Abstract
Translation termination is accomplished by proteins of the Class I release factor family (RF) that recognize stop codons and catalyze the ribosomal release of the newly synthesized peptide. Bacteria have two canonical RFs: RF1 recognizes UAA and UAG, RF2 recognizes UAA and UGA. Despite that these two release factor proteins are sufficient for de facto translation termination, the eukaryotic organellar RF protein family, which has evolved from bacterial release factors, has expanded considerably, comprising multiple subfamilies, most of which have not been functionally characterized or formally classified. Here, we integrate multiple sources of information to analyze the remarkable differentiation of the RF family among organelles. We document the origin, phylogenetic distribution and sequence structure features of the mitochondrial and plastidial release factors: mtRF1a, mtRF1, mtRF2a, mtRF2b, mtRF2c, ICT1, C12orf65, pRF1, and pRF2, and review published relevant experimental data. The canonical release factors (mtRF1a, mtRF2a, pRF1, and pRF2) and ICT1 are derived from bacterial ancestors, whereas the others have resulted from gene duplications of another release factor. These new RF family members have all lost one or more specific motifs relevant for bona fide release factor function but are mostly targeted to the same organelle as their ancestor. We also characterize the subset of canonical release factor proteins that bear nonclassical PxT/SPF tripeptide motifs and provide a molecular-model-based rationale for their retained ability to recognize stop codons. Finally, we analyze the coevolution of canonical RFs with the organellar genetic code. Although the RF presence in an organelle and its stop codon usage tend to coevolve, we find three taxa that encode an RF2 without using UGA stop codons, and one reverse scenario, where mamiellales green algae use UGA stop codons in their mitochondria without having a mitochondrial type RF2. For the latter, we put forward a “stop-codon reinvention” hypothesis that involves the retargeting of the plastid release factor to the mitochondrion.
Keywords: release factor, translation termination, mitochondrion, plastid, evolution, genetic code
Introduction
Mitochondria and plastids translate their own genetic material. Even though the number of protein coding genes in these organelles can be quite limited, ranging from three genes in the mitochondria of apicomplexa (Hikosaka et al. 2010) to 273 in the chloroplasts of Pinus koraiensis (Noh et al. 2007), their translation involves many molecular players—rRNAs, tRNAs, aminoacyl-tRNA synthetases, ribosomal protein subunits and translation initiation, elongation, and termination factors—with at least 150 proteins having been implicated in translating human mitochondrial mRNAs (Rötig 2011). One protein group that is essential for translation is the Class I Release Factor family. These recognize the stop codon at the ribosomal A-site, upon which they hydrolyze the ester-bond that connects the nascent polypeptide to the last tRNA in the ribosomal P-site, thus releasing the newly synthesized protein (Petry et al. 2008). Although cytosolic translation involves a single peptide chain release factor—eRF1—of archaeal origin (Moreira et al. 2002) that decodes all three stop codons (Frolova et al. 1994); organellar translation termination, just like bacterial translation termination, employs two codon-specific release factors: RF1 recognizes UAA and UAG, and RF2 recognizes UAA and UGA (Scolnick et al. 1968). Mitochondrial and plastidial versions of RF1 and RF2—mtRF1a, mtRF2a, pRF1 and pRF2—have been described and some (mtRF1a and pRF2) have been functionally characterized (Meurer et al. 2002; Soleimanpour-Lichaei et al. 2007). But besides these, five other eukaryotic protein families have been recognized as putative members of the organellar release factor family: mtRF1, mtRF2b, mtRF2c, ICT1, and C12orf65 (Raczynska et al. 2006; Chrzanowska-Lightowlers et al. 2011).
Assigning proteins to the release factor family has mostly been done automatically, based on their homology to known RFs, and, with the exception of ICT1, the molecular functions of the noncanonical RFs remain unknown. Nevertheless, the individual domains and sequence motifs within the RFs have been experimentally well characterized. Bona fide release factors exhibit two catalytic domains: the Codon Recognition (CR) domain, composed of the helix alpha-5 and the “anticodon tripeptide motif”—PxT in RF1 and SPF in RF2—; and the peptidyl-tRNA hydrolase (PTH) domain, characterized by its universally conserved GGQ motif (Seit-Nebi et al. 2001). Extrapolating the function of a protein based on the presence/absence patterns of these domains was successful in the case of ICT1. This protein lacks the CR domain but contains the PTH one, and accordingly, was experimentally shown to have a codon-independent release factor activity (Richter et al. 2010).
Tightly linked to translation termination is the genetic code, particularly the identity of the nonsense codons used to stop translation. This is especially important for organellar genomes, because many of them exhibit deviations from the standard genetic code (reviewed in Sengupta et al. 2007; Ohama et al. 2008; Watanabe 2010). The most common deviations involve a nonsense codon reassignment, in which a stop codon (most frequently TGA, but there are also a few reports of TAA [Jacob et al. 2009] and TAG [Hayashi-Ishimaru et al. 1996]) is reassigned to code for an amino acid or is simply not used at all (reviewed in Knight et al. 2001). The first case of such a reassignment was reported in 1979 for the human mitochondrion, whose TGA codes for a tryptophan (Barrell et al. 1979). In time, as more mitogenome sequences got published, this emerged to be the standard mitochondrial genetic code, not only for animals but also for fungi and most green algae and protists (Sengupta et al. 2007). Nevertheless, accurately predicting a genome’s genetic code, and specifically its stop codons is not trivial. In fact, the genetic code of the human mitochondrion has been fully resolved only in 2010 (Temperley et al. 2010), whereas that of many other organisms still remains unknown.
Nearly one decade after the discovery of the mitochondrial TGA reassignment, Lee et al. (1987) published the first report of the coevolution of the mitochondrial genetic code with its termination factors, reporting that the lack of usage of UGA as a stop codon in the rat’s mitochondrion coincided with the absence of a mitochondrial type RF2. Since then, similar trends have been noted in other organisms (Askarian-Amiri et al. 2000; Meurer et al. 2002; Heidel and Glöckner 2008), adding to the hypothesis that the presence of codon-specific release factors in the organelle has coevolved with its genetic code (Jukes and Osawa 1990). However, no systematic studies to corroborate this theory have been done so far, and at least one instance has been reported, in the social amoeba Dictyostelium fasciculatum, where RF2 is retained and expressed, despite the lack of TGA stop codons (Heidel and Glöckner 2008), offering an interesting evolutionary scenario that could represent a transition state in switching between genetic codes. The mechanisms responsible for these reassignments have not been unequivocally established, but it has been proposed that the stop codons’ scarcity (used only once per gene) together with the possibility of fast changes in release factors—for example, if a RF is deleted as a result of genomic streamlining or if a mutation inactivates it—might play an important role (Osawa et al. 1992).
There are very few studies characterizing the organellar members of the RF protein family. Most reports focus either on the prokaryotic proteins or describe a particular organellar RF (e.g., mtRF1a, ICT1, C12orf65, pRF2) (Meurer et al. 2002; Soleimanpour-Lichaei et al. 2007; Antonicka et al. 2010; Richter et al. 2010). A large-scale systematic analysis of the whole RF protein family across all eukaryotes and for all organellar types, allowing the detection of general trends in organellar RF evolution has not been published. Similarly, most studies correlating the RFs with the organellar genetic code have focused on the metazoan mitochondrial genetic code (Knight et al. 2001), leaving this coevolution hypothesis largely untested for most other taxon groups and other organelle types. Here, we classify and describe the nine distinct subfamilies of organellar release factors by combining large-scale phylogenetic analyses with protein function and localization data, the genetic code of organellar genomes and empirical knowledge about the role of particular motifs within RF domains. This systematic study and data conjugation allows us to document the established molecular structure and function of each protein subfamily, as well as to trace its phylogenetic origin and evolution throughout the eukaryotic tree of life. Furthermore, we evaluate the phylogenetic distribution of the RF subfamilies and correlate it with the mitochondrial/plastidial genetic code, reporting several instances that clearly illustrate the coevolution of the release factors with the organellar genetic code.
Materials and Methods
Sequence Data Retrieval and Selection
The sequence dataset used was obtained by retrieving all human mtRF1a (GI: 166795303) homologues, using its sequence as query seed for a PSI-BLAST (Altschul et al. 1997) search of the GenBank nr database, restricted to eukaryotic organisms and iterated until convergence.
The results were manually inspected to remove redundant sequences and guarantee the presence of all RF family members. Using as guideline the systematics described by Simpson and Roger (2004), the dataset taxonomic coverage was balanced by removing species from groups that are over-represented in the databases, like the fungi/metazoa, and keeping and/or manually including species from the under-represented taxa like the excavata, alveolata, and stramenopiles. We selected only fully sequenced organisms, preferably with well-annotated organellar genomes. When needed, organism-specific tBLASTn searches were conducted, and the relevant homologues were included.
Prokaryotic homologues of each RF sub-families were collected by conducting a BLASTp search of NCBI’s RefSeq database restricted to bacteria, and the first hit from the 21 main prokaryotic groups, according to (Wu et al. 2009), was included in the dataset (see supplementary table 3, Supplementary Material online, for the accession numbers of the 359 protein sequences used in this study).
Sequence Alignment, Trimming, and Subfamily Classification
Each main subfamily—RF1, RF2, ICT1, and C12orf65—was aligned separately. The considerable sequence divergence present between some subfamilies lead us to test the performance of three alignment algorithms: Muscle (v3.7) (Edgar 2004), MAFFT with L-INS-i iterative refinement option (v6.717b) (Katoh et al. 2005), and ClustalW (v2.0.10) (Thompson et al. 1994). After careful visual inspection of the alignments and its guide trees, the ClustalW alignment was chosen, given that it yielded the best overall alignment of the known functional elements.
For the individual RF1 and RF2 phylogenies, we used BMGE (v1.0) (Criscuolo and Gribaldo 2010) to remove ambiguously aligned positions. A range of parameter settings was tested, and after visual inspection, a 60% gap removal threshold was chosen because it yielded the best results relative to the accurate alignment of the functionally characterized and well-conserved domains, while maintaining an acceptable number of positions for accurate phylogenetic inference. The final RF1 phylogeny contains 148 sequences with 499 aligned positions, and RF2 contains 74 sequences with 541 amino acid positions.
To visualize and classify the multiple RFs from each species, we computed a Neighbor-Joining tree with QuickTree (v1.1) (Howe et al. 2002) using all 313 eukaryotic full-length sequences. These were aligned through profile to profile sequential alignment of the individual subfamilies’ alignments using ClustalW (v2.0.10) (Thompson et al. 1994) followed by one last round of alignment refinement with Muscle’s refine option. Finally, the whole RF family alignment was inspected and manually adjusted. All alignment visual inspections were performed using Jalview (v2.7) (Waterhouse et al. 2009). All alignment data have been deposited in the Dryad repository: doi:10.5061/dryad.2br48.
Phylogenetic Analysis
The presence of paralogs in the RF1 and RF2 subfamilies (3 and 4, respectively) led us to compute individual Bayesian phylogenies to clarify their phylogenetic relationships. These were computed using PhyloBayes (v3.2e) (Lartillot et al. 2009). Two independent chains were run for RF1 and RF2, using a C20 empirical profile mixture model of amino acid substitution and 4 discrete-rate categories Gamma distribution (C20+G4). Convergence of the phylogenies was assessed following the guidelines provided with PhyloBayes (maximum difference observed across bipartitions between the chains <0.1; maximum discrepancy <0.1; and minimum effective size >100 for the variables estimated). The final majority-rule posterior consensus tree was obtained with a burnin value of 1,000, using every-other tree.
An individual ICT1 plus C12orf65 phylogeny was not calculated given that the alignment between these two proteins would not yield enough confidently aligned positions to obtain a reliable phylogeny (no convergence for a Bayesian phylogeny could be obtained).
Organellar Genetic Code Analysis
A customized set of Perl scripts was developed to analyze the organellar genetic codes. For that, the GenBank files of all available mitochondrion and plastid genomes (total of 2,431 files) were retrieved and all relevant information regarding the number, identity and neighborhood of the stop codons predicted for every ORF was parsed and summarized. For sequenced but unannotated mitochondrial genomes, we used FACIL to predict the genetic code (Dutilh et al. 2011).
Subcellular Localization Data
To complement our bioinformatics analysis, we conducted a scrupulous manual literature search for experimental localization data on all release factor family proteins. We gathered public large-scale localization datasets from several model organisms, namely, Arabidopsis thaliana (Heazlewood et al. 2004; Dunkley et al. 2006; Zybailov et al. 2008; Olinares et al. 2010), Caenorhabditis elegans (Li et al. 2009), Homo sapiens (Pagliarini et al. 2008), Mus musculus (Kislinger et al. 2006), Saccharomyces cerevisiae (Huh et al. 2003), and Schizosaccharomyces pombe (Matsuyama et al. 2006), which we examined for localization information about the RF family proteins (table 1).
Table 1.
Overview of the Organellar Release Factor Family, Summarizing and Comparing Structural and Functional Data about the Nine Members of the RF Family.
| RF Protein | Human/Arabidopsis Gene | Location | Alpha5 Helix | PxT/SPF | GGQ | Function | Recognized Stop Codons | Phylogenetic Origin | Experimental Studies |
|---|---|---|---|---|---|---|---|---|---|
| mtRF1a | MTRF1L/AT2G47020 | Mt | Y | PxT | Y | Codon-specific release factor | UA (A/G) | Alphaprot | Homo sapiens,a Caenorhabditis elegans,b Schizosaccharomyces pombec |
| mtRF1 | MTRF1/— | Mt | Y | PExGxS | Y | ? | None | Euk mtRF1a | H. sapiensa |
| ICT1 | ICT1/AT1G62850 | Mt | N | N | Y | Noncodon-specific release factor | Nonselective | Alphaprot | H. sapiens,d,e C. elegans,b S. pombec |
| C12orf65 | C12orf65/— | Mt | N | N | Y | Hyp: recycling abortive peptidyl-tRNAs | ? | Euk (ICT1?) | H. sapiens,f Saccharomyces cerevisiaeg |
| mtRF2a | —/AT1G56350 | ? | Y | SPF | Y | Codon-specific release factor | U(A/G)A | Alphaprot | None |
| mtRF2b | —/AT3G57190 | ? | N | N | N | ? | ? | ? | None |
| mtRF2c | —/AT1G33330 | Pt | N | N | Y | ? | ? | Euk (pRF2?) | Arabidopsis thalianah |
| pRF1 | —/AT3G62910 | Pt | Y | PxT | Y | Codon-specific release factor | UA(A/G) | Cyanobact | A. thalianah,i |
| pRF2 | —/AT5G36170 | Pt | Y | SPF | Y | Codon-specific release factor | U(A/G)A | Cyanobact | A. thalianah,j |
Note.—Human and Arabidopsis were chosen as representative of mitochondria and plastid containing species for which the gene names are shown. ?, unknown; —, not applicable; Y, yes; N, no; Mt, mitochondrial; Pt, plastidial; Alphaprot, Alphaproteobacteria; Cyanobact, Cyanobacteria.
References for experimental studies: a(Soleimanpour-Lichaei et al. 2007).
b(Li et al. 2009).
g(Huh et al. 2003).
For proteins without experimental localization data, we predicted their subcellular targeting using the method implemented in ConLoc (Park et al. 2009), whose outcome is based on the consensus result of 13 on-line localization prediction servers.
Molecular Modeling
All models were built using the YASARA molecular modeling package (Krieger et al. 2002). The high-resolution structures of RF1 bound to the ribosome of Thermus thermophilus (PDB entries 3D5A, 3D5B [Laurberg et al. 2008] and PDB entries 3MR8 and 3MS1 [Korostelev et al. 2010]) were used as modeling templates. Loops were modeled by scanning a nonredundant subset of the PDB (>8,000 structures) for fragments with matching anchor points, a minimal number of bumps, and maximal sequence similarity. Side chains were added with YASARA’s implementation of SCWRL (Canutescu et al. 2003), and then the model was subjected to an energy minimization with the YASARA2 force field as described elsewhere (Krieger et al. 2009). WHAT CHECK (Hooft et al. 1996) validation scores were used to score and rank the final models.
C12orf65 C-Terminal Extension Analysis
The observation that both C12orf65 and ICT1 shared a basic-residue rich C-terminal extension, together with the recent experimental elucidation of the functional role of this extra domain in ICT1’s bacterial ortholog YaeJ (Gagnon et al. 2012) (see ICT1 section for a detailed discussion) led us to analyze the relationship between these extensions. To confirm the homology between these domains and predict C12orf65’s structure, we used HHpred (Söding et al. 2005) (data not shown), confirming that these terminal extensions are indeed homologous. Moreover, C12orf65’s C-terminal extension is predicted to be an alpha-helix, mirroring the setting in ICT1’s bacterial ortholog.
Results and Discussion
To provide an overview of the organellar release factor family, we first calculated a simple, yet comprehensive and illustrative tree of the nine distinct subfamilies (fig. 1). The figure shows congruence between tree topology, domain architecture, and the presence of functionally relevant motifs allowing the classification of each organisms’ RFs.
Fig. 1.
Full release factor family neighbor-joining tree. This figure presents an overview of the nine RF subfamilies roughly separated by the NJ algorithm, recapturing the pattern of sequence motifs characteristic of each protein. It summarizes, in one image, the main sequence features presented by individual organisms. Each subfamily branch is highlighted with a different color following the exterior labels. Well-resolved branches from well-established taxa were collapsed to improve readability. In these collapsed branches, a representative domain and motif structure is displayed, slightly enlarged in order to stand out from other individual results. The following species were chosen as models for these representative domains: viridiplantae and land plants—Arabidopsis thaliana; metazoa, vertebrates, and mammals—Homo sapiens; insects—Drosophila melanogaster; and fungi—Saccharomyces cerevisiae. (Legend: Pfam domains displayed in front of each leaf: green hexagon—PCRF (peptide chain release factor) and dark-blue arrow—RF-1. Superimposed on the Pfam domains are the functionally characterized motifs: purple diamond—alpha5 helix; cyan oval—PxT motif, yellow oval—SPF motif, red oval—PExGxS motif; red diamond—RT insert; green diamond—GGQ motif; pink hexagon—C-terminal helix; and orange rectangle—ICT1-specific helix.)
Release Factor Family Classification: Subcellular Localization, Structural Characterization, and Phylogenetic Origin
Three widespread organellar release factor protein families have been classified via orthology to their eubacterial counterparts: the two bona fide release factors RF1 and RF2, and the release factor-like ICT1. Although the last one is only present in the mitochondrion, RF1 and RF2 include both the mitochondrial and the plastidial forms, termed mtRF1a, mtRF2a, pRF1, and pRF2a, respectively. C12orf65 is another frequent mitochondrial release factor-like protein. Furthermore, vertebrates possess yet another RF1 homologue in the mitochondrion, named mtRF1, and land plants present two other RF2 homologues, mtRF2b and mtRF2c, amounting in total to nine distinct subfamilies.
Canonical Release Factors: RF1 and RF2
Release factor 1 proteins specifically recognize the stop codons UAA and UAG, while release factor type 2 proteins recognize UAA and UGA. Consistent with their codon-specific peptidyl-tRNA hydrolytic function, both RF1 and RF2 display all three functionally described structural features: the codon-recognition (CR) domain with its alpha-5 helix and codon-discriminator tripeptide motif—PxT in RF1 and SPF in RF2—and the peptidyl-hydrolase (PTH) domain containing the universally conserved GGQ motif (table 1).
Despite having the same domain composition, sharing the same molecular function and the significant sequence similarity—48% sequence identity between mitochondrial and plastidial RF1s and 55% for their RF2 counterparts (calculated using the consensus sequences of each subfamily divided by their average length)—each subfamily can be distinguished by its different phylogenetic origin and subcellular localization.
mtRF1a and pRF1
mtRF1a is the most widespread of all organellar release factors. Every eukaryotic organism with a mitochondrial genome, harbors a mitochondrial type RF1 encoded in the nucleus (supplementary table 1, Supplementary Material online). Consistent with the origin of this organelle, this protein evolved from an alphaproteobacterial ancestor, as clearly demonstrated in figure 2 by the highly supported clustering of the alphaproteobacterium Rhodospirillum rubrum at the basis of the eukaryotic mtRF1a branch, to the exclusion of all other nonalphaproteobacteria prokaryotic sequences. This protein has been experimentally well characterized, particularly the human ortholog. It is a bona fide peptide release factor that localizes in the mitochondrion and specifically releases UAA/UAG, both in vitro and in vivo (Soleimanpour-Lichaei et al. 2007; Nozaki et al. 2008).
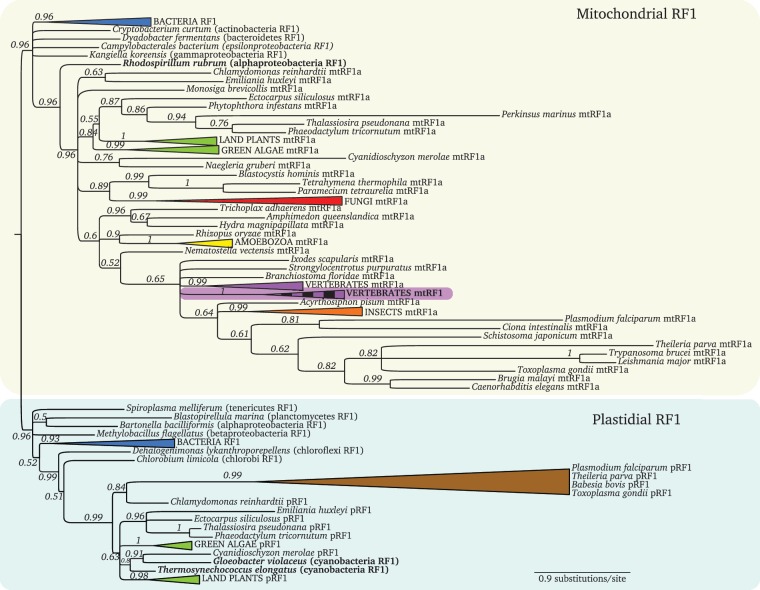
Fig. 2.
Bayesian RF1 phylogeny. The two main branches separate the mitochondrial proteins (yellow box) from the plastidial ones (green box). The mtRF1 branch nested within mtRF1a is highlighted in purple with a vertical striped pattern. Well-supported branches from well-established taxa were collapsed to improve readability (full noncollapsed tree in supplementary fig. 1, Supplementary Materials online). Alphaproteobacteria and cyanobacteria are highlighted in bold. (Colors for collapsed taxa: Blue—bacteria; green—Viridiplantae; red—fungi; yellow—amoebozoa; purple—vertebrates; orange—insects; and brown—apicomplexa.)
pRF1 is ubiquitous in plastid-bearing species: archaeplastida (plants, red, and green algae), rhizaria, diatoms, apicomplexa, and brown algae (supplementary table 1, Supplementary Material online). Deciphering this protein’s phylogenetic origin is not trivial, mainly because, unlike the mitochondrion that has been the result of a single endosymbiotic event, plastids have been acquired several times independently. There were at least two single primary endosymbioses of a cyanobacterium (e.g., red algae’s rhodoplasts, land plant’s, and green algae’s chloroplasts from a beta-cyanobacterium [Reyes-Prieto et al. 2007], and Paulinella’s chromatophores from an alpha-cyanobacterium [Marin et al. 2005; Yoon et al. 2009]); two secondary endosymbiosis of algae (a red algae gave rise to, for example, apicomplexan apicoplasts and stramenopiles’ plastids, whereas two green algae gave rise to the plastids of euglenophytes and chlorarachniophytes’ [Baurain et al. 2010; Janouskovec et al. 2010]) and even tertiary endosymbiosis of haptophytes and diatoms in some plastid-bearing dinoflagellates (for recent reviews see Keeling 2010; Archibald 2012).
As such, one would expect these multiple origins to be, at least partially, recaptured in the phylogeny of the plastidial RF1. Indeed, the two beta-cyanobacterial RF1 orthologs, from Gloeobacter violaceus and Thermosynechococcus elongatus, cluster together in a strongly supported branch, with the land plants, green and red algae and a group of other plastid bearing organisms (fig. 2).
On the other hand, the phylogenetic signal in the pRF1 alignment does not seem to be strong enough to recapitulate the red algal secondary origin of the apicomplexan plastids, because these groups confidently with the green algae Chlamydomonas reinhardtii and not with the red algae Cyanidioschyzon merolae. The same holds true for the diatoms, brown algae and Emiliania, which cluster with each other excluding the red algae (fig. 2).
Several experimental studies have been published regarding pRF1’s localization and function. Two independent reports show its plastidial localization (Zybailov et al. 2008; Olinares et al. 2010), and the molecular function of A. thaliana’s pRF1 has been experimentally characterized in vivo. Not only it is essential for appropriate chloroplast development, but also successfully rescues the temperature-sensitive phenotype of Escherichia coli RF1 mutants, proving that this protein is indeed a functional translation release factor (Motohashi et al. 2007).
mtRF2a and pRF2
The mitochondrial mtRF2a has a relatively narrow phylogenetic distribution, when compared to its mtRF1a counterpart. It has been lost at least five times during the eukaryotic evolution (fig. 4), coevolving together with the mitochondrial genetic code (see section II. Release factors and the evolution of the genetic code). It is only consistently found in streptophytes (land plants), red algae, dictyosteliida, and some stramenopiles (namely in brown algae, oomycetes and Blastocystis). It is absent from animals, fungi and excavata, with the exception of the heterolobosean Naegleria gruberi (supplementary table 1, Supplementary Material online).
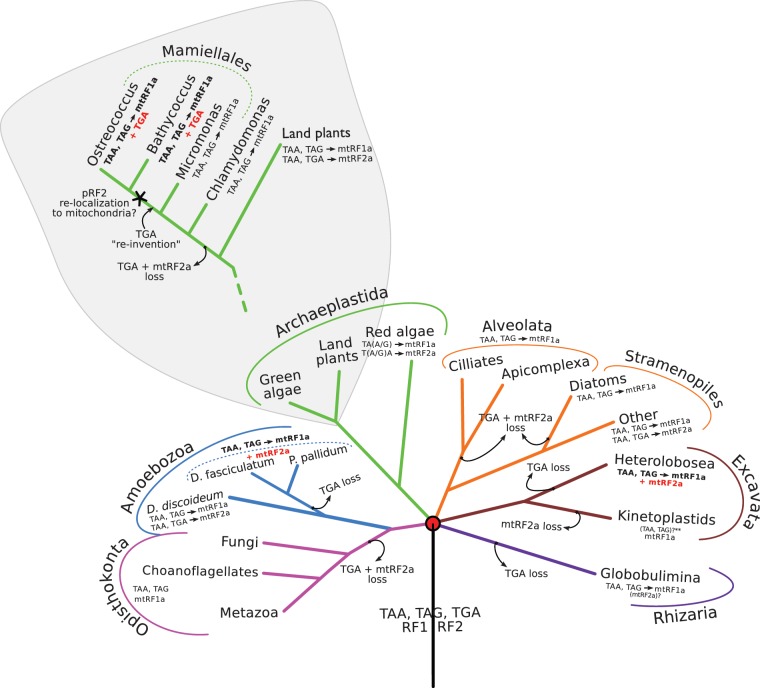
Fig. 4.
Schematic eukaryotic phylogeny displaying, per lineage, the coevolution of the mitochondrial genetic code with the codon-specific RFs. The red circle indicates the unique primary endosymbiosis event that originated the mitochondrion. The green algae stop-codon reinvention hypothesis is detailed in the gray “zoom-in” area. Species relationships were assembled from two studies: the main tree is based on the consensus tree depicting the six main eukaryotic groups from Simpson and Roger (2004) and the green algae lineage is based on the 18S rRNA gene tree published by Worden et al. (2009). Branching order is meaningful, but not branch length. Red font highlights the exceptions to the coevolution discussed in the text. The star marks the “TGA-stop reinvention” with pRF2 relocalization hypothesis in the green lineage. Question marks are used for uncertain data, and two asterisks indicate no mitochondrial genome available. (See supplementary methods, Supplementary Materials online for details about the species used in making this figure).
The expected alphaproteobacterial ancestry of this protein, given the endosymbiotic origin of mitochondria, cannot be unequivocally established from our RF2 phylogeny (fig. 3). Most prokaryotic sequences present in this dataset do not form a monophyletic group, being instead all grouped in an unresolved branch, containing also most eukaryotic mitochondrial RF2s (fig. 3).
Fig. 3.
Bayesian RF2 phylogeny. The top branch, highlighted in yellow, groups known mitochondrial proteins (mtRF2a), several nonresolved bacterial RF2s and, highlighted with a checkerboard pattern, the mtRF2c branch (which has been experimentally shown to localize in the chloroplast). The bottom branch, highlighted in green, clusters the plastidial RF2s and mtRF2b (indicated by a vertical strip pattern). Alphaproteobacteria and cyanobacteria are highlighted in bold. (Colors for collapsed taxa: blue—bacteria; pink—alphaproteobacteria; green—viridiplantae; yellow—amoebozoa; brown—apicomplexa; and cyan—cyanobacteria). (Full noncollapsed tree available in supplementary figure 2, Supplementary Materials online.)
There are no experimental data on this protein’s molecular function and localization in eukaryotes. Nevertheless, its E. coli ortholog has been thoroughly studied and shown to terminate translation by decoding UAA and UAG, both in vitro and in vivo (Scolnick et al. 1968; Mora et al. 2003).
The plastidial RF2’s phylogenetic distribution overlaps perfectly with its RF1 counterpart (with the exception of Toxoplasma gondii, see below), being ubiquitously present both in the primary plastids of land plants, red and green algae, and in the secondary plastids of apicomplexan parasites, diatoms, and brown algae (supplementary table 1, Supplementary Material online).
As mentioned earlier, the multiple origins of plastids challenge the task of tracing the phylogenetic origin of these organellar proteins. The cyanobacterial origin of primary plastids’ RF2 is recaptured by the strongly supported grouping of the two cyanobacteria within the plastidial branch of this phylogeny (fig. 3). No strong conclusions can be drawn regarding the origin of apicomplexan, diatom and brown algae secondary plastids given the unresolved phylogenetic branches comprising these organisms.
Contrasting with the lack of published functional data about the mitochondrial RF2, the chloroplastidial localization of A. thaliana’s pRF2 has been experimentally determined (Meurer et al. 2002; Zybailov et al. 2008), and its function has been shown to be primarily in the termination of UGA stop codons, but also in the regulation of chloroplastidial protein synthesis and stability of UGA-containing mRNAs (Meurer et al. 2002).
Noncanonical RFs
The remaining five RF subfamilies have lost one or both of the structural features that characterize bona fide release factors. Their origins are more diverse and, except for ICT1, their phylogenetic distribution is not as uniform. Most have not been characterized experimentally and for some, their subcellular localization has not been established (table 1).
mtRF1
mtRF1 is probably the most studied nonclassical release factor, and yet its molecular function fails to be determined. It is the longest protein of the RF family—the human protein is 445 amino acids long, while mtRF1a is only 380. Its C-terminal is remarkably similar to bona fide RFs, presenting an analogous PTH domain harboring the ultra conserved GGQ, but it shows some differences within the codon recognition domain that set it apart from other canonical RFs. Most notably, it lacks the characteristic PxT motif, displaying instead PExGxS (most commonly PEVGLS) (table 1). Another intriguing sequence feature is a distinctive RT insert within the alpha-5 helix that extends the recognition loop without disrupting the overall domain architecture (discussed later).
This is a vertebrate-specific mitochondrial protein (Soleimanpour-Lichaei et al. 2007) and it has been reported to have originated by duplication of the mtRF1a gene at the root of this clade (Young, Edgar, Murphy, et al. 2010). In our figure 2 phylogeny, we observe this protein’s branch in an unresolved cluster with the vertebrate mtRF1a branch and several other metazoa and earlier branching eukaryotes. Notwithstanding, its high sequence conservation together with its ubiquitous and exclusive distribution within vertebrates, leave no doubt that this protein arose by mtRF1a duplication at the root of the vertebrate lineage.
Young, Edgar, Murphy, et al. (2010) have suggested that mtRF1 could be responsible for decoding the nonstandard mitochondrial stop codons, AGG and AGA, predicted to terminate numerous vertebrate mitochondrial orfs. A number of observations contribute to this hypothesis. First, it possesses the canonical domains involved in peptidyl-tRNA hydrolysis and in stop codon recognition (although not with the classical tripeptide motif). Second, this protein’s origin at the root of the vertebrate lineage coincides with the origin of AGG/AGA stop codons, hinting that their roles might be functionally connected. Finally, despite the lack of in vitro release activity in response to any potential stop codon (Soleimanpour-Lichaei et al. 2007; Nozaki et al. 2008), mtRF1 has been argued to possess several structural features capable of recognizing adenine as the first base of a stop codon (Young, Edgar, Murphy, et al. 2010), linking again AGG/AGA codons with this protein.
Nevertheless, this hypothesis has never been experimentally confirmed. Moreover, Temperley et al. (2010) demonstrated that, at least in human, AGG and AGA codons do not function as stop. Instead, they promote a -1 frameshift in both genes containing AGG and AGA (ND6 and CO1), yielding a standard TAG stop codon, hence bypassing the need for an extra RF protein. On the other hand, Young, Edgar, Murphy, et al. (2010) remark that some vertebrates’ mitogenomes present a cytb and/or COI gene that do not possess a T immediately before these “frameshifting codons”. In such cases, a -1 frameshift does not generate a standard termination codon, leaving unexplained the mechanism of termination of these genes.
To investigate this matter, we used all 1,604 complete vertebrate mitochondrial genomes deposited at the time of our analysis in the NCBI’s organellar genomes database, to systematically evaluate both the origin of AGG/AGA terminated orfs and verify to what extent the postulated -1 frameshift mechanism would not originate a canonical TAG stop.
First, our findings show that the mitochondrial orfs terminated with AGG/AGA indeed arose at the root of the vertebrates, and are not present in any other eukaryotes, which use them to code for arginine. Second, there were 1,535 orfs predicted to stop with AGG/A, from 947 distinct species. From those, a TGA stop arising from a -1 frameshift could account for 395 orfs, leaving 1,140 orfs from 808 different vertebrate species unable to terminate translation with a standard stop codon. (We also examined if a -2 frameshift, creating a TAA stop, could hypothetically solve the issue of “nonterminated” orfs, but only an extra 188 orfs would be terminated.)
Additionally, to gain more insight on this protein’s putative function, in a separate publication manuscript, we describe the results from a molecular model analysis conducted on mtRF1 3D structure. We predict that it is unlikely that mtRF1 recognizes any codon at all, as amino acid substitutions and insertions at the codon recognition domain of mtRF1 create additional hydrophobic bulk that is highly unlikely to tolerate any mRNA in the A site of the ribosome (Huynen et al. 2012).
ICT1
ICT1 (immature colon carcinoma transcript-1) is much shorter than any of the canonical release factors—206 residues in human, compared to the 380 residues of mtRF1a. It is an experimentally confirmed mitochondrial protein that has lost both structural elements responsible for the stop codon recognition (the C-terminal alpha-5 helix and the tripeptide motif), while retaining the GGQ PTH domain (table 1). Consistent with this domain composition, Richter et al. (2010) have demonstrated that this protein indeed functions as a stop-codon independent PTH. Also, they have shown that, in human, ICT1 is incorporated into the mitoribosome’s large subunit, leading to the suggestion that it was recruited there in the course of eukaryotic evolution. However, recently it has been reported that E. coli’s ICT1 ortholog, YaeJ, is already part of the bacterial ribosome, indicating that the ribosomal location of the orthologous group precedes the origin of eukaryotes (Handa et al. 2011).
Mouse’s ICT1 catalytic domain structure, comprising the loop containing the GGQ, has been determined by NMR spectroscopy, showing an overall topology and structural framework similar to Class I RFs PTH domain, confirming its analogous hydrolytic activity. There is nevertheless a distinguishing feature that sets this domain apart from the one found in canonical Class I release factors. Handa et al. (2010) describe a groove formed by an ICT1-specific alpha-helix inserted between two conserved beta-strands, and they propose that this element might be a site related to this protein’s specific catalytic activity.
Also, it has been noted that ICT1 presents a C-terminal extension rich in basic-residues, characteristic of many ribosomal proteins (Brodersen et al. 2002), agreeing with its ribosomal location. A recent crystal structure of the bacterial ICT1 ortholog YeaJ (Gagnon et al. 2012) reveals that this C-terminal extension acts as a sensor to detect stalled ribosomes, based on the occupancy of the mRNA channel in the ribosome. Upon recognition of an empty mRNA channel, the catalytic GGQ motif of YeaJ can bind in the peptidyl-transferase center, resulting in subsequent release of the nascent peptide chain. Based on these recent findings, it is tempting to speculate that ICT1 performs a similar function in mitochondria.
ICT1’s widespread eubacterial distribution (Handa et al. 2011) suggests that this protein is of ancient origin and not from an RF1 or RF2 gene duplication. Apart from mtRF1a, this is the only subfamily present in all eukaryotic phyla analyzed (with a few notable exceptions, namely C. merolae, Neurospora crassa, Sclerotinia sclerotiorum, and Phytophthora infestans as shown in supplementary table 1, Supplementary Material online). This broad taxonomical distribution is in accordance with its reported essentiality in human (Richter et al. 2010).
C12orf65
The C12orf65 orthologous group provides a similar example of loss of the two stop-codon recognition functional elements, while retaining the catalytic GGQ motif. C12orf65 misses the ICT1-specific alpha-helix, and accordingly it has been reported that, contrary to ICT1, this is a mitochondrial soluble matrix protein that does not exhibit ribosomal-specific PTH activity (Antonicka et al. 2010). Note however, that despite this obvious functional divergence, ICT1 over-expression partially rescues the biochemical defect presented by C12orf65 mutated cells, hinting that both proteins must have at least partially overlapping functions in the mitochondrion. Further evidence for a similar function between the two proteins comes from the observation that C12orf65 has a (predicted) C-terminal alpha-helix that is homologous to a recently described ICT1 C-terminal helix. In ICT1 E. coli ortholog YaeJ this C-terminal helix, as described in the previous section, functions in sensing an empty mRNA channel in the ribosome (Gagnon et al. 2012). The homology between ICT1 and C12orf65 includes the conservation of basic residues that in YaeJ interact with ribosomal proteins and the ribosomal SSU rRNA.
Since only 21 bacterial species from 5 (out of 28) bacterial groups (BLAST results not shown) harbor a C12orf65 homologue, it is likely that this protein is a eukaryotic invention derived from a duplication of a canonical RF. The fact that C12orf65 and ICT1 have lost, relative to canonical RFs, the same stop-codon recognizing domains, and that both share the C-terminal alpha-helix that is absent from canonical RFs provide a strong argument that C12orf76 is derived from ICT1, which has a wider phylogenetic distribution. Nevertheless, our phylogenetic analyses based on the positions that could confidently be aligned between all organellar release factors did not show strong support for a direct relationship between C120rf65 and ICT1.
C12orf65 is notably absent from viridiplantae (land plants and green algae), being present in all other eukaryotic taxa (supplementary table 1, Supplementary Material online). The most parsimonious scenario to explain this absence is that it likely originated at the root of the eukaryotes, and was subsequently lost in the green lineage.
mtRF2c and mtRF2b
Land plants (embryophytes) present two extra RF-like proteins that are not present in any other organism: mtRF2c and mtRF2b. These two proteins have not been experimentally studied and their domain divergence and rearrangement allows only educated guesses regarding their molecular function.
mtRF2c is much shorter than other plant RF2s (only 257 residues in A. thaliana), and has lost both the alpha-5 helix and the stop codon recognizing motif, keeping only the GGQ hydrolyzing tripeptide. This protein has never been functionally characterized.
No rigorous phylogenetic interpretation regarding mtRF2c’s origin can be made from the RF2 phylogeny presented (fig. 3). Not only is its branch nested within the unresolved mtRF2a cluster, but also the very long-branch length precludes any significant conclusion.
Contrary to the “mitochondrial localization” suggested by its name, mtRF2c has been found experimentally in the chloroplast of A. thaliana (Zybailov et al. 2008). Therefore, we propose this protein to be renamed from mtRF2c to pRF2c, to correctly express its subcellular localization, following the convention used for the other release factors.
mtRF2b represents a unique type of release factor given its loss of both RF signature-motifs, that is, the GGQ tripeptide and the stop-codon recognizing motif. Despite its sequence divergence and absence of these two features, this protein has retained the overall structure of the two release factor family domains (Pfam names RF1 and PCRF) (fig. 1), suggesting that this is a genuine member of this family. Also, corresponding EST sequences for several land plants are present in NCBI’s EST database, confirming that this protein is indeed expressed and not a pseudogene.
Despite its “mitochondrial naming,” there are no experimental localization data about this protein. Also, localization prediction analysis using ConLoc (Park et al. 2009) gave no unambiguous results. Nevertheless, it has been described that proteins interacting with organellar multi-subunit complexes tend to inherit the subcellular localization of their ancestral protein (Szklarczyk and Huynen 2010). The strongly supported clustering of mtRF2b’s branch within the plastidial branch of our RF2 phylogeny (fig. 3), indicates not only that this protein has originated from a duplication of the land plants’ plastidial RF2, but also suggests that mtRF2b might be plastidial. Further localization studies are required to corroborate this prediction. Given the loss of the GGQ motif from mtRF2b, it is tempting to speculate that this protein will not present hydrolytic capabilities.
Release Factors and the Evolution of the Genetic Code
The coevolution of the genetic code with the release factors has been proposed by Jukes and Osawa over 20 years ago (Jukes and Osawa 1990). Nevertheless, its universality has never been assessed, and many interesting questions remain unanswered: was RF2 lost before or after the stop codon reassignment; was it lost once in the common ancestor or several times in independent lineages; is it present in species that do not use TGA as stop codon, and if so, does it (apart from the redundant recognition of UAA) have any other function in these organisms?
We performed a systematic analysis of the organellar genetic code and the presence of mitochondrial and plastidial RF2 (figs. 4 and 5). Based on 95 currently sequenced nuclear genomes of organisms with annotated organellar genomes, the mitochondrial-type RF2 has been lost five times in evolution: in kinetoplastids, diatoms, alveolates, at the root of the opisthokonta and in the green algae lineage, whereas the usage of TGA as stop codon in mitochondrial genomes has been lost seven times: not only in the same diatoms, alveolata, opisthokonta, and green algae, but also in heterolobosea, rhizaria, and some amoebozoa (fig. 4).
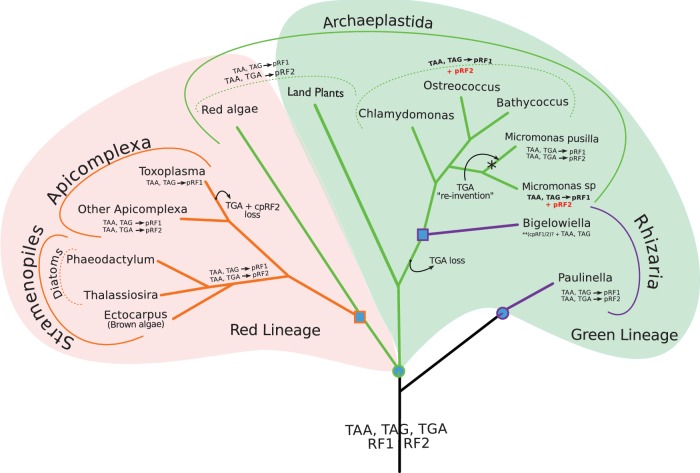
Fig. 5.
Coevolution of the plastidial RFs with the plastidial genetic code. The red and green backgrounds mark the red and green plastid lineages, respectively. The branching order is based on Marin et al. (2005) and Keeling (2010). Blue circles indicate a primary endosymbiosis and blue squares represent a secondary endosymbiosis event. Red font highlights the cases that represent exceptions to the coevolution of the RF with the plastidial genetic code, where the pRF2 is present in the genome, but TGA stop codons are not used. The star marks the possible “TGA-stop reinvention.” Question marks are used for uncertain data and two asterisks indicate no whole genome available. (See supplementary methods, Supplementary Materials online for details about the species used in making this figure.)
Plastidial RF2s and genomes show less volatility than mitochondrial ones. pRF2 has only been lost once (in T. gondii) while TGA as a stop-codon was lost twice: in T. gondii (agreeing with a previous report [Denny et al. 1998]) and at the root of the green algae lineage (fig. 5).
In almost all cases, the RF2 loss coincides with the lack of usage of TGA stop codons, supporting the coevolution hypothesis. For example, Phaeodactylum tricornutum and Thalassiosira pseudonana have lost mtRF2a and they both lack mitochondrial genes terminated by TGA; T. gondii has lost pRF2 and accordingly none of its 26 plastidial orfs is predicted to stop with a TGA codon (supplementary table 4, Supplementary Material online). Nevertheless, 4 exceptions were found, which are briefly discussed in the following two sub-sections.
RF2 Without UGA
Most archaeplastida organisms use the same standard genetic code in both organelles. Green algae represent the exception to this pattern. Despite maintaining pRF2, C. reinhardtii, Ostreococcus tauri, Micromonas sp., and Bathycoccus sp. do not use TGA as a stop codon in their chloroplast genome (fig. 5). In our dataset, only Micromonas pusilla retains one gene (PsbL) that is terminated with TGA, hence likely using its plastidial RF2.
Another such case is observed in the mitochondrion of the heterolobosean N. gruberi (fig. 4). This species still has a mitochondrial RF2, despite not using TGA stop codons in any of its 46 mitochondrially encoded genes.
Particularly intriguing is the scenario displayed by the three social amoebae included in our study (fig. 4). While all three encode mtRF2a, only Dictyostelium discoideum has retained the usage of TGA stop codons (in the two nonhypothetical orfs rpl5 and rpl16). On the other hand, D. fasciculatum and Polysphondylium pallidum do not possess any TGA-terminated mitochondrial genes, and P. pallidum’s mitogenome is even predicted to use TGA encoded tryptophan in five protein coding sequences (rps3, rpl16, rps8, orf919, and orf83). If P. pallidum’s mitogenome truly uses TGA to code for W, this would be an exceptional setting where the same codon could be decoded both by a cognate tRNA and a release factor.
To evaluate the plausibility of this scenario, we compared the sequences of the five peptides containing TGA encoded tryptophan to their orthologous sequences in closely related species, and found no strong arguments that this would be the case. First, TGA codons are only used in five orfs, only once per orf, and in all of them the codon is located near the predicted termination codon—13 amino acids from TAA in rps3, 7 residues from TAG in rps8, immediately before TAA in rpl16, 5 and 2 amino acids from TAA in orf83 and orf919, respectively. Second, for the three nonhypothetical orfs, neither the tryptophan nor the small protein “extensions” (created by including W and the following residues until the annotated stop codon) are conserved in the orthologous proteins from other dictyosteliida species. Together, these observations suggest that P. pallidum uses TGA stop codons, hence making use of its mtRF2a specificity, mirroring what is observed in D. discoideum. Moreover, as long as RF2 is not lost, the nonsense codon reassignment to tryptophan would be hard to establish having to compete with the release factor. This is in line with the idea that as long as a stop codon is recognized by a RF, it cannot become reassigned to code for an amino acid (Osawa et al. 1992).
These repeated instances of TGA loss without loss of RF2, together with the significant TGA-stop reduction in most organisms (supplementary table 4, Supplementary Material online), suggests that alternative genetic codes might arise more commonly by disappearance of the stop-codon first, and only then the loss of its respective release factor.
UGA without RF2
The mitochondrial genome of mamiellales algae display the opposite scenario: UGA stop codons have been retained, but there is no mtRF2a to decode them (fig. 4). Ostreococcus tauri has six predicted orfs ending with TGA (two of which are nonhypothetical proteins—Rpl5 and Rps8) that would be extended by 25 and 45 amino acids, respectively, if they were to use the next in frame non-TGA stop codon. These potential “extensions” are not present in any other members of the Rpl5 and Rps8 gene families (data not shown). Bathycoccus sp has one nonhypothetical open reading frame (Rpl16) that ends in TGA, while its Rps8 gene terminates with a TAA that perfectly aligns with the TGA in Ostreococcus’ Rps8. The most parsimonious scenario to explain this TGA-stop reusage in Ostreococcus and Bathycoccus, would be that early in the evolution of green algae the mitochondrial RF2 was lost concomitantly with the usage of TGA stop codons in the mitogenome, followed by a later “reinvention” of TGA as stop in those two species (fig. 4).
Favoring this hypothesis is the fact that, even though the usage of TGA stop codons was lost earlier in green algae evolution, this codon has not been reassigned as a sense-codon in the earlier branching green algae—TGA is simply not used in the mitochondrial genomes of Chlamydomonas and Micromonas—facilitating the reversion to the ancestral state. Nevertheless, this TGA-stop reusage requires the presence of a release factor capable of recognizing it, and mtRF2a has been lost at the root of the green algae, leaving open the question: how can these algae decode TGA stop codons in their mitochondria?
One possibility would be to retarget the plastidial RF2 to the mitochondrion. This retargeting would explain not only the ability to decode TGA stops in the mitochondrion but also the conservation of pRF2 in green algae, which do not use TGA stop codons in their chloroplast genome (see previous section). Other multiple subcellular targeting examples have been described in organisms with multiple genome-containing organelles, like the apicomplexa (e.g., Plasmodium falciparum and T. gondii [Pino et al. 2007; Ralph 2007]) and A. thaliana (Duchêne et al. 2009).
Noncanonical Motifs
RF1 and RF2 protein family members have been primarily classified based on the identity of the two experimentally characterized tripeptide motifs—PxT in RF1 and SPF in RF2—which confer their distinct codon specificity. Despite their nearly universal conservation, we came across 13 nonclassical motifs: 10 noncanonical PxT and 3 noncanonical SPF (fig. 1 and supplementary table 2, Supplementary Material online).
The three nonclassical RF2s have a SPY motif (Babesia bovis, Ectocarpus siliculosus, and Erythrobacter litoralis), which is rare in organellar RFs but is present in nearly one-third of eubacteria (data not shown), immediately suggesting that this variability does not affect its function. Also, this phenylalanine (F) to tyrosine (Y) change in the third position of the motif is not disruptive given that the amino acid directly involved in the discriminatory role of RF2 is the first residue from the tripeptide (serine) and not the third (observation of SBN).
From the 10 noncanonical PxT motifs, 9 are PxN and 1 is PTS (supplementary table 2, Supplementary Material online). Most of the PxN motifs (7 out of 9) sit on a 2 amino acid shorter recognition loop that, despite its unusual features, has been experimentally tested in C. elegans, displaying full UAA/UAG-specific release activity, both in vitro and in vivo (Young, Edgar, Poole, et al. 2010). This, together with the fact that this novel shorter loop has arisen at least three times independently in evolution—in metazoa, stramenopiles and apicomplexa—suggests that it might represent a viable alternative conformation.
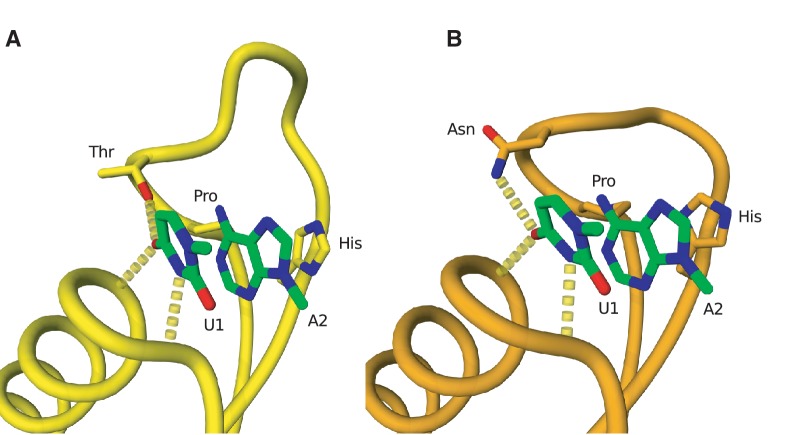
To better understand the retained functionality of this alternative loop conformation, we have built a molecular model of C. elegans’ mtRF1a (with its PVN motif and shorter recognition loop). To do so, we used T. thermophilus’ crystal structure of RF1 bound to a ribosome with a UAA stop codon in the A-site (fig. 6A). Our model clearly shows that, despite the shortened recognition loop, the tripeptide’s asparagine (N) is still able to make the crucial hydrogen bonding interaction to the first nucleotide of the stop codon (fig. 6B), just like the threonine in the canonical PxT motif, which determines selectivity over other nucleotides (Korostelev et al. 2008; Laurberg et al. 2008).
Fig. 6.
Molecular modeling of the noncanonical PxT motif from Caenorhabditis elegans’ mtRF1a. (A) Hydrogen bonding interaction between the first nucleotide of the UAA stop codon (U1) and the threonine of the PxT motif (labeled Thr) of the reading head of RF1 in the Thermus thermophilus crystal structure (PBD entry 3D5A [Laurberg et al. 2008]). (B) Molecular model of the reading head conformation in the C. elegans RF1. The asparagine of the PxN motif is capable of making a similar hydrogen bonding interaction to the first nucleotide of the stop codon as a result of a two amino acid deletion in the recognition loop. The first two stop codon nucleotides (U1 and U2) are shown in green in both panels.
Despite the over-representation of shorter nonclassical loops, there were two PxN motifs in proteins with full-sized recognition loops, that is, without any post-motif deletions: P. falciparum’s pRF1 (PKN) and Cryptococcus neoformans’ mtRF1a (PAN). Again we computed a molecular model for this alternative structure (not shown), this time using Cryptococcus’s mtRF1a sequence. In the model, we unequivocally observe the H-bond between the tripeptide’s asparagine (N) and the first U from the stop codon. This explains the published experimental evidence that full-length recognition loops with a noncanonical tripeptide PxN are also capable of codon-specific release activity in a C. elegans bacterial chimeric system (Young, Edgar, Poole, et al. 2010).
The only non-PxN motif found in our dataset belongs to B. bovis, which has a PTS tripeptide motif in a full-length recognition loop. Based on their biochemical and structural similarity, this threonine to serine substitution in the third position of the motif, most likely, maintains the same intra-molecular interactions, representing a nondisruptive substitution (observation of SBN).
One “Extra” ICT1 without GGQ
The phylogenetic distribution of RF proteins can provide valuable clues about putative functional interactions. We analyzed all cases where a particular species displayed a RF presence/absence pattern that deviates from the trend of the taxonomical group. Despite the several interesting cases found (for details see supplementary table 1, Supplementary Material online), there is just one for which we could find convincing evidence that the departure might be functionally relevant, that is, where the same pattern has been found in related species, and is thus unlikely a sequencing error or a pseudogene.
Ixodes scapularis, the black-legged tick, seems to have gained an additional ICT1-related protein, while loosing C12orf65. This extra ICT1-like protein is approximately the same size as the canonical one (171 vs. 166 residues, respectively) but has lost the GGQ motif, setting it apart from classical ICT1s, and possibly conferring a different molecular function. Also the full-length peptide is expressed in I. scapularis and other Ixodidae family members (e.g., I. ricinus, Rhipicephalus microplus, and Tetranychus urticae) (data from NCBI’s EST database) further supporting its credibility as a “real protein.” Further experimental studies are needed to shed some light on this putative “novel” ICT-like protein.
Conclusion
Organellar translation termination is still far from understood. Although cytosolic translation employs a single release factor—eRF1—belonging to a highly conserved protein family, the organellar release factors comprise nine subfamilies. This protein family seems to be particularly prone to undergo genetic expansion and functional divergence. In fact, this trend can also be observed in bacteria. Apart from RF1, RF2, and YaeJ (ICT1’s bacterial ortholog), at least one other bacterial RF duplicated gene—E. coli’s prfH—has been documented and proposed to be one more member of the Class I release factor family in bacteria (Baranov et al. 2006).
Despite the loss and/or departure from canonical motifs in some RFs, these subfamilies can still be recognized as release factors, suggesting conservation of structure and a possible interaction with the ribosome. Nevertheless, experimental characterization of each subfamily’s specific function is paramount. For example, it would be interesting to experimentally assess the molecular function of the RFs that have lost all functionally characterized motifs—as the mtRF2b plant subfamily or the ICT1-like protein from I. scapularis—to evaluate the effects of such sequence divergence on translation termination. Also, it is necessary to evaluate how comparable this process is between organelles and between the same organelle in different species, given their dissimilar RF content.
Here, we have paved the way for this experimental characterization by classifying and highlighting the most striking attributes of each main RF subfamily. We have clarified, as far as possible, RF1 and RF2 phylogenetic origins and have shown that most organellar release factors tend to keep their ancestral subcellular localizations—mitochondrial RFs derive either from alphaproteobacteria (mtRF1a) or from duplications of canonical mitochondrial RFs (e.g., mtRF1 in Vertebrates); and RFs from primary plastids originated from cyanobacteria (pRF1 and pRF2) or duplications of plastidial proteins (mtRF2b proposed to be renamed pRF2b), following the observed trend that irrespective of the relocalization of the genes, proteins from organellar multi-subunit complexes and their interacting partners tend to continue to function in their original compartment (Szklarczyk and Huynen 2010).
Also, we have explored the tight connection between the organellar RFs and the identity of the stop codons used, revealing a picture of dynamic ongoing evolution within this protein family. The complementarity observed in green algae organelles (where the plastid still retains a RF2 without any gene predicted to terminate with TGA, and the mitochondria has lost the RF2 but still uses TGA stop codons) presents a fascinating scenario that lead us to propose a stop-codon “reinvention” with pRF2 relocalization to the mitochondrion in the green algae lineage. Notably, this would be an exception to the general pattern that proteins that function in a complex maintain their ancestral subcellular localization. Despite the elegance of this hypothesis, it requires experimental validation before any further conclusions can be drawn from such a mechanism.
Overall, our comprehensive classification of the organellar release factor family should serve as a starting point for prioritization of experimental efforts such that, for each of the nine orthologous groups, the subcellular location is unequivocally established, and the effects of knockouts/knockdowns or site-specific mutagenesis on translation termination are measured, better clarifying this essential cellular process.
Supplementary Material
Supplementary methods, figures S1–S2, and tables 1–4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank John van Dam, Radek Szklarczyk, and Robin van der Lee for stimulating discussions and priceless help. This work was supported by the Portuguese Fundação para a Ciência e Tecnologia – FCT (SFRH/BD/32966/2006 to R.M. and SFRH/BD/32959/2006 to I.D.), Bolsas Rui Tavares 2010 (RT2010 to I.D.), the Netherlands Organization for Scientific Research – NWO (VENI grant 700.58.410 to S.B.N.), and by the European Community’s Seventh Framework Programme (FP7/2009 under grant agreement no: 241955, SYSCILIA to M.H.).
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonicka H, Ostergaard E, Sasarman F, et al. (11 co-authors) Mutations in C12orf65 in patients with encephalomyopathy and a mitochondrial translation defect. Am J Hum Genet. 2010;87:115–122. doi: 10.1016/j.ajhg.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald JM. Organelle genetics—plastid origins. Berlin (Germany): Springer-Verlag; 2012. [Google Scholar]
- Askarian-Amiri ME, Pel HJ, Guévremont D, McCaughan KK, Poole ES, Sumpter VG, Tate WP. Functional characterization of yeast mitochondrial release factor 1. J Biol Chem. 2000;275:17241–17248. doi: 10.1074/jbc.M910448199. [DOI] [PubMed] [Google Scholar]
- Baranov PV, Vestergaard B, Hamelryck T, Gesteland RF, Nyborg J, Atkins JF. Diverse bacterial genomes encode an operon of two genes, one of which is an unusual class-I release factor that potentially recognizes atypical mRNA signals other than normal stop codons. Biol Direct. 2006;1:28. doi: 10.1186/1745-6150-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell BG, Bankier AT, Drouin J. A different genetic code in human mitochondria. Nature. 1979;282:189–194. doi: 10.1038/282189a0. [DOI] [PubMed] [Google Scholar]
- Baurain D, Brinkmann H, Petersen J, Rodríguez-Ezpeleta N, Stechmann A, Demoulin V, Roger AJ, Burger G, Lang BF, Philippe H. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol Biol Evol. 2010;27:1698–1709. doi: 10.1093/molbev/msq059. [DOI] [PubMed] [Google Scholar]
- Brodersen DE, Clemons WM, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J Mol Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- Canutescu AA, Shelenkov AA, Dunbrack RL. A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrzanowska-Lightowlers ZM, Pajak A, Lightowlers RN. Termination of protein synthesis in mammalian mitochondria. J Biol Chem. 2011;286:34479–34485. doi: 10.1074/jbc.R111.290585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny P, Preiser P, Williamson D. Evidence for a single origin of the 35 kb plastid DNA in Apicomplexans. Protist. 1998;149:51–59. doi: 10.1016/S1434-4610(98)70009-4. [DOI] [PubMed] [Google Scholar]
- Duchêne A-M, Pujol C, Maréchal-Drouard L. Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria. Curr Genet. 2009;55:1–18. doi: 10.1007/s00294-008-0223-9. [DOI] [PubMed] [Google Scholar]
- Dunkley TPJ, Hester S, Shadforth IP, et al. (13 co-authors) Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci U S A. 2006;103:6518–6523. doi: 10.1073/pnas.0506958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilh BE, Jurgelenaite R, Szklarczyk R, et al. (13 co-authors) FACIL: fast and accurate genetic code inference and logo. Bioinformatics. 2011;27:1929–1933. doi: 10.1093/bioinformatics/btr316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, Philippe M. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- Gagnon MG, Seetharaman SV, Bulkley D, Steitz TA. Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome. Science. 2012;335:1370–1372. doi: 10.1126/science.1217443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa Y, Hikawa Y, Tochio N, et al. (11 co-authors) Solution structure of the catalytic domain of the mitochondrial protein ICT1 that is essential for cell vitality. J Mol Biol. 2010;404:260–273. doi: 10.1016/j.jmb.2010.09.033. [DOI] [PubMed] [Google Scholar]
- Handa Y, Inaho N, Nameki N. YaeJ is a novel ribosome-associated protein in Escherichia coli that can hydrolyze peptidyl-tRNA on stalled ribosomes. Nucleic Acids Res. 2011;39:1739–1748. doi: 10.1093/nar/gkq1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Ishimaru Y, Ohama T, Kawatsu Y, Nakamura K, Osawa S. UAG is a sense codon in several chlorophycean mitochondria. Curr Genet. 1996;30:29–33. doi: 10.1007/s002940050096. [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH. Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. The Plant Cell. 2004;16:241–256. doi: 10.1105/tpc.016055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidel AJ, Glöckner G. Mitochondrial genome evolution in the social amoebae. Mol Biol Evol. 2008;25:1440–1450. doi: 10.1093/molbev/msn088. [DOI] [PubMed] [Google Scholar]
- Hikosaka K, Watanabe Y-I, Tsuji N, et al. (12 co-authors) Divergence of the mitochondrial genome structure in the apicomplexan parasites, Babesia and Theileria. Mol Biol Evol. 2010;27:1107–1116. doi: 10.1093/molbev/msp320. [DOI] [PubMed] [Google Scholar]
- Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- Howe K, Bateman A, Durbin R. QuickTree: building huge Neighbour-Joining trees of protein sequences. Bioinformatics. 2002;18:1546–1547. doi: 10.1093/bioinformatics/18.11.1546. [DOI] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Huynen MA, Duarte I, Chrzanowska-Lightowlers ZMA, Nabuurs SB. Structure based hypothesis of a mitochondrial ribosome rescue mechanism. Biol Direct. 2012;7:14. doi: 10.1186/1745-6150-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob JEM, Vanholme B, Van Leeuwen T, Gheysen G. A unique genetic code change in the mitochondrial genome of the parasitic nematode Radopholus similis. BMC Res Notes. 2009;2:192. doi: 10.1186/1756-0500-2-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouskovec J, Horák A, Oborník M, Lukes J, Keeling PJ. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci U S A. 2010;107:10949–10954. doi: 10.1073/pnas.1003335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes TH, Osawa S. The genetic code in mitochondria and chloroplasts. Experientia. 1990;46:1117–1126. doi: 10.1007/BF01936921. [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K-i, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ. The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci. 2010;365:729–748. doi: 10.1098/rstb.2009.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislinger T, Cox B, Kannan A, et al. (14 co-authors) Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- Knight RD, Freeland SJ, Landweber LF. Rewiring the keyboard: evolvability of the genetic code. Nat Rev Genet. 2001;2:49–58. doi: 10.1038/35047500. [DOI] [PubMed] [Google Scholar]
- Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci U S A. 2008;105:19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Zhu J, Asahara H, Noller HF. Recognition of the amber UAG stop codon by release factor RF1. EMBO J. 2010;29:2577–2585. doi: 10.1038/emboj.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins. 2009;77(Suppl 9):114–122. doi: 10.1002/prot.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA—a self-parameterizing force field. Proteins. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- Lee CC, Timms KM, Trotman CN, Tate WP. Isolation of a rat mitochondrial release factor. Accommodation of the changed genetic code for termination. J Biol Chem. 1987;262:3548–3552. [PubMed] [Google Scholar]
- Li J, Cai T, Wu P, et al. (12 co-authors) Proteomic analysis of mitochondria from Caenorhabditis elegans. Proteomics. 2009;9:4539–4553. doi: 10.1002/pmic.200900101. [DOI] [PubMed] [Google Scholar]
- Marin B, Nowack ECM, Melkonian M. A plastid in the making: evidence for a second primary endosymbiosis. Protist. 2005;156:425–432. doi: 10.1016/j.protis.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Matsuyama A, Arai R, Yashiroda Y, et al. (12 co-authors) ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- Meurer J, Lezhneva L, Amann K, Gödel M, Bezhani S, Sherameti I, Oelmüller R. A peptide chain release factor 2 affects the stability of UGA-containing transcripts in Arabidopsis chloroplasts. Plant Cell. 2002;14:3255–3269. doi: 10.1105/tpc.006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora L, Heurgué-Hamard V, Champ S, Ehrenberg M, Kisselev LL, Buckingham RH. The essential role of the invariant GGQ motif in the function and stability in vivo of bacterial release factors RF1 and RF2. Mol Microbiol. 2003;47:267–275. doi: 10.1046/j.1365-2958.2003.03301.x. [DOI] [PubMed] [Google Scholar]
- Moreira D, Kervestin S, Jean-Jean O, Philippe H. Evolution of eukaryotic translation elongation and termination factors: variations of evolutionary rate and genetic code deviations. Mol Biol Evol. 2002;19:189–200. doi: 10.1093/oxfordjournals.molbev.a004071. [DOI] [PubMed] [Google Scholar]
- Motohashi R, Yamazaki T, Myouga F, et al. (13 co-authors) Chloroplast ribosome release factor 1 (AtcpRF1) is essential for chloroplast development. Plant Mol Biol. 2007;64:481–497. doi: 10.1007/s11103-007-9166-7. [DOI] [PubMed] [Google Scholar]
- Noh EW, Lee JS, Choi YI, Han MS, Yi YS, Han SU. Complete Nucleotide Sequence of Pinus koraiensis. 2007. Direct Submission to Genbank: Biotechnology Division, Korea Forest Research Institute. Accession No. AY228468. [Google Scholar]
- Nozaki Y, Matsunaga N, Ishizawa T, Ueda T, Takeuchi N. HMRF1L is a human mitochondrial translation release factor involved in the decoding of the termination codons UAA and UAG. Genes Cells. 2008;13:429–438. doi: 10.1111/j.1365-2443.2008.01181.x. [DOI] [PubMed] [Google Scholar]
- Ohama T, Inagaki Y, Bessho Y, Osawa S. Evolving genetic code. Proc Jpn Acad Ser B Phys Biol Sci. 2008;84:58–74. doi: 10.2183/pjab.84.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinares PDB, Ponnala L, van Wijk KJ. Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol Cell Proteomics. 2010;9:1594–1615. doi: 10.1074/mcp.M000038-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa S, Jukes TH, Watanabe K, Muto A. Recent evidence for evolution of the genetic code. Microbiol Rev. 1992;56:229–264. doi: 10.1128/mr.56.1.229-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, et al. (16 co-authors) A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Yang J-S, Jang SK, Kim S. Construction of functional interaction networks through consensus localization predictions of the human proteome. J Proteome Res. 2009;8:3367–3376. doi: 10.1021/pr900018z. [DOI] [PubMed] [Google Scholar]
- Petry S, Weixlbaumer A, Ramakrishnan V. The termination of translation. Curr Opin Struct Biol. 2008;18:70–77. doi: 10.1016/j.sbi.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Pino P, Foth BJ, Kwok L-Y, Sheiner L, Schepers R, Soldati T, Soldati-Favre D. Dual targeting of antioxidant and metabolic enzymes to the mitochondrion and the apicoplast of Toxoplasma gondii. PLoS Pathog. 2007;3:e115. doi: 10.1371/journal.ppat.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczynska KD, Le Ret M, Rurek M, Bonnard G, Augustyniak H, Gualberto JM. Plant mitochondrial genes can be expressed from mRNAs lacking stop codons. FEBS Lett. 2006;580:5641–5646. doi: 10.1016/j.febslet.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Ralph SA. Subcellular multitasking—multiple destinations and roles for the Plasmodium falcilysin protease. Mol Microbiol. 2007;63:309–313. doi: 10.1111/j.1365-2958.2006.05528.x. [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A, Weber APM, Bhattacharya D. The origin and establishment of the plastid in algae and plants. Annu Rev Genet. 2007;41:147–168. doi: 10.1146/annurev.genet.41.110306.130134. [DOI] [PubMed] [Google Scholar]
- Richter R, Rorbach J, Pajak A, Smith PM, Wessels HJ, Huynen MA, Smeitink JA, Lightowlers RN, Chrzanowska-Lightowlers ZM. A functional peptidyl-tRNA hydrolase, ICT1, has been recruited into the human mitochondrial ribosome. EMBO J. 2010;29:1116–1125. doi: 10.1038/emboj.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rötig A. Human diseases with impaired mitochondrial protein synthesis. Biochim Biophys Acta. 2011;1807:1198–1205. doi: 10.1016/j.bbabio.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Scolnick E, Tompkins R, Caskey T, Nirenberg M. Release factors differing in specificity for terminator codons. Proc Natl Acad Sci U S A. 1968;61:768–774. doi: 10.1073/pnas.61.2.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seit-Nebi A, Frolova L, Justesen J, Kisselev L. Class-1 translation termination factors: invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res. 2001;29:3982–3987. doi: 10.1093/nar/29.19.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Yang X, Higgs PG. The mechanisms of codon reassignments in mitochondrial genetic codes. J Mol Evol. 2007;64:662–688. doi: 10.1007/s00239-006-0284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AGB, Roger AJ. The real ‘kingdoms’ of eukaryotes. Curr Biol. 2004;14:R693–696. doi: 10.1016/j.cub.2004.08.038. [DOI] [PubMed] [Google Scholar]
- Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimanpour-Lichaei HR, Kühl I, Gaisne M, et al. mtRF1a is a human mitochondrial translation release factor decoding the major termination codons UAA and UAG. Mol Cell. 2007;27:745–757. doi: 10.1016/j.molcel.2007.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk R, Huynen MA. Mosaic origin of the mitochondrial proteome. Proteomics. 2010;10:4012–4024. doi: 10.1002/pmic.201000329. [DOI] [PubMed] [Google Scholar]
- Temperley R, Richter R, Dennerlein S, Lightowlers RN, Chrzanowska-Lightowlers ZM. Hungry codons promote frameshifting in human mitochondrial ribosomes. Science. 2010;327:301. doi: 10.1126/science.1180674. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K. Unique features of animal mitochondrial translation systems. The non-universal genetic code, unusual features of the translational apparatus and their relevance to human mitochondrial diseases. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86:11–39. doi: 10.2183/pjab.86.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden AZ, Lee J-H, Mock T, et al. (51 co-authors) Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- Wu D, Hugenholtz P, Mavromatis K, et al. (34 co-authors) A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Nakayama T, Reyes-Prieto A, Andersen RA, Boo SM, Ishida K-I, Bhattacharya D. A single origin of the photosynthetic organelle in different Paulinella lineages. BMC Evol Biol. 2009;9:98. doi: 10.1186/1471-2148-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DJ, Edgar CD, Murphy J, Fredebohm J, Poole ES, Tate WP. Bioinformatic, structural, and functional analyses support release factor-like MTRF1 as a protein able to decode nonstandard stop codons beginning with adenine in vertebrate mitochondria. RNA. 2010;16:1146–1155. doi: 10.1261/rna.1970310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DJ, Edgar CD, Poole ES, Tate WP. The codon specificity of eubacterial release factors is determined by the sequence and size of the recognition loop. RNA. 2010;16:1623–1633. doi: 10.1261/rna.2117010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ. Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE. 2008;3:e1994. doi: 10.1371/journal.pone.0001994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.