Abstract
Reporter genes are important tools for assessing vector pharmacology in vivo. Although useful, current systems are limited by (1) the need to generate a new vector for each different reporter, (2) the inability to package reporter genes in small vectors, and (3) variations in reporter gene feedback due to variations in cell-to-cell vector copy number. To circumvent these problems, we have used Cre recombinase as a “cat's paw” to activate reporter genes embedded in transgenic mice. The small Cre gene was introduced into self-complementary adeno-associated viral (scAAV) vectors with limited packaging capacity. Injection of scAAV-Cre vectors into mice with loxP-inactivated luciferase enabled in vivo imaging distributions comparable to the signal observed after AAV-luciferase injection. When injected into mT/mG mice, AAV-Cre converted ubiquitous expression of red fluorescent protein (RFP) to green fluorescent protein (GFP) expression only where the vectors transduced cells. Injection into F1 hybrid luciferase and mT/mG mice enabled simultaneous three-reporter tracking. This system was able to discriminate cell-specific transduction in all organs tested, with particular usefulness for detecting AAV serotype-specific transduction in the liver, kidney, and muscle. Given that F1 mice bear exactly one copy of luciferase and one copy of RFP-GFP, each reporter gene is either “on” or “off” in a cell. The Cre system therefore provides a unique quantum method to quantify vector delivery that can be applied when vector capacity is limited.
Hillestad and colleagues use self-complementary adeno-associated viral vectors encoding the Cre recombinase gene (scAAV-Cre) to activate reporter genes embedded in transgenic Cre reporter mouse strains. scAAV-Cre injected into mice containing loxP-inactivated luciferase resulted in luciferase expression similar to AAV-Luciferase vectors. In mice in which Cre-mediated recombination converts ubiquitous red fluorescent protein (RFP) expression to green fluorescent protein (GFP) expression, GFP was detected only in scAAV-Cre-transduced cells. Hybrid mice of these strains enabled simultaneous cell-specific three-reporter tracking.
Introduction
Reporter genes encode proteins that are straightforward to measure and distinguish from the endogenous cellular background. When developing gene delivery vectors, reporters provide the needed feedback for assessing tropism, expression kinetics, biodistribution, and so on. Optical reporter proteins that emit bioluminescence or fluorescence are particularly popular in small-animal studies as they provide quantitative and qualitative feedback, respectively. Of the known bioluminescent reporters, firefly luciferase (Luc) is particularly useful in small-animal models (Rabinovich et al., 2008; Wu et al., 2008). Of the fluorescent reporters, enhanced green fluorescent protein (EGFP) is among the brightest and can be used at necropsy to identify transduced cell populations. Ideally, vector studies in small animals would allow both in vivo imaging in living animals as well as cell-specific detection of vector gene expression in tissue sections or by flow cytometry. Although dual-reporter cassettes have been developed, these can be impossible to package in small vectors and can suffer from reductions in activity (Day et al., 1998).
For self-complementary adeno-associated viral (scAAV) vectors, packaging even one reporter gene can be difficult. scAAV vectors are interesting vehicles because, in vivo, they transduce cells 10 to 15 times more efficiently than their single-stranded counterparts (ssAAV) (McCarty et al., 2001, 2003). However, this boost in gene expression comes at the cost of reduced packaging capacity, with only ∼2.2 kilobase pairs (kbp) available to accommodate foreign DNA. This means large reporter genes such as luciferase (1.65 kbp) can be packaged only in the context of smaller enhancer/promoter sequences that usually have lower activity than larger ones (Pañeda et al., 2009).
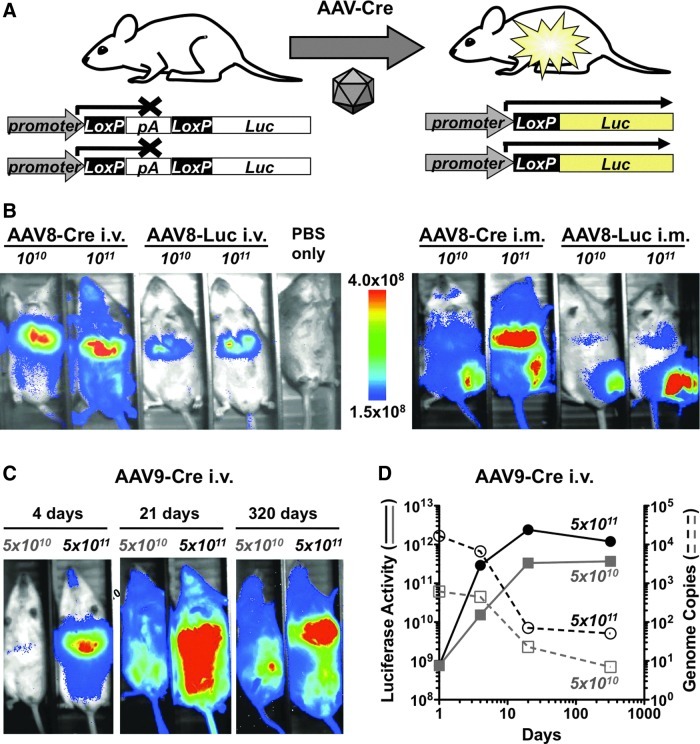
To circumvent the constraints imposed by limited packaging capacity, we generated scAAV vectors armed with the 1-kbp Cre recombinase gene rather than with a larger reporter gene such as luciferase. Cre mediates recombination at loxP sites (Sauer, 1987; Sauer and Henderson, 1988). If two loxP sites in the same orientation flank a sequence, Cre will excise the intervening sequence, leaving only a single loxP site (see Fig. 1A).
FIG. 1.
Comparing luciferase and Cre vectors in mice. (A) Diagram of luciferase activation in L-S-L Luc mice by Cre-expressing vector. Two luciferase cassettes are shown to depict the presence of two target genes per cell in homozygous mice. (B) Comparison of luciferase imaging in female L-S-L Luc mice injected by the intravenous or intramuscular route with ssAAV8-Luc or scAAV8-Cre. Pseudocolor of the day 6 image scale bar is shown. (C and D) Luciferase expression after intravenous injection of scAAV8-Cre into female L-S-L Luc mice. The total photon flux over 10 min per mouse is denoted for both doses by solid lines; relative copy number of the Cre–transgene (dashed lines) was measured by qPCR of total DNA isolated from livers of mice harvested on days 1, 4, 21, and 320 postadministration. The copy number of the reference gene RNase P was used to correct for variation in the amounts of DNA. The final results are expressed as n-fold differences in the Cre–transgene gene copy number relative to the RNase P gene (n=4).
To use Cre as a transgene to track virus transduction, we have applied vectors expressing Cre in mice that are transgenic for Cre-activated reporter genes. In these mice, the reporter genes are inactive because of the presence of a loxP-flanked (floxed) polyadenylation [poly(A)] cassette between a constitutively active promoter and the reporter gene's start methionine. In the absence Cre, these poly(A) sequences prematurely terminate the mRNA before reaching the reporter genes. When Cre is provided to the cell by breeding to a Cre-expressing mouse or in our case by a Cre-expressing vector, the recombinase deletes the floxed poly(A) to activate the downstream reporter gene.
We demonstrate the usefulness of this approach with small scAAV-Cre vectors in transgenic luciferase mice, in transgenic dual-fluorescent protein mice, and in F1 crosses of these mice providing three-way reporter gene detection, using one small vector platform.
Materials and Methods
Cell culture
Human embryonic kidney cells (293) were maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum (Gibco, Carlsbad, CA) and penicillin–streptomycin at 100 U/ml (Gibco).
Plasmids
Adenoviral helper genes for AAV vector production were provided by pHelper (Stratagene/Agilent, La Jolla, CA). pRC-2/8 and pRC-2/9 express the replication gene of AAV2 and the capsid genes of AAV8 and AAV9, respectively (Gao et al., 2005). A codon-optimized Cre recombinase was designed with the DNA Builder Program (UT Southwestern) and was build by overlapping PCR from twenty-eight 60-mer oligonucleotides. The scAAV plasmid expressing codon-optimized Cre, pSC-CMV-Cre, was generated from pSC(TTR-mvm-hFIX) (Wu et al., 2008) by replacing the expression cassette with a cytomegalovirus (CMV) promoter, chimeric intron (Promega, Madison, WI), and Cre recombinase gene. The ssAAV plasmid pAAV-CMV-Fluc was generated by transferring Fluc (Clontech, Palo Alto, CA) into pAAV(CMV-MCS) (Stratagene/Agilent).
AAV vector production
AAV vectors were generated by triple transfection of pHelper, pRC-2/8(9), and either pAAV-CMV-Fluc or pSC-CMV-Cre, using 25-kDa linear polyethylenimine (l-PEI) (Polysciences, Warrington, PA) by adaptation of the methods described by Reed and colleagues (2006). Briefly, first, 7 ml of PEI (1 mM) was diluted in 75 ml of NaCl (150 mM). Separately, 2 mg of the three plasmids at a 1:1:1 molar ratio was also diluted in 75 ml of NaCl (150 mM). Second, diluted PEI was added dropwise to the diluted DNA under constant agitation. After a 10-min incubation, contents were transferred to a 10-chamber CellSTACK (Corning Life Sciences, Lowell, MA) in which 293T cells were at ∼70% confluency. After 12–16 hr, the medium was exchanged and the cells were incubated for 96 hr. The cells were trypsinized and pelleted. Pellets were resuspended in 20 ml of phosphate-buffered saline (PBS: 137 mM NaCl, 2.7 mM KCl, 8.0 mM Na2HPO4, 2.0 mM KH2PO4) and cells were lysed by three rounds of freeze–thawing. Vector particles were released from the lysates, using DNase I (10 μg/ml) for 30 min at 37°C. Lysates were clarified by centrifugation for 15 min at 7000 rpm. Each lysate was then divided into four Quick-Seal Ultra-Clear (25×89 mm) centrifuge tubes (Beckman Coulter, Brea, CA). Iodixanol gradients were formed by underlying the lysate with 10 ml of 15%, 8 ml of 25%, 6 ml of 40%, and 4 ml of 54% iodixanol in 1× PBS (Zolotukhin et al., 1999). The gradients were centrifuged for 1 hr at 300,000×g, using a Ti-70 fixed angle rotor (Beckman coulter). The 40–60% gradient interface was harvested and the iodixanol was removed by multiple rinses on an Amicon Ultra-15 (100 kDa) concentrator (EMD Millipore, Billerica, MA). AAV vectors expressing non-codon-optimized Cre under the control of the CAG promoter (AAV2/1.CMV.PI.Cre.rBG, AAV2/8.CMV.PI.Cre.rBG, and AAV2/9.CMV.PI.Cre.rBG) were also purchased from the University of Pennsylvania Vector Core (Philadelphia, PA).
qPCR of AAV
AAV genomes were quantitated by qPCR on an ABI PRISM 7900HT sequence detection system (Applied Biosystems, Warrington, UK), using CMV-specific primers: RT-CMV-F (5′-CAAGTGTATCATATGCCAAGTACGCCCC-3′) and RT-CMV-R (5′-CCCCGTGAGTCAAACCGCTATCCAC GCC-3′). Yields ranged from 1×1011 to 1×1013 genome copies (GC)/ml.
Mice
mT/mG mice [also known as Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J mice] and L-S-L-Luc mice [also known as FVB.129S6(B6)-Gt(ROSA)26Sortm1(Luc)Kael/J] bearing a Cre-inducible reporter were purchased from Jackson Laboratory (Bar Harbor, ME). All studies involving animals were carried out according to the provisions of the Animal Welfare Act, Public Health Service Animal Welfare Policy, and Guide for the Care and Use of Laboratory Animals (National Research Council).
In vivo bioluminescence imaging
Mice were anesthetized with ketamine–xylazine and injected intraperitoneally with 100 μl of d-luciferin (20 mg/ml; Molecular Imaging Products, Bend, OR). Images were taken with a Lumazone imaging system (Photometrics/Roper Scientific, Tucson, AZ) over 10 min with 1×1 binning, using no filters or photo multiplication. Photons were quantitated with the Lumazone imaging software to determine total light intensity per mouse (photons/10 min).
Relative GC quantification in liver
Total DNA from liver tissue was isolated, using a Maxwell 16 mouse tail DNA purification kit (cat. no. AS1120; Promega) and detected with Universal ProbeLibrary probes (Roche, Indianapolis, IN). Combinations of primers (900 nM per reaction) and probes (100 nM) were designed online with Roche ProbeFinder software as follows: CoOpt.Cre, probe 105, 5′-GTCAGGACATCCGGAACCT-3′ and 5′-GCGATCC TCAGCAGGGTA3′; RNase P (calibrator), probe 56, 5′-CGA GCCTTATAACTTCATATCAACC3′ and 5′-CAGAAAAG CTTGGGCTACCA3′. In the 384-well format, 100 ng of DNA per reaction (final volume, 20 μl) was amplified in triplicate with FastStart universal probe master (Rox) (cat. no. 04913957001; Roche) on an Applied Biosystems 7900HT instrument as follows: 95°C for 10 min; and 40 cycles of 95°C for 15 sec, 60°C for 60 sec. Calculations were performed with ABI SDS2.4 and RQ manager 1.2.1 software, using RNase P as the sample calibrator.
Tissue sectioning and confocal microscopy
Organs were fixed for 4 hr in 4% paraformaldehyde (PFA)–PBS at room temperature and cryoprotected by overnight immersion in 30% sucrose–PBS. Trimmed organs were then flash frozen by liquid nitrogen-cooled isopentane in optimal cutting temperature (O.C.T.) medium (Sakura Finetek USA, Torrance, CA). Cryosections (thickness, 7 μm) were prepared with a Leica CM-850 cryostat and mounted on slides (Superfrost Plus; Thermo Fisher Scientific, Waltham, MA) with VECTASHIELD with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA), and CytoSeal-60 coverslip sealant (Thermo Fisher Scientific). Confocal imaging was performed at the Optical Morphology Core facility at Mayo Clinic Rochester (Rochester, MN), using a Zeiss Axiovert LSM510 laser confocal microscope (Carl Zeiss Jena, Jena, Germany) at 1 Airy unit pinhole with objective C-Apochromat ×40/1.2 W Corr M27.
Immunofluorescence staining
Kidneys were fixed and cryoprotected as described previously. Before flash freezing low-temperature antigen retrieval was performed by incubating trimmed kidney organ in 150 mM Tris-HCl at pH 9.0 for 5 min, followed by heating at 70°C for 15 min (Inoue and Wittbrodt, 2011). After heating, the kidney tissues were recryoprotected in 30% sucrose at 4°C overnight and flash frozen as described in the previous section. Cryosection (thickness, 18 μm) slides were equilibrated in 1× PBS (pH 7.4) and incubated at 4°C overnight with primary antibodies diluted 1:50 in PBS–incubation buffer (1% bovine serum albumin [BSA], 0.3% Triton X-100, and 1% calf serum). To stain endothelial cells, sections were conjugated with rat anti-CD31 (cat. no. 550274; BD Biosciences, San Jose, CA), for podocytes with rabbit anti-synaptopodin antibody (cat. no. ab101883; Abcam, Cambridge, UK), and for proximal tubules with rabbit anti-EpCAM (epithelial cell adhesion molecule) antibody (cat. no. ab32392; Abcam). Secondary antibodies Alexa Fluor 647-conjugated chicken anti-rat (cat. no. A21472; Invitrogen, Carlsbad, CA) and Alexa Fluor 647-conjugated anti-rabbit (A21245; Invitrogen) were both used at 1:200 dilution in incubation buffer.
Statistical analysis
GraphPad Prism (GraphPad Software, San Diego, CA) was used to perform the Student t test and one-way analysis of variance (Newman–Keuls post-test).
Results
Generation of scAAV vectors expressing codon-optimized Cre recombinase
Large reporter genes such as the 1.65-kbp firefly luciferase are too large to package in scAAV vectors, which contain a robust but large CMV promoter, an intron, and polyadenylation sequences. To circumvent this problem, we generated scAAV vectors expressing Cre recombinase rather than the reporter gene itself. Cre recombinase is a site-specific topoisomerase from P1 bacteriophage that recognizes and cleaves loxP sites. To maximize the efficiency of Cre for use in mammalian systems, the 1030-base pair (bp) bacteriophage sequence was codon-optimized for human tRNA bias and cloned into a self-complementary AAV2 vector under the control of the CMV promoter. This self-complementary vector was then packaged into AAV2/8 and AAV2/9 particles, generating the vectors scAAV8-Cre and scAAV9-Cre.
In vivo activation of genomic luciferase by AAV-Cre
To evaluate the use of Cre as a pseudoreporter gene, AAV vectors were first injected into L-S-L Luc mice (Safran et al., 2003). In these mice, expression of firefly luciferase is blocked by a loxP-STOP fragment (polyadenylation signal)-loxP (i.e., L-S-L) between the luciferase gene and the promoter in the (ROSA)26Sor locus (Zambrowicz et al., 1997). After Cre recombination, the polyadenylation sequence is deleted, allowing luciferase to be expressed from the ROSA26 promoter (Soriano, 1999) (Fig. 1A).
Self-complementary scAAV8-Cre was first compared with single-stranded AAV8 expressing firefly luciferase (ssAAV8-Luc), because luciferase was too large to be accommodated in a CMV scAAV vector. Therefore, this comparison was not intended to compare expression levels between Cre and luciferase vectors. Rather, the luciferase vector was used to compare vector tropism. scAAV8-Cre and ssAAV8-Luc vectors (1010 and 1011 GC) were injected by intravenous and intramuscular routes into L-S-L Luc mice (Fig. 1B). Imaging 6 days after injection revealed that both vectors generated luciferase bioluminescence that correlated to dose and delivery route. Intravenous injection of scAAV9-Cre into L-S-L mice also produced robust luciferase expression (Fig. 1C). The kinetics and stability of scAAV9-Cre-induced luciferase expression were monitored for 320 days (Fig. 1C and D). In this case, luciferase expression peaked around day 14 and remained high through 320 days. To compare vector genome persistence with expression, qPCR for AAV was performed on select mice at 1, 4, 14, and 320 days (Fig. 1D). In contrast to Cre-activated luciferase expression, scAAV-Cre genomes were highest on day 1 and decayed 100-fold over the same 320-day time period. These data indicate that the Cre vector and loxP host system mediates persistent changes to the host genome that mediate long-lasting expression beyond the persistence of the vector itself.
In vivo activation and deactivation of multiple reporter genes by Cre vectors
Our results so far suggested that inducing luciferase expression through vector-mediated delivery of Cre in L-S-L Luc mice produced a luciferase expression “fingerprint” similar to that generated by a vector carrying luciferase itself. Although this is useful, tracking only one reporter gene is insufficient to validate virus tropism. One must also determine virus tropism at the cellular level by either tissue sectioning or by flow cytometry to better understand the cell specificity of vectors. Unfortunately, luciferase bioluminescence is not generally sensitive enough in sections or by cytometry. In contrast, vectors carrying genes for fluorescent proteins such as green fluorescent protein (GFP), red fluorescent protein (RFP), or β-galactosidase are proven to assess cell specificity. Although these additional reporter genes may be optimal under different situations, testing of each also necessitates the production of an entirely different set of vectors with another set of genes.
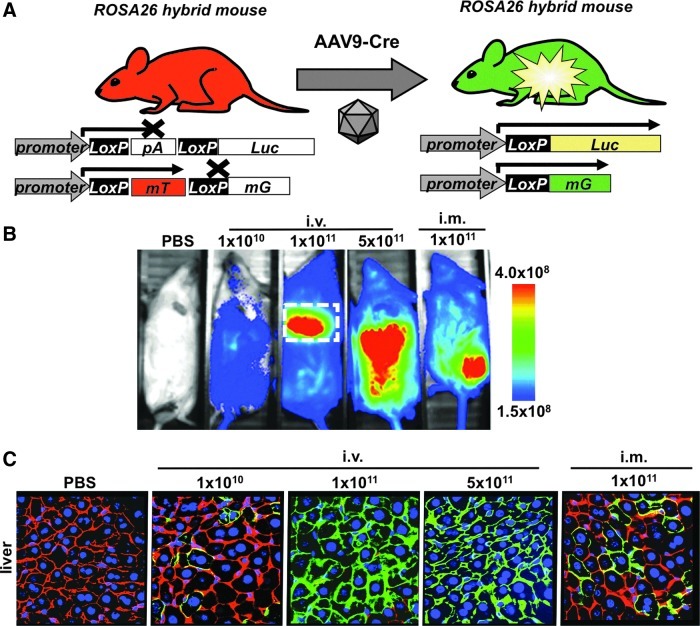
Fortunately, a Cre-vectored system circumvents this problem and requires that only one vector be generated. This is feasible because in addition to L-S-L Luc mice, there is a series of alternative, Cre-activatable mice that can be used with Cre vectors. For example, Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J mice (abbreviated here as mT/mG mice) are a useful double-fluorescent Cre-activatable mouse model for assessing cell specificity when bred to a Cre recombinase-expressing strain (Muzumdar et al., 2007). In the absence of Cre, cells express a membrane-targeted red fluorescent protein called mTomato or “mT” (Fig. 2A) that is flanked by two loxP sites. After Cre-mediated excision of these loxP sites, a downstream membrane-targeted GFP (“mG”) is expressed. Therefore, before Cre delivery, the membranes of all cells in the mice are red fluorescent and after excision the membranes become green fluorescent. Membrane targeting of the fluorescent proteins not only provides striking definition for each cell, but also ensures the proteins are not lost during fixation.
FIG. 2.
Vector transduction monitored by Cre modification of three reporter genes. (A) Diagram of Cre-induced reporter activation in hybrid mice after vector administration. One luciferase and one mT/mG cassette depict the presence of exactly one of each target gene per cell in these heterozygous mice. (B) Luciferase imaging and signal quantification in female hybrid mice injected by the intravenous route with the indicated amounts of scAAV9-Cre 18 days after injection. (C) Confocal images of liver tissue sections from the same mice harvested on day 18. Tissues are counterstained for nuclei with DAPI. Original magnification, ×630.
This mouse therefore provides a unique on-and-off dual signal wherein mTomato is turned off (one signal) and mGFP is turned on (a second signal). Only when both signals are present would a cell be flagged as being legitimately transduced. When combined with Cre-activated luciferase imaging, this provides a three-reporter vector detection system with three separate signals to confirm vector specificity (Fig. 2A).
To test the feasibility of this three-reporter vector detection approach, mT/mG mice were crossed with L-S-L Luc mice, generating F1 hybrids. Given that both transgene cassettes are inserted into the same Rosa26 locus, these mice are genetically determined to have exactly one copy of mTomato/mGFP and one copy of luciferase (Fig. 2A).
These hybrid mice were tested by injecting them by intramuscular and intravenous routes with scAAV9-CMV-Cre at different doses. Eighteen days after injection mice were anesthetized and imaged for luciferase expression (Fig. 2B). Consistent with AAV8- and AAV9-Cre data in L-S-L mice (Fig. 1), increasing the doses of AAV9-Cre also produced increasing bioluminescence output after intravenous injection in the hybrid mice. After intramuscular injection, AAV9-Cre mediated strong muscle expression with some distribution elsewhere in the body.
Immediately after luciferase imaging, the mice were killed and their livers were sectioned and visualized for red and green fluorescence by confocal microscopy (Fig. 2C). In PBS-treated mice, liver cells showed exclusive red fluorescence at their membranes. With increasing doses of scAAV9-Cre by the intravenous route, increasing numbers of cells were observed that had green fluorescent membranes. At the highest intravenous doses, all liver cells appeared to have been converted from red to green fluorescence. Where red and green cells were observed, the two fluorescent membranes appeared adjacent to each other rather than in the same location, consistent with the “on/off” genetics of the mT/mG single gene locus. Liver sections from the intramuscularly injected mice also showed a mixture of red and green fluorescent liver cells, with lower numbers of cells converted to mGFP expression. When luciferase expression and GFP-positive cells were compared, there was good concordance between the signals provided by the two reporter proteins after Cre activation.
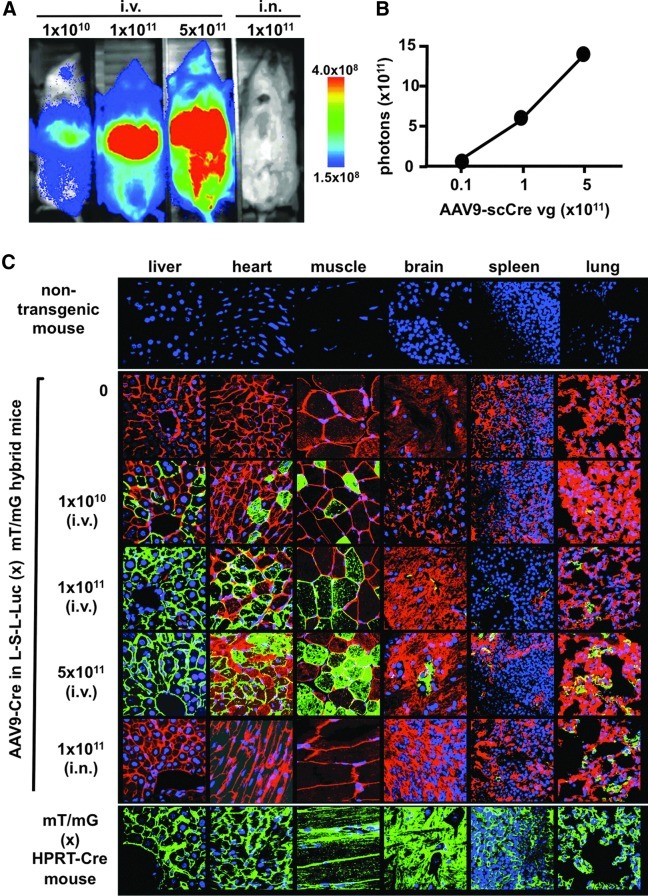
Three-way reporter detection in multiple tissues after scAAV9-Cre injection
To more fully assess the system, L-S-L Luc:mT/mG hybrid male mice were injected with scAAV9-Cre by the intravenous route and also by the intranasal route, and multiple tissues were analyzed for reporter conversion 18 days later (Fig. 3). With increasing intravenous doses, luciferase expression increased (Fig. 3A and B). The percentage of green-fluorescing cells in mice injected intravenously at doses from 1×1010 to 5×1011 GC increased from 45 to 100% in the liver, from 20 to 60% in the heart, from 20 to 60% in skeletal muscle, from 0 to 0.5% in the brain, from 0 to 0.5% in the spleen, and from 2 to 30% in the lungs (Fig. 3C). Interestingly, vector activity was observed in the testes after intravenous injection of scAAV9-Cre (Supplementary Fig. 1; supplementary data are available online at www.liebertpub.com/hum). Under closer microscopic examination, no Cre-mediated activation of green fluorescence was observed in germ cells in the seminiferous tubules. In contrast, reporter activation was observed in the interstitial space occupied by Leydig cells. For the mouse receiving scAAV9-Cre by the intranasal route, green fluorescence was observed only in the lungs and spleen. In this case, as much as 35% of lung cells were modified by AAV9-Cre whereas only scattered spleen cell modification was observed.
FIG. 3.
Three-reporter gene monitoring in multiple tissues after Cre vector injection. (A) Imaging for luciferase activity in male hybrid mice injected by the intravenous or intranasal route with the indicated amounts of scAAV9-Cre 20 days postinjection. (B) Quantification of luciferase activity in the same mice by measuring total photon flux over 10 min per mouse. (C) Confocal images of select tissue sections from mice injected with the indicated amounts of scAAV9-Cre by the indicated routes. Tissues are counterstained for nuclei with DAPI. Nontransgenic mice do not express fluorescent proteins and show only DAPI stain. Positive control mice encode only mG, which is constitutively expressed. Original magnifications are 400x.
These vector-mediated modifications to the mouse genome contrast well with those in positive control mice produced by crossing mT/mG mice with HPRT-Cre mice. In these control mice, mT is converted to mG expression in all cells of the mouse, due to ubiquitous Cre expression early in development. They therefore show only mG expression throughout the tissues in contrast to the varied conversion of mT to mG mediated by scAA9-Cre (Fig. 3). These data demonstrate that this vector–host system enables quantitative assessment of vector pharmacology by tracking three separate reporters using only a single Cre vector.
Comparison of various AAV serotypes for kidney transduction
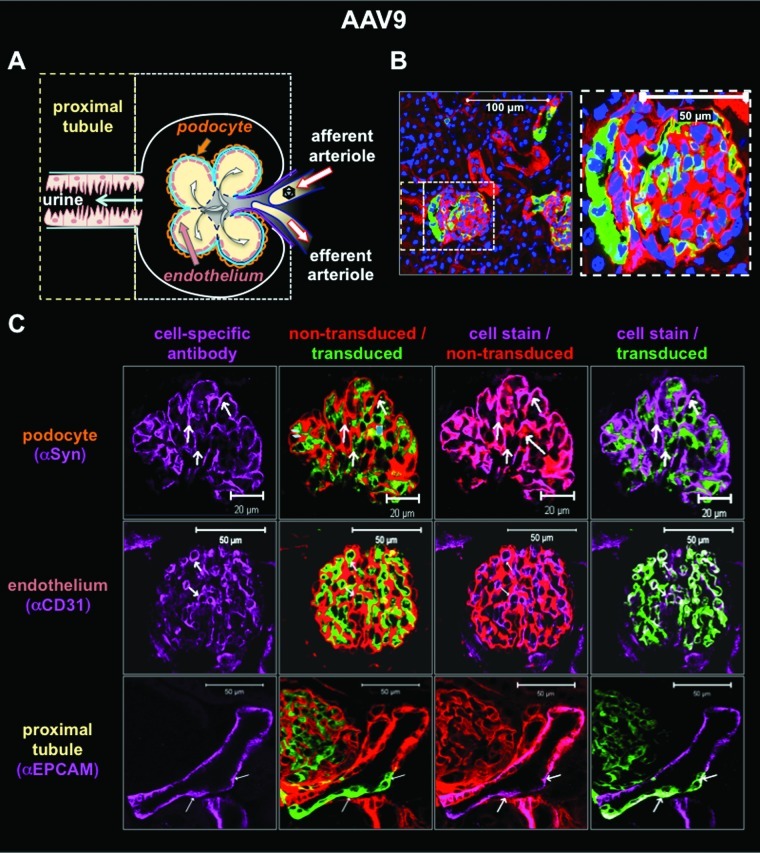
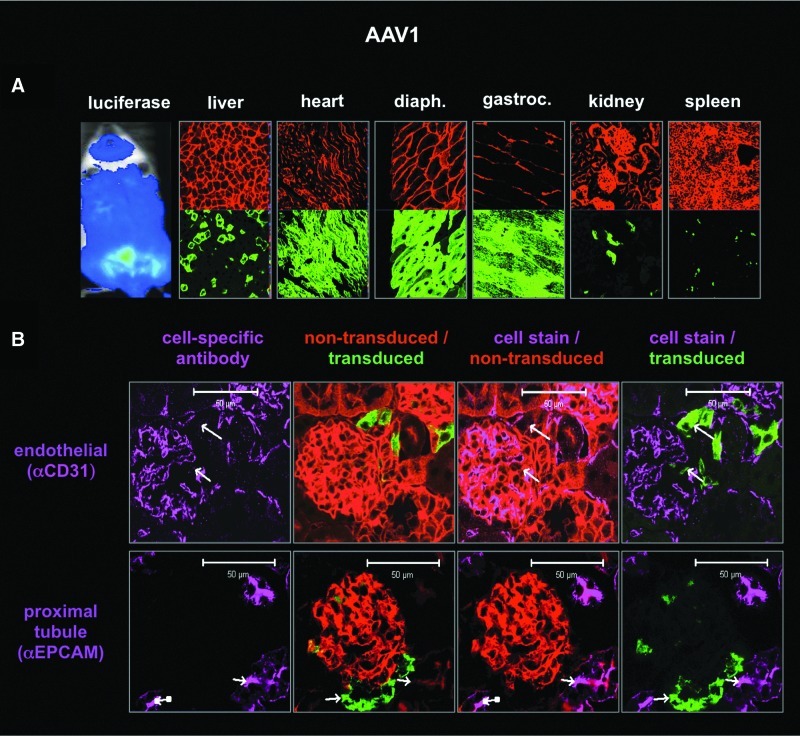
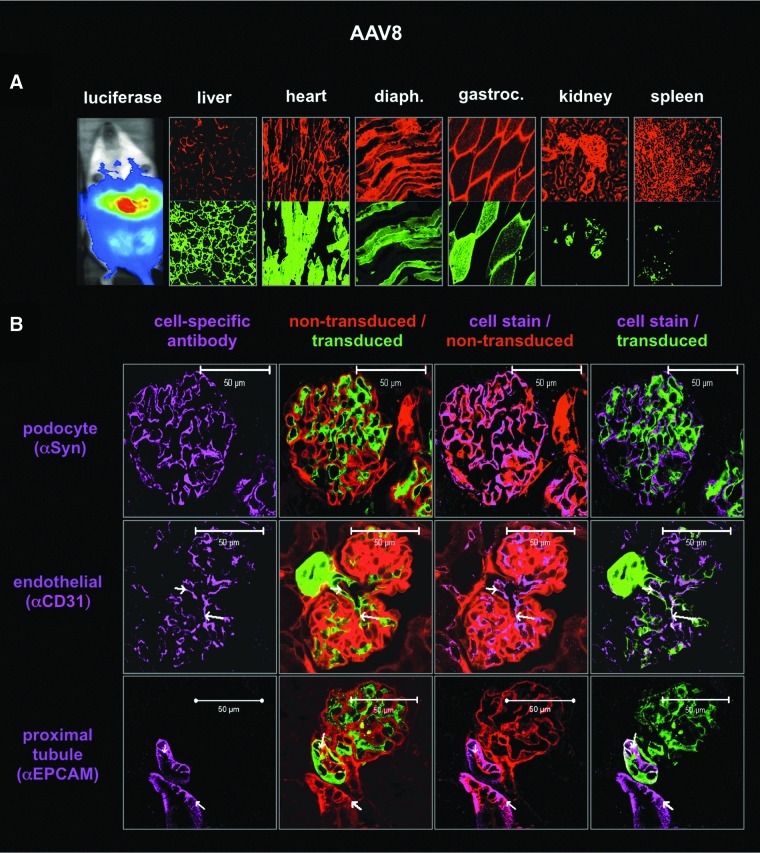
The hybrid mouse system and “on/off” nature of the Cre system were tested for their ability to detect cell-specific transduction in the kidney after intravenous injection. scAAV1, scAAV8, and scAAV9 vectors expressing wild-type Cre were injected into hybrid mice for luciferase imaging and confocal microscopy (Figs. 4–6). The kidneys of mice injected with scAAV9-Cre were sectioned and stained with kidney cell-specific antibodies (Fig. 4). Nontransduced red cells were compared with transduced green cells with and without staining for podocytes (with anti-synaptopodin), for endothelial cells (with anti-CD31), or for proximal tubules with anti-EpCAM). Comparison of red/green fluorescence of an individual glomerulus (Fig. 4B) with nephron structure (Fig. 4A) demonstrated that intravenous injection of AAV9 mediated genetic modification of a subset of kidney cells. When separate sections were stained with the cell-specific antibodies, podocytes did not appear to be transduced. In contrast, AAV9 appeared to transduce some, but not all CD31+endothelial cells and EpCAM+ proximal tubules (Fig. 4C). AAV8 did not transduce podocytes, but did modify endothelial cells as well as cells in the proximal tubules (Fig. 5). AAV1 tropism was markedly different than those of AAV8 and AAV9 (Figs. 1–6). AAV1 predominantly modified skeletal and cardiac muscle cells with markedly less transduction of the liver (Fig. 6A) than AAV8 and AAV9. AAV1-Cre marked a subset of cells in the kidney with GFP (Fig. 6A). Confocal microscopy of these kidneys indicated that AAV1 did not transduce endothelial cells or tubules, as did AAV8 and AAV9 (Fig. 6B). These data demonstrate the usefulness of the Cre-activated vector–host system for analyzing virus tropism at macroscopic, microscopic, and cell-specific levels.
FIG. 4.
Transduction profile of scAAV9-Cre in the glomeruli of female hybrid mice by immunofluorescence staining 21 days after intravenous injection with 1×1012 GC. (A) Diagram depicting structural relationship of blood flow, podocytes, endothelial cells, and proximal tubules in kidney. (B) Confocal image of PFA-fixed kidney cryosections (thickness, 7 μm) at magnifications of ×100 and ×400 with DAPI-stained nuclei (405 nm emission, blue), transduced cells (488 nm emission, green), and nontransduced cells (563 nm emission, red). (C) Immunofluorescence confocal imaging and overlays of PFA-fixed kidney cryosections (thickness, 18 μm) for denoted cell types (647 nm emission, purple); DAPI counterstain not shown. EpCAM, epithelial cell adhesion molecule; Syn, synaptopodin.
FIG. 6.
Transduction profile of scAAV1-Cre in female hybrid mice 21 days after intravenous injection with 1×1012 GC. PFA-fixed kidney cryosections. (A) Luciferase image and confocal images (400×) of cryosections taken from various tissues (thickness, 7 μm) at ×100 and ×400 magnification with DAPI-stained nuclei (405 nm emission, blue), transduced cells (488 nm emission, green), and nontransduced cells (563 nm emission, red). (B) Immunofluorescence imaging by confocal of kidney tissue sections (thickness, 18 μm) for denoted cell types (647 nm emission, purple); DAPI counterstain not shown. EpCAM, epithelial cell adhesion molecule.
FIG. 5.
Transduction profile of scAAV8-Cre in female hybrid mice 21 days after intravenous injection with 1×1012 GC. PFA-fixed kidney cryosections. (A) Luciferase image and confocal images ( ×400) of cryosections taken from various tissues (thickness, 7 μm) at ×100 and ×400 magnification with DAPI-stained nuclei (405 nm emission, blue), transduced cells (488 nm emission, green), and nontransduced cells (563 nm emission, red). (B) Immunofluorescence imaging by confocal of kidney tissue sections (thickness, 18 μm) for denoted cell types (647 nm emission, purple); DAPI counterstain not shown. EpCAM, epithelial cell adhesion molecule; Syn, synaptopodin.
Discussion
In this study we demonstrate that the Cre–lox system can be used as an efficient approach to assess vector pharmacology in a quantitative fashion with confirmation by tracking multiple reporter genes simultaneously. Previous studies have used viral vectors carrying Cre to induce reporter expression in ROSA26(LacZ) mice. For example, ssAAV2-Cre was shown to induce β-galactosidase expression in tissues when administered intratracheally (Rossiter et al., 2008; Liu et al., 2009), intracranially (Kaspar et al., 2002; Ahmed et al., 2004), and by transcoronary injection (Iwatate et al., 2003; Lee et al., 2008). These studies provided good proof of principle for the use of Cre, but did not leverage this potent transgene for in vivo luciferase imaging and for fluorescent protein detection.
In this study, we originally applied the small Cre gene to circumvent problems in packaging capacity in scAAV vectors. This approach also has practical benefit, because only one vector needs to be produced to track many different reporter genes, because many useful Cre reporter strains have already been generated (i.e., luciferase, GFP, RFP, YFP, β-galactosidase, alkaline phosphatase, etc.). In contrast, if one wanted to track each of these reporters with their unique features, this would require generating a battery of parallel vectors.
This approach also looks promising to assess vector pharmacology by allowing many different flavors of reporter to be sampled. When applied in a hybrid such as the L-S-L Luc:mT/mG mouse, vector tropism is sampled in parallel by different techniques. Each activation or deactivation of a given reporter gene can also be used to validate the signals of the other reporters in the same mouse. These data therefore suggest that Cre-expressing vectors may be useful to assess vector pharmacology in preclinical models. Given the burgeoning number of Cre-activated and Cre-inactivated animal models that have been provided by our colleagues in transgenic mouse research, the approach is likely to provide even more utility to the gene therapy field.
Supplementary Material
Acknowledgments
The authors thank Torey Batts, Mary Barry, Anthony J. Croatt, and James E. Tarara for excellent technical assistance. This work was supported by a grant to M.A.B. from the NIH (R01-CA136945) and by a grant from the Muscular Dystrophy Association. It was also supported by a grant to K.A.N. from the NIH (R37-DK047060). M.L.H. was supported by the Kidney Disease Research Training Program (T32-DK007013).
Author Disclosure Statement
M.L.H., A.J.G., K.A.N., and M.A.B. have no competing financial interests.
References
- Ahmed B.Y. Chakravarthy S. Eggers R., et al. Efficient delivery of Cre-recombinase to neurons in vivo and stable transduction of neurons using adeno-associated and lentiviral vectors. BMC Neurosci. 2004;5:4. doi: 10.1186/1471-2202-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day R.N. Kawecki M. Berry D. Dual-function reporter protein for analysis of gene expression in living cells. Biotechniques. 1998;25:848–850. doi: 10.2144/98255bt02. 852–844, 856. [DOI] [PubMed] [Google Scholar]
- Gao G. Vandenberghe L.H. Wilson J.M. New recombinant serotypes of AAV vectors. Curr. Gene Ther. 2005;5:285–297. doi: 10.2174/1566523054065057. [DOI] [PubMed] [Google Scholar]
- Inoue D. Wittbrodt J. One for all—a highly efficient and versatile method for fluorescent immunostaining in fish embryos. PLoS One. 2011;6:e19713. doi: 10.1371/journal.pone.0019713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatate M. Gu Y. Dieterle T., et al. In vivo high-efficiency transcoronary gene delivery and Cre–LoxP gene switching in the adult mouse heart. Gene Ther. 2003;10:1814–1820. doi: 10.1038/sj.gt.3302077. [DOI] [PubMed] [Google Scholar]
- Kaspar B.K. Vissel B. Bengoechea T., et al. Adeno-associated virus effectively mediates conditional gene modification in the brain. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2320–2325. doi: 10.1073/pnas.042678699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N. Robitz R. Zurbrugg R.J., et al. Conditional, genetic disruption of ciliary neurotrophic factor receptors reveals a role in adult motor neuron survival. Eur. J. Neurosci. 2008;27:2830–2837. doi: 10.1111/j.1460-9568.2008.06298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. Luo M. Guo C., et al. Analysis of adeno-associated virus progenitor cell transduction in mouse lung. Mol. Ther. 2009;17:285–293. doi: 10.1038/mt.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty D.M. Monahan P.E. Samulski R.J. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 2001;8:1248–1254. doi: 10.1038/sj.gt.3301514. [DOI] [PubMed] [Google Scholar]
- McCarty D.M. Fu H. Monahan P.E., et al. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. 2003;10:2112–2118. doi: 10.1038/sj.gt.3302134. [DOI] [PubMed] [Google Scholar]
- Muzumdar M.D. Tasic B. Miyamichi K., et al. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Pañeda A. Vanrell L. Mauleon I, et al. Effect of adeno-associated virus serotype and genomic structure on liver transduction and biodistribution in mice of both genders. Hum. Gene Ther. 2009;20:908–917. doi: 10.1089/hum.2009.031. [DOI] [PubMed] [Google Scholar]
- Rabinovich B.A. Ye Y. Etto T., et al. Visualizing fewer than 10 mouse T cells with an enhanced firefly luciferase in immunocompetent mouse models of cancer. Proc. Natl. Acad. Sci. U.S.A. 2008;105:14342–14346. doi: 10.1073/pnas.0804105105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S.E. Staley E.M. Mayginnes J.P., et al. Transfection of mammalian cells using linear polyethylenimine is a simple and effective means of producing recombinant adeno-associated virus vectors. J. Virol Methods. 2006;138:85–98. doi: 10.1016/j.jviromet.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Rossiter H.B. Scadeng M. Tang K., et al. Doxycycline treatment prevents alveolar destruction in VEGF-deficient mouse lung. J. Cell Biochem. 2008;104:525–535. doi: 10.1002/jcb.21643. [DOI] [PubMed] [Google Scholar]
- Safran M. Kim W.Y. Kung A.L., et al. Mouse reporter strain for noninvasive bioluminescent imaging of cells that have undergone Cre-mediated recombination. Mol. Imaging. 2003;2:297–302. doi: 10.1162/15353500200303154. [DOI] [PubMed] [Google Scholar]
- Sauer B. Functional expression of the Cre–lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1987;7:2087–2096. doi: 10.1128/mcb.7.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B. Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. U.S.A. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Wu Z. Sun J. Zhang T., et al. Optimization of self-complementary AAV vectors for liver-directed expression results in sustained correction of hemophilia B at low vector dose. Mol. Ther. 2008;16:280–289. doi: 10.1038/sj.mt.6300355. [DOI] [PubMed] [Google Scholar]
- Zambrowicz B.P. Imamoto A. Fiering S., et al. Disruption of overlapping transcripts in the ROSA βgeo 26 gene trap strain leads to widespread expression of β-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin S. Byrne B.J. Mason E., et al. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.