Abstract
In many invertebrate and vertebrate species, cell fates are assigned through the cellular inheritance of differentially localized maternal determinants. Whether mammalian embryogenesis is also regulated by deterministic mechanisms is highly controversial. The caudal domain transcription factor CDX2 has been reported to act as a maternal determinant regulating cell fate decisions in mouse development. However, this finding is contentious because of reports that maternal Cdx2 is not essential for development. Notably, all of the previously published studies of maternal Cdx2 relied on injected RNA interference constructs, which could introduce experimental variation. Only deletion of the maternal gene can unambiguously resolve its requirement in mouse development. Here, we genetically ablated maternal Cdx2 using a Cre/lox strategy, and we definitively establish that maternal Cdx2 is not essential for mouse development.
Keywords: Blastocyst, Maternal-effect gene, Trophectoderm, Caudal
INTRODUCTION
In animals, early embryonic development is regulated by maternal genes, which are transcribed in the female germline during oogenesis. The extent to which maternal genes participate in embryo patterning varies among species. For example, in fruit fly embryos, differentially localized maternal factors define body axes, whereas in frog and nematode, maternal factors, in addition to information from the site of sperm entry, regulate early cell fate decisions. In mice, maternal factors have been identified that are essential for embryo viability (Li et al., 2010), but mouse early embryos are thought to undergo regulative, rather than mosaic, development (Johnson, 2009; Rossant and Tam, 2009; Yamanaka et al., 2006). Therefore, the contribution of maternal factors to embryo patterning remains speculative.
Mouse Cdx2, the ortholog of the Drosophila maternal-effect gene caudal (Frohnhöfer and Nüsslein-Volhard, 1986; Mlodzik et al., 1985), is essential for early development, and loss of zygotic Cdx2 disrupts trophectoderm development, leading to preimplantation lethality (Strumpf et al., 2005). Whether maternal Cdx2 is also required for development is unresolved. Studies of the requirement for maternal Cdx2 in development have yielded conflicting results (Jedrusik et al., 2010; Wang et al., 2010a; Wu et al., 2010). Notably, these reports relied on RNA interference (RNAi) to reduce Cdx2 levels, leaving open the possibility that the differing phenotypes resulted from differing degrees of gene inactivation. To resolve unambiguously the requirement for maternal Cdx2 in development, Cdx2 must be deleted from the oocyte prior to fertilization using a conditional null allele. We use this approach to show that Cdx2 is not a maternal-effect gene in mouse.
MATERIALS AND METHODS
Mouse strains
The following alleles or transgenes were maintained in an outbred (CD1) background: Cdx2tm1Fbe (a null allele) (Chawengsaksophak et al., 1997), Tg(Zp3-cre)93Knw (de Vries et al., 2000) and Cdx2fl (a new conditional allele). All animal work conformed to the guidelines and regulatory standards of the University of California Santa Cruz Institutional Animal Care and Use Committee.
Generation of the Cdx2 conditional allele
To generate the Cdx2 conditional allele, the targeting construct was created by PCR amplification of 129X1/SvJ genomic DNA using PfuTurbo Hotstart DNA polymerase (Stratagene) to amplify regions of the Cdx2 locus. Amplified regions were assembled in ploxPF1pneo plasmid [gift of Dr James Shayman (Hiraoka et al., 2006)]. PvuI-linearized plasmid was then electroporated into R1 ES cells. NheI-digested genomic DNA from a total of 480 neo-resistant clones was then screened by Southern blot using a probe complementary to the 3′ region. Ten positive clones were then digested with ScaI and screened by Southern blot using the 5′ probe and by PCR using primers P1 and P2 (5′-GAATACGTCGTGTAATTAGCA-3′ and 5′-CAAAGCCAACAACT GGAC-3′). A single correctly targeted clone was selected for injection into C57BL/6J blastocysts to produce chimeric mice. Germline transmission was observed in 4/11 male chimeras, which were then bred to establish founder Cdx2fl/+ mice. Subsequently, the neo cassette was removed by crossing founder lines to the B6;SJL-Tg(ACTFLPe)9205Dym/J mouse (Rodríguez et al., 2000).
Embryo genotyping
Embryo genotyping was performed blind, without prior knowledge of phenotypes. Genomic DNA was extracted from individual embryos using the Red Extract-N-Amp Kit (Sigma) in a final volume of 10 μl extraction/neutralization buffers. Subsequently, 0.5-1 μl lysate was used for PCR detection of the various alleles using the following primers (5′-3′): wild type and Cdx2tm1Fbe, AGGGACTATTCAAACTACAGGAG, TAAAAGTCAACTGTGTTCGGATCC and ATATTGCTGAAGAGCTTGGCGGC; Zp3Cre, GCGGTCTGGCAGTAAAAACTATC and GTGAAACAGCATTGCTGTCACTT; and wild type, Cdx2fl and Cdx2del, P1 and P2 (see above).
Immunofluorescence, embryo culture and microscopy
Embryos were collected from timed natural matings by flushing dissected oviducts or uteri with M2 medium (Millipore). Embryos were either fixed and stained as previously described (Ralston and Rossant, 2008) or cultured in KSOM (Millipore) at 37°C and 6% CO2 to monitor morphological development. Primary antibodies included mouse anti-CDX2 (Biogenex CDX-88), rabbit anti-NANOG (Reprocell), rat anti-CDH1 (Sigma) and rat anti-KRT8 (Troma-1; R. Kemler, Developmental Studies Hybridoma Bank, developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biology, Iowa City, IA 52242, USA). Secondary antibodies, nuclear stain (Draq5) and confocal microscopy methods were as described (Ralston and Rossant, 2008), using a Leica SP5 confocal microscope and 20×, 0.7 NA objective.
Real-time PCR
To obtain oocytes, female mice were superovulated by subcutaneous injections of 5 IU each pregnant mare serum (PMS) and human chorionic gonadotropin (HCG) (Sigma), 46 hours apart. MII oocytes were collected 23 hours after HCG injection from dissected ampullae. Oocytes were then denuded of cumulus cells by incubation and gentle pipetting in 300 μg/ml bovine type IV-S hyaluronidase (Sigma) in M2 medium. RNA was extracted from ~20 pooled oocytes or individual blastocysts using the PicoPure RNA Isolation Kit (Arcturus) according to the manufacturer's instructions. As previously described (Ralston et al., 2010), cDNA was prepared and amplified by SYBR Green-based relative quantification PCR using a Roche LightCycler 480. For each primer pair, the PCR efficiency was empirically determined from a standard curve, and this was used to calculate measurements using the ΔCT method using Roche software. Primers were (5′-3′): Actb, CTGAACCCTAAGGCCAACC and CCAGAGGCATACAGGGACAG; Cdx2, AAACCTGTGCGAGTGGATG and TCTGTGTACACCACCCGGTA.
RESULTS AND DISCUSSION
Oocytes express lower levels of Cdx2 than do embryonic stem cells
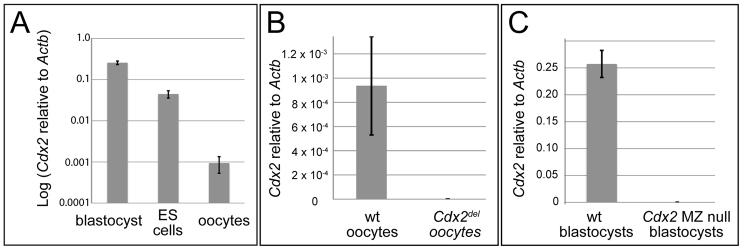
As a first step toward determining whether maternal Cdx2 is important for development, several groups have characterized the level of protein (CDX2) and mRNA (Cdx2) present in mouse oocytes and ovaries. By immunohistochemistry, CDX2 was undetectable in mouse ovaries (Beck et al., 1995). Similarly, CDX2 was not detected in mouse oocytes by mass spectrometry (Wang et al., 2010b). Post-fertilization, CDX2 is not detectable at the 2- or 4-cell stages by immunofluorescence (Ralston and Rossant, 2008), suggesting that maternal Cdx2 is not a major source of CDX2 in the embryo. Cdx2 mRNA levels were reportedly low, but detectable, in oocytes (Jedrusik et al., 2010; Wang et al., 2010a). Both these studies reported lower levels of Cdx2 in oocytes than in blastocysts, but neither group examined Cdx2 levels in a cell type in which Cdx2 is functionally irrelevant. For example, embryonic stem (ES) cells do not require Cdx2 (Chawengsaksophak et al., 2004) and could be used as a negative control. We therefore compared levels of Cdx2 mRNA in blastocysts, oocytes and ES cells by reverse transcription and relative quantification real-time PCR (qPCR). Consistent with previous reports, we detected higher levels of Cdx2 mRNA in blastocysts than in oocytes. However, Cdx2 mRNA levels were more than an order of magnitude lower in oocytes than in ES cells (Fig. 1A). This suggests that the level of oocyte Cdx2 mRNA is functionally irrelevant.
Fig. 1.
Relative quantification of Cdx2 levels by qPCR. (A) Average Cdx2 levels, normalized to β-actin (Actb), in E3.5 blastocysts, oocytes and ES cells. Averages were calculated from three biological replicate measurements: three wild-type (wt) blastocysts, three ES cell lines (R1, E14 and G4), and oocytes from three mice. (B) Levels of Cdx2, relative to Actb mRNA, in wild-type and Cdx2 M null oocytes (average of three biological replicates for each genotype). (C) Average levels of Cdx2, relative to Actb mRNA, in single wild-type blastocysts (n=4) and Cdx2 MZ null blastocysts (n=5) at E3.5. Error bars indicate s.d. of biological replicates.
Generation of viable mice lacking maternal Cdx2
Our qPCR analysis indicated that Cdx2 is unlikely to be required for development, and we next sought a means to test this hypothesis. Several groups have examined the requirement for maternal Cdx2 by injecting siRNA constructs into oocytes and zygotes (Jedrusik et al., 2010; Wang et al., 2010a; Wu et al., 2010). Curiously, these groups obtained contradictory findings, even after injection of identical constructs (Wu and Schöler, 2011). Groups led by Wu and Wang reported that maternal Cdx2 is not required for development (Wang et al., 2010a; Wu et al., 2010), whereas Jedrusik et al. (Jedrusik et al., 2010) reported that maternal Cdx2 is required for development. Since different methods of injection might lead to different experimental outcomes (Morris, 2011), we used a non-invasive, genetic strategy to remove Cdx2 from the maternal germline.
First, we created a conditional allele of Cdx2 (supplementary material Fig. S1A-D). In this allele, Cre-mediated recombination between loxP sites leads to deletion of the Cdx2 transcription start site and introduction of a nonsense frame shift. We then used this allele to create adult females carrying Cdx2 null oocytes using the female germline-specific Zp3-Cre (de Vries et al., 2000) (supplementary material Fig. S1E). Zp3-Cre is expressed specifically in oocytes, where it causes recombination of floxed alleles (de Vries et al., 2000) very early in oogenesis (Lan et al., 2004). We first confirmed that Cdx2 mRNA was ablated in oocytes from Cdx2 germline null (Cdx2fl/Cdx2fl or del; Zp3-Cre/+) females by qPCR (Fig. 1B).
We then bred Cdx2 germline null females to wild-type males, with the expectation that if maternal Cdx2 were required for development then no viable offspring would result. However, we obtained comparable numbers of viable offspring from Cdx2 germline null and control females (7.3±1.5 and 7.0±0.8 pups/litter, respectively). We examined whether Cre-mediated excision had occurred in these crosses by PCR genotyping pups and embryos collected from Cdx2 germline null females. This confirmed that Cre-mediated excision was highly penetrant, as the excised allele was detected in almost all progeny examined (6/6 pups and 70/72 blastocysts). We also determined that, although only around half of the progeny inherit the Zp3-Cre transgene (11/25 blastocysts), excision occurs in all progeny (25/25 blastocysts), consistent with excision prior to the first meiotic division (de Vries et al., 2000; Lan et al., 2004). We infer that germline Cdx2 is dispensable for female fertility and embryo development.
Maternal Cdx2 is not required for trophectoderm specification
Jedrusik et al. (Jedrusik et al., 2010) suggested that maternal Cdx2 is required for trophectoderm cell polarization and cell fate specification prior to the establishment of inner and outer cell populations because zygotic Cdx2 knockdown, which presumably interfered with both maternal and zygotic Cdx2, led to phenotypes that were more severe than that of the Cdx2 zygotic (Z) null (Jedrusik et al., 2010). Cdx2 maternal-zygotic (MZ) knockdown phenotypes included prolonged cell cycle during cleavage stages, lethality prior to the blastocyst stage in over half of the embryos and formation of a blastocyst with a reduced number of trophectoderm cells in the remaining embryos. In addition, molecular defects were noted in Cdx2 MZ knockdown embryos, including failure to express cell polarity genes, including E-cadherin (Cdh1) and keratin 8 (Krt8), and lineage markers such as Nanog (Jedrusik et al., 2010). By contrast, another group reported that knockdown of Cdx2 in the zygote did not disrupt formation of the blastocyst, but phenocopied Cdx2 Z null embryos (Wu et al., 2010). Accordingly, Cdx2 MZ knockdown blastocysts expressed CDH1 and KRT8, whereas NANOG was ectopically expressed in the trophectoderm, consistent with the Cdx2 Z null phenotype (Ralston and Rossant, 2008; Strumpf et al., 2005). Cdx2 MZ knockdown blastocysts then collapsed around implantation stage (Wu et al., 2010), as do Cdx2 Z null blastocysts (Strumpf et al., 2005). Subsequently, differences among phenotypes resulting from Cdx2 MZ knockdown have been discussed in the literature (Bruce, 2011; Johnson, 2011; Wu and Schöler, 2011), but no consensus has been reached to explain the differing phenotypes.
To resolve the debate, we bred Cdx2 germline null (Cdx2fl/fl or fl/del; Zp3-Cre/+) females with Cdx2null/+ (Chawengsaksophak et al., 1997) males to generate embryos lacking both maternal and zygotic Cdx2 (Cdx2 MZ null). In contrast to the study by Jedrusik et al. (Jedrusik et al., 2010) and consistent with studies by Wu et al. and Wang et al. (Wu et al., 2010; Wang et al., 2010a), we found that Cdx2 MZ null embryos reached the blastocyst stage and then collapsed around the time of implantation (11/11 embryos; data not shown). We confirmed that Cdx2 mRNA was ablated in Cdx2 MZ null blastocysts (Fig. 1C). Thus, Cdx2 MZ null embryos morphologically phenocopied Cdx2 Z null embryos. In fact, MZ null and Z null embryos cultured side by side from 1-cell to implantation stages underwent cleavages, compaction, cavitation, expansion and collapse within the same time frame (n=11 MZ null and n=10 Z null cultured embryos; data not shown), indicating no difference in cell cycle length or morphology between Cdx2 MZ null and Z null embryos. Importantly, we observed no phenotypic differences among Cdx2del/del, Cdx2del/null and Cdx2null/null embryos, and none of these maintained an expanded blastocoel or hatched from the zona, indicating that deleted and null alleles are functionally equivalent.
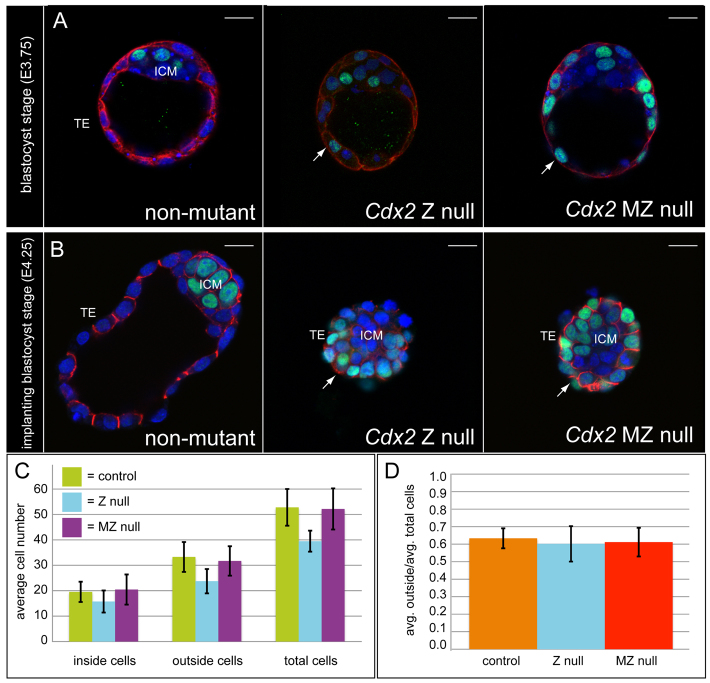
To examine the consequences of simultaneous loss of maternal and zygotic Cdx2 on a molecular level, we examined the expression of CDH1, KRT8 and NANOG in Cdx2 MZ null blastocysts. We found that these proteins were all still detectable in Cdx2 MZ null blastocysts (Fig. 2A,B). Notably, NANOG was ectopically expressed in trophectoderm cells of Cdx2 MZ null blastocysts and Z null blastocysts (Fig. 2A,B), as previously reported for Cdx2 Z null (Strumpf et al., 2005) and knockdown (Wu et al., 2010) blastocysts. In addition, KRT8 was expressed at a slightly lower level in both Cdx2 MZ null and Z null blastocysts (Fig. 2A), consistent with previous studies of Cdx2 Z null (Ralston and Rossant, 2008) and knockdown (Wu et al., 2010) blastocysts. At later time points, when the embryos had collapsed, CDH1 localization appeared identical in Cdx2 Z null and MZ null embryos (Fig. 2B), consistent with disrupted trophectoderm cell polarity in the absence of Cdx2 (Strumpf et al., 2005; Wu et al., 2010). Finally, we did not detect a significant difference in the proportion of trophectoderm cells in Cdx2 MZ null embryos compared with Cdx2 Z null or control blastocysts (Fig. 2C,D). Thus, neither trophectoderm cell polarity nor initial cell fate depends on maternal Cdx2. The findings presented here therefore show that maternal Cdx2 is not required for mouse development.
Fig. 2.
Loss of maternal Cdx2 does not worsen the Cdx2 zygotic null phenotype. (A) Expression of KRT8 (red) and NANOG (green) in confocal transverse sections of preimplantation mouse blastocysts at E3.75 (nuclei, blue). Images are representative of n=20 control (Cdx2+/− or wild type), n=6 Z null, n=5 MZ null blastocysts. In control blastocysts, KRT8 is restricted to the trophectoderm (TE) and NANOG is restricted to the inner cell mass (ICM). In Cdx2 Z null and MZ null blastocysts, KRT8 is still expressed in the TE and NANOG is ectopically expressed in the TE (arrows). (B) Expression of CDH1 (red) and NANOG (green) in implantation stage blastocysts at E4.25 (nuclei, blue). Control blastocysts are expanded and Cdx2 Z null and MZ null blastocysts are collapsed. CDH1 and NANOG are detectable in Cdx2 Z null and MZ null blastocysts and NANOG is ectopically expressed in the TE of both mutants (arrows). Representative of n=7 control, n=7 Z null, n=9 MZ null blastocysts. (C) Average numbers of inside, outside and total cells in control (Zp3-Cre/+; n=28), Cdx2 Z null (n=4) and Cdx2 MZ null (n=22) blastocysts at E3.5. Inside and outside cells were counted on the basis of morphological position in the blastocyst. (D) Data from C showing the average proportion of outside cells per embryo, indicating no difference in the proportion of TE cells for any genotype (P>0.05, t-tests). Error bars indicate s.d. Scale bars: 20 μm.
Supplementary Material
Acknowledgements
We thank Yojiro Yamanaka, Brian J. Cox, William Sullivan, David Feldheim, Michael Halbisen and Needhi Bhalla for discussion; Lane Sharon, Tiffany Medina, Eryn Wicklow for assistance with mouse breeding; and Armen Shamamian and the UCSC Transgenic Facility for assistance with oocyte collection.
Footnotes
Funding
This study was supported by UCSC Committee on Research grants to A.R.; a California Institute for Regenerative Medicine Major Facilities Grant [FA1-00617-1] to UCSC; and a National Institutes of Health (NIH) grant [5R01 CA082223] to E.F.; S.B. and T.F. were supported by the NIH [T32 GM008646]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.086025/-/DC1
References
- Beck F., Erler T., Russell A., James R. (1995). Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev. Dyn. 204, 219-227 [DOI] [PubMed] [Google Scholar]
- Bruce A. W. (2011). What is the role of maternally provided Cdx2 mRNA in early mouse embryogenesis? Reprod. Biomed. Online 22, 512-515 [DOI] [PubMed] [Google Scholar]
- Chawengsaksophak K., James R., Hammond V. E., Köntgen F., Beck F. (1997). Homeosis and intestinal tumours in Cdx2 mutant mice. Nature 386, 84-87 [DOI] [PubMed] [Google Scholar]
- Chawengsaksophak K., de Graaff W., Rossant J., Deschamps J., Beck F. (2004). Cdx2 is essential for axial elongation in mouse development. Proc. Natl. Acad. Sci. USA 101, 7641-7645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries W. N., Binns L. T., Fancher K. S., Dean J., Moore R., Kemler R., Knowles B. B. (2000). Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis 26, 110-112 [PubMed] [Google Scholar]
- Frohnhöfer H. G., Nüsslein-Volhard C. (1986). Organization of anterior pattern in the Drosophila embryo by the maternal gene bicoid. Nature 324, 120-125 [Google Scholar]
- Hiraoka M., Abe A., Lu Y., Yang K., Han X., Gross R. W., Shayman J. A. (2006). Lysosomal phospholipase A2 and phospholipidosis. Mol. Cell. Biol. 26, 6139-6148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrusik A., Bruce A. W., Tan M. H., Leong D. E., Skamagki M., Yao M., Zernicka-Goetz M. (2010). Maternally and zygotically provided Cdx2 have novel and critical roles for early development of the mouse embryo. Dev. Biol. 344, 66-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. (2011). Decisions, decisions: how are they made in the early embryo – and does it matter? Reprod. Biomed. Online 22, 509-511 [DOI] [PubMed] [Google Scholar]
- Johnson M. H. (2009). From mouse egg to mouse embryo: polarities, axes, and tissues. Annu. Rev. Cell Dev. Biol. 25, 483-512 [DOI] [PubMed] [Google Scholar]
- Lan Z. J., Xu X., Cooney A. J. (2004). Differential oocyte-specific expression of Cre recombinase activity in GDF-9-iCre, Zp3cre, and Msx2Cre transgenic mice. Biol. Reprod. 71, 1469-1474 [DOI] [PubMed] [Google Scholar]
- Li L., Zheng P., Dean J. (2010). Maternal control of early mouse development. Development 137, 859-870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzik M., Fjose A., Gehring W. J. (1985). Isolation of caudal, a Drosophila homeo box-containing gene with maternal expression, whose transcripts form a concentration gradient at the pre-blastoderm stage. EMBO J. 4, 2961-2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris S. A. (2011). Cell fate in the early mouse embryo: sorting out the influence of developmental history on lineage choice. Reprod. Biomed. Online 22, 521-524 [DOI] [PubMed] [Google Scholar]
- Ralston A., Rossant J. (2008). Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev. Biol. 313, 614-629 [DOI] [PubMed] [Google Scholar]
- Ralston A., Cox B. J., Nishioka N., Sasaki H., Chea E., Rugg-Gunn P., Guo G., Robson P., Draper J. S., Rossant J. (2010). Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 137, 395-403 [DOI] [PubMed] [Google Scholar]
- Rodríguez C. I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A. F., Dymecki S. M. (2000). High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25, 139-140 [DOI] [PubMed] [Google Scholar]
- Rossant J., Tam P. P. (2009). Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development 136, 701-713 [DOI] [PubMed] [Google Scholar]
- Strumpf D., Mao C. A., Yamanaka Y., Ralston A., Chawengsaksophak K., Beck F., Rossant J. (2005). Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093-2102 [DOI] [PubMed] [Google Scholar]
- Wang K., Sengupta S., Magnani L., Wilson C. A., Henry R. W., Knott J. G. (2010a). Brg1 is required for Cdx2-mediated repression of Oct4 expression in mouse blastocysts. PLoS ONE 5, e10622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Kou Z., Jing Z., Zhang Y., Guo X., Dong M., Wilmut I., Gao S. (2010b). Proteome of mouse oocytes at different developmental stages. Proc. Natl. Acad. Sci. USA 107, 17639-17644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Schöler H. R. (2011). Role of mouse maternal Cdx2: what's the debate all about? Reprod. Biomed. Online 22, 516-518 [DOI] [PubMed] [Google Scholar]
- Wu G., Gentile L., Fuchikami T., Sutter J., Psathaki K., Esteves T. C., Araúzo-Bravo M. J., Ortmeier C., Verberk G., Abe K., et al. (2010). Initiation of trophectoderm lineage specification in mouse embryos is independent of Cdx2. Development 137, 4159-4169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka Y., Ralston A., Stephenson R. O., Rossant J. (2006). Cell and molecular regulation of the mouse blastocyst. Dev. Dyn. 235, 2301-2314 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.