Abstract
The prominent role of Horizontal Gene Transfer (HGT) in the evolution of bacteria is now well documented, but few studies have differentiated between evolutionary events that predominantly cause genes in one lineage to be replaced by homologs from another lineage (“replacing HGT”) and events that result in the addition of substantial new genomic material (“additive HGT”). Here in, we make use of the distinct phylogenetic signatures of replacing and additive HGTs in a genome-wide study of the important human pathogen Streptococcus pyogenes (SPY) and its close relatives S. dysgalactiae subspecies equisimilis (SDE) and S. dysgalactiae subspecies dysgalactiae (SDD). Using recently developed statistical models and computational methods, we find evidence for abundant gene flow of both kinds within each of the SPY and SDE clades and of reduced levels of exchange between SPY and SDD. In addition, our analysis strongly supports a pronounced asymmetry in SPY–SDE gene flow, favoring the SPY-to-SDE direction. This finding is of particular interest in light of the recent increase in virulence of pathogenic SDE. We find much stronger evidence for SPY–SDE gene flow among replacing than among additive transfers, suggesting a primary influence from homologous recombination between co-occurring SPY and SDE cells in human hosts. Putative virulence genes are correlated with transfer events, but this correlation is found to be driven by additive, not replacing, HGTs. The genes affected by additive HGTs are enriched for functions having to do with transposition, recombination, and DNA integration, consistent with previous findings, whereas replacing HGTs seen to influence a more diverse set of genes. Additive transfers are also found to be associated with evidence of positive selection. These findings shed new light on the manner in which HGT has shaped pathogenic bacterial genomes.
Keywords: bacterial evolutionary genomics, recombination, Streptococcus pyogenes, Streptococcus dysgalactiae
Introduction
During the last 20 years, it has become increasingly apparent that Horizontal Gene Transfer (HGT) has played a major role in genomic evolution, particularly among bacteria and other microbes (Smith et al. 1992; Maynard-Smith et al. 1993; Ochman et al. 2000; Koonin et al. 2001). Indeed, it is likely that HGT has been sufficiently prevalent throughout evolutionary history that no present-day gene can trace an unbroken history of vertical descent to a common ancestor for all species (Zhaxybayeva and Doolittle 2011), meaning that no single phylogenetic marker can be used to reconstruct a universal tree of life.
For HGT to occur in bacteria, DNA must first be transferred from a donor to a recipient cell by transformation, transduction, or conjugation, then must be integrated into the recipient cell's genome (setting aside the case of replication-proficient plasmids). This typically occurs in one of two major ways: either the new sequence replaces a homologous sequence through the process of homologous recombination (similar to “gene conversion” in sexually reproducing organisms) or it is acquired through an additive (nonreplacing) integration process (Thomas and Nielsen 2005). Although the mechanisms underlying these “replacing” and “additive” events overlap (e.g., additive HGTs can be associated with homologous recombination at flanking regions), replacing and additive HGTs nevertheless leave distinct molecular evolutionary signatures (fig. 1). The rates at which these processes occur depend on many factors, including the co-occurrence of bacterial species in the environment, the degree of competence of recipient cells, sequence similarity between the donor and recipient, and barriers to conjugation such as surface exclusion. In addition, the persistence of horizontally transferred DNA segments in present-day species depends strongly on the subsequent effects of natural selection. Not surprisingly, rates of HGT seem appear to vary considerably across groups of bacteria (e.g., Feil et al. 2001).
Fig. 1.

Replacing and additive HGT. (A) Foreign DNA can be added to a recipient genome by a replacing HGT (left) or an additive HGT (right). (B) These types of transfers produce distinct phylogenetic signatures. In a replacing HGT, a segment of DNA is effectively overwritten by a homologous segment from another species, which causes a lineage in the phylogeny to be replaced by a transferred lineage. This type of transfer can be identified in gene-tree/species-tree reconciliation by the appearance of coinciding transfer and loss events (left tree). An additive HGT, on the other hand, leads to a transfer event that is not paired with a loss event (right tree). Of course, parallel losses can cause an additive HGT to appear similar to a replacing HGT. Therefore, in our analysis, we conservatively require that at least one descendant species (here, B) contains genes that both descend from (b1) and do not descend from (b2) a transferred gene when inferring an additive HGT event. It is worth noting that similar biological processes may contribute to both types of events—for example, an additive HGT may be associated with homologous recombination in flanking regions. Our interest here is in distinguishing between evolutionary processes that do (additive HGT) and do not (replacing HGT) tend to alter the size and gene composition of a genome.
HGT in the Streptococcus genus is of particular interest. This group of Gram-positive bacteria contains several human pathogens, including Streptococcus pyogenes (SPY; the cause of Group A streptococcal infections, including pharyngitis, impetigo, cellulitis, necrotizing fasciitis, and rheumatic fever), S. pneumoniae (bacterial pneumonia), S. agalactiae (neonatal sepsis), S. mutans (dental caries), and S. dysgalactiae ssp. equisimilis (SDE; cellulitis, peritonitis, pneumonia, and other infections), as well as agricultural pathogens, such as S. uberis and S. dysgalactiae ssp. dysgalactiae (SDD; mastitis in cows, ewes, and goats), S. equi ssp. equi (strangles in horses), and S. canis (various infections in dogs and other animals). Many of these species are concentrated in the predominantly beta-hemolytic pyogenic division (Facklam 2002). These species have adapted to a remarkable variety of distinct ecological niches and display high rates of recombination and HGT, as well as substantial evidence of positive selection (Feil et al. 2001; Marri et al. 2006; Anisimova et al. 2007; Lefébure and Stanhope 2007; Suzuki et al. 2011). Complete genome sequences are now available for several species in the pyogenic division (e.g., Ferretti et al. 2001; Holden et al. 2009; Shimomura et al. 2011; Suzuki et al. 2011), providing a rich resource for the study of the genomic evolution of pathogenic bacteria.
Several studies have focused in particular on the issue of HGT between the human-colonizing species SPY and SDE. Until recently, SDE was considered to be primarily a commensal organism (Vandamme et al. 1996), unlike SPY, but it has become increasingly apparent that it also has an important pathogenetic role, with a disease spectrum similar to that of SPY (Brandt and Spellerberg 2009). Shared virulence genes are well documented between the two species (e.g., Davies, McMillan, Beiko et al. 2007), genetic exchange presumably having been enabled by both their close evolutionary relationship and shared ecological niches. In recent years, evidence has emerged for more widespread SPY–SDE gene flow (Kalia and Bessen 2004; Davies et al; 2005, Davies, McMillan, Beiko et al. 2007; Davies, McMillan, Domselaar et al. 2007; Ahmad et al. 2009; McMillan et al. 2010; Jensen and Kilian 2012). An early study of seven housekeeping genes used for multilocus sequence typing (MLST) (Kalia et al. 2001) additionally claimed a pronounced bias in the direction of gene flow, favoring the SPY-to-SDE direction. However, this study was eventually retracted due to the authors' inability to replicate their findings and concerns about sample contamination (Kalia et al. 2009). Subsequent analyses of the same loci have not supported SPY-to-SDE gene flow (Ahmad et al. 2009; McMillan et al. 2010), and examples have been found of gene flow in the opposite (SDE-to-SPY) direction (Sachse et al. 2002). In any case, these studies have all been limited to small numbers of loci, and questions remain open about genome-wide patterns of gene flow between SPY and SDE. These issues are of particular interest because of the possibility that increasing virulence of SDE may derive, at least in part, from gene flow from SPY.
Most analyses of HGT have been based on relatively simple phylogenetic methods that make use of incongruity of inferred gene trees across loci (e.g., Kalia and Bessen 2004; Ahmad et al. 2009). These methods are sensitive to errors in phylogenetic reconstruction and ignore information from branch lengths and phylogenetic correlation at adjacent loci. Recently, alternative, model-based approaches have been proposed for statistical inference of replacing HGTs, assumed to derive from homologous recombination (Didelot and Falush 2007; Didelot et al. 2010). Using Bayesian principles and Markov chain Monte Carlo (MCMC) techniques, these methods allow for uncertainty in the phylogeny, make use of branch lengths and correlations across loci, and allow for inference not only of individual recombination events but also of parameters describing global rates and patterns of recombination. In addition, several recent methods have been introduced to detect HGT in a phylogenetic framework, by parsimoniously reconciling reconstructed gene trees with a given species tree, allowing for duplication, transfer, and loss events (Merkle et al. 2010; David and Alm 2011; Doyon et al. 2011; Doyon et al. 2012). These new model- and parsimony-based methods are complementary in many respects, and could be particularly useful in combination.
In this article, we take a fresh look at the issue of HGT in the pyogenic division, making use of newly available complete genome sequences for SPY, SDE, and the related species SDD, as well as new statistical and parsimony-based methods for analysis. The combined use of these methods allows us to examine both replacing and additive HGTs and compare their genome-wide effects. We find evidence for abundant gene flow within SPY, within SDE, and between SPY and SDE and for greatly reduced exchange between SPY and SDD. In addition, our genome-wide analysis supports a pronounced preference for the SPY-to-SDE direction in SPY–SDE gene flow. We find that this property is much more evident for replacing than for additive transfers. We also examine correlations of gene transfer events with functional categories and positive selection. Our results have been made available as tracks in our recently released Streptococcus genome browser.
Materials and Methods
Genome Sequences and Alignments
Our primary analysis was based on five complete genome sequences, including two representatives of SPY (accessions NC_004070 [Beres et al. 2002; SPY1] and NC_008024 [Beres et al. 2006; SPY2]), two of SDE (CP002215 [Suzuki et al. 2011; SDE1] and NC_012891 [Shimomura et al. 2011; SDE2]), and one of SDD (CM001076 [Suzuki et al. 2011]). In addition, we used S. equi ssp. equi (SEE) strain 4047 (NC_012471 [Holden et al. 2009]) as an outgroup in our parsimony-based analysis (supplementary table S1, Supplementary Material online). Our primary data set includes only 2 of the 14 publicly available genome sequences for SPY. The reason for this restriction is that our evolutionary analyses require a well-defined “clonal frame,” or species phylogeny, that applies genome wide and, therefore, is not well suited for simultaneous analysis of multiple genomes from the same species (assuming non-negligible levels of intraspecies recombination). Instead, we tested the robustness of our results to the choice of SPY genomes by repeating several of our analyses using 10 different pairs of genomes in place of SPY1 and SPY2, making sure to include representatives of all 11 serotypes for which genome sequences are publicly available (see Results).
For the model-based analysis of replacing transfers, we obtained high-quality alignments of SPY1, SPY2, SDE1, SDE2, and SDD using the pipeline detailed by Didelot and coworkers (Didelot and Falush 2007; Didelot et al. 2010). Briefly, we first aligned these five genomes using progressiveMauve v2.3.1 (Darling et al. 2004, 2010) with default options (supplementary fig. 1, Supplementary Material online), and then used stripSubsetLCBs (from the ClonalOrigin package; Didelot et al. 2010) to identify 276 blocks of sequence alignments that exceed a length threshold of 1,500 bp. This conservative threshold was selected to avoid biases from boundary effects in short alignment blocks based on a preliminary analysis. Two blocks containing long gaps were excluded from the analysis. The final set of 274 alignment blocks contained 1,155,016 columns with relatively few gaps (0.2% of characters) and covered 1,106 of the 1,951 genes in the SPY1 genome. Note that this alignment procedure effectively focused on the “core genome” by discarding regions that were not conserved among the five bacterial individuals in question. The same pipeline was applied to the 10 alternative pairs of SPY genomes with similar results.
For the parsimony-based analysis, we began with 3,408 gene families representing all six genomes (including SEE), as described by Suzuki et al (2011). Approximately one-third of these families (1,094) are represented by a single gene from a single genome. These families do not contribute to transfer events among the analyzed genomes and were thus removed from the analysis. Among the remaining 2,314 gene family clusters, approximately half (1,066) contain a single gene from each of the six genomes, whereas the remaining families ranged from as few as two genes (415 families) to as many as 59 (one family) (supplementary fig. S2, Supplementary Material online). We aligned protein sequences corresponding to each family using MUSCLE (Edgar 2004) and then obtained nucleotide alignments by reverse translation, using the genomic sequences as a guide. Next we identified likely intragenic recombinations using the single breakpoint recombination (SBP) method from the HyPhy package (Kosakovsky Pond et al. 2006). Based on the Akaike Information Criterion, SBP identified at least one topology-altering recombination in 1,094 (47.5%) gene families. These genes were split into two putative nonrecombining gene fragments for subsequent analysis. We later performed a similar analysis in which we allowed for as many as eight breakpoints per gene and observed little change in our results (Supplementary Material online). Note that the alignments described here in are representative of the “pan genome,” including regions that frequently turn over (“dispensable” regions), in addition to the “core genome” analyzed by the model-based approach (e.g., Tettelin et al. 2005; Lefébure and Stanhope 2007).
Model-Based Analysis of Replacing HGTs
ClonalFrame and ClonalOrigin
To study replacing HGTs, we made use of the ClonalOrigin program recently developed by Didelot et al. (2010). Given a set of alignment blocks, ClonalOrigin samples from the posterior distribution of recombinant graphs using a MCMC algorithm. A recombinant graph consists of a pre-estimated rooted phylogeny with branch lengths (a “clonal frame”), augmented by a set of recombinant edges. Each recombinant edge is defined by the two points along the branches of the clonal frame that correspond to the donor and recipient in the hypothesized recombination event and by the start and end of the affected genomic interval. The donor can predate the recipient to allow for a delayed coalescence, but it cannot postdate it (supplementary fig. S3, Supplementary Material online). Another program, called ClonalFrame (Didelot and Falush 2007), can be used to infer the phylogeny that serves as the basis of the recombinant graph. ClonalFrame makes use of a similar, but somewhat simpler, probabilistic model.
Fitting the Model
Following Didelot et al. (2010), we used ClonalFrame (v1.1) to estimate a clonal frame for our five-way alignments, running the algorithm for 104 burn-in iterations followed by another 104 sampling iterations. A series of seven additional sampling runs with random initialization converged to essentially identical estimates, indicating stable convergence. We then used an initial run of ClonalOrigin (subversion r19) to estimate global parameters including the mutation rate, recombination rate, and average recombinant tract length. We applied the method separately to each of the 274 blocks, in all cases using 106 burn-in iterations, 107 sampling iterations, and subsampling every 105 iterations. A global estimate for each parameter was then obtained by taking the block-length-weighted median of the block-specific posterior means, which down-weighed less reliable estimates obtained from shorter alignment blocks.
Sampling Recombinant Graphs
We ran ClonalOrigin a second time on all alignment blocks, this time fixing the clonal frame and global model parameters at their pre-estimated values, and collected samples of recombinant graphs across the genome. For each block, we ran the sampler for 107 burn-in iterations and 108 sampling iterations, subsampling every 105 iterations to obtain 1,001 recombinant graphs per alignment block. We performed a replicate of the two stages of the ClonalOrigin inference to ensure adequate convergence of the sampler.
Summary statistics of recombinant graphs
We used various summary statistics to describe the sampled recombinant graphs. First, we recorded the relative frequencies of the 105 possible rooted trees at each site in the alignments. Second, we recorded the frequencies of sampled recombinant edges, grouping them by the associated donor and recipient branches in the clonal frame (Didelot et al. 2010), and took ratios of these sampled counts with corresponding expected values under the assumed prior distribution. Third, we recorded the frequencies of these edges types per site. Finally, we defined a general, branch-independent recombination intensity at each site by summing the posterior probabilities of all recombinant edges. In some cases, we also computed intensities for particular types of replacing transfers, such as those between the SPY and SDE clades or those producing topologies different from the clonal frame (Supplementary Material online).
Significance Testing
We used simulations to assess the significance of the prior-normalized counts of recombinant edges. First, we generated 100 replicate data sets under the prior model, with the same block number and block lengths as the real data. We then applied our entire inference procedure to these data sets and computed the same ratios of posterior estimates to prior expectations as for the real data. We then compared the ratios estimated from real data with this empirical null distribution. Empirical one-sided P values were computed as the fraction of null estimates greater or less than the values estimated from real data. This approach had the advantage of not only assessing the significance of departures from prior expectations but also correcting for any systematic biases imposed by ClonalOrigin (Supplementary Material online).
To compare the rates of SPY-to-SDE and SDE-to-SPY gene flow, we computed, for each of the 1,001 genome-wide collections of recombinant graphs, the number of recombinant edges from any of the three SPY branches (SPY1, SPY2, and SPY) to any of the three SDE branches (SDE1, SDE2, and SDE) and the number of recombinant edges in the reverse direction. We then took the ratio of these two counts. This gave us 1,001 samples from the posterior distribution of the ratio of SPY-to-SDE and SDE-to-SPY recombinant edges. These posterior samples were compared with the prior expected value of the ratio of SPY-to-SDE to SDE-to-SPY counts.
Parsimony-Based Analysis
Mowgli
To study additive HGTs, we used a parsimony-based method for reconciling gene and species trees called Mowgli (Doyon et al. 2011). Given a gene tree and a species tree, Mowgli finds a reconciliation scenario that minimizes the number of gene duplication, loss, and transfer (DLT) events, considering tree topologies with branching orders (“labeled histories”; Edwards 1970). We estimated unrooted gene trees from the nucleotide alignments for each of our putative nonrecombining gene fragments using RAxML (Stamatakis 2006), then applied Mowgli to each of these trees. We used the tree topology estimated by ClonalFrame, augmented with the SEE outgroup, as the species tree in this analysis. Since Mowgli required rooted gene trees, we ran Mowgli for all possible rootings of each gene tree, and chose the scenario that produced the minimum DLT score. In the case of multiple most parsimonious reconciliations, one was chosen uniformly at random.
Distinguishing between Replacing and Additive HGTs
Although Mowgli's evolutionary model does not distinguish between additive and replacing HGTs, the two kinds of events can be separated, to a degree, in a subsequent step. In the absence of prediction error, this separation could be accomplished by simply assuming a replacing HGT whenever an inferred transfer coincided exactly with an inferred loss in the recipient lineage. However, loss events are difficult to place accurately because they tend to be pushed toward the root of the tree due to errors in phylogeny reconstruction or parallel losses in multiple lineages. Therefore, we classified a transfer event as additive whenever the recipient species or a descendant contained both a gene descended from the transferred copy and a gene not descended from that copy. All other transfers were considered replacing HGTs. This approach is conservative about inferring additive HGTs (supplementary fig. S4, Supplementary Material online).
Statistical Enrichment
As with replacing HGTs, we classified transfer events by donor and recipient branch and recorded the number of inferred additive HGTs of each type. We compared these numbers to expectations based on simulations that assumed constant rates of DLT across the species tree, given the total number of events inferred across all 2,314 gene families divided by the total branch lengths of the trees (Supplementary Material online).
Gene Category Associations and Putative Virulence Genes
We assigned genes to gene ontology (GO) categories by comparing the Streptococcus genes with bacterial proteins from the Uniref90 database using BLASTP and then assigning the same GO classification as the target gene of the uniProt GOA database if the match had an E value of <1.0 × 10−5. Putative virulence genes were identified using the methods described by Suzuki et al. (2011). A gene family was assigned a given classification if any of its genes was assigned that classification. To test for associations with replacing gene transfers we used the recombination intensity estimated for each gene, and for additive gene transfers we used the number of additive transfers inferred for each family. To test for significance, we performed a Mann–Whitney U-test (MWU) of the values (recombination intensities or numbers of transfers) associated with a given category verses the values for the all other genes/families. We used the Benjamini and Hochberg (1995) method to correct for multiple comparisons.
Results
Clonal Frame and Global Parameter Estimates
By applying ClonalFrame to our five-way genomic alignments, we obtained a phylogeny in which the two SPY samples grouped together, as did the two SDE samples, and in which SDD and SDE formed a clade, with SPY as an outgroup (fig. 2). This tree was consistent with previous results from 16S rRNA sequences (Facklam 2002) and with general patterns observed genome wide (Suzuki et al. 2011). The estimated levels of interspecies divergence were substantial, at 0.48 substitution per site for SDD and SDE and 0.66 substitutions per site for SPY and SDD/SDE. An initial analysis with ClonalOrigin (see Materials and Methods) produced an estimated per-site population-scaled mutation rate of θ = 0.081 (interquartile range across blocks: [0.067–0.094]), a per-site population-scaled recombination rate of ρ = 0.012 (0.006–0.019), and an average recombinant tract length of δ = 744 (346–2848) bp. The full distributions of these quantities across alignment blocks are shown in supplementary figure S5, Supplementary Material online. A second run of the MCMC sampler produced nearly identical results. For comparison, Didelot et al. (2010) obtained estimates of θ = 0.044, ρ = 0.017, and δ = 236 for the Bacillus cereus group. Our estimates implied a ratio of ρ/θ = 0.15, considerably lower than the estimate of Didelot et al. (2010) of ρ/θ = 0.405 for B. cereus, suggesting increased rates of mutation and/or decreased rates of recombination after controlling for differences in effective population size. We found that estimates of the average recombinant tract length, δ , were quite sensitive to our threshold for minimum block length, with the inclusion of short alignments producing much larger estimates due to edge effects. We experimented with a range of thresholds and selected a value (1,500 bp) at which the estimates stabilized.
Fig. 2.
Clonal frame inferred for the five genomes. This phylogeny was inferred using the ClonalFrame program (Didelot and Falush 2007). Branch lengths are in units of expected substitutions per site and are drawn to scale in the horizontal dimension. The labels for the ancestral nodes of the tree and the branches immediately ancestral to them (SPY, SDE, and SD) are used throughout the article. The outgroup species, SEE, was not part of the estimated clonal frame but was used in the parsimony-based analysis.
Model-Based Analysis of Replacing HGTs
In a second round of analysis with ClonalOrigin, we obtained samples from an approximate posterior distribution of recombinant graphs along the genome, conditional on our five-way alignments and the previously estimated parameters. (A recombinant graph is modeled by ClonalOrigin as the clonal frame augmented by zero or more recombinant edges; see Materials and Methods.) This distribution indicated that a majority of sites in the genome (66%) were topologically consistent with the clonal frame but that there was also substantial support for various alternative tree topologies (supplementary table S2, Supplementary Material online). The second most frequent tree topology (9.3% of the sites) had SPY and SDE as sister clades and SDD as an outgroup, providing an initial indication of gene transfer between SPY and SDE. No other single tree topology appeared with a frequency of ≥ 4%.
To gain further insight into rates and patterns of HGT, we estimated the rates of occurrence of various types of recombinant edges, grouping them by their donor and recipient edges (see Materials and Methods). There are 81 possible edge pairs associated with our nine-branch rooted phylogeny, but because donor edges cannot postdate recipient edges under the model, 21 of these pairs are prohibited, leaving 60 possible recombinant edge types. Each of these viable edge types corresponds to a class of replacing HGTs, including not only transfers from the donor to the recipient edge but also transfers from any (sufficiently old) descendant of the donor to the recipient (due to the possibility of delayed coalescence). Since all recombinant edges are not equally likely a priori, we summarized the estimated rates by taking ratios with respect to their prior expectations under the ClonalOrigin model. Experiments conducted on simulated data demonstrated that our approach had reasonable power to detect recombination scenarios that deviated from the prior, with some underestimation of the degree of deviation due to the use of the prior in the inference procedure (Supplementary Material online and supplementary figs. S6 and S7).
We summarized these normalized rates in a heatmap (fig. 3A; supplementary tables S3 and S4, Supplementary Material online) and found that most cells fell in the blue-to-white range, corresponding to rates of occurrence less than or equal to what was expected under the prior. A few edge types were strongly under-represented (blue), such as those between the SDD branch and the branches of the SPY clade (P < 0.01 based on simulations; see Materials and Methods), probably due to ecological isolation of human and strictly veterinary pathogens. Edges from the branch at the root were also under-represented, suggesting a deficiency of long delays in coalescence relative to the prior. In contrast, the recombinant edges between the two SDE genomes, and between the two SPY genomes, were significantly over-represented (P < 0.01), consistent with previous evidence for abundant intraspecies recombination in Streptococcus (Feil et al. 2001). In addition, we found a pronounced enrichment for recombinant edges between the SPY and SDE clades, as has been reported previously (Sachse et al. 2002; Kalia and Bessen 2004; Davies, McMillan, Beiko et al. 2007; Davies, McMillan, Domselaar et al. 2007). We also observed a slight enrichment for SDE-to-SDD edges.
Fig. 3.
Heatmaps showing rates of replacing and additive transfers. Each cell of the heat map represents the base-2 logarithm of the ratio of the estimated number of recombination events to its prior expectation, for the corresponding donor (y axis) and recipient (x axis) branches. Cells in black indicate prohibited transfer events. (A) Replacing transfers, as inferred by the model-based approach. The plotted values represent average values across sampled recombinant graphs. The prior considers the clonal frame with branch lengths (fig. 2) and the global estimates of the three population parameters. Prohibited transfer events are ones for which the recipient branch is strictly older than the donor branch. An asterisk indicates statistical significance (P < 0.01). (B) Additive transfers, as inferred by the parsimony-based approach. The plotted log ratios reflect total numbers of inferred additive transfers across all gene families. The prior considers the clonal frame (with branch lengths) and the total number of events of each type inferred by the analysis. Prohibited transfer events are ones for which the recipient and donor branch do not share a common time interval. Significance levels are denoted by asterisks: one for P < 0.01, two for P < 0.005, and three for P < 0.001. See Materials and Methods for details.
Although SPY–SDE edges occurred at elevated rates in both directions, they showed a pronounced directional asymmetry, with edges from SPY to SDE occurring at up to four times the expected rate, whereas those in the reverse direction occurred at only about twice the expected rate. By reanalyzing the sampled recombinant graphs, we were able to obtain a Bayesian posterior mean estimate of 2.0 (95% credible interval: 1.8–2.2) for the ratio of the numbers of SPY-to-SDE to SDE-to-SPY recombinant edges, compared with a ratio of 1.4 implied by the prior (the prior ratio is larger than one because of differences in branch lengths). None of the sampled values was as small as the prior expected ratio, indicating strong support for a preference for the SPY-to-SDE direction. In addition, we counted genes having high probability of recombination (>0.6) for various types of recombinant edges (supplementary table S5, Supplementary Material online), and found that many more genes showed strong evidence of SPY-to-SDE than of SDE-to-SPY recombinant edges (e.g., 59 vs. 11 genes, when the entire SPY and SDE clades are considered).
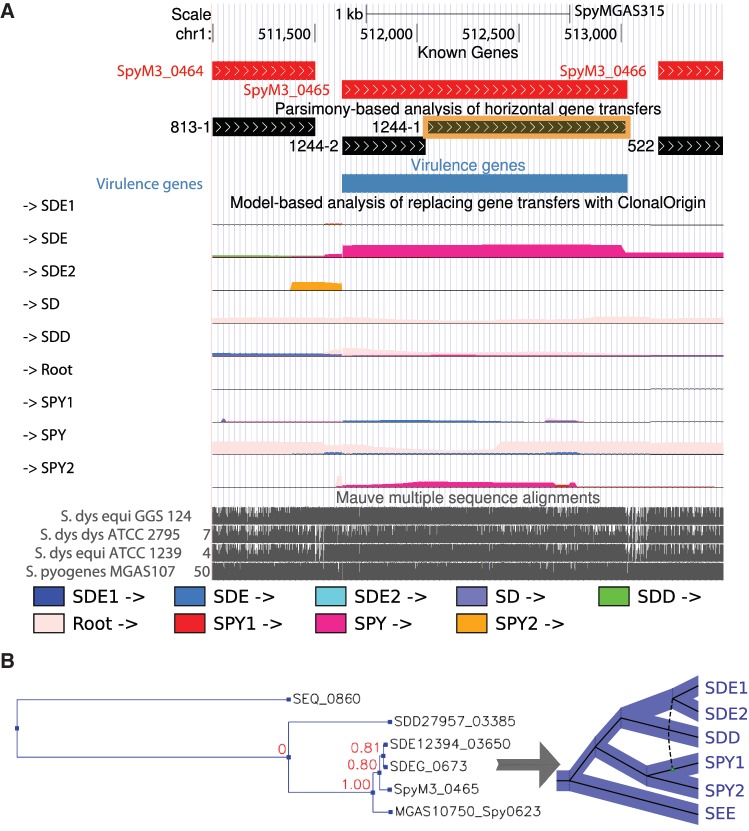
We generated new tracks for our recently released Streptococcus Genome Browser (http://strep-genome.bscb.cornell.edu) that summarize the results of this model-based analysis alongside known genes, alignments, and other annotations (fig. 4). These tracks can be used to inspect loci of interest and to compare the results of our model- and parsimony-based analyses (later). They can also be queried and intersected with other tracks using the University of California, Santa Cruz (UCSC) Table Browser.
Fig. 4.
Genome Browser tracks. Our inferences of replacing and additive horizontal gene transfers are summarized in new tracks in the Streptococcus Genome Browser (Suzuki et al. 2011). (A) The main browser display, with gene annotations for the selected reference genome (S. pyogenes strain MGAS315) shown in red and putative virulence genes highlighted in blue. The third track from top (in black) shows the putative non-recombining gene fragments analyzed by Mowgli. The next series of tracks shows the results of the ClonalOrigin analysis of replacing gene transfers. Each track in this series corresponds to a recipient lineage in the phylogeny (fig. 2) and describes the posterior probabilities along the genome of recombinant edges from all possible donor lineages (shown in different colors; see key). Here the putative virulence gene SpyM3_0465, a dipeptidase, shows strong evidence of a recombinant edge from the SPY to the SDE lineage, as well as some evidence of a SPY → SPY2 edge. The genome-wide multiple alignment obtained with Mauve is shown at bottom. (B) The gene tree and its reconciliation displayed after clicking on the gene fragment highlighted in orange. Note that the most parsimonious reconciliation of the tree estimated by RAxML involves a single SPY → SDE replacing HGT; however, this tree is also consistent with the combined influence of SPY → SDE and SPY → SPY2 replacing transfers, as inferred under the richer statistical model of ClonalOrigin.
Parsimony-Based Analysis
To gain further insight into HGT in Streptococcus, we estimated gene trees for 2,314 gene families from six genomes (including the SEE outgroup), obtained parsimonious reconciliations of these gene trees with the clonal frame (fig. 2) using the Mowgli program (Doyon et al. 2011), and partitioned the inferred transfers into replacing and additive HGTs (see Materials and Methods). This analysis produced estimated numbers of five types of events for each branch of the phylogeny (gene duplications, losses, replacing and additive transfers, and “appearances,” most of which are probably transfers from phylogenetically distant species) and an estimated number of genes at each ancestral node (fig. 5). We observed large numbers of appearances on all branches of the tree, suggesting a steady influx of genes into the clade by HGT. Indeed, appearance events accounted for 1,957 of the 3,432 (57%) gene additions (by duplication, transfer, or appearance; loss events are excluded in this study). The somewhat reduced numbers of genes in SPY (particularly in SPY1) were primarily explained by reduced rates of gene appearance, rather than by increased loss or other factors. The estimated numbers of ancestral genes tended to decrease toward the root of the tree, but this likely reflects under estimation due to parallel losses of some genes (especially on the branches beneath the root) rather than a true increase in gene number over evolutionary time. An exception was the branch leading to the ancestor of the two SPY individuals, which was enriched for gene loss events, perhaps associated with niche adaptation. High rates of loss also occurred on the branches to SDE1, SDE2, and SDD. Duplications were relatively infrequent overall, but, as has been noted previously (Marri et al. 2006), they were substantially enriched on external branches of the phylogeny.
Fig. 5.
Distribution of inferred gene duplication, loss, and transfer events across the six-species phylogeny. Numbers at nodes represent gene counts for extant species and estimated counts for ancestral species. Numbers on branches indicate inferred gene appearances (A*), duplications (D*), losses (L*), additive transfers (T*), and replacing transfers (R*). Transfer events are recorded on recipient branches (donors are not indicated). Singleton families (containing a single gene) are included as appearance events on external branches of the phylogeny.
We conservatively identified 290 of the 1,218 (non appearance) transfers (23.8%) as additive HGTs. As with the replacing HGTs evaluated in the model-based analysis, these predicted additive transfers were significantly enriched within the SDE clade, and they were weakly enriched within the SPY clade (fig. 3B and supplementary fig. S8, Supplementary Material online). They were also significantly enriched between SDE and SDD, with a preference for the SDE-to-SDD direction, and significantly depleted between SDD and SPY. However, unlike the replacing HGTs from the previous section, these additive transfers were not significantly enriched between SPY and SDE. They also did not exhibit a pronounced directional asymmetry between SPY and SDE, except between the ancestral SPY and SDE branches, where they were significantly depleted in both directions, but more strongly from SDE-to-SPY. It is worth noting, however, that this apparent asymmetry in depletion could in fact reflect an asymmetry in enrichment, as the normalization in this case reflects average rates across the tree, and an inflated average could cause a global shift in the heatmap toward depletion (blue). Other differences also make it difficult to compare the model- and parsimony-based analyses—for example, the first considered the core genome only, whereas the second considered the pan genome; and the two types of analysis might have quite different power for events affecting different portions of the phylogeny. Nevertheless, we find much clearer evidence that homologous recombination, rather than additive transfer, has driven the apparent gene flow between SPY and SDE, particularly in the SPY-to-SDE direction. Notably, we compared the replacing HGTs identified in the parsimony-based analysis with those from the model-based analysis, and found reasonable concordance, despite substantial differences between the data sets and methods (Supplementary Material online).
Functional Categories of Transferred Genes
To gain insight into the functional impact of HGT, we assigned genes to various functional categories and looked for statistical associations between these categories and predicted gene transfer events (see Materials and Methods). First, we partitioned all genes into “virulence” and “nonvirulence” categories, considering genes whose homologs in other genera of bacteria exhibit putative virulence phenotypes (based on VFDB) to be putative virulence genes (see Suzuki et al. 2011). Among gene families for which we inferred additive HGTs, putative virulence genes were significantly enriched (P = 1.33 × 10−11). Moreover, putative virulence genes associated with additive HGTs showed a pronounced enrichment for SPY-to-SDE events (25.0% of the events compared with 12.1% for all genes at the level of the entire SPY and SDE clades, P = 0.01, Fisher's exact test; supplementary table S6, Supplementary Material online). Transfers in the reverse direction (SDE-to-SPY) were also over represented (17.5% vs. 10.6%) but not significantly (P = 0.11). Replacing gene transfers, on the other hand, were not significantly enriched for putative virulence genes, and those putative virulence genes that did exhibit strong evidence of replacing transfers were not enriched for SDE/SPY transfers (supplementary table S5, Supplementary Material online). We also found a significant association between putative virulence genes and gene duplications (P = 7.13 × 10−3). Not surprisingly, families associated with bacteriophages (based on an annotated “phage” product for least one gene in the family) were strongly enriched for additive HGTs; 88 of 288 (30.6%) of phage families exhibited additive HGTs compared with only 269 of all 3,408 (7.8%) families (P < 1.6 × 10−33).
Next, we assigned GO terms to all genes and searched for significant associations with transfer events. Several GO categories having to do with transposition, recombination, and DNA integration were significantly associated with additive gene transfers (table 1), consistent with observations in other bacterial groups (e.g., Liu et al. 2009). Essentially the same categories were enriched when phage genes were excluded. Replacing transfers, on the other hand, showed much weaker, and quite different, functional associations compared with additive transfers. These associations were similar for replacing transfers inferred by the model- and parsimony-based methods (supplementary tables S7 and S8, Supplementary Material online).
Table 1.
GO Enrichments for Additive Gene Transfers.
| Pa | qb | Countc | GO Term | Description |
|---|---|---|---|---|
| 7e-61 | 3e-58 | 67 | GO:0006313 | Transposition, DNA mediated |
| 3e-58 | 7e-56 | 59 | GO:0004803 | Transposase activity |
| 1e-30 | 2e-28 | 52 | GO:0015074 | DNA Integration |
| 1e-28 | 1e-26 | 23 | GO:0032196 | Transposition |
| 1e-13 | 1e-11 | 527 | GO:0003677 | DNA binding |
| 2e-12 | 2e-10 | 86 | GO:0006310 | DNA recombination |
| 1e-11 | 7e-10 | 197 | GO:0003676 | Nucleic acid binding |
| 3e-09 | 1e-07 | 36 | GO:0016987 | Sigma factor activity |
| 3e-09 | 1e-07 | 36 | GO:0006352 | Transcription initiation |
| 3e-08 | 1e-06 | 142 | GO:0043565 | Sequence-specific DNA binding |
| 4e-07 | 1e-05 | 12 | GO:0008170 | N-methyltransferase activity |
| 1e-06 | 4e-05 | 12 | GO:0019867 | Outer membrane |
| 2e-06 | 5e-05 | 25 | GO:0007059 | Chromosome segregation |
| 1e-05 | 4e-04 | 15 | GO:0006306 | DNA methylation |
| 2e-05 | 6e-04 | 15 | GO:0006470 | Protein amino acid dephosphorylation |
| 2e-04 | 4e-03 | 76 | GO:0005618 | Cell wall |
| 3e-04 | 7e-03 | 25 | GO:0008236 | Serine-type peptidase activity |
| 3e-04 | 7e-03 | 65 | GO:0009986 | Cell surface |
| 5e-04 | 1e-02 | 10 | GO:0003872 | 6-phosphofructokinase activity |
| 1e-03 | 2e-02 | 11 | GO:0009307 | DNA restriction–modification system |
| 2e-03 | 5e-02 | 84 | GO:0006974 | Response to DNA damage stimulus |
P value based on a Mann–Whitney U-test (see Materials and Methods).
Corresponding false discovery rate estimated by the Benjamini–Hochberg method. All categories having at least 10 genes and q ≤ 0.05 are displayed.
Number of genes assigned to category.
Positive Selection and Gene Transfer
We tested for evidence of positive selection within the gene families and for associations between positive selection and various types of events. We performed a likelihood ratio test using the “sites models” M1a and M2b from the Phylogenetic Analysis by Maximum Likelihood (PAML) package (Yang 2007) on each of the 2,991 gene fragment families with three or more genes. After controlling for multiple comparisons, 31 distinct families showed evidence of positive selection (false discovery rate < 5%). Among high-confidence gene trees (bootstrap values of > 90% on every branch), families with duplication events were slightly enriched for evidence of positive selection (P < 0.046, MWU) as were families with additive transfers (P < 0.002, MWU). These results held even when additional recombination break points were inferred (Supplementary Material online). Of the 31 families with significant positive selection, 13 families were part of the core genome analyzed by ClonalOrigin. Replacing transfers from SDE to SPY and from SPY to SDE were slightly enriched for positive selection (P < 0.005 and P < 0.01, MWU). For 11 of the 13 (85%) families showing evidence of positive selection, the SPY-to-SDE direction had greater intensity than the SDE-to-SPY direction, consistent with overall directional bias (83.3%). The same directional bias was not seen for additive transfers between the two clades.
Comparison with Previous Results for Housekeeping Genes
Several previous studies have considered the issue of gene flow between SPY and SDE based on MLST data for small numbers of housekeeping genes (e.g., Ahmad et al. 2009; McMillan et al. 2010; Jensen and Kilian, 2012). We compared our results with those of Ahmad et al. (2009), who performed one of the few MLST studies to address the issue of directionality in gene flow. Six of the seven genes examined by Ahmad et al. (2009) (gki, gtr, murI, mutS, recP, and xpt) were included in both our model- and parsimony-based analyses, whereas the seventh (yqiL; atoB in SDE) was excluded from our model-based analysis, owing to elevated sequence divergence between SPY and SDE.
We observed general concordance with Ahmad et al. (2009) in the genes predicted to have undergone HGT. Specifically, we found a strong signal for SPY–SDE HGT in mutS, recP, and xpt, a weaker signal in gki, and no evidence for HGT in gtr and murI, in agreement with Ahmad et al. (2009). (Two of the genes showing evidence of HGT, gki, and mutS, are annotated as putative virulence genes.) Our model- and parsimony-based analyses were also concordant for all of these genes. The recombinant edges sampled by ClonalOrigin in gki and mutS (and, to a lesser extent, xpt) showed a clear bias for the SPY-to-SDE direction, whereas both directions were sampled at similar rates in recP. In contrast, Ahmad et al. (2009) found some evidence for gene flow in the opposite direction, from SDE to SPY. However, this discrepancy may be due to limitations in methods by Ahmad et al. (see Discussion).
Alternative SPY Genomes
To examine the robustness of our results to the choice of SPY genomes, we repeated our model-based analysis 10 additional times, in each case substituting two different SPY genomes for SPY1 and SPY2, whereas leaving SDE1, SDE2, and SDD unchanged. These experiments made use of 12 of the 14 publicly available SPY genomes and covered all 11 represented serotypes (supplementary tables S9 and S10, Supplementary Material online). For each replicated data set, we applied our entire analysis pipeline, including multiple alignment, processing and filtering of alignment blocks, estimation of a clonal frame, and two rounds of ClonalOrigin analysis (see Materials and Methods). The estimated alignments were qualitatively similar in all cases, and all clonal frames showed a high degree of concordance, as did the global parameters estimated by ClonalOrigin (supplementary table S10 and supplementary fig. S9, Supplementary Material online). In addition, the heatmaps summarizing the model-based analysis all strongly resembled the one in figure 3A (supplementary fig. S10, Supplementary Material online). Because the finding of directional asymmetry in SPY/SDE gene flow was of particular interest, we repeated our analysis to estimate the ratio of the numbers of SPY-to-SDE to SDE-to-SPY recombinant edges under both the prior and posterior distributions for all replicate data sets. We found the resulting estimates to be highly consistent with prior means ranging from 1.37 to 1.47 and posterior means from 1.81 to 2.05 (supplementary table S11, Supplementary Material online). In all cases, all posterior samples of this ratio exceeded the expected value under the prior, indicating strong evidence in the data for a bias in gene flow favoring the SPY-to-SDE direction. Notably, because these replicate analyses involved a reapplication of the entire analysis pipeline, they allow to a degree for statistical uncertainty in the estimation of the alignment and the clonal frame, as well as in the ClonalOrigin analysis. We performed a similar validation of our parsimony-based analysis, substituting two alternative SPY genomes for SPY1 and SPY2 (supplementary fig. S11, Supplementary Material online), and also observed general agreement with the primary analysis in this case.
Discussion
There has been a great deal of interest for decades in the process of HGT and the manner in which it has influenced phylogenetic relationships, particularly among bacteria (Koonin et al. 2001). Most previous studies, however, have either simply examined patterns of phylogenetic discordance, without differentiating between additive and replacing HGTs (e.g., Lerat et al. 2005), or have focused specifically on the process of homologous recombination, which is primarily associated with replacing HGTs (Didelot and Falush 2007; Didelot et al. 2010). To our knowledge, this is the first study to examine both processes on a genome-wide scale, using both model- and parsimony-based methods.
We have focused our analysis on SPY and two of its close relatives, SDE and SDD, a human commensal-like organism and a strict veterinary pathogen, respectively. We find strong evidence of gene flow both within and between the SPY and SDE groups. Events between SPY and SDE are more prominent among inferred replacing than among inferred additive transfers, suggesting that they may be driven by homologous recombination, although we acknowledge several challenges in comparing inferences from the model- and parsimony-based analyses. We also find strong support for an asymmetry in replacing gene transfers between SPY and SDE, with a preference for the SPY-to-SDE direction. The situation with SDD is more complex, with somewhat elevated rates of SDE-to-SDD transfer (more pronounced for additive than for replacing HGTs), mixed evidence for SDD-to-SDE transfer, and generally reduced rates of transfer between SDD and SPY, perhaps due to a combination of genetic divergence and different ecological niches. These findings hold regardless of the particular SPY genomes selected for the analyses. We also find that putative virulence genes are significantly associated with additive, but not replacing, gene transfers, as are genes under positive selection. Genes that have undergone replacing transfers between SPY and SDE are also enriched for positive selection. A central component of our study is the use of simulations to demonstrate that our methods have good power and accuracy in the detection of both types of transfer events.
In comparing replacing and additive HGTs, it is worth bearing in mind that these events are likely to be strongly correlated with the core and dispensable portions of the genome. In part, this reflects an ascertainment bias in the inference procedures used to detect these events. Methods for detecting recombination (replacing transfers) tend to rely on large-scale alignments spanning multiple syntenic loci to gain power. As a result, these methods are effectively limited to application in the core genome, even though homologous recombination probably also occurs between genes that are less widely shared across species. In contrast, phylogenetic reconciliation methods can be applied to the entire pan genome, as in this study. In addition, true additive transfer events will be enriched in the dispensable genome, by definition, and it is likely that homologous recombination occurs at higher rates in the core genome (which is enriched for close homologs between species). For all these reasons, it is likely that some apparent differences between the two types of transfer events simply reflect general differences between the core and dispensable portions of the genome, rather than properties specifically associated with modes of gene transfer.
Our main goal in this study was to statistically characterize global patterns of HGT in the pyogenic division, and our power for reconstructing the histories of individual genes was necessarily limited by the small number of genomes analyzed. Nevertheless, we were able to compare our results for six genes with those of Ahmad et al. (2009) in some detail. We observed remarkably good concordance with Ahmad et al. (2009) in predictions of which genes had experienced HGT but poorer agreement in the inferred directionality of gene flow. However, Ahmad et al. (2009) based their conclusions on a simple clustering analysis that used pairwise comparisons of SPY and SDE sequences. Our model-based approach, which additionally made use of the SDD outgroup and phylogenetic signals at flanking sites (as well as in the genes themselves), might offer improved power in addressing the issue of directionality, despite our limited number of samples. Of course, it is also possible that the collection of genomes we examined and the one analyzed by Ahmad et al. (2009) simply had quite different histories of HGT at the few genes at which a comparison was possible.
The question of the mechanism responsible for the asymmetric gene flow between SPY and SDE remains open. Possible mechanisms include differences in the sizes or demographic structure of commingling populations of cells or differences in barriers to gene transfer (Thomas and Nielsen 2005). One possibility that has been raised is that SPY is inherently less capable than SDE of acquiring DNA by homologous recombination, for example, due to differences in the restriction modification system (RMS), heteroduplex formation, and/or mismatch repair. Interestingly, although most of SPY genome contain both types I and II RMSs, the two SDE genomes studied here is contain only type II RMSs (Roberts et al. 2010).
As sequence data become available for many more closely and distantly related organisms, it will become increasingly important to devise improved methods that consider both additive and replacing HGT. As noted earlier, the current statistical models of recombination, such as ClonalOrigin, are effectively limited to considering orthologs, with one copy per species, in the core genome. The phylogenetic reconciliation methods, on the other hand, fail to allow for multilocus gene transfer events and ignore information from branch lengths. We have shown that it is possible, to a degree, to extend these methods to distinguish between additive and replacing transfers, but our methods are limited by their dependency on heuristic rules and parsimony assumptions. It may be possible to develop integrated statistical models that consider both types of gene transfers and produce unbiased estimates of the relative rates at which these events occur. Another extension worth considering is direct modeling of incomplete lineage sorting (ILS), which is likely to be increasingly important as larger phylogenies are considered, especially given the large effective population sizes of many bacteria. ILS has recently been integrated into models for duplication and loss (Rasmussen and Kellis 2012) but has yet to be considered together with gene transfer. We expect that the combination of improved models and richer data sets will allow for a much more detailed understanding of the important process of HGT.
Supplementary Material
Supplementary material, tables S1–S11, and figures S1–S11 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Xavier Didelot for assistance with ClonalOrigin. This study was funded by the National Institute of Allergy and Infectious Disease, US National Institutes of Health, under grant number AI073368-01A2 (to M.J.S. and A.S.). Additional support was provided by National Science Foundation CAREER Award DBI-0644111 and a David and Lucile Packard Fellowship for Science and Engineering (to A.S.).
References
- Ahmad Y, Gertz RE, Li Z, et al. (11 co-authors) Genetic relationships deduced from emm and multilocus sequence typing of invasive Streptococcus dysgalactiae subsp. equisimilis and S. canis recovered from isolates collected in the United States. J Clin Microbiol. 2009;47:2046–2054. doi: 10.1128/JCM.00246-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Bielawski J, Dunn K, Yang Z. Phylogenomic analysis of natural selection pressure in Streptococcus genomes. BMC Evol Biol. 2007;7:154. doi: 10.1186/1471-2148-7-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B. 1995;57:289–300. [Google Scholar]
- Beres SB, Richter EW, Nagiec MJ, Sumby P, Porcella SF, DeLeo FR, Musser JM. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc Natl Acad Sci U S A. 2006;103:7059–7064. doi: 10.1073/pnas.0510279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres SB, Sylva GL, Barbian KD, et al. (16 co-authors) Genome sequence of a serotype M3 strain of Group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc Natl Acad Sci U S A. 2002;99:10078–10083. doi: 10.1073/pnas.152298499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt CM, Spellerberg B. Human infections due to Streptococcus dysgalactiae subspecies equisimilis. Clin Infect Dis. 2009;49:766–772. doi: 10.1086/605085. [DOI] [PubMed] [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Alm EJ. Rapid evolutionary innovation during an archaean genetic expansion. Nature. 2011;469:93–96. doi: 10.1038/nature09649. [DOI] [PubMed] [Google Scholar]
- Davies MR, McMillan DJ, Beiko RG, Barroso V, Geffers R, Sriprakash KS, Chhatwal GS. Virulence profiling of Streptococcus dysgalactiae subspecies equisimilis isolated from infected humans reveals 2 distinct genetic lineages that do not segregate with their phenotypes or propensity to cause diseases. Clin Infect Dis. 2007;44:1442–1454. doi: 10.1086/516780. [DOI] [PubMed] [Google Scholar]
- Davies MR, McMillan DJ, Domselaar GHV, Jones MK, Sriprakash KS. Phage 3396 from a Streptococcus dysgalactiae subsp. equisimilis pathovar may have its origins in Streptococcus pyogenes. J Bacteriol. 2007;189:2646–2652. doi: 10.1128/JB.01590-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MR, Tran TN, McMillan DJ, Gardiner DL, Currie BJ, Sriprakash KS. Inter-species genetic movement may blur the epidemiology of streptococcal diseases in endemic regions. Microbes Infect. 2005;7:1128–1138. doi: 10.1016/j.micinf.2005.03.018. [DOI] [PubMed] [Google Scholar]
- Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Lawson D, Darling A, Falush D. Inference of homologous recombination in bacteria using whole-genome sequences. Genetics. 2010;186:1435–1449. doi: 10.1534/genetics.110.120121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon JP, Hamel S, Chauve C. An efficient method for exploring the space of gene tree/species tree reconciliations in a probabilistic framework. IEEE/ACM Trans Comput Biol Bioinform. 2012;9:26–39. doi: 10.1109/TCBB.2011.64. [DOI] [PubMed] [Google Scholar]
- Doyon JP, Ranwez V, Daubin V, Berry V. Models, algorithms and programs for phylogeny reconciliation. Brief Bioinformatics. 2011;12:392–400. doi: 10.1093/bib/bbr045. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AWF. Estimation of the branch points of a branching diffusion process. J Roy Stat Soc B. 1970;32:155–174. [Google Scholar]
- Facklam R. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin Microbiol Rev. 2002;15:613–630. doi: 10.1128/CMR.15.4.613-630.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil EJ, Holmes EC, Bessen DE, et al. (12 co-authors) Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc Natl Acad Sci U S A. 2001;98:182–187. doi: 10.1073/pnas.98.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti JJ, McShan WM, Ajdic D, et al. (23 co-authors) Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci U S A. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden MTG, Heather Z, Paillot R, et al. (26 co-authors) Genomic evidence for the evolution of Streptococcus equi: host restriction, increased virulence, and genetic exchange with human pathogens. PLoS Pathog. 2009;5:e1000346. doi: 10.1371/journal.ppat.1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen A, Kilian M. Delineation of Streptococcus dysgalactiae, its subspecies, and its clinical and phylogenetic relationship to Streptococcus pyogenes. J Clin Microbiol. 2012;50:113–126. doi: 10.1128/JCM.05900-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia A, Bessen DE. Natural selection and evolution of streptococcal virulence genes involved in tissue-specific adaptations. J Bacteriol. 2004;186:110–121. doi: 10.1128/JB.186.1.110-121.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia A, Enright MC, Spratt BG, Bessen DE. (Retracted) Directional gene movement from human-pathogenic to commensal-like streptococci. Infect Immun. 2001;69:4858–4869. doi: 10.1128/IAI.69.8.4858-4869.2001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kalia A, Enright MC, Spratt BG, Bessen DE. Retraction. directional gene movement from human-pathogenic to commensal-like streptococci. Infect Immun. 2009;77:4688. doi: 10.1128/IAI.00966-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Aravind L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu Rev Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW. GARD: a genetic algorithm for recombination detection. Bioinformatics. 2006;22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- Lefébure T, Stanhope MJ. Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome Biol. 2007;8:R71. doi: 10.1186/gb-2007-8-5-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E, Daubin V, Ochman H, Moran NA. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 2005;3:e130. doi: 10.1371/journal.pbio.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Siezen RJ, Nauta A. In silico prediction of horizontal gene transfer events in Lactobacillus bulgaricus and Streptococcus thermophilus reveals protocooperation in yogurt manufacturing. Appl Environ Microbiol. 2009;75:4120–4129. doi: 10.1128/AEM.02898-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marri PR, Hao W, Golding GB. Gene gain and gene loss in Streptococcus: is it driven by habitat? Mol Biol Evol. 2006;23:2379–2391. doi: 10.1093/molbev/msl115. [DOI] [PubMed] [Google Scholar]
- Maynard-Smith J, Smith NH, O'Rourke M, Spratt BG. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan DJ, Bessen DE, Pinho M, Ford C, Hall GS, Melo-Cristino J, Ramirez M. Population genetics of Streptococcus dysgalactiae subspecies equisimilis reveals widely dispersed clones and extensive recombination. PLoS One. 2010;5:e11741. doi: 10.1371/journal.pone.0011741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle D, Middendorf M, Wieseke N. A parameter-adaptive dynamic programming approach for inferring cophylogenies. BMC Bioinformatics. 2010;11(Suppl 1):S60. doi: 10.1186/1471-2105-11-S1-S60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Rasmussen MD, Kellis M. A unified model of gene duplication, loss, and coalescence using a locus tree. Genome Res. 2012 doi: 10.1101/gr.123901.111. In revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse S, Seidel P, Gerlach D, Günther E, Rödel J, Straube E, Schmidt KH. Superantigen-like gene(s) in human pathogenic Streptococcus dysgalactiae subsp equisimilis: genomic localisation of the gene encoding streptococcal pyrogenic exotoxin G (speG(dys)) FEMS Immunol Med Microbiol. 2002;34:159–167. doi: 10.1111/j.1574-695X.2002.tb00618.x. [DOI] [PubMed] [Google Scholar]
- Shimomura Y, Okumura K, Murayama SY, Yagi J, Ubukata K, Kirikae T, Miyoshi-Akiyama T. Complete genome sequencing and analysis of a lancefield Group G Streptococcus dysgalactiae subsp. equisimilis strain causing streptococcal toxic shock syndrome (STSS) BMC Genomics. 2011;12:17. doi: 10.1186/1471-2164-12-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MW, Feng DF, Doolittle RF. Evolution by acquisition: the case for horizontal gene transfers. Trends Biochem Sci. 1992;17:489–493. doi: 10.1016/0968-0004(92)90335-7. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Lefébure T, Hubisz MJ, Bitar PP, Lang P, Siepel A, Stanhope MJ. Comparative genomic analysis of the Streptococcus dysgalactiae species group: gene content, molecular adaptation, and promoter evolution. Genome Biol Evol. 2011;3:168–185. doi: 10.1093/gbe/evr006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, et al. (46 co-authors) Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.”. Proc Natl Acad Sci U S A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- Vandamme P, Pot B, Falsen E, Kersters K, Devriese LA. Taxonomic study of lancefield streptococcal groups C, G, and L (Streptococcus dysgalactiae) and proposal of S. dysgalactiae subsp. equisimilis subsp. nov. Int J Syst Bacteriol. 1996;46:774–781. doi: 10.1099/00207713-46-3-774. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhaxybayeva O, Doolittle WF. Lateral gene transfer. Curr Biol. 2011;21:R242–R246. doi: 10.1016/j.cub.2011.01.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.