol Biol. 2006;(Unit 21):11. doi: 10.1002/0471142727.mb2111s74. [DOI] [PubMed] [Google Scholar]
- 43.Elgin SC. DNAase I-hypersensitive sites of chromatin. Cell. 1981;27:413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- 44.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 45.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 46.Hon GC, Hawkins RD, Ren B. Predictive chromatin signatures in the mammalian genome. Hum Mol Genet. 2009;18:R195–R201. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 48.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 49.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5' ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Stoep N, Quinten E, van den Elsen PJ. Transcriptional regulation of the MHC class II trans-activator (CIITA) promoter III: identification of a novel regulatory region in the 5'-untranslated region and an important role for cAMP-responsive element binding protein 1 and activating transcription factor-1 in CIITA- promoter III transcriptional activation in B lymphocytes. J Immunol. 2002;169:5061–5071. doi: 10.4049/jimmunol.169.9.5061. [DOI] [PubMed] [Google Scholar]
- 51.Accolla RS. Human B cell variants immunoselected against a single Ia antigen subset have lost expression of several Ia antigen subsets. J Exp Med. 1983;157:1053–1058. doi: 10.1084/jem.157.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown JA, He XF, Westerheide SD, Boss JM. Characterization of the expressed CIITA allele in the class II MHC transcriptional mutant RJ2.2.5. Immunogenetics. 1996;43:88–91. doi: 10.1007/BF00186611. [DOI] [PubMed] [Google Scholar]
- 53.DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 54.Dekker J. The three 'C' s of chromosome conformation capture: controls, controls, controls. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 55.Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 56.Eberharter A, Becker PB. ATP-dependent nucleosome remodelling: factors and functions. J Cell Sci. 2004;117:3707–3711. doi: 10.1242/jcs.01175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Restriction site accessibility
All restriction sites assessed for 3C were equally accessible to digestion by HindIII irrespective of the cell line or the condition used. A. A schematic of CIITA region analyzed by 3C assays is shown with Hind III sites, and primers that were used to assess HindIII accessibility. Nuclei from crosslinked cells prepared for 3C was digested with Hind III or left undigested. 30 ng of the purified DNA from the above reactions was used to perform PCR with indicated primers. Agarose gel electrophoresis used to assess each of the PCR products with (B) comparing BCL1 and P3X cells; and (C) undifferentiated and 3 day ex vivo differentiated primary splenic B cells.