Abstract
The nuclear receptor (NR) superfamily of ligand-regulated transcription factors directs ligand- and tissue-specific transcriptomes in myriad developmental, metabolic, immunological, and reproductive processes. The NR signaling field has generated a wealth of genome-wide expression data points, but due to deficits in their accessibility, annotation, and integration, the full potential of these studies has not yet been realized. We searched public gene expression databases and MEDLINE for global transcriptomic datasets relevant to NRs, their ligands, and coregulators. We carried out extensive, deep reannotation of the datasets using controlled vocabularies for RNA Source and regulating molecule and resolved disparate gene identifiers to official gene symbols to facilitate comparison of fold changes and their significance across multiple datasets. We assembled these data points into a database, Transcriptomine (http://www.nursa.org/transcriptomine), that allows for multiple, menu-driven querying strategies of this transcriptomic “superdataset,” including single and multiple genes, Gene Ontology terms, disease terms, and uploaded custom gene lists. Experimental variables such as regulating molecule, RNA Source, as well as fold-change and P value cutoff values can be modified, and full data records can be either browsed or downloaded for downstream analysis. We demonstrate the utility of Transcriptomine as a hypothesis generation and validation tool using in silico and experimental use cases. Our resource empowers users to instantly and routinely mine the collective biology of millions of previously disparate transcriptomic data points. By incorporating future transcriptome-wide datasets in the NR signaling field, we anticipate Transcriptomine developing into a powerful resource for the NR- and other signal transduction research communities.
Keywords: nuclear receptors, steroid hormone receptors, transcription, transcriptomes, database, genome-wide expression profiling, expression microarray
nuclear receptors (NRs) comprise a large superfamily of conserved ligand-regulated transcription factors that regulate target genes controlling essential biological processes in development, immunity, reproduction, and metabolism (2). Included in this family are receptors for endocrine steroids (i.e., corticosteroids, progesterone, androgens, and estrogens), retinoic acid, vitamin D3, thyroid hormone, fatty acids, oxysterols, bile acids, and numerous xenobiotic molecules derived from the diet and the environment.
NR signaling in target cells occurs primarily through the interaction of ligands with cognate NRs. NRs contain a domain that mediates interaction with ligand (the ligand binding domain, or LBD), a zinc finger-containing DNA binding domain (DBD) that binds to hormone response elements (HREs) in the vicinity of target genes, and a dimerization domain that mediates homodimerization or heterodimerization with other NRs. Ligand-bound NRs effect transcriptional regulation by recruitment of coactivators and the subsequent assembly of a transcriptional preinitiation complex at target gene promoters. In contrast, in the absence of ligand, certain NRs are capable of exerting repressive effects on target gene promoters by recruitment of corepressor complexes. Abundant evidence supports the notion that posttranslational modifications catalyzed by coregulator-associated enzymes target a variety of substrates, including histones, and contribute to specific epigenomic marks across target promoters that are critical determinants of their transcriptional status. In addition to direct transcriptional effects, certain physiological ligands are known to engage, in both NR-dependent and non-NR-dependent fashions, kinase-mediated signaling pathways that also ultimately contribute to the overall transcriptional response to NR signaling (19). Occupying a pivotal position in cellular signaling, NRs encompass one of the most successful groups of drug targets for the treatment of a broad range of therapeutic indexes, including obesity, diabetes, cancer, and a variety of cardiovascular, metabolic, toxic/environmental, senescent, and reproductive diseases.
Given the primary function of NRs as transcription factors, the NR field has generated a large number of genome-wide expression analysis datasets, mostly in the form of expression microarrays and, increasingly, on next-generation sequencing (NGS) platforms such as RNA-Seq. However, despite the development of community-based standards for reporting genome-wide expression datasets and the existence of public repositories of expression datasets such as the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO), and the European Bioinformatics Institute (EBI) ArrayExpress, public deposition of array datasets remains far from routine (31). Gene lists in published articles are inconsistently presented in an unpredictable variety of electronic formats, ranging from spreadsheets to PDFs, text, and graphics documents or lack basic fold-change metadata - such as P values and time of ligand treatment - that are essential for meaningful data mining. Moreover, investigators frequently report only those data points relevant to their experimental hypothesis, leaving thousands of potentially useful expression fold changes unreported and inaccessible to bench researchers. Even when publically available in repositories, datasets are accessible only as raw downloads, presenting a prohibitive barrier to their analysis by many scientists. The net result is that unlike genomics research, which has thrived upon enthusiastic universal archiving of sequence data in publically funded repositories, NR transcriptomic data remain largely underreported and underutilized by bench researchers. By aggregating public cancer microarray data from disparate sources for analysis in a single location, the Oncomine database has had a powerful impact in its field (33). Reasoning that a deeply annotated, comprehensive public database of NR transcriptomes would be a similarly valuable resource for data validation and hypothesis generation, as well as a potential catalyst for future drug discovery efforts in this field, we set out to collect, annotate, and expose this universe of data points to the community through a new data-mining tool, Transcriptomine.
MATERIALS AND METHODS
Data Acquisition and Processing
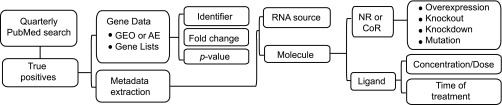
Studies were identified as previously described (32). In brief, a three-part Perl script (Supplementary File S1) was used with NCBI's eUtils to identify and download journal abstracts from studies investigating NR, NR ligand, and coregulator-dependent gene expression specifically in the context of genome-wide technologies such as microarray and RNA-Seq (see Supplementary File S2 for a list of molecule terms).1 Fold changes were extracted preferentially from public high-throughput database repositories containing full datasets (NCBI GEO and EBI ArrayExpress) or, when these were unavailable, were manually retrieved from investigator-curated gene lists published in journal articles and supplementary files (Fig. 1).
Fig. 1.
Transcriptomine data and metadata curation strategy. See text for details.
Expression data obtained from GEO and ArrayExpress are the investigator-provided summarized and normalized array feature expression intensities available in the “series matrix” or “processed” files, respectively. The full set of processed and normalized sample expression values provided by the investigator was extracted and processed in the statistical program R (version 2.13; Supplementary File S3) (16). To calculate differential gene expression for investigator-defined experimental contrasts, we used the linear modeling functions from the Bioconductor limma analysis package (38). Initially, a linear model was fitted to a group-means parameterization design matrix defining each experimental variable. Subsequently, we fitted a contrast matrix that recapitulated the sample contrasts of interest as defined in the study, producing fold-change and significance values for each array feature present on the array. P values obtained from Limma analysis were not corrected for multiple comparisons. In cases where a given gene was represented on an array by more than one probe-set, data from individual probe-sets were generated separately and fold-change values were not pooled across array features.
Where the full raw dataset was unavailable (i.e., had not been deposited in a public repository), fold-change and significance values were transcribed directly from journal and supplementary tables as reported by the investigator. For both sources of gene expression data, individual identifiers provided by the investigator or on the array were mapped to the current Entrez Gene identifier.
Dataset Annotation
In addition to basic annotations such as species (human, mouse, or rat), gene fold-change value, direction, significance, and regulating molecule, experiments were annotated for a number of variables known to be critical determinants of the transcriptional response to NRs and their ligands (See Fig. 1).
Ligand concentration.
The transcriptional function of NRs is known to be closely linked to the amount of ligand available for binding, and receptor-ligand pairs have been postulated to act as rheostats, which function to fine tune the transcriptional response in a given tissue (36). Moreover, many endocrine-disrupting chemicals (EDCs) such as bisphenol A are biologically active only at concentrations several orders of magnitude higher than natural agonists for the NR in question. As such, the ligand concentration at which regulation of a specific gene is observed is a critical experimental descriptor.
Length of ligand exposure.
Experimental systems yield distinct transcriptional profiles based on the length of time they are exposed to NR ligand. Rapid nongenomic signaling by 17β-estradiol, for example, elicits transcriptional responses within minutes that differ fundamentally from those elaborated after multiple hours of exposure, when the genomic effects of liganded estrogen receptors are manifest (44).
RNA source.
Tissue and cell line identity are important determinants of transcriptional and functional endpoints of NR signaling pathways. For example, the breast cancer drug Tamoxifen, while an estrogen receptor (ER) antagonist in the breast, has partial ER agonist activity in the uterus and ER agonist activity in the skeleton (37) and induction or repression of the same gene by glucocorticoid receptor (GR) agonists has been shown to be a function of cell type (3, 11). To align our RNA source annotations with published standards, we used the EFO Experimental Factor Ontology (24) and the CL cell type ontology (5) where possible. All data and annotations are stored in the Nuclear Receptor Signaling Atlas (NURSA) website Oracle database.
Maintenance and All-trans-retinoic Acid-induced Differentiation of Embryonic Stem Cells
Wild-type R1 embryonic stem (ES) cells (a gift from Andras Nagy, Mount Sinai Hospital, Toronto, Ontario, Canada) were maintained on 0.1% gelatinized plates in Dulbecco's modified Eagle's medium supplemented with 15% fetal bovine serum, 100 mM nonessential amino acids, 2 mM glutamine, 100 U of penicillin-streptomycin/ml (Invitrogen), 0.55 μM β-mercaptoethanol (Sigma, St. Louis, MO), and 1,000 U/ml of leukemia inhibitor factor (LIF, Millipore). Cells were rinsed twice with 1× phosphate-buffered saline, treated with 0.25% trypsin, 0.5 mM EDTA (Invitrogen) for 2–3 min, and then split. We plated ∼100,000 cells on a gelatinized 60 mm culture dish and differentiated them by withdrawal of LIF from ES medium and addition of 10 μM all-trans-retinoic acid (ATRA) (Sigma) with daily medium changes.
Quantitative Reverse Transcription Polymerase Chain Reaction Analysis
Total RNA was extracted from ES cells with TRIzol reagent (Invitrogen). cDNA was generated using the Super Script III First Strand Synthesis Kit (Invitrogen) with Oligo dT primers following the manufacturer's protocol. Quantitative expression of endogenous genes was carried out using QuantiFast SYBR Green PCR (Qiagen, Valencia, CA; http://www.qiagen.com) on a Step 1 Plus Real Time PCR System (Applied Biosystems, Carlsbad, CA; https://products.appliedbiosystems.com). Target gene expression was normalized to GAPDH expression in all experiments. Quantitative reverse transcription polymerase chain reaction (q-RT-PCR) primers: Gas1-F, ACTGCGGCAAGCTTTTCAACGG; Gas1-R, CTCTTTGACCGATTCGCAGATGG; Greb1L-F, AGATGGTGAGCACCCTCTTGGA; Greb1L-R, CATAGTGGTGAGAGCCATCAAGG; Foxg1-F, TACTACCGCGAGAACAAGCAGG; Foxg1-R, GAGCATCCAGTAGTTGCCCTTG; Gata3-F, CCTCTGGAGGAGGAACGCTAAT; Gata3-R, GTTTCGGGTCTGGATGCCTTCT; Gapdh-F, GTCTCCTCTGACTTCAACAGCG; Gapdh-R, ACCACCCTGTTGCTGTAGCCAA; Ncoa3-F, 5′GGAGAATTACGGTGCCAACATGG-3′; Ncoa3-R, 5′-GACTTGATGCCAGAGGCTCCAT-3′.
RESULTS
Data Querying Strategies
To maximize its utility to a broad audience, the Transcriptomine user interface allows for flexible query construction. The form is designed with five sections, through which the user navigates to assemble their query of choice (Table 1) using any combination of the criteria below.
Table 1.
Query options in Transcriptomine
| Query Parameter | Options | Strategy |
|---|---|---|
| Gene(s) of interest | any gene | return any gene that matches the query parameters |
| single gene (AC) | specify a single gene of interest | |
| GO term (AC) | specify a gene ontology biological process, cellular component, or molecular function as a surrogate for gene(s) of interest | |
| disease term (AC) | specify a disease term as a surrogate for gene(s) of interest | |
| upload gene list | upload a custom gene list (≤5,000 genes) | |
| Regulation | fold change | options: ≥twofold (default); enter custom value |
| direction | options: up- or downregulated (default); upregulated; downregulated | |
| significance | options: any (default); P < 0.05; enter custom value | |
| Regulating molecule | single molecule (AC) | specify a regulating NR ligand, NR or coregulator; if a ligand is entered, Time Treated becomes available [options: ≤1 h (default); >1 h-≤4 h; >4 h-≤24 h; >24 h] |
| RNA source | species | options: all (default), human; mouse; rat |
| tissue/cell line (AC or select from list) | specify individual tissues (e.g., liver) or cell lines (e.g., MCF-7) or all tissues and cell lines within a given tissue type or organ (e.g., adipose tissue, all tissues and cell lines) |
AC, autocomplete text field.
Gene(s) of interest.
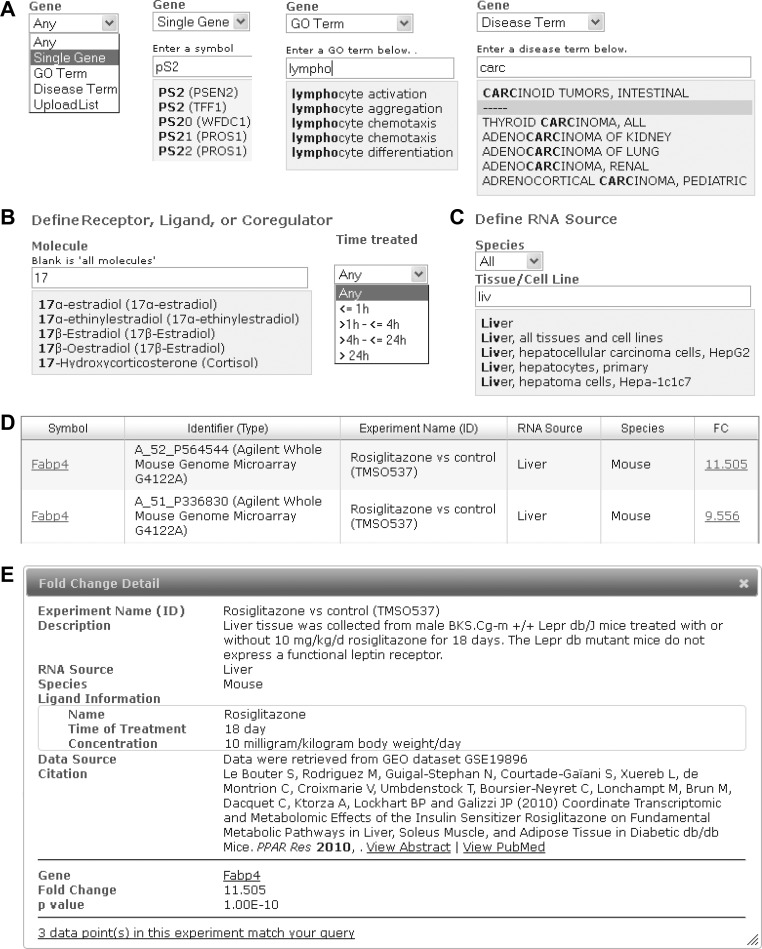
See Fig. 2A.
Fig. 2.
Schematic of Transcriptomine v. 1.0 user interface features. A: gene query options - any, single, GO term, disease term and gene list upload. B: regulating molecule, including treatment time option for ligands. C: RNA source. D: search results page. E: fold change detail window.
any gene.
Returns any gene fulfilling the criteria in the query.
single gene.
As users enter text, an autosuggest functionality displays suggested symbols based on the text entered along with the corresponding official symbol. To simplify query entry, only the human symbol for a given gene is displayed in the autosuggest drop-down, and fold changes for all orthologs (human, mouse, and rat) are returned by default in the search results. Users can override this setting and retrieve orthologs for a given species by specifying this in the “RNA Source” section of the form.
Gene Ontology term.
The Gene Ontology (GO) initiative provides a standard, universal vocabulary of biological attributes of individual genes and their products. To provide a platform for the exploration of higher-level regulation of cellular biology by NR signaling pathways, users can specify a GO term (biological process, cellular component, or molecular function) as a surrogate for a specific gene or list of genes.
disease term.
Members of the NR superfamily are well-characterized therapeutic targets in a broad range of disease states, and filtering based on a specific disease or group of diseases is an essential query option. Accordingly, we utilized the NCBI Online Mendelian Inheritance in Man (OMIM) database (13), which maps Entrez Gene IDs to human genetic diseases.
gene list.
To accommodate users wishing to query custom gene lists against the database, we provide for uploading of .CSV files of official gene symbols or Entrez GeneIDs.
Regulation.
This subsection provides for filtering search results based on the amplitude (fold change), direction (induction or repression) and significance.
fold change.
The default value is 2.
direction.
NR signaling pathways effect induction or repression of target gene networks in a context-dependent manner, and Transcriptomine provides for filtering upregulated genes, downregulated genes, or both. Most experiments in Transcriptomine are “gain-of-function” (GOF) experiments, in which perturbation of a system - usually with a ligand (agonist or antagonist) or overexpression of a NR or coregulator, is compared with a control. A small number are “loss of function” (LOF) experiments, in which the effect of removal of an entire molecule is examined, namely knockout vs. control experiments in animal models or knockdown vs. control in cultured cells. In LOF experiments, the fold-change direction of regulated genes is opposite to that in GOF experiments. For example if gene A is positively regulated by a NR in a given system, it will have a negative fold change when that receptor is knocked out or knocked down in that system, and vice versa. In these experiments, we modified the normal logic of the query form such that in LOF contrasts, fold changes of an opposite direction to that specified in the query form are returned, i.e., if a user specifies genes upregulated by a given NR (direction = “upregulated”), their results will include downregulated fold changes from LOF experiments involving that NR. LOF contrasts are clearly defined in the descriptions for such fold changes so that users understand why fold changes of the opposite value anticipated are being displayed.
significance.
The default P value is P < 0.05. P values in Transcriptomine fall into one of two categories. 1) P values from deposited datasets. Where datasets were deposited in a public gene expression data repository such as GEO or ArrayExpress, P values are calculated from the data in the deposited files using the Bioconductor limma package for R. 2) P values from publications. The majority of gene expression experiments are not deposited in public repositories (31) and in these cases P values associated with a specific fold change are presented in Transcriptomine as reported in the original publication without modification or reanalysis. In a surprising percentage of publications, no P values are reported by the investigator and the significance of a specific fold change cannot be clearly determined; these P values are displayed as “not reported”.
Molecule.
See Fig. 2B. Version 1 of Transcriptomine contains transcriptomes for a broad variety of NR signaling molecules, including NRs, their ligands, and coregulators (Fig. 3). The user next defines the regulating molecule, NR ligand, NR, or coregulator, for which the default value is “any”. While NRs and coregulators have been assigned official gene symbols by the organizations overseeing human, mouse, and rat gene nomenclatures, numerous familiar names for these molecules remain in everyday discourse in the field (30). NURSA website curators maintain official and unofficial synonyms for all active NRs, coregulators, and ligands defined on the NURSA website. To accommodate as many end users as possible, investigators can enter their preferred symbol synonym, which Transcriptomine maps to a specific molecule using terms curated by NURSA Bioinformatics Resource curators. For example, the search terms p/CIP, ACTR, TRAM-1, AIB1, RAC3, SRC-3 all map to the molecule defined by the official symbol NCOA3. Selecting anyone of these terms will retrieve data relevant from Transcriptomine that map to NCOA3.
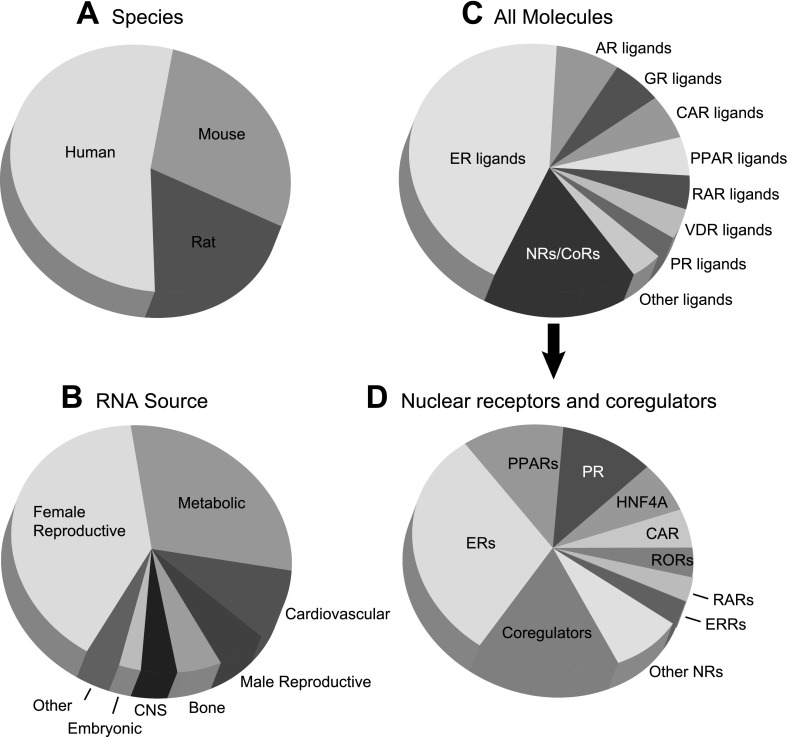
Fig. 3.
Data coverage of Transcriptomine 1.0. >700 individual experiments representing a total of >10,000,000 data points are broken down by species (A), RNA source (B), and molecule (C, D). A: species. B: RNA Source. Metabolic tissues: liver, muscle, adipose tissue, pancreas, gastrointestinal, kidney. Female reproductive: mammary gland, uterus, endometrium, ovary, vagina, cervix. Male reproductive: prostate, testis. Connective: skin, cartilage. Embryonic: embryonal carcinoma cells, Mullerian duct, whole embryo, embryonic fibroblasts. Other tissues: skin, cartilage, dendritic cells, thyroid gland, lung, eye, mesenchyme. C: all molecules. Other ligands: ligands for TRs, RXRs, ERRs, RORs, LXRs, FXR, and PXR. D: nuclear receptors and coregulators - other NRs: TRs, AR, SHP, RXRs, DAX1, FXR, GR. Coregulators: AIB1, BAF60a, BRCA1, CAV1, E6AP, GRIP1, PGC1α, PGC1β, RIP140, SMRT, SRC-1. Due to space constraints, RNA sources in this figure represent broader categories of the cell lines and tissues in Transcriptomine.
RNA source.
See Fig. 2C. Version 1 of Transcriptomine has broad tissue and cell line coverage in a variety of species (Fig. 3). RNA Source is defined as a specific combination of species and organ or tissue/cell line using a controlled vocabulary. To allow users to query fold changes across entire organs we mapped individual RNA Sources to catch-all terms. For example, with species = “all,” selecting “liver, all tissues, and cell lines” will encompass fold changes from all liver-related experiments in all species, whether in cultured or primary cell lines, or the broader tissue-/organ-level analyses typically carried out in animal experiments. Terms can be entered by autosuggest-assisted free text entry or by selecting from a list of currently curated RNA Sources in a pop-up window.
Search Results
See Fig. 2D. The fold changes and P values reported by Transcriptomine are derived either from .CEL files in publically deposited datasets or directly from investigator-supplied gene lists reported in publications. Expression fold-change values are viewable in one of two options: 1) by symbol, in which official gene symbols are sorted alphabetically by default along with the identifier type (e.g., array probe-set, Ref-seq ID), identifier, experiment name, RNA Source, species, and fold change. Fold changes are reported according to the convention 2.0 = twofold induction (upregulation) and 0.5 = twofold repression (downregulation); and 2) by experiment, in which individual experiments are listed, along with a link to the number of fold changes within that experiment that match the query criteria. In Symbol view, more information on individual fold changes can be viewed by clicking on the fold-change value hyperlink, which displays additional information such as the original data source, P value, supporting publication, and other relevant metadata in a pop-up window (Fig. 2E). Data can be downloaded as Excel files for downstream analysis.
Use Cases
We next demonstrate the utility of Transcriptomine as research tool in a series of four in silico and experimental use cases that represent the kind of questions that can be asked of Transcriptomine when seeking information with which to generate or validate a biological hypothesis.
Use case 1: identification of previously uninvestigated 1,25-dihdroxyvitamin D3-regulated genes.
An investigator wishes to investigate previously uncharacterized targets of 1,25-dihdroxyvitamin D3 in tissues of the cardiovascular system.
query.
Gene, any gene; fold change, twofold; Direction, up or down; Significance, P < 0.05; Regulatory molecule, 1,25-dihydroxyvitamin D3; Time of treatment, any; RNA source, cardiovascular, all tissues, all cell lines.
results.
See Supplementary File S4. The large number of fold-change data returned by this search can be accommodated by computational methods but is more than a user unexperienced in bioinformatics can readily analyze. To reduce the number of results, this user can click on the “modify query” button and adjust the significance cut-off to a more stringent confidence level (P < 1E-5) to reduce the number of fold changes to ∼1,000, corresponding to three separate studies (Supplementary File S4). The regulation of previously characterized targets of 1,25-dihydroxyvitamin D3 signaling, such as CYP24A1 and several members of the integrin family (ITGA4 & ITGAM), serve as in silico internal “positive controls” to increase confidence in previously uncharacterized genetic feature fold changes in the dataset. The search also returns data points for genes not previously characterized as targets of VDR signaling, and whose biology is consistent with known physiological effects of 1,25-dihydroxyvitamin D3. The cross-validation that arises from the consistent regulation of a group of these genes in multiple studies serves as an additional source of confidence in their identity as “true” 1,25-dihydroxyvitamin D3 target genes. One of these genes, CHST15, encodes N-acetylgalactosamine 4-sulfate 6-o-sulfotransferase. Null deletion of this gene results in decreased synthesis of chondroitin sulfate, an inhibitor of NFKB activation and an important structural component of cartilage that is used as a dietary supplement in the treatment of osteoarthritis (43). Another, CES1, encodes monocyte esterase, deficiency of which is associated with reduced tumor cell killing by monocytes and which is underexpressed in non-Hodgkin lymphoma and B-cell lymphocytic leukemia (25).
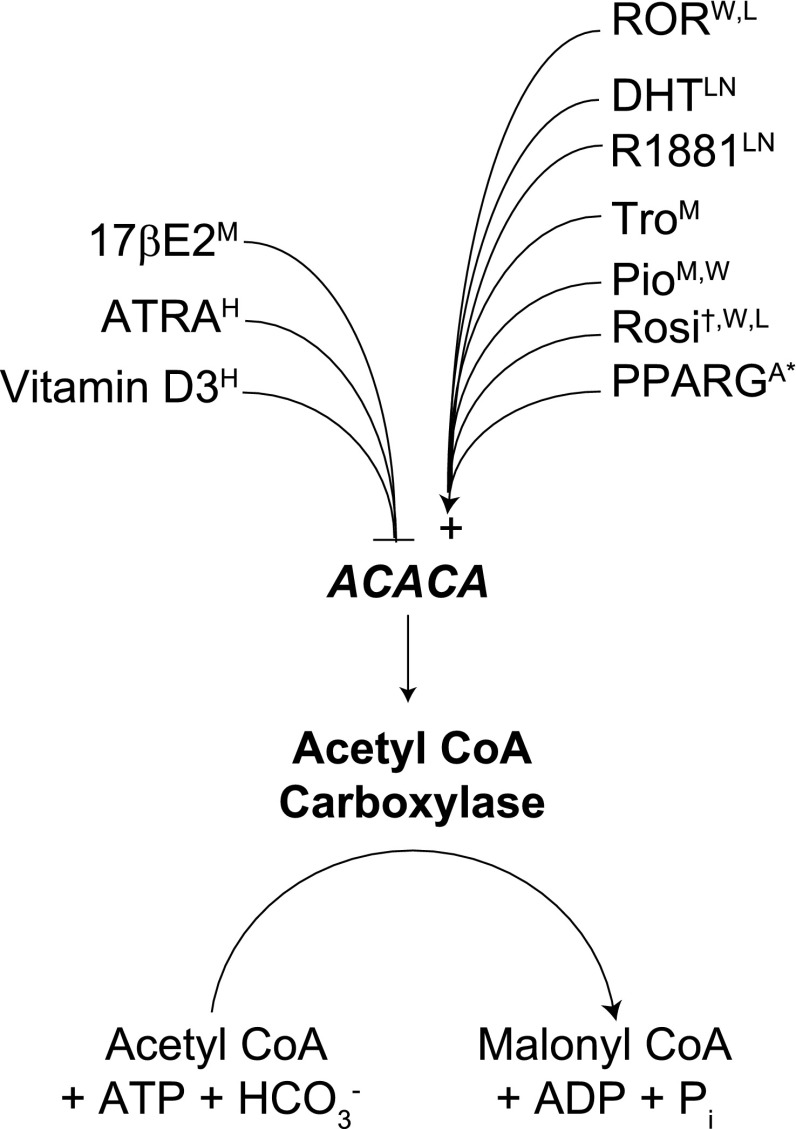
Use case 2: convergent regulation of the acetyl CoA carboxylase gene by multiple NR signaling pathways.
In this use case, the user has a specific gene in which they are interested. Acetyl CoA carboxylase A (ACACA) is a key control site in saturated long-chain fatty acid synthesis in the liver, catalyzing the formation of malonyl CoA from acetyl CoA, bicarbonate, and ATP. It is controlled at the protein level through tandem opposing effects of glucagon and insulin. Low blood sugar stimulates the release of glucagon, which initiates a phosphorylation cascade that represses ACACA activity. Insulin, a signal that energy sources are abundant, has the opposite effect, restoring ACACA activity. While ACACA is known to be regulated at the transcriptional level by SREBP1, to date regulation of ACACA gene expression by NR signaling pathways has been essentially uninvestigated. Given the abundant data linking NR signaling to regulation of fat metabolism, the following question arises: is ACACA subject to control by one or more metabolic NR signaling pathways? Transcriptomine can be used to guide a researcher toward potential regulatory pathways.
query.
Gene: single gene, ACACA; Fold change, twofold; Direction, up or down; Significance, P < 0.05; Regulatory molecule, any; RNA source, any.
results.
See Supplementary File S5 and Fig. 4. Consistent with its role in fatty acid synthesis, ACACA is strongly induced in adipogenesis in 3T3L1 cells, as well as being upregulated in the Zucker fa/fa rat. In agreement with the improved insulin sensitivity characteristic of RORα-deficient staggerer mice (18), ACACA is induced in the white adipose tissue of these mice, indicating its repression by RORα. Consonant with the insulin-sensitizing effect of PPARγ agonists, a total of seven independent experiments show that ACACA is induced by three different PPARγ agonists in muscle, white adipose tissue, and liver (Fig. 3 and Supplementary Fig. S5a). Moreover, adjustment of the fold-change cut-off to 1.9 adds an additional data point showing downregulation of ACACA in PPARγ siRNA-treated adipocytes, supporting its identity as a direct target of PPARγ. By connecting corroborating data points from studies with different ligands for a given NR, as well as LOF experiments involving that NR, Transcriptomine can therefore provide the user with a high confidence level of regulation of a specific gene by an NR pathway.
Fig. 4.
Transcriptomine implicates multiple NR signaling pathways in regulation of the acetyl CoA carboxylase gene (ACACA) in metabolic tissues and cancer cell lines. All data points are ≥2 (except * = 1.9) and P <0.05 and are previously uninvestigated (except †, see Ref. 17) in the research literature. See Supplementary File S5 for data. Superscript denotes RNA Source: A3T3-L1 adipocytes, HHL-60 cells, Lliver, LNLNCaP cells, Mmuscle, Wwhite adipose tissue.
The ACACA use case also demonstrates the ability of Transcriptomine to compile entirely unrelated data points that collectively prompt a hypothesis not related to that on which the original search was predicated. Elevated rates of fatty acid synthesis in tumor growth were first characterized over 60 years ago (26), and the importance of fatty acid synthesis in supporting growth of a variety of tumors, including prostate cancer (42) and leukemia (34), has been well documented. Moreover, studies have shown that silencing of ACACA gene expression by RNA interference (8) or chemical inhibition of the enzyme (6) causes growth inhibition and induction of cell death in prostate cancer cells. Consistent with the androgen-dependent growth of many prostate cancers, a total of six independent experiments in Transcriptomine demonstrate that both DHT and R1881 induce ACACA (see Supplementary File S5). Conversely, the robust repression of ACACA by ATRA and vitamin D3 in the HL-60 acute promyelocytic leukemia cell line reported by Transcriptomine (see Supplementary File S4), reflects the prominence of these NR ligands in the treatment of leukemia. Indeed, the case of ATRA provides a transcriptional basis for the observation nearly three decades ago that decreased ACACA enzymatic activity is associated with HL-60 cells exposed to retinoic acid (12). Intriguingly, further mining suggests that the effect of NR ligands on ACACA expression may be only one component of a coordinated regulation of the fatty acid biosynthetic pathway. A search for a gene encoding another important enzyme in fatty acid synthesis, FASN, shows that it too is subject to induction by DHT and R1881 in prostate cancer cells and profound downregulation by ATRA and 1,25-dihydroxyvitamin D3 in leukemia cell lines (Supplementary File S5a). Collectively, these data points posit the hypothesis that regulation of fatty acid biosynthesis by NR signaling pathways is an important component of their effects on the growth of various cancers. In a wider context they demonstrate how a user can leverage Transcriptomine to quickly and efficiently assemble in silico models tying regulation of gene expression into NR physiology.
Use case 3: coordinate regulation of de novo purine base biosynthesis by NR signaling pathways.
In many cases, a user of Transcriptomine might not have a specific gene or NR signaling pathway in mind but, rather, might wish to ascertain the effect of NR signaling as a whole on multiple genes involved in a certain physiological process, for which the GO query option can be used. Nucleotide biosynthesis plays fundamental roles in the cell, generating activated precursors that drive DNA and RNA synthesis in cell division, provide energy to drive cellular processes, and function as important regulators of cellular signaling pathways. While feedback inhibition of nucleotide biosynthetic enzymes has been intensively studied, regulation of this process at the transcriptional level, particularly by NR signaling pathways, has been relatively poorly characterized. The following search was run to broadly determine the role of NRs in regulation of genes involved in purine biosynthesis.
Gene: GO term, purine base biosynthetic process; Fold change: twofold; Direction: up or down; Significance: P < 0.05; Regulatory molecule: any; RNA source: any.
results.
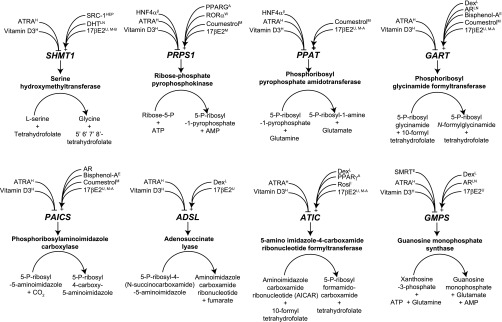
See Supplementary File S6 and Fig. 5. Several of the genes in the pathway are positively regulated by dexamethasone, consistent with historical effects of positive regulation by glucocorticoids of hepatic purine synthesis (10). Strikingly, multiple independent studies report that eight genes encoding enzymes in the purine biosynthetic pathway (SHMT1, PRPS1, PPAT, GART, PAICS, ADSL, ATIC, and GMPS) are all induced by 17β-estradiol in MCF-7 breast cancer cells and repressed by ATRA and 1,25-dihydroxyvitamin D3 (Fig. 5) in HL-60 human promyelocytic leukemia cells. The induction by 17βE2 of the specific activities of enzymes involved in purine and DNA synthesis in MCF-7 cells, and the attendant increases in [3H]thymidine uptake and cell proliferation, have been well documented (1), but to date no transcriptional regulation of these enzymes by 17βE2-signaling has been documented. Moreover, the repression by ATRA and 1,25-dihydroxyvitamin D3 of genes encoding enzymes in this pathway is consistent with the growth inhibitory and prodifferentiative effect of these ligands in hematological malignancies. Interestingly, coumestrol, an endocrine-disrupting chemical with estrogenic activity (28), induces five of the six genes.
Fig. 5.
Coordinate regulation by NR signaling pathways of genes encoding enzymes in the de novo purine base biosynthesis pathway. All fold changes are ≥2 and P < 0.05 and are previously uninvestigated in the research literature. See Supplementary File S6 for data. Superscript denotes RNA Source: β, pancreatic β-cells; A, 3T3-L1 adipocytes; E, endometrial Ishikawa cells; H, HL-60 cells; HEP, liver HepG2 cells; L, liver; LN, LNCaP cells; M, MCF-7 cells; M-A, MCF-7/AKT cells; M-B, MCF-7/BUS cells; U, uterus; W, white adipose tissue.
Coregulators were also implicated in the regulation of purine biosynthesis: SRC-1 knockout results in downregulation of SHMT1, positing this coactivator as a regulator of the reaction that is the single largest source of one-carbon units in the cell. Moreover, SMRT is a corepressor for GMPS, thereby overseeing the branch-point for synthesis of adenine or guanine nucleotides (Fig. 5 and Supplementary File S6). Collectively, these data imply that orchestrated regulation of multiple steps of the purine biosynthetic pathway is effected by diverse NR signaling pathways.
Use case 4: validation of ATRA-regulated genes in mouse ES cells.
The final use case demonstrates how Transcriptomine can be used to corroborate the results of a wet bench experiment and guide the design of subsequent bench studies. ATRA is one of the principle active metabolites of vitamin A (retinol) that, in concert with members of the retinoic acid receptor (RAR) family, directs transcriptional programs in a spectrum of critical developmental and physiological processes, one of which is the induction of differentiation in ES cells (39). Our laboratory is interested in identifying new transcription factors and transcriptional coregulators that play roles in ES cell ATRA-differentiation. Accordingly, we carried out transcriptome-wide expression profiling of ATRA-treated ES cells (data not shown). To assist us in focusing on targets for validation and follow-up study we queried Transcriptomine for any genes regulated by ATRA in any tissue or cell line.
Gene: any; Fold change, twofold; Direction, up or down; Significance, P < 0.05; Regulatory molecule, all-trans retinoic acid; RNA source, any.
results.
See Fig. 6. This query generated >11,000 fold changes and a supplementary file has not been included. Of the multiple genes returned in our search, we focused on five (GREB1L, GAS1, GATA3, FOXG1, and NCOA3) based on 1) their induction in the original array dataset, 2) their involvement in transcription, and 3) the lack of previous data implicating them in ES cell differentiation. FOXG1 encodes a member of the Forkhead Box family of transcription factors and has recently been shown to be capable of generating neural precursor cells when overexpressed in mouse embryonic fibroblasts (21). Another transcription factor, GATA3, is a member of the GATA family of transcriptional regulators with well-characterized roles in the development of T lymphocytes (27). It has previously been shown to be transcriptionally regulated by the global chromatin organizer and regulator of gene expression, SATB1 (29), an essential factor in stem cell differentiation (35). Consistent with the function of ATRA in prodifferentiative signaling, GAS1 encodes a protein that inhibits entry into S-phase (9). GREB1L encodes a virtually uncharacterized protein that we have recently identified as corepressor for members of the NR superfamily (unpublished data). Finally, the NR coregulator SRC-3/NCOA3 has been shown to be required for the efficient induction of genes targeted by a number of different signaling pathways (23).
Fig. 6.
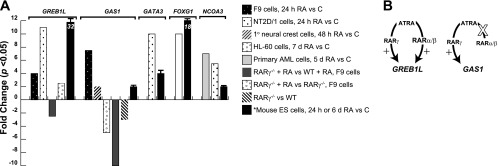
Mining of Transcriptomine identifies previously uninvestigated ATRA-responsive genes in silico. A: data points were retrieved from Transcriptomine except * obtained from Q-PCR analysis of mRNA from ATRA-treated mouse ES cells. Original citations for Transcriptomine data points: F9 cells (41), NT2D/1 cells (14), primary neural crest cells (46), HL-60 cells (15), and primary AML cells (40). All TM data points P < 0.05 except Ref. 15, P values not reported. B: differential regulation by ATRA of GREB1L and GAS1.
Transcriptomine reported induction by ATRA of the following: GREB-1L in mouse F9 cells and human NT2D teratocarcinoma cells (Fig. 6A); GAS1 in primary neural crest cells; GATA3 in NT2D/1 cells; FOXG1 in human promyelocytic leukemia HL-60 cells [note that this is in contrast to its repression by ATRA as reported by Li et al. (20) in human ES cells]; and NCOA3/SRC-3 in both primary AML cells and NT2D/1 cells (Fig. 6A). Additional evidence for direct regulation of GREB1L and GAS1 was gleaned from data points in knockout studies of RARγ in F9 cells. GAS1 was downregulated in RAR−/− F9 cells vs. WT F9 cells, and both genes were downregulated in ATRA-treated RARγ−/− F9 cells relative to ATRA-treated WT F9 cells. Interestingly, while there was some residual induction of GREB1L in ATRA-treated RARγ−/− cells relative to untreated RARγ−/− cells, no such induction was observed for GAS1 (Fig. 6A). Collectively these studies suggest a model in which ATRA can signal through RARα or RARβ to regulate GREB1L in the absence of RARγ but signals exclusively through RARγ to regulate GAS1 (Fig. 6B).
Based on the corroborative evidence gleaned from Transcriptomine we next validated regulation of these genes by Q-PCR analysis in the original RNA samples. These experiments confirmed induction in ES cells of Greb1l (32-fold), Gas1 (twofold), and Gata3 (fourfold) in response to 24 h treatment with ATRA and induction of Foxg1 (10-fold) and Ncoa3 (fourfold) in response to 6 days ATRA treatment (Fig. 6A). In summary, we observed broad agreement for all five genes between the original array dataset, the in silico fold changes in Transcriptomine, and the experimentally validated Q-PCR fold changes, indicating that they indeed represent previously uninvestigated ATRA-responsive genes.
Comparison of Transcriptomine With Other Search Strategies
For each use case we compared the Transcriptomine search strategy with those involved in retrieving similar information through Google, PubMed, and GEO (Supplementary File S7). In all four cases, after reasonable effort on our part, none of the alternative search strategies was able to reproduce the quality and quantity of the Transcriptomine search results, nor the efficiency and convenience with which those results were obtained.
DISCUSSION
Despite the presence of a peer review system and the existence for nearly a decade of the MIAME standard for reporting microarray data and metadata (7), the quality of annotation and the existence of publically deposited transcriptomic datasets remain sporadic and unpredictable (31). While Google, PubMed, and GEO are routinely used as information retrieval strategies, the high rate of false positives that confound these search tools reflects a substantial disconnect between the intended logic of the query and the quality of the information retrieved. Moreover, the dearth of consistent data organization and metadata descriptors prevents the retrieval of “like” datasets in any one search. Even in the event of results being returned, the level of organization of these data required to identify meaningful biological trends and patterns across multiple datasets assumes a solid working knowledge of bioinformatics on the part of the user. Collectively, these factors have combined to profoundly limit the utility of a vast number of public transcriptomic data points in the NR signaling field.
We have described here the development of Transcriptomine, an innovative data resource for both the NR signaling field and the wider field of transcriptional regulation. The value of this resource to the community lies in its aggregation of all publically available transcriptomic datasets in the NR signaling field; the value it adds to these datasets through deep, consistent reannotation using controlled vocabularies; a user-friendly user interface that provides for logical query construction and intuitive browsing of search results; and the availability of comprehensively annotated data points for browsing or downloading for downstream analysis by advanced users. The Transcriptomine user interface guides the user toward gene expression fold changes that occur consistently across multiple related datasets, allowing for cross-validation and insight into the biological significance of these expression changes in specific regulatory contexts. Similar fold changes that are individually suggestive construct a more convincing argument when viewed side by side. For example, Transcriptomine can pull together available multiple independent data points showing regulation of a given gene in both cell lines and native tissue, or that knockout or knockdown of an NR affects expression of a gene regulated by its cognate ligand. Through this accretion of evidence from different sources, the user derives a higher level of confidence that a given gene is a true target of the ligand or NR and would therefore triage this gene for validation and functional studies.
Google searching did achieve some success in use case 4 in identifying studies showing ATRA induction of two of our candidate genes (4, 45), and GEO returned several positive results in the same use case (Supplementary File S7), but these were the exceptions. Overall, Transcriptomine consistently outperformed free-text searches both in ease and speed of searching, as well as quantity and quality of results, thereby validating our approach of combining accurate, manual annotation with menu-driven queries to virtually eliminate search result noise. Moreover, the use cases, particularly use cases 2 and 3, demonstrate a powerful consequence of the uniform structure of the data and metadata, namely the ability of the Transcriptomine to compile previously unconnected data points to generate unanticipated hypotheses on the fly, thereby opening up novel and potentially profitable lines of enquiry to the bench researcher.
Transcriptomine is not designed to reanalyze or call into question any investigator's use of a particular normalization method. In any case, accurate descriptions of the precise normalization steps employed are typically absent from publications, making it impossible to precisely replicate fold-change values from any given study. For this reason we preferentially generate data points from publically deposited raw files using an industry-standard data-processing pipeline or, absent these, gene lists retrieved from the published paper. In this way we enable users of Transcriptomine to retrieve fold-change values upon which the study authors based their original biological interpretation of the datasets.
Data points in Transcriptomine are annotated as completely and uniformly as possible given the information supplied by the authors at the time of publication. In some cases, however, critical metadata for a given experiment were either incomplete or absent altogether, and to preserve the quality of the data in Transcriptomine, we were unable to include them. The advent of NGS data platforms offers a renewed opportunity to establish standards for reporting and archiving genome-wide transcriptome analysis datasets, and we call upon authors, publishers, and funding organizations alike to collaborate upon establishing and ensuring the adoption of these standards.
Future versions of the database will provide for complex queries, in which two queries can be combined to identify an intersecting set of genes that fulfill both sets of regulatory criteria. Additional features will also include expanding upon the range of options for querying the database, to encompass canonical signaling pathways, protein domains, as well as other biological ontologies and disease-to-gene databases. The utility of Transcriptomine can also be enhanced by incorporation of additional contextual data layers. For example, while Transcriptome can furnish the user with functional evidence for regulation of a given gene by a specific NR signaling pathway, an additional level of confidence in determining direct regulation of the gene can be attained by evaluating NR DNA binding at the genomic locus of that gene in a given tissue in a specific signaling context. The cistrome for a given NR is defined as its set of cognate cis regulatory elements in a given cell type in a specific signaling context (22) and can be determined on a variety of experimental platforms, such as chromatin immunoprecipitation (ChIP)-chip and ChIP-Seq. A natural extension of Transcriptomine is therefore to integrate genomic coordinates from the growing number of cistromic studies profiling DNA binding by NRs in a variety of cell types and signaling contexts. We also plan to incorporate data visualization options into the user interface, such as heat maps, and adding pathway annotations available from Cytoscape and other resources. Finally, the development of similar databases for other families of transcription factors would illuminate cross talk at the transcriptional level between NRs and the myriad other signal transduction pathways that impinge upon cell function, growth, and differentiation. By bringing existing and future transcriptomic datasets to the fingertips of researchers in this field, we anticipate that Transcriptomine will catalyze the development of novel hypotheses and biological models of NR signaling pathways.
GRANTS
This work was supported in part by the NURSA U19DK-62434 to N. J. McKenna and the National Cancer Institute Cancer Center Support Grant P30-CA-125123 (to L. B. Becnel).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: S.A.O., C.M.W., A.M., Y.F.D., A.J.C., D.L.S., L.B.B., and N.J.M. conception and design of research; S.A.O. and A.J.C. analyzed data; S.A.O., L.B.B., and N.J.M. edited and revised manuscript; S.A.O., C.M.W., A.M., X.X., Y.F.D., M.D.D., A.J.C., D.L.S., L.B.B., and N.J.M. approved final version of manuscript; X.X. and M.D.D. performed experiments; A.J.C. interpreted results of experiments; N.J.M. prepared figures; N.J.M. drafted manuscript.
Supplementary Material
ACKNOWLEDGMENTS
Transcriptomine is available at http://www.nursa.org/transcriptomine.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1. Aitken SC, Lippman ME. Effect of estrogens and antiestrogens on growth-regulatory enzymes in human breast cancer cells in tissue culture. Cancer Res 45: 1611–1620, 1985. [PubMed] [Google Scholar]
- 2. Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev 81: 1269–1304, 2001. [DOI] [PubMed] [Google Scholar]
- 3. Badrinarayanan R, Rengarajan S, Nithya P, Balasubramanian K. Corticosterone impairs the mRNA expression and activity of 3beta- and 17beta-hydroxysteroid dehydrogenases in adult rat Leydig cells. Biochem Cell Biol 84: 745–754, 2006. [DOI] [PubMed] [Google Scholar]
- 4. Bai A, Lu N, Zeng H, Li Z, Zhou X, Chen J, Liu P, Peng Z, Guo Y. All-trans retinoic acid ameliorates trinitrobenzene sulfonic acid-induced colitis by shifting Th1 to Th2 profile. J Interferon Cytokine Res 30: 399–406, 2010. [DOI] [PubMed] [Google Scholar]
- 5. Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biol 6: R21, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beckers A, Organe S, Timmermans L, Scheys K, Peeters A, Brusselmans K, Verhoeven G, Swinnen JV. Chemical inhibition of acetyl-CoA carboxylase induces growth arrest and cytotoxicity selectively in cancer cells. Cancer Res 67: 8180–8187, 2007. [DOI] [PubMed] [Google Scholar]
- 7. Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet 29: 365–371, 2001. [DOI] [PubMed] [Google Scholar]
- 8. Brusselmans K, De Schrijver E, Verhoeven G, Swinnen JV. RNA interference-mediated silencing of the acetyl-CoA-carboxylase-alpha gene induces growth inhibition and apoptosis of prostate cancer cells. Cancer Res 65: 6719–6725, 2005. [DOI] [PubMed] [Google Scholar]
- 9. Del Sal G, Ruaro ME, Philipson L, Schneider C. The growth arrest-specific gene, gas1, is involved in growth suppression. Cell 70: 595–607, 1992. [DOI] [PubMed] [Google Scholar]
- 10. Feigelson M, Gross PR, Feigelson P. Early effects of cortisone on nucleic acid and protein metabolism of rat liver. Biochim Biophys Acta 55: 495–504, 1962. [DOI] [PubMed] [Google Scholar]
- 11. Feltus FA, Cote S, Simard J, Gingras S, Kovacs WJ, Nicholson WE, Clark BJ, Melner MH. Glucocorticoids enhance activation of the human type II 3beta-hydroxysteroid dehydrogenase/Delta5-Delta4 isomerase gene. J Steroid Biochem Mol Biol 82: 55–63, 2002. [DOI] [PubMed] [Google Scholar]
- 12. Fischkoff SA, Papuchis GC, Nickols WA. Decreased activity of acetyl-CoA carboxylase during chemically induced neutrophilic differentiation of human promyelocytic leukemia cells. J Cell Biochem 26: 75–81, 1984. [DOI] [PubMed] [Google Scholar]
- 13. Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 33: D514–D517, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heim KC, White KA, Deng D, Tomlinson CR, Moore JH, Freemantle SJ, Spinella MJ. Selective repression of retinoic acid target genes by RIP140 during induced tumor cell differentiation of pluripotent human embryonal carcinoma cells. Mol Cancer 6: 57, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang S, Eichler G, Bar-Yam Y, Ingber DE. Cell fates as high-dimensional attractor states of a complex gene regulatory network. Phys Rev Lett 94: 128701, 2005. [DOI] [PubMed] [Google Scholar]
- 16. Ihaka R, Gentleman RR. R: a language for data analysis and graphics. J Computational Graph Stat 5: 299–314, 1996. [Google Scholar]
- 17. Kadegowda AK, Bionaz M, Piperova LS, Erdman RA, Loor JJ. Peroxisome proliferator-activated receptor-gamma activation and long-chain fatty acids alter lipogenic gene networks in bovine mammary epithelial cells to various extents. J Dairy Sci 92: 4276–4289, 2009. [DOI] [PubMed] [Google Scholar]
- 18. Lau P, Fitzsimmons RL, Pearen MA, Watt MJ, Muscat GE. Homozygous staggerer (sg/sg) mice display improved insulin sensitivity and enhanced glucose uptake in skeletal muscle. Diabetologia 54: 1169–1180, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Levin ER. Minireview: Extranuclear steroid receptors: roles in modulation of cell functions. Mol Endocrinol 25: 377–384, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li XJ, Zhang X, Johnson MA, Wang ZB, Lavaute T, Zhang SC. Coordination of sonic hedgehog and Wnt signaling determines ventral and dorsal telencephalic neuron types from human embryonic stem cells. Development 136: 4055–4063, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lujan E, Chanda S, Ahlenius H, Sudhof TC, Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc Natl Acad Sci USA 109: 2527–2532, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lupien M, Eeckhoute J, Meyer CA, Krum SA, Rhodes DR, Liu XS, Brown M. Coactivator function defines the active estrogen receptor alpha cistrome. Mol Cell Biol 29: 3413–3423, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lydon JP, O'Malley BW. Minireview: steroid receptor coactivator-3: a multifarious coregulator in mammary gland metastasis. Endocrinology 152: 19–25, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Malone J, Holloway E, Adamusiak T, Kapushesky M, Zheng J, Kolesnikov N, Zhukova A, Brazma A, Parkinson H. Modeling sample variables with an Experimental Factor Ontology. Bioinformatics 26: 1112–1118, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Markey GM. Carboxylesterase 1 (Ces1): from monocyte marker to major player. J Clin Pathol 64: 107–109, 2011. [DOI] [PubMed] [Google Scholar]
- 26. Medes G, Thomas , Weinhouse S. Metabolism of neoplastic tissue. IV. A study of lipid synthesis in neoplastic tissue slices in vitro. Cancer Res 13: 27–29, 1953. [PubMed] [Google Scholar]
- 27. Naito T, Tanaka H, Naoe Y, Taniuchi I. Transcriptional control of T-cell development. Int Immunol 23: 661–668, 2011. [DOI] [PubMed] [Google Scholar]
- 28. Ndebele K, Graham B, Tchounwou PB. Estrogenic activity of coumestrol, DDT, and TCDD in human cervical cancer cells. Int J Environ Res Public Health 7: 2045–2056, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Notani D, Gottimukkala KP, Jayani RS, Limaye AS, Damle MV, Mehta S, Purbey PK, Joseph J, Galande S. Global regulator SATB1 recruits beta-catenin and regulates T(H)2 differentiation in Wnt-dependent manner. PLoS Biol 8: e1000296, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. O'Malley BW, McKenna NJ. Coactivators and corepressors: what's in a name? Mol Endocrinol 22: 2213–2214, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ochsner SA, Steffen DL, Stoeckert CJ, Jr, McKenna NJ. Much room for improvement in deposition rates of expression microarray datasets. Nat Meth 5: 991, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ochsner SA, Watkins CM, LaGrone BS, Steffen DL, McKenna NJ. Research resource: Tissue-specific transcriptomics and cistromics of nuclear receptor signaling: a web research resource. Mol Endocrinol 24: 2065–2069, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, Varambally S, Ghosh D, Chinnaiyan AM. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9: 166–180, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Samudio I, Harmancey R, Fiegl M, Kantarjian H, Konopleva M, Korchin B, Kaluarachchi K, Bornmann W, Duvvuri S, Taegtmeyer H, Andreeff M. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J Clin Invest 120: 142–156, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Savarese F, Davila A, Nechanitzky R, De La Rosa-Velazquez I, Pereira CF, Engelke R, Takahashi K, Jenuwein T, Kohwi-Shigematsu T, Fisher AG, Grosschedl R. Satb1 and Satb2 regulate embryonic stem cell differentiation and Nanog expression. Genes Dev 23: 2625–2638, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schwimmer LJ, Rohatgi P, Azizi B, Seley KL, Doyle DF. Creation and discovery of ligand-receptor pairs for transcriptional control with small molecules. Proc Natl Acad Sci USA 101: 14707–14712, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Smith CL, O'Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev 25: 45–71, 2004. [DOI] [PubMed] [Google Scholar]
- 38. Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3, 2004. [DOI] [PubMed] [Google Scholar]
- 39. Soprano DR, Teets BW, Soprano KJ. Role of retinoic acid in the differentiation of embryonal carcinoma and embryonic stem cells. Vitam Horm 75: 69–95, 2007. [DOI] [PubMed] [Google Scholar]
- 40. Stegmaier K, Ross KN, Colavito SA, O'Malley S, Stockwell BR, Golub TR. Gene expression-based high-throughput screening(GE-HTS) and application to leukemia differentiation. Nat Genet 36: 257–263, 2004. [DOI] [PubMed] [Google Scholar]
- 41. Su D, Gudas LJ. Gene expression profiling elucidates a specific role for RARgamma in the retinoic acid-induced differentiation of F9 teratocarcinoma stem cells. Biochem Pharmacol 75: 1129–1160, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Swinnen JV, Vanderhoydonc F, Elgamal AA, Eelen M, Vercaeren I, Joniau S, Van Poppel H, Baert L, Goossens K, Heyns W, Verhoeven G. Selective activation of the fatty acid synthesis pathway in human prostate cancer. Int J Cancer 88: 176–179, 2000. [DOI] [PubMed] [Google Scholar]
- 43. Vallieres M, du Souich P. Modulation of inflammation by chondroitin sulfate. Osteoarthritis Cartilage 18, Suppl 1: S1–S6, 2010. [DOI] [PubMed] [Google Scholar]
- 44. Vasudevan N, Pfaff DW. Membrane-initiated actions of estrogens in neuroendocrinology: emerging principles. Endocr Rev 28: 1–19, 2007. [DOI] [PubMed] [Google Scholar]
- 45. Wang SY. A retinoic acid-inducible GATA-binding protein binds to the regulatory region of J6 serpin gene. J Biol Chem 269: 607–613, 1994. [PubMed] [Google Scholar]
- 46. Williams SS, Mear JP, Liang HC, Potter SS, Aronow BJ, Colbert MC. Large-scale reprogramming of cranial neural crest gene expression by retinoic acid exposure. Physiol Genomics 19: 184–197, 2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.