Abstract
Angiogenesis is the formation of new blood vessels from pre-existing microvessels. Excessive and insufficient angiogenesis have been associated with many diseases including cancer, age-related macular degeneration, ischemic heart, brain, and skeletal muscle diseases. A comprehensive understanding of angiogenesis regulatory processes is needed to improve treatment of these diseases. To identify proteins related to angiogenesis, we developed a novel integrative framework for diverse sources of high-throughput data. The system, called GeneHits, was used to expand on known angiogenesis pathways to construct the angiome, a protein-protein interaction network for angiogenesis. The network consists of 478 proteins and 1,488 interactions. The network was validated through cross validation and analysis of five gene expression datasets from in vitro angiogenesis assays. We calculated the topological properties of the angiome. We analyzed the functional enrichment of angiogenesis-annotated and associated proteins. We also constructed an extended angiome with 1,233 proteins and 5,726 interactions to derive a more complete map of protein-protein interactions in angiogenesis. Finally, the extended angiome was used to identify growth factor signaling networks that drive angiogenesis and antiangiogenic signaling networks. The results of this analysis can be used to identify genes and proteins in different disease conditions and putative targets for therapeutic interventions as high-ranked candidates for experimental validation.
Keywords: bioinformatics, systems biology, interactome, endothelial cell, tubulogenesis
angiogenesis, the formation of new blood vessels from pre-existing microvessels, is required for both normal physiological development such as wound healing and exercise, and it accompanies pathological progression of diseases such as cancer and age-related macular degeneration (32). An angiogenic cascade is necessary for tumor growth and metastasis. Without the stable blood supply provided by angiogenesis, tumors are unable to grow past a critical volume of ∼1 mm3 in size or to metastasize (7). Angiogenesis is not determined by a single gene such as vascular endothelial growth factor (VEGF-A), one of the first identified molecular determinants of angiogenesis (7), but by many interacting components. To understand the relationships between these interacting components, we have investigated the systems biology of VEGF (21), the cross talk of protein interaction networks (PINs) including type IV collagens, CXC chemokines, and thrombospondin domain-containing proteins (28), and identified novel and missing angiogenesis annotations (29). Other studies investigated the signaling pathways in angiogenesis (16), identified the responsive gene modules in inflammation and angiogenesis (12) by merging time-series microarray data and genome-wide PINs, and constructed a gene functional network to model in vitro angiogenesis regulation by network perturbation analysis (6). These advances indicate the need for a global analysis of angiogenesis regulatory processes.
The global analysis presented here relies on data integration to identify genes associated with angiogenesis. Several groups have used gene-gene association networks to combine diverse biological datasets, providing new biological understanding and generating testable hypotheses (24, 26). Methods that integrate these gene-gene association networks as kernels, essentially representing each dataset by pairwise relationships between genes, have proven to be some of the most effective for identifying genes with related processes. Early methods transferred gene functions from direct neighbors to annotate genes. Extensions to these methods allowed more distant annotations through shortest paths (38) and graph diffusion (3, 20). Others make use of diverse machine learning techniques including support vector machines (19) and Bayesian networks (37).
To identify the network of angiogenesis-related proteins, we developed a method called GeneHits that combines graph diffusion kernels from protein interactions and pairwise associations from protein domain occurrence to construct a global angiogenesis PIN, the angiome. We then investigated the structural properties of angiome and validated the role of angiome proteins in angiogenesis through analysis of existing microarray datasets. Functional enrichment analysis was also applied for both angiogenesis-annotated and associated proteins in the angiome. We used a two-stage approach to construct the angiome. We first constructed a network with 478 proteins and then extended it to 1,233 proteins. The process allowed us to analyze the results in stages, despite the large network size. The global angiome integrates the current knowledge of angiogenesis.
MATERIALS AND METHODS
Missing gene annotations can be identified from numerous sources of biological data. Proteins that physically interact are more likely to participate in the same biological process or pathway. In addition, proteins that share similar domains are more likely to have similar molecular functions. The integration of these and other sources of data can improve the annotation of proteins. To integrate the attributes of protein physical interactions and domain similarity, we have constructed a method named GeneHits and implemented it as a search engine, as described below.
GeneHits for heterogeneous data integration.
We set up a gene search engine called GeneHits at http://sysbio.bme.jhu.edu. Unlike other methods for heterogeneous data integration, GeneHits explicitly learns weights for network-gene combinations. Let K, Q, and P be the set of networks, query genes, and all proteins, respectively. GeneHits results are broken up into two sections. The first section describes the weighted combination of networks that best discriminate between query and nonquery genes. The second section uses the weights from the first section in a linear combination to score all other genes by their likelihood of association with the query genes. We use the Lasso framework to avoid colinearity and overfitting. For adaptive GeneHits, we learn a vector x of weights with each value representing the influence of a dataset-gene combination. In equal GeneHits, all dataset-gene combinations are presumed to contribute equally. As the gold standard, the vector b indicates the partition between query and nonquery proteins. Entries in b are 1 if the associated protein is a query and zero otherwise. Let k, q, and n be the number of kernels, queries, and proteins, respectively. For each submitted query we solve the following convex optimization problem:
| 1 |
where A is a matrix of size n by k×q, b is a column vector of length n, x is a column vector of length k×q, and λ is the shrinkage coefficient. For protein d in P, e in Q, and f in K, the element Ad,e×f contains the value of the association between d and e from network f. Equation 1 is the standard Lasso objective. The objective contains two parts: the first term is standard multiple linear regression, while the second term penalizes any nonzero entries in x, making x sparse. The selected features correspond to nonzero values in x. In this method, the features we consider are gene and dataset pairs. The scalar parameter λ controls the number of features. A large value of λ will allow fewer features to be selected. We disallow anticorrelation by requiring nonnegative values in the vector x. The objective leads to an additive model for predicting gene associations. For each protein g, we assign the weight sg of the gene to be
| 2 |
In the current study, we evaluated the contribution of two datasets: physical interaction diffusion and protein domain similarity. Higher weights indicate a greater chance of functional association. Perturbation testing and shuffling of these query proteins are also implemented on our website to evaluate the significance of queries.
Example of the adaptive optimization procedure in GeneHits.
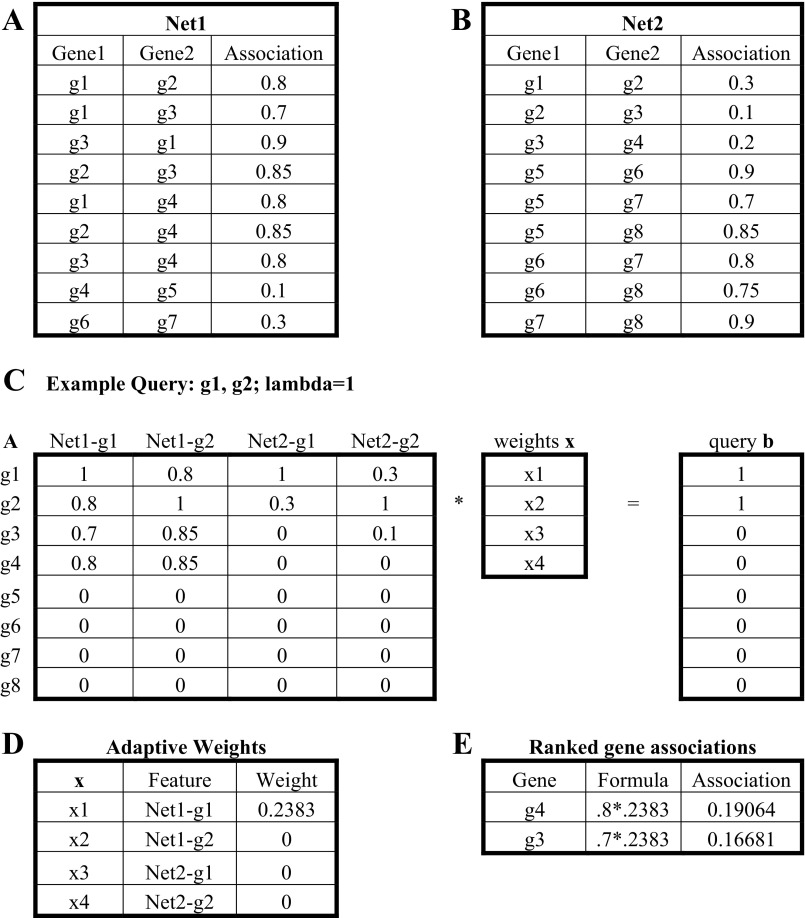
A toy example of the GeneHits adaptive computation is shown in Fig. 1. Figure 1, A and B, shows example network datasets detailing the relationships between pairs of genes (g1-g8). Self-associations are assumed to have weight 1, and other unlisted interactions are assumed to have weight 0. If we take g1 and g2 as an example query, matrices A and b can be constructed as shown in Fig. 1C. If we assume λ = 1, solving for the matrix x gives the feature Net1-g1 a nonzero weight in Fig. 1D. This indicates that the dataset Net1 in association with gene g1 is sufficient to separate queries from nonqueries. Using the weighted feature Net1-g1, we can identify other genes that may be related to the query.
Fig. 1.
A toy example of adaptive weighting of GeneHits. A and B: example network datasets detailing the relationships between pairs of genes (g1–g8). C: examply query: g1, g2: lambda = 1. D: adaptive weights. E: ranked gene associations.
Construction and analysis of angiome.
The MiMI database (9) integrates 11 protein interaction data sources (BIND, CCSB, DIP, GRID, HPRD, IntAct, KEGG, MDC, MINT, PubMed, and Reactome). We included all the 11 databases from MiMI plugin 3.0.1 on Cytoscape 2.8 (34) to construct the angiome. We computed functional enrichment of the genes in the angiome using BiNGO (22) on Cytoscape 2.8 (34). All network parameters in this study are defined in the form of undirected graphs. Network parameters are analyzed by NetworkAnalyzer (1) on Cytoscape 2.8 (34).
Microarray analysis.
We use the software from gene set enrichment analysis studies (36) to compute the q value of the enrichment of angiogenesis-associated proteins in a ranked list of the most perturbed gene expression transcripts. We used packages in Bioconductor to complete this task, including Affy (10) and Limma (35).
RESULTS
The set of angiogenesis-annotated genes.
A list of angiogenesis-annotated genes was compiled from three sources: SABiosciences (84 genes), Gene Ontology (GO) (370 genes) and GeneCards (1,244 genes). The Venn diagram in Fig. 2 shows that 82 of 84 proteins from SABiosciences (Table 1) overlap with GeneCards (Supplementary Table S1; see supplementary files) or GO (Supplementary Table S2).1 Because of the high overlap (∼97.6%) between SABiosciences and the two public databases, we used the 84 genes in the SABiosciences set as the seeds to construct the angiome.
Table 1.
84 genes from SABiosciences
| Angiogenic factors | |
| Growth factors and receptors | ANGPT1, ANGPT2, ANPEP, ECGF1, EREG, FGF1, FGF2, FIGF, FLT1, JAG1, KDR, LAMA5, NRP1, NRP2, PGF, PLXDC1, STAB1, VEGFA, VEGFC |
| Adhesion molecules | ANGPTL3, BAI1, COL4A3, IL8, LAMA5, NRP1, NRP2, STAB1 |
| Proteases, inhibitors and other matrix proteins | ANGPTL4, PECAM1, PF4, PROK2, SERPINF1, TNFAIP2 |
| Transcription factors and others | HAND2, SPHK1 |
| Other factors involved in angiogenesis | |
| Cytokines and chemokines | CCL11, CCL2, CXCL1, CXCL10, CXCL3, CXCL5, CXCL6, CXCL9, IFNA1, IFNB1, IFNG, IL1B, IL6, MDK, TNF |
| Other growth factors and receptors | EDG1, EFNA1, EFNA3, EFNB2, EGF, EPHB4, FGFR3, HGF, IGF1, ITGB3, PDGFA, TEK, TGFA, TGFB1, TGFB2, TGFBR1 |
| Adhesion molecules | CCL11, CCL2, CDH5, COL18A1, EDG1, ENG, ITGAV, ITGB3, THBS1, THBS2 |
| Proteases, inhibitors and other matrix proteins | LECT1, LEP, MMP2, MMP9, PLAU, PLG, TIMP1, TIMP2, TIMP3 |
| Transcription factors and others | AKT1, HIF1A, HPSE, ID1, ID3, NOTCH4, PTGS1 |
Comparison with other topological annotation methods.
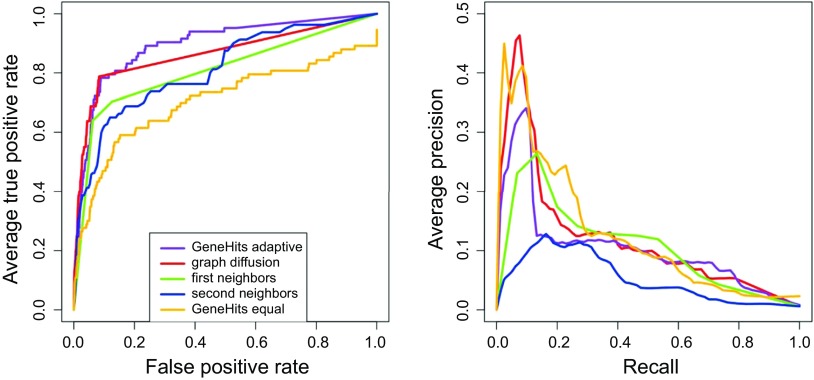
To evaluate the performance of GeneHits, we compared GeneHits to graph diffusion, first neighbor, and second neighbor methods that also predict angiogenesis annotations. We performed a leave-one-out cross-validation (LOOCV) procedure. In Fig. 3, we show the receiver operating characteristic (ROC) and precision recall curves. Graph diffusion is a recent method for functional annotation by counting paths of all lengths between all pairs of vertices in a graph, and adding these path counts to give kernel values for all vertex pairs (27). Detailed mathematical formulation is presented in Refs. 27, 28. The first neighbor method ranks proteins by the number of direct interactions with annotated proteins. The second neighbor method ranks proteins by the number of annotated proteins reachable by paths of length one or two. These unweighted methods are appropriate for the unweighted set of protein-protein interactions used in this comparison.
Fig. 3.
Cross validation by ROC and PR curves. The receiver operating characteristic (ROC) and precision recall (PR) curves display the prediction accuracy of angiogenesis-annotated seeds from leave-one-out cross validation. The areas under the curve (AUC) by GeneHits with adaptive weighting, graph diffusion, first and second neighbor method, and GeneHits with equal weighting are 0.895, 0.864, 0.817, 0.811, and 0.705, respectively.
We measure performance of these approaches by computing the area under the ROC curve (AUC). The AUC of dynamic GeneHits achieves 0.895, compared with 0.864, 0.817, and 0.811 by graph diffusion, first neighbor, and second neighbor methods, respectively. We observe from the ROC curve that GeneHits recovers more angiogenesis-related seeds in LOOCV, or higher recall than first and second neighbor methods under the same false-positive rate. GeneHits adaptively weights dataset and gene combinations. To determine the contribution of this adaptive approach, we compared GeneHits with adaptive weighting to GeneHits with static weighting. In Fig. 3, we show that adaptive weighting results in an AUC of 0.895, while equal weighting results in an AUC of 0.705. We also noticed that GeneHits with static weights performs well in the high-precision, low-recall range; however, GeneHits with adaptive weights performs better overall. The algorithm comparison in Fig. 3 was graphed using the ROCR package (33). Based on this analysis, we constructed the network using GeneHits with adaptive weighting.
Angiogenesis network expansion.
We used the GeneHits method to expand the set of 84 genes (Supplementary Table S3). The method integrated the diffusion kernel for the physical PIN (9, 27) and the protein domain associations (8). Using this method, we scored all the genes by their association to angiogenesis. The threshold for annotating additional angiogenesis proteins was set using the median LOOCV score, representing 50% recall of angiogenesis-annotated proteins. Each angiogenesis-annotated gene in the initial set is assigned an angiogenesis-association score using a model built from all other angiogenesis-annotated genes. The median of angiogenesis-association scores is taken to be the threshold for the angiome. The selection threshold for annotation is needed to strike a balance between false positives and false negatives. With this threshold, it is estimated that the number of false negatives will be <50% of all angiogenesis-related proteins. The false-negative bound indicates that the number of true angiogenesis-related genes not found by our study is estimated to be <50%. For a false-negative rate (i.e., recall) of <50%, the ROC curve in Fig. 3 indicates that adaptive gene hits are likely to have a false-positive rate of <10%.
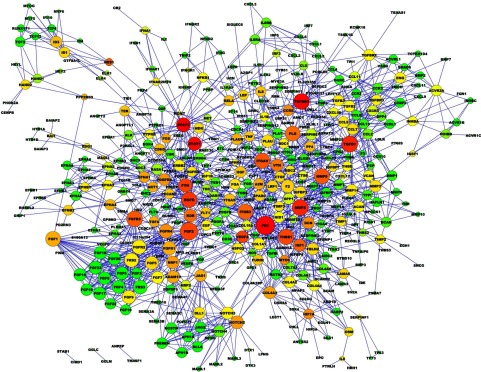
The angiome is composed of 478 proteins and 1,488 interactions as shown in Fig. 4. Larger node size indicates higher degree. The colors from red to green represent the betweenness centrality of each node in descending order. Betweenness centrality measures the fraction of shortest paths that pass through a particular protein. Proteins with high betweenness centrality may be important conduits of information in the network. Details of proteins and interactions in the angiome are listed in Supplementary Tables S4 and S5, respectively. In Supplementary Table S4, 77 proteins from 84 seeds on SABiosciences in Table 1 are angiogenesis-annotated proteins, and the other 401 (= 478 − 77) proteins are angiogenesis-associated proteins.
Fig. 4.
Graphic representation of angiome. Using graph diffusion and MiMI, we constructed the angiome with 478 proteins and 1,488 interactions. Larger nodes indicate higher degrees. Color shifts from red to green indicate the betweenness centrality of each node from larger to lower, respectively. Details of these proteins and interactions are in Supplementary Tables S4 and S5, respectively.
Functional characterization of the angiogenesis network.
Receptor-ligand interactions in endothelial cells drive angiogenesis. To identify pathways and biological processes associated with angiogenesis, we computed functional enrichment of genes in the angiome using BiNGO (22) in Supplementary Table S6, which ranked GO terms by P values. The functional enrichment analysis of genes in the angiome includes most of the molecular and cellular mechanisms of angiogenesis (7). For example, we identify 60 proteins in growth factor activity, 34 proteins in heparin binding, 27 proteins in cytokine binding, 11 proteins in collagen binding, 22 proteins in metallopeptidase activity, and 43 proteins in calcium ion binding in angiogenesis PIN. We will repeat the same procedure of functional analysis of the extended angiome below.
Structure and topological properties of angiome.
The concept of biological networks could integrate the gene regulations, protein interactions, and metabolic networks (2). To compare the entire human interactome with angiome, we measured structural and topological parameters of the angiome using the same mathematical definitions as previous studies (1, 2). The descriptions of these parameters that were discussed in this study are shown in Table 2. The results of the comparison between the entire human interactome and angiome are given in Table 3.
Table 2.
Definition of network parameters
| Parameter | Definition |
|---|---|
| Betweenness centrality | Cb(n) = Σs≠n≠t [σst(n)/σst], where s and t are nodes in the network different from n, σst denotes the number of shortest paths from s to t, and σst(n) is the number of shortest paths from s to t that n lies on |
| Clustering coefficient | Cn = 2en/kn(kn − 1), where en is the number of connected pairs between all neighbors of n, and kn is the number of neighbors of n |
| Degree | number of links to a node |
| Network diameter | largest distance between any 2 nodes in the network |
| Network density | normalized average of degree of nodes in the network |
| Shortest path length | length of the shortest path between 2 nodes n and m |
| Topological coefficient | Tn = avg[J(n,m)]/kn, where J(n,m) is defined for all nodes m that share at least 1 neighbor with n, and kn is the number of neighbors of node n |
Table 3.
Measurement of network parameters in the entire human protein interaction network and the angiome
| Entire | Angiome | |
|---|---|---|
| a [P(k) = ak−γ] | 14594 | 235.23 |
| γ [P(k) = ak−γ] | 1.76 | 1.388 |
| R2 of the power-law fitting | 0.923 | 0.92 |
| Number of nodes | 13,584 | 478 |
| Number of edges | 85,083 | 1,488 |
| Average number of neighbors | 11.27 | 6.226 |
| Clustering coefficient | 0.109 | 0.237 |
| Average shortest path length | 4.086 | 3.972 |
| Network diameter | 11 | 9 |
| Network density | 0.001 | 0.013 |
Network parameters in entire human PIN are directly cited from Ref. 18.
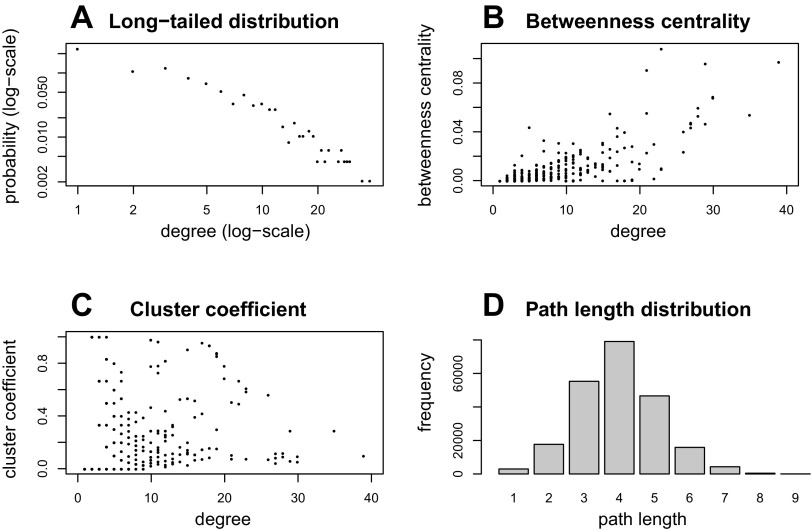
Several features of the parameters of the angiome are worth mentioning. First, the degree distribution has a long tail, but downward curvature on a log-log plot indicates that the degree distribution is not a power law (Fig. 5A). Second, there are 21 proteins with degree ≥15 among the top 24 proteins (∼5%) with the highest value of betweenness centrality. We can also observe in Fig. 4 that nodes with higher degree (bigger node size) are usually associated with higher betweenness centrality (red-colored nodes). Degree and betweenness centrality are positively correlated, as shown in Fig. 5B, which has been observed before for long-tailed networks (17). Third, the clustering coefficient does not correlate with degree of nodes (2), as shown in Fig. 5C. The greater value of clustering coefficient of the angiome than the human interactome in Table 3 indicates that the angiome is more densely connected than the entire human interactome. Fourth, Fig. 5D shows that 4 is the most frequent shortest path length. Fifth, none of the five VEGF ligands VEGFA, VEGFB, VEGFC, VEGFD (FIGF), and PIGF (PGF) and none of the five VEGF receptors VEGFR1 (FLT1), VEGFR2 (KDR), VEGFR3 (FLT4), NRP1, and NRP2 are in the top list, but all the 10 VEGF ligands and receptors (21) are included in the angiome. This suggests that many proteins such as fibroblast growth factor (FGF) family in addition to VEGF ligands and receptors play important roles in the development, maintenance, and remodeling of the vasculature.
Fig. 5.
Network properties of angiome. A: long-tailed degree distribution, with both axes on logarithmic scale. B: plot of degree vs. betweenness centrality. C: plot of degree vs. clustering coefficient. D: bar graph of the distribution of path lengths.
Validation by in vitro time course transcriptional profiling.
We used five published gene expression datasets to verify a relationship between genes in the angiome and angiogenesis. We hypothesized that angiogenesis-associated proteins would have a higher level of expression perturbation on average during angiogenesis than other proteins. We tested the set of angiogenesis-annotated and associated proteins from the angiome using three separate time series microarray sets of VEGF-stimulated endothelial cells (11, 23, 31). Schweighofer et al. (31) measured gene expressions in VEGF-stimulated HUVEC (human umbilical vein endothelial cells) at 0 min, 30 min, 1 h, and 2.5 h. Glesne et al. (11) measured transcripts during tubulogenesis and proliferation on HMVEC (human microvascular endothelial cells) stimulated with VEGF and measured gene expressions at 30 min and 1, 2, 4, and 8 h. Mellberg et al. (23) cultured TIME cells (telomerase-immortalized human microvascular endothelial cells) on fibronectin matrix and 3D collagen gels, stimulated with VEGF and made gene expression measurements at 15 min and 1, 3, 6, 9, 12, 18, and 24 h.
We ranked transcripts by the maximum differential expression observed during the time course relative to an untreated control. We tested the null hypothesis that the distribution of angiogenesis-associated proteins was uniform throughout the ranked list of transcripts. We determined the significance of the enrichment using the Kolmogorov-Smirnov test. The significant false discovery rate (FDR) q value indicates that the angiogenesis-associated proteins were disproportionally ranked at either the head or tail of the ranked list of transcripts (36).
The results of the external gene expression analysis are summarized in Table 4. We found that the set of angiogenesis-annotated proteins was significantly perturbed (FDR, q<0.05) in HMVEC tubulogenesis, HMVEC combination of tubulogenesis and proliferation, TIME collagen, TIME fibronectin, and TIME combination of collagen and fibronectin. The five sets of angiogenesis-associated proteins and both annotated and associated proteins on HMVEC (GDS2039; GDS: Gene Expression Omnibus datasets) and TIME cells (Mellberg et al., Ref. 23) except HUVEC (GDS3567) were significantly perturbed (q < 0.05). We found that the combined set of proteins was more significantly perturbed than the set of angiogenesis-annotated proteins alone in HUVEC (GDS3567) and HMVEC proliferation (GDS2039).
Table 4.
Enrichment of angiogenesis-annotated and -associated proteins in a ranked list of the most perturbed gene expression transcripts during angiogenesis
| FDR (q value) | Angiogenesis-annotated Proteins Only | Angiogenesis-associated Proteins Only | Angiogenesis-annotated and -associated Proteins |
|---|---|---|---|
| HUVEC (GDS3567) | 0.147 | 0.104 | 0.122 |
| HMVEC proliferation (GDS2039) | 0.33 | 0.032 | 0.029 |
| HMVEC tubulogenesis (GDS2039) | 0.0001 | 0.012 | 0.005 |
| HMVEC combination (GDS2039) | 0.004 | 0.006 | 0.003 |
| TIME collagen (Ref. 23) | 0.0001 | 0.005 | 0.001 |
| TIME fibronectin (Ref. 23) | 0.0001 | 0.0001 | 0.0001 |
| TIME combination (Ref. 23) | 0.0001 | 0.001 | 0.0001 |
Biological analysis of angiogenesis-associated proteins.
Out of the 478 proteins, there are 235 that are not included in any of the three databases in Fig. 2. We used ToppGene http://toppgene.cchmc.org/enrichment.jsp (5) to analyze GO molecular functions and biological processes for angiogenesis-annotated and angiogenesis-associated proteins in Supplementary Table S7, with P value cutoff 0.05 and Bonferroni correction. The top five features (P < 1.33E-65) of GO biological processes in angiogenesis-annotated proteins include vasculature development, angiogenesis, blood vessel development, blood vessel morphogenesis, and cardiovascular system development. The top five features (P < 1.47E-31) of GO biological processes in angiogenesis-associated proteins include enzyme-linked receptor protein signaling pathway, locomotion, transmembrane receptor protein tyrosine kinase signaling pathway, response to wounding, and response to external stimulus. Many of the associated proteins linked to angiogenesis are involved in the regulation of many biological processes of endothelial cells, including angiogenesis, cell proliferation, cell migration, cell adhesion, cell division, cell motility, cell differentiation, and cell communication.
Fig. 2.
Overlap of angiogenesis genes from SABiosciences, GO and GeneCards. There are 84, 370, and 1,244 angiogenesis-related genes in SABiosciences, Gene Ontology (GO), and GeneCards, respectively. Of the 84 proteins from SABiosciences, 82 overlap with GO or GeneCards. Because of the high overlap (∼97.6%) between SABiosciences and 2 different public databases, we used the 84 genes from the SABiosciences website as the angiogenesis-annotated seeds to construct the angiome.
Extended angiome.
GeneCards and GO contain 810 (= 1,288 − 478) proteins that are absent from the angiome. To ameliorate the effects of missing data, we constructed an extended angiome by adding the original angiome and the genes in the union of the three databases shown in Fig. 2, and all molecular interactions from MiMI (9). The extended angiome is composed of 1,233 proteins and 5,726 interactions. Supplementary Table S8 lists the proteins in this extended angiome ranked by the degree of nodes. The top 20 genes with the highest degree values in this extended angiome are JUN, SRC, MYC, GRB2, TP53, EGFR, EP300, PIK3R1, MAPK1, CTNNB1, SHC1, ESR1, STAT3, FYN, FN1, CREBBP, AKT1, RELA, PTPN11, and MAPK3. These proteins play an important role in cancer and human interactomes, or cross talk between different signaling pathways and angiogenesis, but may not be specific to angiogenesis.
Use of gene expression data sets.
Integration of gene coexpression matrix based on Pearson correlation coefficients (PCC) of each gene pair and protein-protein interactions could provide biological interpretations of interactions under different experimental conditions. We calculated the PCC of each interaction among 5,726 links from the five independent time series in vitro gene expression data (11, 23, 31) as shown in Supplementary Table S9. The coexpression matrix shows the divergence of gene expression profiles of each protein-protein interaction pair among five different experimental conditions. PCC rank could reveal gene-to-gene functional relationships (25). PCC values have relevance in identifying whether interaction partners of a protein are simultaneously or independently expressed, often described as party and date hubs (14).
PubMed search for newly identified angiogenesis-related genes.
To further validate our results, we searched for “angiogenesis protein novel” and compiled newly identified angiogenesis-related proteins that have been reported in PubMed from January 1, 2012 to June 30, 2012. Supplementary Table S10 lists 30 novel angiogenesis-related proteins and references. Six proteins (CXCL12, DCN, ENG, IGFBP7, SDC2, and SEMA3A) are included in both angiome and extended angiome. Ten proteins (CDH1, CDKN2A, DKK1, ERG, MAPK14, MCAM, NES, SEMA4D, SHC1, and TRAF6) are not included in angiome but are included in extended angiome. Nine proteins (BAG3, DLK1, FABP4, GOLGA2, HTRA1, NF2, NOSTRIN, STUB1, and TFCP2) are not included in either angiome or extended angiome but are the first neighbors linked to proteins in angiome or extended angiome. The remaining five proteins (HSPB6, CYTH2, MMRN2, NRN1, and TMIGD2) are not linked to any proteins in angiome or extended angiome. Except for HSPB6, there are no protein-protein interaction records of these unlinked angiome or extended angiome proteins in MiMI (9). These results further demonstrate that the proteins and neighbors in the angiome and extended angiome could be a rich resource for discovery of novel angiogenesis-related proteins.
DISCUSSION
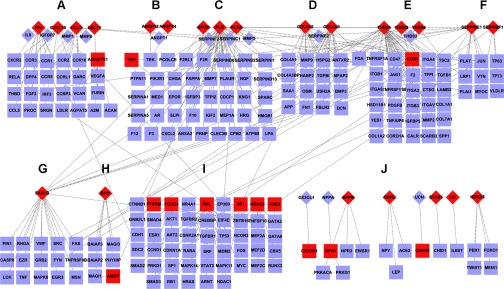
In 2000 Hagedorn and Bikfalvi (13) summarized the receptor-ligand interactions responsible for angiogenesis. The presented network of angiogenesis-related proteins could be used as a foundation to display additional growth factor signaling pathways and their interactions. To elucidate all diverse growth factors and interactions with receptors, we used BiNGO (22) to analyze functional enrichment of 171 genes enriched with growth factor-related annotations in Supplementary Table S11. In Fig. 6, we present the major extracellular regulators of angiogenesis and their receptors. We divided these regulator proteins into 11 categories: (A) vascular endothelial growth factor signaling, (B) fibroblast growth factor signaling, (C) epidermal growth factor signaling, (D) transforming growth factor signaling, (E) insulin-like growth factor signaling, (F) platelet-derived growth factor signaling, (G) inflammation and immune response, (H) other cytokines, (I) focal adhesion, (J) Wnt signaling pathway, (K) others. Figure 6 illustrates the cross talk and redundancy inherent in angiogenesis-related receptor-ligand signaling.
Fig. 6.
Angiogenesis regulators and associated proteins. Based on Supplementary Tables S8 and S9, we divided these 171 angiogenesis regulator proteins into 11 categories: (A) vascular endothelial growth factor signaling, (B) fibroblast growth factor signaling, (C) epidermal growth factor signaling, (D) transforming growth factor signaling, (E) insulin-like growth factor signaling, (F) platelet-derived growth factor signaling, (G) inflammation and immune response, (H) other cytokines, (I) focal adhesion, (J) Wnt signaling pathway, (K) others.
To validate the angiogenesis regulator proteins shown in Fig. 6 and Supplementary Table S11, we used ToppGene (5) as an independent resource of gene list enrichment analysis other than BiNGO (22). The top 10 GO molecular functions (P < 2.72E-24) are growth factor activity, receptor binding, growth factor binding, growth factor receptor binding, cytokine receptor binding, transmembrane receptor protein kinase activity, cytokine activity, fibroblast growth factor receptor binding, platelet-derived growth factor receptor binding, and transmembrane receptor protein tyrosine kinase activity; these results corroborate Fig. 6 and Supplementary Table S11.
Angiogenesis inhibitors, particularly polypeptides or endogenous peptides, may become the safest and least toxic therapy for diseases associated with abnormal angiogenesis. These peptides have been derived from thrombospondin, collagens, chemokines, coagulation cascade proteins, growth factors, and other classes of proteins and target different receptors (30). From the extended angiome, we selected the 36 proteins (marked as red in Fig. 7) that were also annotated for negative regulation of angiogenesis by BINGO analysis of the extended angiome, and extracted their interaction partners (Supplementary Table S12). Nodes shown as diamonds and rectangles represent the proteins in extracellular and intracellular or membrane regions, respectively. We generally divided negative regulators of angiogenesis proteins into 10 categories: (A) chemokines, (B) angiopoietin, (C) urokinase plasiminogen, (D) collagen family, (E) thrombospondin, (F) serpins, (G) Fas ligand, (H) brain-specific angiogenesis inhibitors, (I) transcription factor activities, (J) others. These proteins cover most of endogenous antiangiogenesis proteins that were discussed in Rosca et al. (30).
Fig. 7.
Negative regulators of angiogenesis and associated proteins. From the extended angiome, we selected 36 proteins (nodes shown in red) annotated as negative regulation of angiogenesis by BINGO analysis and found their 210 interactive proteins. Diamond- and rectangular-shaped nodes represent the proteins in extracellular and intracellular or membrane regions, respectively. We generally divided negative regulators of angiogenesis proteins into 10 categories: (A) chemokines, (B) angiopoietin, (C) urokinase plasiminogen, (D) collagen family, (E) thrombospondin, (F) serpins, (G) Fas ligand, (H) brain-specific angiogenesis inhibitors, (I) transcription factor activities, (J) others.
We also compared the angiome with CancerGenes (15), a gene selection resource for cancer genome projects. There are only 42 genes from the gene list on http://cbio.mskcc.org/CancerGenes/Select.action as listed in Supplementary Table S13. Many genes would be missed if researchers use one single information source, providing an incomplete view of the proteins involved in a specific process. When comparing the angiome to the gene functional association network constructed by Chen et al. (6) for antiangiogenic kinase inhibitor activity assessment, we found a number of known angiogenesis-related processes missing. Some of these processes include the platelet-derived growth factor receptor binding, ephrin receptor activity, fibroblast growth factor receptor binding, metalloendopeptidase activity, interleukin-1 receptor activity, chemokine receptor binding, and Notch binding. Therefore, our results significantly expand the protein network of biological importance for angiogenesis.
In summary, we constructed the most complete network of proteins that have been demonstrated as angiogenesis related or have the potential to be angiogenesis related. This network should be viewed as a resource for discovery that balances selectivity and sensitivity, and some of the proteins identified by our analysis (Supplementary Tables S8 and S9) may not be involved in angiogenesis. Top candidates from this analysis may be validated in experimental studies in various applications. We can envision some of the applications of the angiome networks. Angiogenesis is a key component of over 70 diseases, including cancer, wet age-related macular degeneration, pre-eclampsia, atherosclerosis, pathological obesity, asthma, diabetes, endometriosis, autoimmune diseases such as Crohn's disease, and ischemic diseases such as coronary and peripheral artery diseases (4). In addition to pathophysiological, disease conditions angiogenesis plays fundamental role in physiological processes such as development, exercise, and aging. In many of these physiological and pathophysiological conditions a vast amount of genomic, epigenomic, and proteomic information is becoming available and accessible via public databases. The angiome provides us with a platform to identify those genes and proteins in the databases that are associated with angiogenesis by comparing the disease- or condition-specific data to the angiome. Examples may include identification of specific angiogenesis-associated genes that are up- or downregulated or mutated in disease conditions. This information will be new and not readily identifiable by other means. Some of these genes and proteins may then be considered as putative targets for therapeutic interventions that, ultimately, will need to be experimentally validated.
GRANTS
This work was supported by National Institutes of Health Grants R01 CA-138264 and R01 HL-101200 (A. S. Popel), and U54 RR-020839 and the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation (J. S. Bader).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: L.-H.C. conception and design of research; L.-H.C. and C.G.R. performed experiments; L.-H.C. analyzed data; L.-H.C. and C.G.R. interpreted results of experiments; L.-H.C. and C.G.R. prepared figures; L.-H.C. and C.G.R. drafted manuscript; L.-H.C., C.G.R., A.S.P., and J.S.B. edited and revised manuscript; L.-H.C., C.G.R., A.S.P., and J.S.B. approved final version of manuscript.
Supplementary Material
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Assenov Y, Ramirez F, Schelhorn SE, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics 24: 282–284, 2008. [DOI] [PubMed] [Google Scholar]
- 2.Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet 5: 101–113, 2004. [DOI] [PubMed] [Google Scholar]
- 3.Borgwardt KM, Ong CS, Schonauer S, Vishwanathan SV, Smola AJ, Kriegel HP. Protein function prediction via graph kernels. Bioinformatics 21, Suppl 1: i47–i56, 2005. [DOI] [PubMed] [Google Scholar]
- 4.Carmeliet P. Angiogenesis in life, disease and medicine. Nature 438: 932–936, 2005. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37: W305–W311, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, Wei T, Yan L, Lawrence F, Qian HR, Burkholder TP, Starling JJ, Yingling JM, Shou J. Developing and applying a gene functional association network for anti-angiogenic kinase inhibitor activity assessment in an angiogenesis co-culture model. BMC Genomics 9: 264, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Figg WD, Folkman J. Angiogenesis an integrative approach from science to medicine. New York: Springer, 2008. [Google Scholar]
- 8.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res 36: D281–D288, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao J, Ade AS, Tarcea VG, Weymouth TE, Mirel BR, Jagadish HV, States DJ. Integrating and annotating the interactome using the MiMI plugin for cytoscape. Bioinformatics 25: 137–138, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315, 2004. [DOI] [PubMed] [Google Scholar]
- 11.Glesne DA, Zhang W, Mandava S, Ursos L, Buell ME, Makowski L, Rodi DJ. Subtractive transcriptomics: establishing polarity drives in vitro human endothelial morphogenesis. Cancer Res 66: 4030–4040, 2006. [DOI] [PubMed] [Google Scholar]
- 12.Gu J, Chen Y, Li S, Li Y. Identification of responsive gene modules by network-based gene clustering and extending: application to inflammation and angiogenesis. BMC Syst Biol 4: 47, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagedorn M, Bikfalvi A. Target molecules for anti-angiogenic therapy: from basic research to clinical trials. Crit Rev Oncol Hematol 34: 89–110, 2000. [DOI] [PubMed] [Google Scholar]
- 14.Han JD, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, Dupuy D, Walhout AJ, Cusick ME, Roth FP, Vidal M. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 430: 88–93, 2004. [DOI] [PubMed] [Google Scholar]
- 15.Higgins ME, Claremont M, Major JE, Sander C, Lash AE. CancerGenes: a gene selection resource for cancer genome projects. Nucleic Acids Res 35: D721–D726, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y, Li S. Detection of characteristic sub pathway network for angiogenesis based on the comprehensive pathway network. BMC Bioinformatics 11, Suppl 1: S32, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang YJ, Hang D, Lu LJ, Tong L, Gerstein MB, Montelione GT. Targeting the human cancer pathway protein interaction network by structural genomics. Mol Cell Proteomics 7: 2048–2060, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kar G, Gursoy A, Keskin O. Human cancer protein-protein interaction network: a structural perspective. PLoS Comput Biol 5: e1000601, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanckriet GR, De Bie T, Cristianini N, Jordan MI, Noble WS. A statistical framework for genomic data fusion. Bioinformatics 20: 2626–2635, 2004. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Chen H, Li J, Zhang Z. Gene function prediction with gene interaction networks: a context graph kernel approach. IEEE Trans Inf Technol Biomed 14: 119–128, 2010. [DOI] [PubMed] [Google Scholar]
- 21.Mac Gabhann F, Popel AS. Systems biology of vascular endothelial growth factors. Microcirculation 15: 715–738, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449, 2005. [DOI] [PubMed] [Google Scholar]
- 23.Mellberg S, Dimberg A, Bahram F, Hayashi M, Rennel E, Ameur A, Westholm JO, Larsson E, Lindahl P, Cross MJ, Claesson-Welsh L. Transcriptional profiling reveals a critical role for tyrosine phosphatase VE-PTP in regulation of VEGFR2 activity and endothelial cell morphogenesis. FASEB J 23: 1490–1502, 2009. [DOI] [PubMed] [Google Scholar]
- 24.Murali TM, Wu CJ, Kasif S. The art of gene function prediction. Nat Biotechnol 24: 1474–1475, 2006. [DOI] [PubMed] [Google Scholar]
- 25.Obayashi T, Kinoshita K. Rank of correlation coefficient as a comparable measure for biological significance of gene coexpression. DNA Res 16: 249–260, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pena-Castillo L, Tasan M, Myers CL, Lee H, Joshi T, Zhang C, Guan Y, Leone M, Pagnani A, Kim WK, Krumpelman C, Tian W, Obozinski G, Qi Y, Mostafavi S, Lin GN, Berriz GF, Gibbons FD, Lanckriet G, Qiu J, Grant C, Barutcuoglu Z, Hill DP, Warde-Farley D, Grouios C, Ray D, Blake JA, Deng M, Jordan MI, Noble WS, Morris Q, Klein-Seetharaman J, Bar-Joseph Z, Chen T, Sun F, Troyanskaya OG, Marcotte EM, Xu D, Hughes TR, Roth FP. A critical assessment of Mus musculus gene function prediction using integrated genomic evidence. Genome Biol 9 Suppl 1: S2, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi Y, Suhail Y, Lin YY, Boeke JD, Bader JS. Finding friends and enemies in an enemies-only network: a graph diffusion kernel for predicting novel genetic interactions and co-complex membership from yeast genetic interactions. Genome Res 18: 1991–2004, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rivera CG, Bader JS, Popel AS. Angiogenesis-associated crosstalk between collagens, CXC chemokines, and thrombospondin domain-containing proteins. Ann Biomed Eng 39: 2213–2222, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rivera CG, Mellberg S, Claesson-Welsh L, Bader JS, Popel AS. Analysis of VEGF-A regulated gene expression in endothelial cells to identify genes linked to angiogenesis. PLoS One 6: e24887, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosca EV, Koskimaki JE, Rivera CG, Pandey NB, Tamiz AP, Popel AS. Anti-angiogenic peptides for cancer therapeutics. Curr Pharm Biotechnol 12: 1101–1116, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schweighofer B, Testori J, Sturtzel C, Sattler S, Mayer H, Wagner O, Bilban M, Hofer E. The VEGF-induced transcriptional response comprises gene clusters at the crossroad of angiogenesis and inflammation. Thromb Haemost 102: 544–554, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seaman S, Stevens J, Yang MY, Logsdon D, Graff-Cherry C, St Croix B. Genes that distinguish physiological and pathological angiogenesis. Cancer Cell 11: 539–554, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics 21: 3940–3941, 2005. [DOI] [PubMed] [Google Scholar]
- 34.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27: 431–432, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3, 2004. [DOI] [PubMed] [Google Scholar]
- 36.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Troyanskaya OG, Dolinski K, Owen AB, Altman RB, Botstein D. A Bayesian framework for combining heterogeneous data sources for gene function prediction (in Saccharomyces cerevisiae). Proc Natl Acad Sci USA 100: 8348–8353, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou X, Kao MC, Wong WH. Transitive functional annotation by shortest-path analysis of gene expression data. Proc Natl Acad Sci USA 99: 12783–12788, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.