Abstract
Gibbons (Hylobatidae) are small, arboreal apes indigenous to Southeast Asia that diverged from other apes ∼15–18 Ma. Extant lineages radiated rapidly 6–10 Ma and are organized into four genera (Hylobates, Hoolock, Symphalangus, and Nomascus) consisting of 12–19 species. The use of short interspersed elements (SINEs) as phylogenetic markers has seen recent popularity due to several desirable characteristics: the ancestral state of a locus is known to be the absence of an element, rare potentially homoplasious events are relatively easy to resolve, and samples can be quickly and inexpensively genotyped. During radiation of primates, one particular family of SINEs, the Alu family, has proliferated in primate genomes. Nomascus leucogenys (northern white-cheeked gibbon) sequences were analyzed for repetitive content with RepeatMasker using a custom library. The sequences containing Alu elements identified as members of a gibbon-specific subfamily were then compared with orthologous positions in other primate genomes. A primate phylogenetic panel consisting of 18 primate species, including 13 gibbon species representing all four extant genera, was assayed for all loci, and a total of 125 gibbon-specific Alu insertions were identified. The resulting amplification patterns were used to generate a phylogenetic tree. We demonstrate significant support for Symphalangus as the most basal lineage within the family. Our findings also place Nomascus as a derived lineage, sister to Hoolock, with the Nomascus–Hoolock clade sister to Hylobates. Further, our analysis groups N. leucogenys and Nomascus siki as sister taxa to the exclusion of the other Nomascus species assayed. This study represents the first use of SINEs to determine the genus level phylogenetic relationships within the family Hylobatidae. These relationships have been resolved with robust support at most internal nodes, demonstrating the utility of SINE-based phylogenetic analysis. We postulate that hybridization and rapid radiation may have contributed to the complex and contradictory findings of the previous studies. Our findings will aid in the conservation of these threatened primates and inform future studies of the biogeographical history and distribution of modern gibbon species.
Keywords: primate, ape, phylogeny, retrotransposon, SINE, mobile elements
Introduction
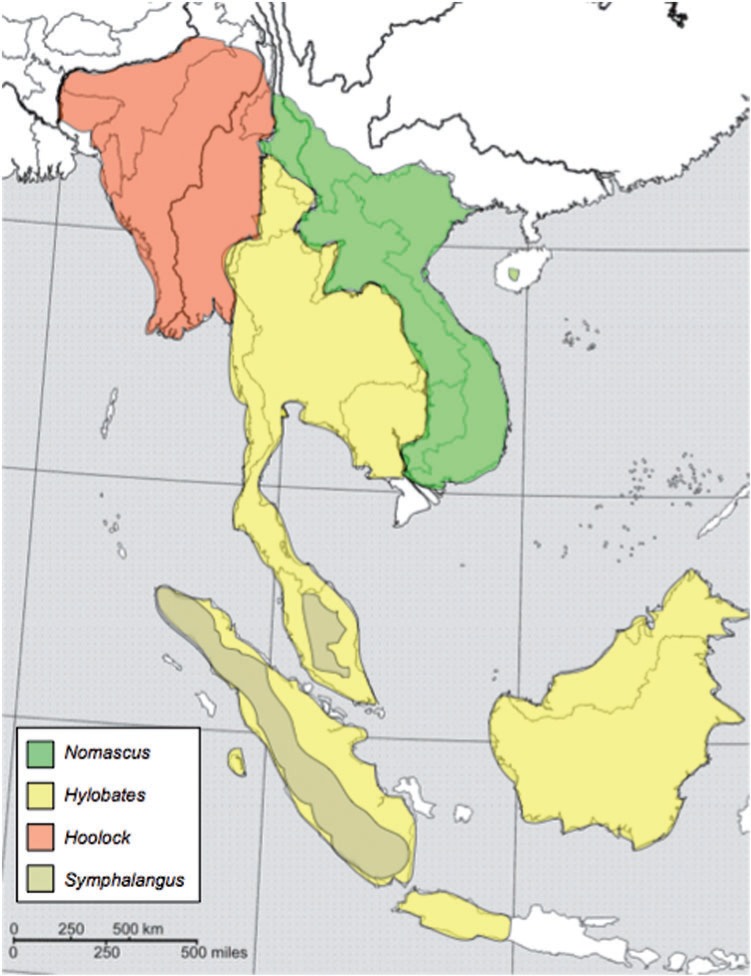
Gibbons (Hylobatidae) are small, arboreal apes indigenous to Southeast Asia. They occupy a range stretching from northeastern India, south to Sumatra, Borneo, and Java, and north into the southernmost parts of China, with ranges among genera being largely separated by major rivers (fig. 1) (Geissmann 1995). All are threatened or endangered due to human activities, some critically (Mootnick 2006). Gibbons are by far the most speciose group of apes (Hominoidea) (Groves 2001), with most of the radiation of extant lineages occurring rapidly between 6 and 10 Ma (Matsudaira and Ishida 2010). They are organized into four genera (Hylobates, Hoolock, Symphalangus, and Nomascus) primarily based on diploid chromosome number, and there are between 12 and 19 extant species, according to various classification schemes (Roos and Geissmann 2001; Chatterjee 2006; Mootnick 2006; Carbone et al. 2009; Thinh et al. 2010a, 2010b). The Hylobatidae represents a valuable perspective on the phylogenetic link between the great apes and the Old World monkeys (Cercopithecoidea) as well as a useful position just before the radiation of great apes. The Old World monkeys diverged from the apes ∼25 Ma (Gibbs et al. 2007; Han et al. 2007), whereas the first divergence within the great apes that of the orangutan lineage occurred ∼14 Ma (Locke et al. 2011). The current estimates show that gibbons diverged from the other apes ∼15–18 Ma (Carbone et al. 2009).
Fig. 1.
Geographic distribution of the four gibbon genera. The Mekong River divides the Nomascus and mainland Hylobates groups. The Hoolock and mainland Hylobates groups are divided by the Salween River. Sympatry exists between Symphalangus and some members of the Hylobates group with conspecific breeding likely avoided due to significant size differences between these taxa. Map after Geissmann (1995) (http://www.gibbons.de/main/system/system.html).
Although clearly delineated from other apes, questions about the complex systematic relationships between gibbon genera and species remain contentious. Numerous phylogenies based on the behavior, morphology, geographic distribution, karyotype, and genetic analyses have been constructed, often leading to more questions than answers (Garza and Woodruff 1992; Geissmann 1995; Roos and Geissmann 2001; Muller, Hollatz, and Wienberg 2003; Takacs et al. 2005; Chatterjee 2006; Matsudaira and Ishida 2010; Thinh et al. 2010a; Kim et al. 2011). Especially debatable are the divergence dates of individual genera and the determination of which lineage is most basal.
The studies of mitochondrial markers have recently begun to achieve some statistical support for various phylogenetic hypotheses. A study by Takacs et al. (2005) of sequence from the mitochondrial ND3–ND4 region was able to resolve species relationships within genera, but it was unable to resolve relationships between genera. Another more recent mitochondrial study of complete cytochrome b gene sequences also failed to robustly resolve the branching patterns among the four genera (Thinh et al. 2010a). However, analysis of complete mitochondrial sequences found Nomascus supported as the most basal group within Hylobatidae (Chan et al. 2010; Matsudaira and Ishida 2010). In addition, a surprising frequency of chromosomal rearrangement within Hylobatidae, resulting in a diverse range of diploid chromosome numbers (Nomascus, 2n = 52; Symphalangus, 2n = 50; Hoolock, 2n = 38; and Hylobates, 2n = 44), has contributed to the confusion over the relationships between gibbons (Muller et al. 2003; Carbone et al. 2009).
The use of retrotransposon insertions as phylogenetic and population genetic markers has seen an increasing popularity in recent years (Okada 1991; Minghetti and Dugaiczyk 1993; Novick et al. 1993; Batzer et al. 1994; Stoneking et al. 1997; Hamdi et al. 1999; Nasidze et al. 2001; Romualdi et al. 2002; Salem et al. 2003; Watkins et al. 2003; Roos et al. 2004; Shedlock et al. 2004; Ray et al. 2005; Schmitz et al. 2005; Nikaido et al. 2006; Witherspoon et al. 2006; Herke et al. 2007; Xing et al. 2007a; Osterholz et al. 2008; Li et al. 2009; Osterholz et al. 2009; Nilsson et al. 2010; Roos et al. 2011; Suh et al. 2011). Short interspersed elements (SINEs) are <500 bp in length and generate new copies of themselves through reverse transcription of an RNA intermediate into a new position in the genome (Luan et al. 1993; Luan and Eickbush 1995; Cost et al. 2002). This pattern of mobilization results in an increase in the copy number of most SINEs within their host genomes. The copies of these elements produce many important impacts on the structure and function of the genome, especially when their insertion disrupts coding or regulatory regions (Callinan and Batzer 2006; Belancio et al. 2009; Cordaux and Batzer 2009; Belancio et al. 2010; Konkel and Batzer 2010; Konkel et al. 2010; Beck et al. 2011). However, coding and regulatory regions comprise only ∼5% of the genome (Lander et al. 2001), so that most new SINE insertions are likely to cause no disruption during their insertion (Cordaux et al. 2006; Konkel and Batzer 2010). This subset of insertions into neutrally evolving regions of the genome has a number of characteristics that make them desirable as phylogenetic markers. Genetic markers are said to be homoplasious if the phylogenetic tree inferred from them is not the true tree. SINEs are nearly homoplasy-free markers because the ancestral state of any locus is known to be the absence of the element (Batzer and Deininger 1991; Murata et al. 1993; Batzer et al. 1994; Shedlock and Okada 2000; Okada et al. 2004; Ray et al. 2006; Ray 2007; Xing et al. 2007b). The presence of a SINE insertion at a specific location within the genomes of two or more lineages is evidence that the insertion occurred at a point in time when the lineages shared a common ancestor. Potentially homoplasious events, such as precise deletion and parallel independent insertion, have been shown to be exceptionally rare, with most suspected cases being easily resolved by sequencing the suspicious loci (Ray et al. 2006; Xing et al. 2007a; Li et al. 2009). However, most loci are quickly and easily genotyped using a combination of polymerase chain reaction (PCR) and gel electrophoresis.
The expansion of one particular family of SINEs, the Alu family, began ∼65 Ma and has dominated within the radiation of primates ever since (Batzer and Deininger 2002; Deininger and Batzer 2002; Kriegs et al. 2007; Konkel et al. 2010; Perelman et al. 2011). Alu elements are the most successful lineage of mobile elements in primate genomes, being present in ∼1.2 million copies in the human genome and having been found in every primate sequenced thus far (Lander et al. 2001; Chimpanzee Sequencing and Analysis Consortium 2005; Gibbs et al. 2007; Locke et al. 2011). An Alu element is ∼300 bp long and is nonautonomous; hence, it does not encode the enzymatic machinery necessary for its own reverse transcription. Instead, Alu elements hijack the enzymatic machinery of a long interspersed element, L1 (Schmid 2003). The evolution of Alu elements within the primate radiation has resulted in a large number of subfamilies of elements, identifiable by the diagnostic mutations that they share with their progenitor copies (reviewed in Cordaux and Batzer 2009; Konkel et al. 2010). The mobilization activity of these subfamilies varies with time, allowing researchers to tailor their assays to specific subfamilies active only in the lineages and during the time periods in which they are interested (Churakov et al. 2010). Because of this, phylogenetic studies of primates using Alu elements as markers have been increasingly popular, helping to elucidate the relationships in many primate taxa, including those within the Homininae (Salem et al. 2003), Catarrhini (Xing et al. 2005, 2007a), Platyrrhini (Ray et al. 2005; Osterholz et al. 2009), and Strepsirrhini (Roos et al. 2004) clades, as well as the more detailed relationships within the genus Macaca (Li et al. 2009) and the Colobinae (Osterholz et al. 2008; Roos et al. 2011) and the affiliation of the genus Tarsius to other primates (Zietkiewicz et al. 1999; Schmitz et al. 2001).
In this study, we resolve relationships within gibbons using Alu elements as phylogenetic markers. We computationally screened the available genomic sequences of Nomascus leucogenys that have been generated as a part of the ongoing gibbon genome project. PCR assays in a panel of 18 primate species, including 13 gibbon species, produced 125 gibbon-specific Alu insertions that were used to generate a phylogeny of the Hylobatidae.
Materials and Methods
Computational Analysis of Candidate Loci
Genomic sequence generated by the gibbon sequencing consortium for N. leucogenys (northern white-cheeked gibbon) in the form of a large number of sequenced bacterial artificial chromosome (BAC) clones was obtained from the Ensembl database system (Hubbard et al. 2009). A local installation of RepeatMasker was then used to scan the sequences on the sensitive setting to classify all identifiable repetitive sequences (Smit et al. 1996–2010). Next, in-house Perl scripts were used to filter the RepeatMasker output, keeping only those sequences that contained Alu elements classified by RepeatMasker in the AluYc3 subfamily. However, further inspection revealed these elements to have been misidentified. They were instead members of the AluYd3a1_gib subfamily, which was not in the RepeatMasker RepBase library available at the time of analysis (Jurka 2000). This yielded a total of 9,701 candidate sequences that contained putatively gibbon-specific Alu insertions.
These computationally derived candidates were then queried against four available outgroup genomes, the human (hg19), chimpanzee (panTro2), orangutan (ponAbe2), and rhesus macaque (rheMac2), using BLAT searches (Kent 2002) via the UCSC Genome Bioinformatics website (http://genome.ucsc.edu/). Only those sequences whose putatively gibbon-specific Alu insertions were not present at the orthologous loci in the outgroups were retained for further analysis. In addition, the position of each orthologous locus in the human genome was noted for each candidate sequence that passed our BLAT analysis. Since at the time of analysis, the Ensembl data set represented an unassembled genome, many of the sequences were duplicate reads of the same region. The orthologous human positions of the gibbon-specific Alu insertions were used to identify all sequences in the data set that corresponded to a single locus. A total of 430 gibbon-specific candidate loci were identified using this strategy.
PCR Amplification and DNA Sequencing of Candidate Loci
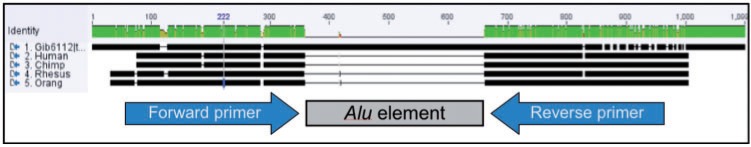
Flanking oligonucleotide primers for PCR amplification of each gibbon-specific Alu element were designed using the CLC Main Workbench v.5 software suite ([cited 2012 Jun 25], http://www.clcbio.com/index.php?id=92). This involved the creation of an alignment of each gibbon sequence to any orthologous sequences obtained from the outgroup genomes. The gibbon-specific Alu element and ∼20 bp of flanking sequence upstream and downstream were designated as the region to amplify (fig. 2). Oligonucleotide primers were selected as close to the element as possible while still amplifying some orthologous flanking sequence. This is because increased amplicon sizes containing excessive flanking sequence into which near parallel independent insertions may occur can potentially confound analyses. This problem is greater for analyses of distantly related species because researchers may need to search further into flanking regions to find sequences sufficiently conserved to allow primer amplification across all species. However, with due diligence and sequencing of questionable loci, this can be overcome. Next, the suggested primers were screened computationally against the available human, chimp, orangutan, and rhesus macaque genomes using the In-Silico PCR function available at the UCSC Genome Bioinformatics website. This was done to reaffirm the established gibbon specificity of the locus to be amplified and to determine whether the primer pairs amplified single loci (i.e., they were not designed within other repeats). Of the 430 candidate loci, only 231 had sufficient unique flanking sequence available to allow primers to be designed that passed in silico PCR analysis.
Fig. 2.
An illustration of how multiple alignments of orthologous loci are used to design oligonucleotide primers for lineage-specific insertions. The multiple alignment was performed using Clustal X. The gibbon sequence is N. leucogenys and contains a gibbon-specific Alu insertion (indicated by the 300 bp region in the center that does not match the other sequences). Also included in this alignment are H. sapiens, P. troglodytes, Pongo abelii, and Macaca mulatta. The gray rectangle shows the location of the Alu insertion in the gibbon. The arrows indicate the orthologous flanking regions in which primers are designed that will match all five species in the alignment.
Because our supply of some gibbon species and individual DNA was limited, and to verify that our primers amplified unambiguous loci and absent sites in non-gibbon primates, we first PCR amplified each Alu insertion locus against a primate panel consisting of one of each of the following species: Homo sapiens, Pan troglodytes, Gorilla gorilla, Pongo pygmaeus, Hylobates lar (PR00715), Nomascus gabriellae (PR00652), Symphalangus syndactylus (KB11539), and Chlorocebus aethiops. Of the 231 primer pairs tested, pairs that did not result in amplification in many species, amplified multiple paralogous fragments in each species, or produced smears were removed from further analysis. The remaining loci were amplified in a larger gibbon panel consisting of 31 individuals representing 18 species as follows: one H. sapiens, one P. troglodytes, one G. gorilla, one P. pygmaeus, four N. leucogenys, one Nomascus siki, one Nomascus annamensis, two N. gabriellae, three Hylobates moloch, one Hylobates agilis, one Hylobates albibarbis, one Hylobates muelleri, two H. lar, two Hylobates pileatus, one Hylobates klossii, three Hoolock leuconedys, three S. syndactylus, and one C. aethiops. A complete list of all DNA samples used in our analyses is available in the supplementary data (supplementary table S1, Supplementary Material online).
Due to the limited quantity of genomic DNA available for some individuals, some samples were subjected to whole genome amplification using the GenomiPhi genome amplification kit (Amersham, Sunnyvale, CA) following the manufacturer's instructions. PCR amplification was performed in the following conditions: 25 μl total volume using 15 ng of template DNA, 200 nM of each primer, 200 μM dNTPs in 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris–HCl (pH 8.4), and 2 units of Taq DNA polymerase. The conditions for PCR were as follows: an initial denaturation at 95°C for 1 min, followed by 32 cycles of denaturation at 95°C, annealing at 55°C, and extension at 72°C for 30 s each, followed by a final extension of 72°C for 1 min. Products were run out on 2% agarose stained with 0.25 μg ethidium bromide and visualized with ultraviolet fluorescence. The complete list of loci and primer sequences are available in the supplementary data (supplementary table S2, Supplementary Material online).
Any loci that indicated contradictory relationships to those shown in the most parsimonious tree were also subjected to automated DNA sequence analysis to verify the computationally derived Alu insertion sequences and to confirm the phylogenetic distribution of the insertions. In the case of these potentially homoplasious loci, representative filled and empty sites from the taxa involved were selected for sequencing analysis to determine the origin of the homoplasious signal. Ten such loci were apparent in our analysis and investigated through DNA sequencing. PCR products from selected individuals were directly sequenced as described previously (Ray et al. 2005). In addition, six critical diagnostic loci that inform the center of our tree (three grouping Nomascus, Hoolock, and Hylobates, and three grouping Nomascus and Hoolock) were subjected to DNA sequencing. All sequences were then aligned against orthologous sequences from other sequenced individuals and those obtained from BLAT analysis. These alignments are available in the supplementary data (Supplementary Material online), and all DNA sequences generated for this project have been deposited in GenBank under accession numbers (JQ611584-JQ611670, JQ861884-JQ861906, and JX034756-JX034761). In all but the one potentially homoplasious instance (locus m37; see Results and Discussion), sequencing allowed the resolution of the contradicting loci. All nine of the critical diagnostic loci reported had the insertions and associated target site duplications (TSDs) and flanking confirmed through sequencing, confirming significant support for these loci (see below).
Phylogenetic Analysis
A total of 125 loci were amplified in a majority of the gibbons in the 31 individual panel as well as in at least 4 of the 5 outgroups and were therefore used in the phylogenetic analysis. Each locus for each individual was scored based on whether it amplified as a filled site (presence of an Alu insertion), an empty site (absence of an insertion), or showed no amplification (unknown insertion status) (fig. 3). Bands in individuals heterozygous for both filled and empty sites were sequenced, and if the filled site was found to be an identical insertion, they were scored as having a filled site in our matrix. Filled sites were scored as a “1,” empty sites were scored as a “0,” and unknown sites were scored as “?.” Scores were entered using the Mesquite program ([cited 2012 Jun 25], http://mesquiteproject.org/.), and all loci were set to Dollo.up for parsimony analysis. The matrix used in the analysis can be found in the supplementary data (supplementary table S3, Supplementary Material online).
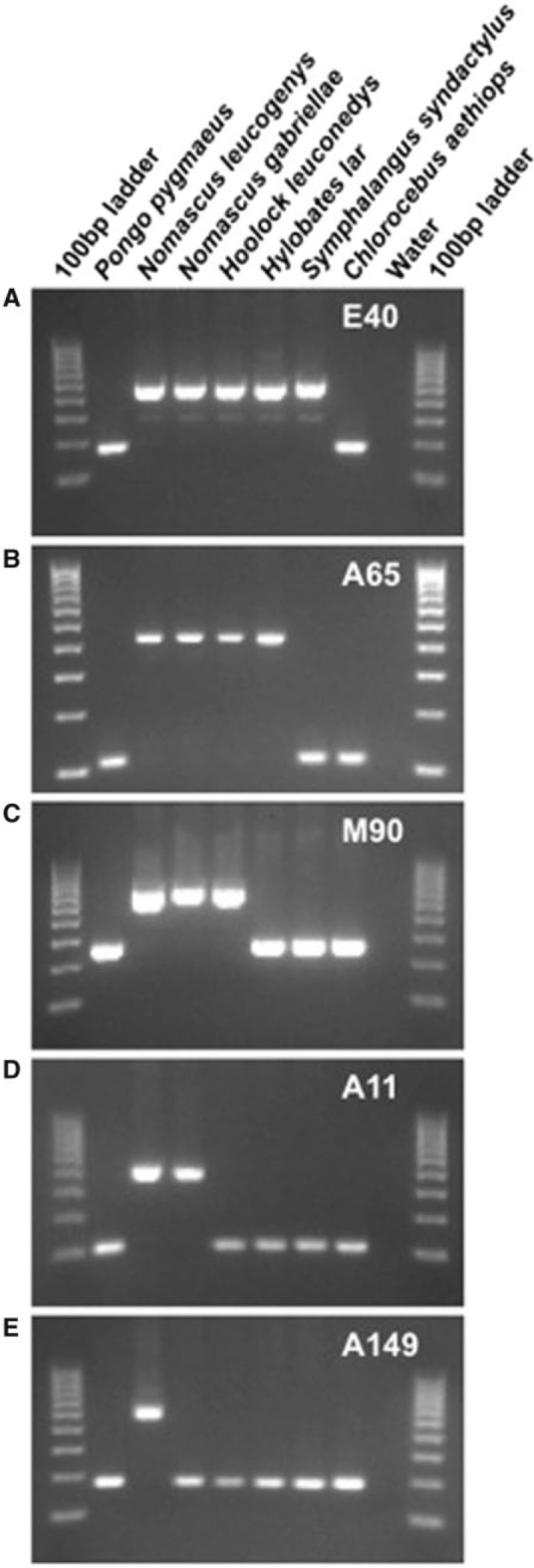
Fig. 3.
PCR amplification assay demonstrating how gibbon-specific Alu insertion polymorphisms can be used to distinguish lineages. The five gel electrophoresis images show the PCR amplification products from five primer sets in seven primate species, including five gibbons, one hominid, and one Old World monkey. The DNA template for each lane is shown at the top, and the locus is indicated in the upper right of each image. Larger bands indicate “filled” sites in which Alu elements have inserted, whereas lower bands indicate “empty” sites containing no Alu insertion. (A) An Alu insertion present in all gibbon species from all four genera. (B) An Alu insertion present only in Nomascus, Hoolock, and Hylobates. (C) An Alu insertion restricted only to Nomascus and Hoolock. (D) An Alu insertion that is Nomascus specific. (E) An Alu insertion that is present only in the species N. leucogenys.
A heuristic search in PAUP* 4.0b10 (Swofford 2000) was performed as described by Xing (2005). Briefly, C. aethiops was set as the outgroup, and all loci were treated as individual insertions following the criteria required for Dollo parsimony analysis. Trees and statistics were generated by PAUP*. Because our loci were harvested from the N. leucogenys genome, no resolution of the relationships within Hylobates was possible, resulting in a large polytomy for the members of this genus. Therefore, in the final tree, only H. lar and H. agilis were included as representatives of the genus. Tree space was limited to 100 trees, and 10,000 bootstrap replicates were run to provide percentage support for each branch under the majority rule tree, which omits bootstrap values less than 50% per branch (126 steps; consistency index [CI] = 0.9921; homoplasy index [HI] = 0.0079; and retention index [RI] = 0.9935). The statistically significant number of SINE insertions required for acceptance is provided under the likelihood model described by Waddell et al. (2001) (fig. 4). The tree was visualized using the FigTree software ([cited 2012 Jun 25], http://tree.bio.ed.ac.uk/software/figtree/).
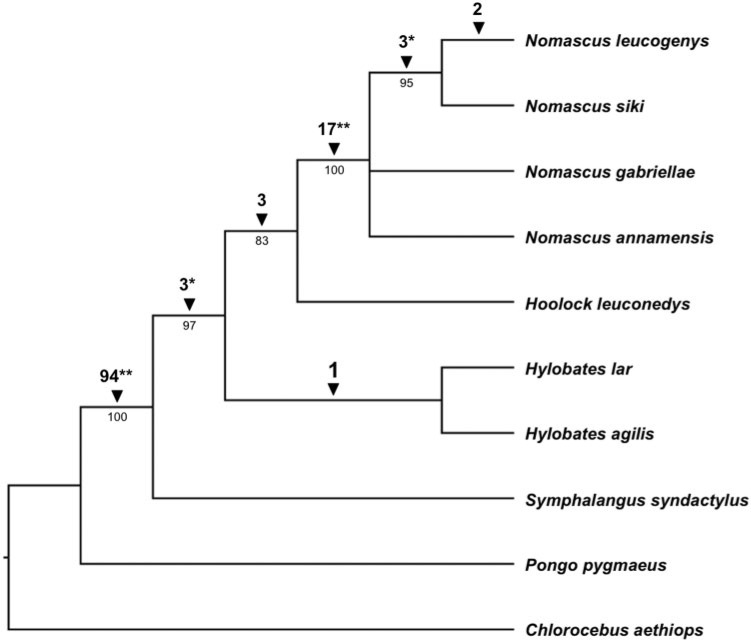
Fig. 4.
Cladogram of Hylobatidae phylogenetic relationships derived from 125 Alu insertion polymorphisms. Amplification patterns of insertion loci were used to construct a Dollo parsimony tree of genus level gibbon phylogenetic relationships using C. aethiops as an outgroup. Also included is P. pygmaeus, the orangutan, which is the hominid that diverged from other hominids relatively soon after gibbons diverged from other hominoids. The numbers below the branches are percentage bootstrap replicates (out of 10,000 iterations) producing trees that include that node. Numbers above the branches indicate the number of Alu insertions supporting the node. The significance level of each node supported by insertions as determined by likelihood testing is indicated by either *P < 0.05 or **P < 0.01.
Results and Discussion
Computational Data Mining of the N. leucogenys Genome
The unassembled sequence data available from the northern white-cheeked gibbon (N. leucogenys) genome sequencing project consists of 750,923 sequences generated via combined whole-genome shotgun plasmid, fosmid, and BAC end sequences. We analyzed the repetitive content of this data set using the RepeatMasker program. We identified 9,701 sequences of interest that contain Alu elements identified as belonging to the AluYc3 subfamily via an in-house Perl script (available on request). These sequences were subjected to BLAT alignment to determine whether the Alu of interest was shared with outgroup species from the Hominidae (human, chimpanzee, and orangutan) or the Old World monkeys (rhesus macaque). We later determined that those Alu insertions that were gibbon specific were not, in fact, AluYc3 elements, but were actually members of a previously reported gibbon-specific subfamily, AluYd3a1_gib. Although this subfamily was present in the RepBase database, it was not included in the repeat library used in the RepeatMasker analysis, explaining the initial misidentification.
The N. leucogenys genome will be a 6x coverage genome once assembly is complete. However, at the time of analysis, the sequence was unassembled, and as a result, many of the sequences analyzed were duplicate reads. Some loci were present in a single copy, but others were present in the data set more than 35 times. In total, 430 unique, computationally derived candidates containing a gibbon-specific Alu element were identified (fig. 2).
PCR Assays of Gibbon-Specific Alu Insertions
Multiple alignments of gibbon and outgroup sequences and automated primer design were attempted for all 430 computational candidates. Some loci were removed from analysis due to a lack of sufficient or unique sequence flanking the Alu insertion in which to design primers. Primers were tested using in silico PCR to determine whether they would amplify single loci in most of the outgroups. A total of 231 loci passed this scrutiny and were then used in PCR verification.
Each locus was first amplified on a panel of primate DNA representing one individual from each of eight species, three of which were gibbons. Those primer pairs that amplified well on this panel and indicated gibbon specificity were then run against the second, larger primate panel. This panel included 31 individuals from 18 species; 26 of these individuals were gibbons from 13 gibbon species including species from each of the 4 gibbon genera.
The 125 loci that amplified unique fragments in a majority of the species on the panel were scored for use in the phylogenetic analysis. Figure 3 demonstrates how lineage-specific insertions can be used to infer the phylogenetic relationships between groups. Loci that produced smears or multiple nonspecific banding patterns were removed from further analysis. Several loci produced double-banding patterns in some species or individuals. These loci may represent a useful set of markers for population genetics studies of these species, as such patterns may be indicative of heterozygous individuals, indicating an insertion that is still polymorphic in the population (Cordaux et al. 2007; Ray et al. 2007). Other possible explanations for such loci include an Alu insertion that occurred after a duplication event generated two paralogous regions or introgression stemming from hybridization events (Xing et al. 2007a; Osterholz et al. 2008).
Phylogenetic Relationships within the Hylobatidae
In total, we used 125 loci in the phylogenetic analysis. A heuristic search was performed using the Dollo parsimony method, with C. aethiops designated as the outgroup taxon. A total of 108 of the loci were found to be parsimony informative, resulting in a single most parsimonious tree (126 steps; CI = 0.9921; HI = 0.0079; RI = 0.9935) (fig. 4). In total, 10,000 bootstrap replicates were performed as well as a likelihood test based on the number of unambiguous Alu insertions supporting each node (Waddell et al. 2001). Bootstrap values, significance levels, and the number of supportive insertions for each node are indicated in figure 4. The likelihood tests of every genus level node except one within the phylogenetic tree were found to be significant.
The topology of our tree clearly defines relationships between the four extant genera within Hylobatidae: Nomascus, Hoolock, Hylobates, and Symphalangus. Previous attempts to reconstruct a genus level tree for Hylobatidae have proven problematic, resulting in many different topologies (Geissmann 1995; Roos and Geissmann 2001; Takacs et al. 2005; Chatterjee 2006; Chan et al. 2010; Matsudaira and Ishida 2010; Thinh et al. 2010a; Kim et al. 2011), often with insufficient support at the genus level nodes. This is likely due to the relatively recent and rapid radiation within the group (Matsudaira and Ishida 2010) leading to incomplete lineage sorting of characters used in phylogenetic analyses. Our finding for the most basal lineage is consistent with some of these previous studies. We demonstrate significant support for Symphalangus as the most basal lineage within the family. This was the finding in a previous study utilizing morphometric data (Creel and Preuschoft 1984) and one cytochrome b analysis (Garza and Woodruff 1992). However, this is inconsistent with other more recent cytochrome b analyses (Chatterjee 2006; Thinh et al. 2010a) as well as analyses of morphological and behavioral characters (Haimoff et al. 1982), chromosomal homology (Muller et al. 2003), and the mitochondrial control region (Roos and Geissmann 2001). Our findings also place Nomascus as a derived lineage, sister to Hoolock, with the Nomascus–Hoolock clade forming a sister to Hylobates. The insertions supporting these affiliations were subjected to DNA sequencing and analysis, and in every case, the shared insertion architecture was confirmed. Some previous studies placed Nomascus as the most basal clade (Roos and Geissmann 2001; Chan et al. 2010; Thinh et al. 2010a), representing a distinct difference in our reconstruction from most previous ones. Within the Nomascus, we found N. leucogenys and N. siki to be more closely related to one another than either was to the other two Nomascus species assayed. This is not surprising given that N. siki was previously classified as a subspecies of N. leucogenys (Garza and Woodruff 1992; Geissmann 1995), and this relationship is consistent with other recent reconstructions (Thinh et al. 2010a; Thinh et al. 2010c). Given the frequency of chromosomal rearrangements among gibbons and the rapid radiation of extant lineages, we favor the Alu-based phylogenetic reconstruction method over the alternatives. This method has allowed us to pull a robust phylogenetic signal out of the data despite the difficulties that other systems have encountered with this group.
Only one locus in the analysis, m37, did not support the most parsimonious tree. This locus is the source of the small amount of homoplasy (HI = 0.0079) observed. The insertion was found in all Nomascus species and all Hylobates species except Hylobates pileatus. However, this insertion was absent in Hoolock and Symphalangus. This locus was among the subset sequenced to verify the computational findings and analyze the sequence architecture of the insertion site in detail. It shows identical TSDs and identical Alu elements across all species sequenced. Possible explanations for these observations are that this locus represents an example of precise parallel insertion and/or precise deletion in the lineages affected or that this locus represents an incomplete lineage sorting event. We favor the latter explanation given the relatively rapid speciation time over which the extant gibbon genera are known to have diverged from one another (6–10 Ma) (Matsudaira and Ishida 2010). This locus affects the likelihood values supporting the Nomascus–Hoolock clade. With three loci supporting this clade, and one supporting a Nomascus–Hylobates grouping, the support for this branch is only P = 0.111 (fig. 4).
The inconsistency with which the topology of the gibbon genus level tree has been resolved in the past, and our own difficulty in obtaining robust support for the Nomascus–Hoolock grouping, is presumably the result of the rapid radiation of extant genera. However, introgression may also play a large role. Gibbon species have been known to hybridize in the wild and generate viable offspring (Myers and Shafer 1979). Furthermore, most of the previous studies of gibbon phylogenetics involve samples taken from either zoo specimens or specimens from captive breeding programs. Gibbons are notoriously difficult to reliably identify to the species level, and hybridization in captivity is known to occur (Mootnick 2006). If such hybrids were misidentified as purebred individuals in previous studies, they could potentially confound the true phylogenetic signal (Churakov et al. 2009). In fact, the individual sampled need not be a direct hybrid, as such introgression events could have occurred several generations in the past and still adversely affect identification. However, given the support for our topology and the fact that only one homoplasious locus was identified in our data set, we believe it is unlikely that sufficient introduction of Alu insertion loci from other lineages has occurred via hybridization between members of different gibbon genera to affect our conclusions. In addition, this analysis represents a necessarily single-sided view of gibbon phylogeny, as it is dependent on the complete genome of N. leucogenys. In the future, we hope that complete genomes for members of the Hylobates and Hoolock genera become available to allow a comparison with our results from Nomascus. The Alu insertion markers developed by this, and future analyses will help conservation efforts by providing relatively inexpensive and quick assays to determine the species identity of individuals being selected for breeding programs.
Conclusion
This study represents the first use of SINEs to investigate the phylogenetic relationships within the family Hylobatidae. By using a combination of computational and wet bench techniques, a total of 125 loci containing gibbon-specific Alu insertions were found. These were used to generate a phylogenetic hypothesis of gibbon phylogeny, with statistically significant support for all but one internal branch. The nearly homoplasy-free nature of Alu insertions is ideally suited to the Dollo parsimony method for the reconstruction of well-supported primate phylogenies. We found Symphalangus to be the most basal lineage, followed by a divergence between the Hylobates and a clade consisting of Hoolock and Nomascus. The four Nomascus species analyzed, N. leucogenys, N. siki, N. annamensis, and N. gabriellae, were also well supported as a clade to the exclusion of Hoolock. Furthermore, N. leucogenys and N. siki were grouped as sister species within the Nomascus. The most striking difference between our tree and those produced in the previous analyses is the grouping of Nomascus with Hoolock as the most derived multigenera clade. A single locus in our data set contradicting this relationship may be the result of the hybridization and rapid radiation characteristic of the gibbon radiation. These factors may have contributed to the complex and contradictory findings of previous studies. In addition, our findings may inform future studies of the biogeographical history and distribution of modern gibbon species. It is our hope that the markers developed in this study will provide a means by which individuals may be quickly and inexpensively genotyped by conservation workers, so that hybridization can be avoided in breeding programs.
Supplementary Material
Supplementary tables S1–S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors wish to express their deep sadness for the loss of their colleague and coauthor, Alan R. Mootnick, who died during the preparation of this manuscript. He worked selflessly, establishing the Gibbon Conservation Center in 1976. He oversaw a successful gibbon breeding program there that gave hope that some of these fascinating apes may yet survive their endangered status. In addition, he worked tirelessly with the academic community to promote research into the animals he so loved. He will be greatly missed. They thank the Parc Zoologique de Cleres, the Gladys Porter Zoo, the Los Angeles Zoo, the Fort Wayne Children's Zoo, the Henry Doorly Zoo, and the Louisiana Purchase Zoo for providing some of the gibbon samples used in this study, as well as the Gibbon Genome Sequencing and Analysis Consortium for providing the N. leucogenys genome sequence. The whole genome shotgun sequencing of the Northern white-cheeked gibbon was performed at the Washington University Genome Sequencing Center and the Baylor College of Medicine Human Genome Sequencing Center. They also thank J.A. Walker and M.K. Konkel for their invaluable help throughout this project. This work was supported by National Institutes of Health grant RO1 GM59290 to M.A.B.
References
- Batzer MA, Deininger PL. A human-specific subfamily of Alu sequences. Genomics. 1991;9:481–487. doi: 10.1016/0888-7543(91)90414-a. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Stoneking M, Alegria-Hartman M, et al. (11 co-authors) African origin of human-specific polymorphic Alu insertions. Proc Natl Acad Sci U S A. 1994;91:12288–12292. doi: 10.1073/pnas.91.25.12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Deininger PL, Roy-Engel AM. LINE dancing in the human genome: transposable elements and disease. Genome Med. 2009;1:97. doi: 10.1186/gm97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Roy-Engel AM, Deininger PL. All y'all need to know ‘bout retroelements in cancer. Semin Cancer Biol. 2010;20:200–210. doi: 10.1016/j.semcancer.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callinan PA, Batzer MA. Retrotransposable elements and human disease. Genome Dyn. 2006;1:104–115. doi: 10.1159/000092503. [DOI] [PubMed] [Google Scholar]
- Carbone L, Harris RA, Vessere GM, et al. (12 co-authors) Evolutionary breakpoints in the gibbon suggest association between cytosine methylation and karyotype evolution. PLoS Genet. 2009;5:e1000538. doi: 10.1371/journal.pgen.1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YC, Roos C, Inoue-Murayama M, Inoue E, Shih CC, Pei KJ, Vigilant L. Mitochondrial genome sequences effectively reveal the phylogeny of Hylobates gibbons. PLoS One. 2010;5:e14419. doi: 10.1371/journal.pone.0014419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee HJ. Phylogeny and biogeography of gibbons: a dispersal-vicariance analysis. Int J Primatol. 2006;27:699–712. [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Churakov G, Grundmann N, Kuritzin A, Brosius J, Makalowski W, Schmitz J. A novel web-based TinT application and the chronology of the Primate Alu retroposon activity. BMC Evol Biol. 2010;10:376. doi: 10.1186/1471-2148-10-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churakov G, Kriegs JO, Baertsch R, Zemann A, Brosius J, Schmitz J. Mosaic retroposon insertion patterns in placental mammals. Genome Res. 2009;19:868–875. doi: 10.1101/gr.090647.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Lee J, Dinoso L, Batzer MA. Recently integrated Alu retrotransposons are essentially neutral residents of the human genome. Gene. 2006;373:138–144. doi: 10.1016/j.gene.2006.01.020. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Srikanta D, Lee J, Stoneking M, Batzer MA. In search of polymorphic Alu insertions with restricted geographic distributions. Genomics. 2007;90:154–158. doi: 10.1016/j.ygeno.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. Embo J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creel N, Preuschoft H. Systematics of the lesser apes: a quantitative taxonomic analysis of craniometric and other variables. In: Preuschoft H, Chivers DJ, Broekelman WY, Creel N, editors. The lesser apes: evolutionary and behavioural biology. Edinburgh (United Kingdom): Edinburgh University Press; 1984. pp. 562–613. [Google Scholar]
- Deininger PL, Batzer MA. Mammalian retroelements. Genome Res. 2002;12:1455–1465. doi: 10.1101/gr.282402. [DOI] [PubMed] [Google Scholar]
- Garza JC, Woodruff DS. A phylogenetic study of the gibbons (Hylobates) using DNA obtained noninvasively from hair. Mol Phylogenet Evol. 1992;1:202–210. doi: 10.1016/1055-7903(92)90016-a. [DOI] [PubMed] [Google Scholar]
- Geissmann T. Gibbon systematics and species identification. Int Zoo News. 1995;42:467–501. [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, et al. (176 co-authors) Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Groves C. Primate taxonomy. Washington (DC): Smithsonian Press; 2001. [Google Scholar]
- Haimoff EH, Chivers DJ, Gittins SP, Whitten T. A phylogeny of gibbons (Hylobates spp.) based on morphological and behavioural characters. Folia Primatol (Basel) 1982;39:213–237. doi: 10.1159/000156079. [DOI] [PubMed] [Google Scholar]
- Hamdi H, Nishio H, Zielinski R, Dugaiczyk A. Origin and phylogenetic distribution of Alu DNA repeats: irreversible events in the evolution of primates. J Mol Biol. 1999;289:861–871. doi: 10.1006/jmbi.1999.2797. [DOI] [PubMed] [Google Scholar]
- Han K, Konkel MK, Xing J, et al. (15 co-authors) Mobile DNA in Old World monkeys: a glimpse through the rhesus macaque genome. Science. 2007;316:238–240. doi: 10.1126/science.1139462. [DOI] [PubMed] [Google Scholar]
- Herke SW, Xing J, Ray DA, Zimmerman JW, Cordaux R, Batzer MA. A SINE-based dichotomous key for primate identification. Gene. 2007;390:39–51. doi: 10.1016/j.gene.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Ayling S, et al. (58 co-authors) Ensembl 2009. Nucleic Acids Res. 2009;37:D690–697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Carbone L, Becquet C, Mootnick AR, Li DJ, de Jong PJ, Wall JD. Patterns of genetic variation within and between gibbon species. Mol Biol Evol. 2011;28:2211–2218. doi: 10.1093/molbev/msr033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel MK, Batzer MA. A mobile threat to genome stability: the impact of non-LTR retrotransposons upon the human genome. Semin Cancer Biol. 2010;20:211–221. doi: 10.1016/j.semcancer.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel MK, Walker JA, Batzer MA. LINEs and SINEs of primate evolution. Evol Anthropol. 2010;19:236–249. doi: 10.1002/evan.20283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegs JO, Churakov G, Jurka J, Brosius J, Schmitz J. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 2007;23:158–161. doi: 10.1016/j.tig.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, et al. (249 co-authors) Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Li J, Han K, Xing J, Kim HS, Rogers J, Ryder OA, Disotell T, Yue B, Batzer MA. Phylogeny of the macaques (Cercopithecidae: Macaca) based on Alu elements. Gene. 2009;448:242–249. doi: 10.1016/j.gene.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Hillier LW, Warren WC, et al. (102 co-authors) Comparative and demographic analysis of orangutan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Eickbush TH. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol Cell Biol. 1995;15:3882–3891. doi: 10.1128/mcb.15.7.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- Matsudaira K, Ishida T. Phylogenetic relationships and divergence dates of the whole mitochondrial genome sequences among three gibbon genera. Mol Phylogenet Evol. 2010;55:454–459. doi: 10.1016/j.ympev.2010.01.032. [DOI] [PubMed] [Google Scholar]
- Minghetti PP, Dugaiczyk A. The emergence of new DNA repeats and the divergence of primates. Proc Natl Acad Sci U S A. 1993;90:1872–1876. doi: 10.1073/pnas.90.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootnick AR. Gibbon (Hylobatidae) species identification recommended for rescue or breeding centers. Primate Conservation. 2006;21:103–138. [Google Scholar]
- Muller S, Hollatz M, Wienberg J. Chromosomal phylogeny and evolution of gibbons (Hylobatidae) Hum Genet. 2003;113:493–501. doi: 10.1007/s00439-003-0997-2. [DOI] [PubMed] [Google Scholar]
- Murata S, Takasaki N, Saitoh M, Okada N. Determination of the phylogenetic relationships among Pacific salmonids by using short interspersed elements (SINEs) as temporal landmarks of evolution. Proc Natl Acad Sci U S A. 1993;90:6995–6999. doi: 10.1073/pnas.90.15.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RH, Shafer DA. Hybrid ape offspring of a mating of gibbon and siamang. Science. 1979;205:308–310. doi: 10.1126/science.451603. [DOI] [PubMed] [Google Scholar]
- Nasidze I, Risch GM, Robichaux M, Sherry ST, Batzer MA, Stoneking M. Alu insertion polymorphisms and the genetic structure of human populations from the Caucasus. Eur J Hum Genet. 2001;9:267–272. doi: 10.1038/sj.ejhg.5200615. [DOI] [PubMed] [Google Scholar]
- Nikaido M, Hamilton H, Makino H, Sasaki T, Takahashi K, Goto M, Kanda N, Pastene LA, Okada N. Baleen whale phylogeny and a past extensive radiation event revealed by SINE insertion analysis. Mol Biol Evol. 2006;23:866–873. doi: 10.1093/molbev/msj071. [DOI] [PubMed] [Google Scholar]
- Nilsson MA, Churakov G, Sommer M, Tran NV, Zemann A, Brosius J, Schmitz J. Tracking marsupial evolution using archaic genomic retroposon insertions. PLoS Biol. 2010;8:e1000436. doi: 10.1371/journal.pbio.1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick GE, Gonzalez T, Garrison J, Novick CC, Batzer MA, Deininger PL, Herrera RJ. The use of polymorphic Alu insertions in human DNA fingerprinting. Exs. 1993;67:283–291. doi: 10.1007/978-3-0348-8583-6_26. [DOI] [PubMed] [Google Scholar]
- Okada N. SINEs. Curr Opin Genet Dev. 1991;1:498–504. doi: 10.1016/s0959-437x(05)80198-4. [DOI] [PubMed] [Google Scholar]
- Okada N, Shedlock AM, Nikaido M. Retroposon mapping in molecular systematics. Methods Mol Biol. 2004;260:189–226. doi: 10.1385/1-59259-755-6:189. [DOI] [PubMed] [Google Scholar]
- Osterholz M, Vermeer J, Walter L, Roos C. A PCR-based marker to simply identify Saimiri sciureus and S. boliviensis boliviensis. Am J Primatol. 2008;70:1177–1180. doi: 10.1002/ajp.20606. [DOI] [PubMed] [Google Scholar]
- Osterholz M, Walter L, Roos C. Phylogenetic position of the langur genera Semnopithecus and Trachypithecus among Asian colobines, and genus affiliations of their species groups. BMC Evol Biol. 2008;8:58. doi: 10.1186/1471-2148-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterholz M, Walter L, Roos C. Retropositional events consolidate the branching order among New World monkey genera. Mol Phylogenet Evol. 2009;50:507–513. doi: 10.1016/j.ympev.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Perelman P, Johnson WE, Roos C, et al. (14 co-authors) A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DA. SINEs of progress: mobile element applications to molecular ecology. Mol Ecol. 2007;16:19–33. doi: 10.1111/j.1365-294X.2006.03104.x. [DOI] [PubMed] [Google Scholar]
- Ray DA, Walker JA, Batzer MA. Mobile element-based forensic genomics. Mutat Res. 2007;616:24–33. doi: 10.1016/j.mrfmmm.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Ray DA, Xing J, Hedges DJ, et al. (13 co-authors) Alu insertion loci and platyrrhine primate phylogeny. Mol Phylogenet Evol. 2005;35:117–126. doi: 10.1016/j.ympev.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Ray DA, Xing J, Salem AH, Batzer MA. SINEs of a nearly perfect character. Syst Biol. 2006;55:928–935. doi: 10.1080/10635150600865419. [DOI] [PubMed] [Google Scholar]
- Romualdi C, Balding D, Nasidze IS, Risch G, Robichaux M, Sherry ST, Stoneking M, Batzer MA, Barbujani G. Patterns of human diversity, within and among continents, inferred from biallelic DNA polymorphisms. Genome Res. 2002;12:602–612. doi: 10.1101/gr.214902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos C, Geissmann T. Molecular phylogeny of the major hylobatid divisions. Mol Phylogenet Evol. 2001;19:486–494. doi: 10.1006/mpev.2001.0939. [DOI] [PubMed] [Google Scholar]
- Roos C, Schmitz J, Zischler H. Primate jumping genes elucidate strepsirrhine phylogeny. Proc Natl Acad Sci U S A. 2004;101:10650–10654. doi: 10.1073/pnas.0403852101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos C, Zinner D, Kubatko LS, et al. (16 co-authors) Nuclear versus mitochondrial DNA: evidence for hybridization in colobine monkeys. BMC Evol Biol. 2011;11:77. doi: 10.1186/1471-2148-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem AH, Ray DA, Xing J, Callinan PA, Myers JS, Hedges DJ, Garber RK, Witherspoon DJ, Jorde LB, Batzer MA. Alu elements and hominid phylogenetics. Proc Natl Acad Sci U S A. 2003;100:12787–12791. doi: 10.1073/pnas.2133766100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid CW. Alu: a parasite's parasite? Nat Genet. 2003;35:15–16. doi: 10.1038/ng0903-15. [DOI] [PubMed] [Google Scholar]
- Schmitz J, Ohme M, Zischler H. SINE insertions in cladistic analyses and the phylogenetic affiliations of Tarsius bancanus to other primates. Genetics. 2001;157:777–784. doi: 10.1093/genetics/157.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J, Roos C, Zischler H. Primate phylogeny: molecular evidence from retroposons. Cytogenet Genome Res. 2005;108:26–37. doi: 10.1159/000080799. [DOI] [PubMed] [Google Scholar]
- Shedlock AM, Okada N. SINE insertions: powerful tools for molecular systematics. Bioessays. 2000;22:148–160. doi: 10.1002/(SICI)1521-1878(200002)22:2<148::AID-BIES6>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Shedlock AM, Takahashi K, Okada N. SINEs of speciation: tracking lineages with retroposons. Trends Ecol Evol. 2004;19:545–553. doi: 10.1016/j.tree.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. 1996–2010. [cited 2012 Jun 25]. Available from: http://www.repeatmasker.org.
- Stoneking M, Fontius JJ, Clifford SL, Soodyall H, Arcot SS, Saha N, Jenkins T, Tahir MA, Deininger PL, Batzer MA. Alu insertion polymorphisms and human evolution: evidence for a larger population size in Africa. Genome Res. 1997;7:1061–1071. doi: 10.1101/gr.7.11.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh A, Paus M, Kiefmann M, Churakov G, Franke FA, Brosius J, Kriegs JO, Schmitz J. Mesozoic retroposons reveal parrots as the closest living relatives of passerine birds. Nat Commun. 2011;2:443. doi: 10.1038/ncomms1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP: phylogenetic analysis using parsimony. Sunderland (MA): Sinauer Associates Inc.; 2000. [Google Scholar]
- Takacs Z, Morales JC, Geissmann T, Melnick DJ. A complete species-level phylogeny of the Hylobatidae based on mitochondrial ND3-ND4 gene sequences. Mol Phylogenet Evol. 2005;36:456–467. doi: 10.1016/j.ympev.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Thinh VN, Mootnick AR, Geissmann T, Li M, Ziegler T, Agil M, Moisson P, Nadler T, Walter L, Roos C. Mitochondrial evidence for multiple radiations in the evolutionary history of small apes. BMC Evol Biol. 2010a;10:74. doi: 10.1186/1471-2148-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thinh VN, Mootnick AR, Thanh VN, Nadler T, Roos C. A new species of crested gibbon, from the central Annamite mountain range. Vietn J Primatol. 2010b;1:1–12. [Google Scholar]
- Thinh VN, Rawson B, Hallam C, Kenyon M, Nadler T, Walter L, Roos C. Phylogeny and distribution of crested gibbons (genus Nomascus) based on mitochondrial cytochrome b gene sequence data. Am J Primatol. 2010c;72:1047–1054. doi: 10.1002/ajp.20861. [DOI] [PubMed] [Google Scholar]
- Waddell PJ, Kishino H, Ota R. A phylogenetic foundation for comparative mammalian genomics. Genome Inform Ser Workshop Genome Inform. 2001;12:141–154. [PubMed] [Google Scholar]
- Watkins WS, Rogers AR, Ostler CT, et al. (14 co-authors) Genetic variation among world populations: inferences from 100 Alu insertion polymorphisms. Genome Res. 2003;13:1607–1618. doi: 10.1101/gr.894603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherspoon DJ, Marchani EE, Watkins WS, et al. (13 co-authors) Human population genetic structure and diversity inferred from polymorphic L1(LINE-1) and Alu insertions. Hum Hered. 2006;62:30–46. doi: 10.1159/000095851. [DOI] [PubMed] [Google Scholar]
- Xing J, Wang H, Han K, Ray DA, Huang CH, Chemnick LG, Stewart CB, Disotell TR, Ryder OA, Batzer MA. A mobile element based phylogeny of Old World monkeys. Mol Phylogenet Evol. 2005;37:872–880. doi: 10.1016/j.ympev.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Xing J, Wang H, Zhang Y, Ray DA, Tosi AJ, Disotell TR, Batzer MA. A mobile element-based evolutionary history of guenons (tribe Cercopithecini) BMC Biol. 2007a;5:5. doi: 10.1186/1741-7007-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Witherspoon DJ, Ray DA, Batzer MA, Jorde LB. Mobile DNA elements in primate and human evolution. Am J Phys Anthropol Suppl. 2007b;45:2–19. doi: 10.1002/ajpa.20722. [DOI] [PubMed] [Google Scholar]
- Zietkiewicz E, Richer C, Labuda D. Phylogenetic affinities of tarsier in the context of primate Alu repeats. Mol Phylogenet Evol. 1999;11:77–83. doi: 10.1006/mpev.1998.0564. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.