Abstract
Type 2 diabetes (T2D) can be prevented in pre-diabetic individuals with impaired glucose tolerance (IGT). Here, we have used a metabolomics approach to identify candidate biomarkers of pre-diabetes. We quantified 140 metabolites for 4297 fasting serum samples in the population-based Cooperative Health Research in the Region of Augsburg (KORA) cohort. Our study revealed significant metabolic variation in pre-diabetic individuals that are distinct from known diabetes risk indicators, such as glycosylated hemoglobin levels, fasting glucose and insulin. We identified three metabolites (glycine, lysophosphatidylcholine (LPC) (18:2) and acetylcarnitine) that had significantly altered levels in IGT individuals as compared to those with normal glucose tolerance, with P-values ranging from 2.4 × 10−4 to 2.1 × 10−13. Lower levels of glycine and LPC were found to be predictors not only for IGT but also for T2D, and were independently confirmed in the European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam cohort. Using metabolite–protein network analysis, we identified seven T2D-related genes that are associated with these three IGT-specific metabolites by multiple interactions with four enzymes. The expression levels of these enzymes correlate with changes in the metabolite concentrations linked to diabetes. Our results may help developing novel strategies to prevent T2D.
Keywords: early diagnostic biomarkers, IGT, metabolomics, prediction, T2D
Introduction
Type 2 diabetes (T2D) is defined by increased blood glucose levels due to pancreatic β-cell dysfunction and insulin resistance without evidence for specific causes, such as autoimmune destruction of pancreatic β-cells (Krebs et al, 2002; Stumvoll et al, 2005; Muoio and Newgard, 2008). A state of pre-diabetes (i.e., impaired fasting glucose (IFG) and/or impaired glucose tolerance (IGT)) with only slightly elevated blood glucose levels may precede T2D for years (McGarry, 2002; Tabak et al, 2012). The development of diabetes in pre-diabetic individuals can be prevented or delayed by dietary changes and increased physical activity (Tuomilehto et al, 2001; Knowler et al, 2002). However, no specific biomarkers that enable prevention have been reported.
Metabolomics studies allow metabolites involved in disease mechanisms to be discovered by monitoring metabolite level changes in predisposed individuals compared with healthy ones (Shaham et al, 2008; Newgard et al, 2009; Zhao et al, 2010; Pietilainen et al, 2011; Rhee et al, 2011; Wang et al, 2011; Cheng et al, 2012; Goek et al, 2012). Altered metabolite levels may serve as diagnostic biomarkers and enable preventive action. Previous cross-sectional metabolomics studies of T2D were either based on small sample sizes (Shaham et al, 2008; Wopereis et al, 2009; Zhao et al, 2010; Pietilainen et al, 2011) or did not consider the influence of common risk factors of T2D (Newgard et al, 2009). Recently, based on prospective nested case–control studies with relative large samples (Rhee et al, 2011; Wang et al, 2011), five branched-chain and aromatic amino acids were identified as predictors of T2D (Wang et al, 2011). Here, using various comprehensive large-scale approaches, we measured metabolite concentration profiles (Yu et al, 2012) in the population-based (Holle et al, 2005; Wichmann et al, 2005) Cooperative Health Research in the Region of Augsburg (KORA) baseline (survey 4 (S4)) and follow-up (F4) studies (Rathmann et al, 2009; Meisinger et al, 2010; Jourdan et al, 2012). The results of these cross-sectional and prospective studies allowed us to (i) reliably identify candidate biomarkers of pre-diabetes and (ii) build metabolite–protein networks to understand diabetes-related metabolic pathways.
Results
Study participants
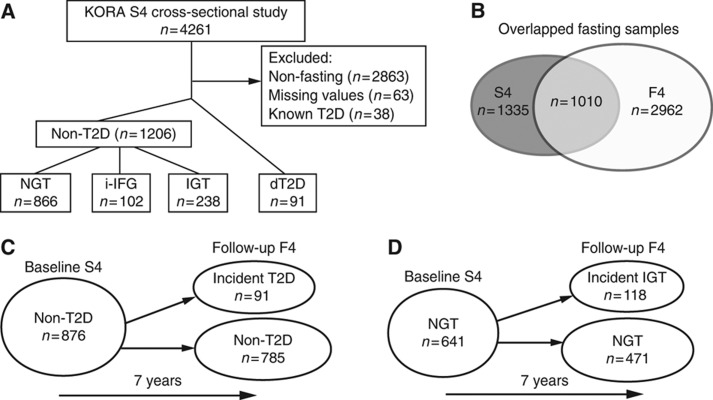
Individuals with known T2D were identified by physician-validated self-reporting (Rathmann et al, 2010) and excluded from our analysis, to avoid potential influence from anti-diabetic medication with non-fasting participants and individuals with missing values (Figure 1A). Based on both fasting and 2-h glucose values (i.e., 2 h post oral 75 g glucose load), individuals were defined according to the WHO diagnostic criteria to have normal glucose tolerance (NGT), isolated IFG (i-IFG), IGT or newly diagnosed T2D (dT2D) (WHO, 1999; Rathmann et al, 2009; Meisinger et al, 2010; Supplementary Table S1). The sample sets include 91 dT2D patients and 1206 individuals with non-T2D, including 866 participants with NGT, 102 with i-IFG and 238 with IGT, in the cross-sectional KORA S4 (Figure 1A; study characteristics are shown in Table I). Of the 1010 individuals in a fasting state who participated at baseline and follow-up surveys (Figure 1B, study characteristics of the KORA F4 survey are shown in Supplementary Table S2), 876 of them were non-diabetic at baseline. Out of these, about 10% developed T2D (i.e., 91 incident T2D) (Figure 1C). From the 641 individuals with NGT at baseline, 18% developed IGT (i.e., 118 incident IGT) 7 years later (Figure 1D). The study characteristics of the prospective KORA S4→F4 are shown in Table II.
Figure 1.
Population description. Metabolomics screens in the KORA cohort, at baseline S4 (A), overlapped between S4 and F4 (B) and prospective (C, D). Participant numbers are shown. Normal glucose tolerance (NGT), isolated impaired fasting glucose (i-IFG), impaired glucose tolerance (IGT), type 2 diabetes mellitus (T2D) and newly diagnosed T2D (dT2D). Non-T2D individuals include NGT, i-IFG and IGT participants.
Table 1. Characteristics of the KORA S4 cross-sectional study sample.
| Clinical and laboratory parameters | NGT | i-IFG | IGT | dT2D |
|---|---|---|---|---|
| N | 866 | 102 | 238 | 91 |
| Age (years) | 63.5±5.5 | 64.1±5.2 | 65.2±5.2 | 65.9±5.4 |
| Sex (female) (%) | 52.2 | 30.4 | 44.9 | 41.8 |
| BMI (kg/m2) | 27.7±4.1 | 29.2±4 | 29.6±4.1 | 30.2±3.9 |
| Physical activity (% >1 h per week) | 46.7 | 35.3 | 39.9 | 36.3 |
| Alcohol intakea (%) | 20.2 | 20.5 | 25.2 | 24.2 |
| Current smoker (%) | 14.8 | 10.8 | 10.9 | 23.1 |
| Systolic BP (mm Hg) | 131.7±18.9 | 138.9±17.9 | 140.7±19.8 | 146.8±21.5 |
| HDL cholesterol (mg/dl) | 60.5±16.4 | 55.7±15.9 | 55.7±15.1 | 50.0±15.8 |
| LDL cholesterol (mg/dl) | 154.5±39.8 | 152.1±37.7 | 155.2±38.6 | 146.1±44.6 |
| Triglycerides (mg/dl) | 120.7±68.3 | 145.0±96.0 | 146.6±80.0 | 170.6±107.1 |
| HbA1c (%) | 5.56±0.33 | 5.62±0.33 | 5.66±0.39 | 6.21±0.83 |
| Fasting glucose (mg/dl) | 95.6±7.1 | 114.2±3.7 | 104.5±9.7 | 133.2±31.7 |
| 2-h Glucose (mg/dl) | 102.1±21.0 | 109.3±18.7 | 163.4±16.4 | 232.1±63.7 |
| Fasting insulin (μU/ml) | 10.48±7.28 | 16.26±9.67 | 13.92±9.53 | 17.70±12.61 |
NGT, normal glucose tolerance; i-IFG, isolated impaired fasting glucose; IGT, impaired glucose tolerance; dT2D, newly diagnosed type 2 diabetes; BP, blood pressure; HDL, high-density lipoprotein; LDL, low-density lipoprotein.
Percentages of individuals or means±s.d. are given for each variable and each group (NGT, i-IFG, IGT and dT2D).
a⩾20 g/day for women; ⩾40 g/day for men.
Table 2. Characteristics of the KORA S4→F4 prospective study samples.
| NGT at baseline (n=589) |
Non-T2D at baseline (n=876) |
|||
|---|---|---|---|---|
| Remained NGT at follow-up | Developed IGT at follow-up | Remained Non-T2D at follow-up | Developed T2D at follow-up | |
| N | 471 | 118 | 785 | 91 |
| Age (years) | 62.4±5.4 | 63.9±5.5 | 62.9±5.4 | 65.5±5.2 |
| Sex (female) (%) | 52.2 | 55.9 | 50.8 | 34.1 |
| BMI (kg/m2) | 27.2±3.8 | 28.2±3.9 | 27.9±4 | 30.2±3.6 |
| Physical activity (% >1 h per week) | 52.9 | 43.2 | 52.2 | 58.2 |
| Alcohol intakea (%) | 19.9 | 20.3 | 20.6 | 19.8 |
| Smoker (%) | 14.6 | 9.3 | 12.0 | 14.3 |
| Systolic BP (mm Hg) | 129.6±18.2 | 134.2±18.7 | 132.4±18.6 | 137.8±19 |
| HDL cholesterol (mg/dl) | 61.3±16.8 | 58.9±16.2 | 60.0±16.5 | 51.9±12.4 |
| LDL cholesterol (mg/dl) | 153.9±38.4 | 156.9±42.7 | 154.5±39.5 | 157.7±41.6 |
| Triglycerides (mg/dl) | 118.1±63.9 | 129.5±79.0 | 125.0±70.0 | 151.2±74.2 |
| HbA1c (%) | 5.54±0.33 | 5.59±0.34 | 5.6±0.3 | 5.8±0.4 |
| Fasting glucose (mg/dl) | 94.7±6.9 | 96.6±7.1 | 97.7±8.8 | 106.1±10.1 |
| 2-h Glucose (mg/dl) | 98.2±20.5 | 109.9±16.8 | 109.3±28 | 145.9±32.3 |
| Fasting insulin (μU/ml) | 9.91±6.48 | 11.79±8.83 | 11.0±7.6 | 16.2±9.6 |
BP, blood pressure; HDL, high-density lipoprotein; LDL, low-density lipoprotein.
Percentages of individuals or means±s.d. are given for each variable and each group.
a⩾20 g/day for women; ⩾40 g/day for men.
Analyses strategies
We first screened for significantly differed metabolites concentration among four groups (dT2D, IGT, i-IFG and NGT) for 140 metabolites with cross-sectional studies in KORA S4, and for 131 metabolites in KORA F4. Three IGT-specific metabolites were identified and further investigated in the prospective KORA S4→F4 cohort, to examine whether the baseline metabolite concentrations can predict incident IGT and T2D, and whether they are associated with glucose tolerance 7 years later. Our results are based on a prospective population-based cohort, which differed from previous nested case–control study (Wang et al, 2011). We also performed analysis with same study design using our data. The obtained results provided clues to explain the differences between the two sets of biomarkers. The three metabolites were also replicated in an independent European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam cohort. Finally, the relevance of the identified metabolites was further investigated with our bioinformatical analysis of protein-metabolite interaction networks and gene expression data.
Identification of novel pre-diabetes metabolites distinct from known T2D risk indicators
To identify metabolites with altered concentrations between the individuals with NGT, i-IFG, IGT and dT2D, we first examined five pairwise comparisons (i-IFG, IGT and dT2D versus NGT, as well as dT2D versus either i-IFG or IGT) in the cross-sectional KORA S4. Based on multivariate logistic regression analysis, 26 metabolite concentrations differed significantly (P-values<3.6 × 10−4) between two groups in at least one of the five comparisons (Figure 2A; odds ratios (ORs) and P-values are shown in Table III). These associations were independent of age, sex, body mass index (BMI), physical activity, alcohol intake, smoking, systolic blood pressure (BP) and HDL cholesterol (model 1). As expected, the level of total hexose H1, which is mainly represented by glucose (Pearson’s correlation coefficient value r between H1 and fasting glucose reached 0.85; Supplementary Table S3), was significantly different in all five comparisons. The significantly changed metabolite panel differed from NGT to i-IFG or to IGT. Most of the significantly altered metabolite concentrations were found between individuals with dT2D and IGT as compared with NGT (Supplementary Table S4A).
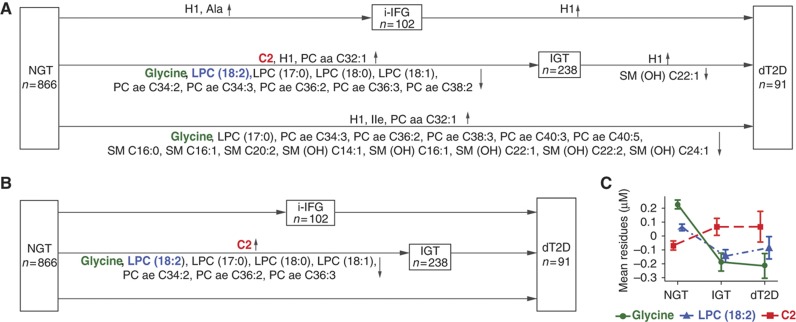
Figure 2.
Differences in metabolite concentrations from cross-sectional analysis of KORA S4. Plots (A, B) show the names of metabolites with significantly different concentrations in multivariate logistic regression analyses (after the Bonferroni correction for multiple testing with P<3.6 × 10−4) in the five pairwise comparisons of model 1 and model 2. Plot (C) shows the average residues of the concentrations with standard errors of the three metabolites (glycine, LPC (18:2) and acetylcarnitine C2) for the NGT, IGT and dT2D groups. Plot (A) shows the results with adjustment for model 1 (age, sex, BMI, physical activity, alcohol intake, smoking, systolic BP and HDL cholesterol), whereas plots (B, C) have additional adjustments for HbA1c, fasting glucose and fasting insulin (model 2). Residuals were calculated from linear regression model (formula: T2D status∼metabolite concentration+model 2). For further information, see Supplementary Table S4.
Table 3. Odds ratios (ORs) and P-values in five pairwise comparisons with two adjusted models in the KORA S4.
| Metabolite | Model 1 |
Model 2 |
||
| OR (95% CI), per s.d. | P-value | OR (95% CI), per s.d. | P-value | |
| 238 IGT versus 866 NGT | ||||
| Glycine | 0.65 (0.53–0.78) | 5.6E-06 | 0.67 (0.54–0.81) | 8.6E-05 |
| LPC (18:2) | 0.58 (0.47–0.7) | 1.3E-07 | 0.58 (0.46–0.72) | 2.1E-06 |
| C2 | 1.37 (1.18–1.59) | 3.8E-05 | 1.38 (1.16–1.64) | 2.4E-04 |
| 91 dT2D versus 866 NGT | ||||
| Glycine | 0.47 (0.33–0.65) | 1.1E-05 | 0.44 (0.22–0.83) | 1.6E-02 |
| LPC (18:2) | 0.62 (0.44–0.85) | 4.1E-03 | 0.61 (0.32–1.07) | 1.1E-01 |
| C2 | 1.17 (0.94–1.45) | 1.5E-01 | 1.71 (1.14–2.52) | 6.8E-03 |
| 91 dT2D versus 234 IGT | ||||
| Glycine | 0.81 (0.61–1.07) | 1.5E-01 | 0.76 (0.51–1.1) | 1.6E-01 |
| LPC (18:2) | 0.91 (0.69–1.19) | 4.8E-01 | 0.84 (0.57–1.22) | 3.7E-01 |
| C2 | 0.93 (0.71–1.2) | 5.9E-01 | 1.27 (0.87–1.86) | 2.2E-01 |
| 102 i-IFG versus 866 NGT | ||||
| Glycine | 0.75 (0.57–0.98) | 3.9E-02 | 0.62a | 1.0E+00 |
| LPC (18:2) | 0.99 (0.77–1.26) | 9.6E-01 | 0.79a | 1.0E+00 |
| C2 | 1.2 (0.99–1.46) | 5.9E-02 | 0.18a | 1.0E+00 |
| 91 dT2D versus 102 i-IFG | ||||
| Glycine | 0.62 (0.43–0.87) | 7.8E-03 | 0.62 (0.4–0.93) | 2.5E-02 |
| LPC (18:2) | 0.62 (0.43–0.89) | 1.1E-02 | 0.54 (0.33–0.84) | 8.9E-03 |
| C2 | 0.92 (0.66–1.27) | 6.2E-01 | 1.23 (0.82–1.85) | 3.1E-01 |
ORs were calculated with multivariate logistic regression analysis with adjustment for age, sex, BMI, physical activity, alcohol intake, smoking, systolic BP and HDL cholesterol in model 1; model 2 includes those variable in model 1 plus HbA1c, fasting glucose and fasting insulin. CI denotes confidence interval.
aFasting glucose values were added as co-variants to the model 2, resulting in a perfect separation between i-IFG and NGT.
To investigate whether HbA1c, fasting glucose and fasting insulin levels mediate the shown associations, these were added as covariates to the regression analysis (model 2) in addition to model 1 (Figure 2B). We observed that, under these conditions, no metabolite differed significantly when comparing individuals with dT2D to those with NGT, suggesting that these metabolites are associated with HbA1c, fasting glucose and fasting insulin levels (r values are shown in Supplementary Table S3). Only nine metabolite concentrations significantly differed between IGT and NGT individuals (Table III; Supplementary Table S4B). These metabolites therefore represent novel biomarker candidates, and are independent from the known risk indicators for T2D. The logistic regression analysis was based on each single metabolite, and some of these metabolites are expected to correlate with each other. To further assess the metabolites as a group, we employed two additional statistical methods (the non-parametric random forest and the parametric stepwise selection) to identify unique and independent biomarker candidates. Out of the nine metabolites, five molecules (i.e., glycine, LPC (18:2), LPC (17:0), LPC (18:1) and C2) were select after random forest, and LPC (17:0) and LPC (18:1) were then removed after the stepwise selection. Thus, three molecules were found to contain independent information: glycine (adjusted OR=0.67 (0.54–0.81), P=8.6 × 10−5), LPC (18:2) (OR=0.58 (0.46–0.72), P=2.1 × 10−6) and acetylcarnitine C2 (OR=1.38 (1.16–1.64), P=2.4 × 10−4) (Figure 2C). Similar results were observed in the follow-up KORA F4 study (Supplementary Figure S1). For instance, when 380 IGT individuals were compared with 2134 NGT participants, these three metabolites were also found to be highly significantly different (glycine, OR=0.64 (0.55–0.75), P=9.3 × 10−8; LPC (18:2), OR=0.47 (0.38–0.57), P=2.1 × 10−13; and C2, OR=1.33 (1.17–1.49), P=4.9 × 10−6) (Supplementary Table S5).
Predict risks of IGT and T2D
To investigate the predictive value for IGT and T2D of the three identified metabolites, we examined the associations between baseline metabolite concentrations and incident IGT and T2D using the prospective KORA S4→F4 cohort (Table II). We compared baseline metabolite concentrations in 118 incident IGT individuals with 471 NGT control individuals. We found that glycine and LPC (18:2), but not C2, were significantly different at the 5% level in both adjusted model 1 and model 2 (Table IV; Supplementary Table S6). Significant differences were additionally observed for glycine and LPC (18:2), but not for C2, at baseline concentrations between the 91 incident T2D individuals and 785 participants who remained diabetes free (non-T2D). Each standard deviation (s.d.) increment of the combinations of the three metabolites was associated with a 33% decreased risk of future diabetes (OR=0.39 (0.21–0.71), P=0.0002). Individuals in the fourth quartile of the combined metabolite concentrations had a three-fold lower chance of developing diabetes (OR=0.33 (0.21–0.52), P=1.8 × 10−5), compared with those whose serum levels were in the first quartile (i.e., combination of glycine, LPC (18:2) and C2), indicating a protective effect from higher concentrations of glycine and LPC (18:2) combined with a lower concentration of C2. With the full adjusted model 2, consistent results were obtained for LPC (18:2) but not for glycine (Supplementary Table S6). When the three metabolites were added to the fully adjusted model 2, the area under the receiver-operating-characteristic curves (AUC) increased 2.6% (P=0.015) and 1% (P=0.058) for IGT and T2D, respectively (Supplementary Figure S2; Supplementary Table S7). Thus, this provides an improved prediction of IGT and T2D as compared with T2D risk indicators.
Table 4. Prediction of IGT and T2D in the KORA cohort.
| Model | Glycine | LPC (18:2) | C2 | Glycine, LPC (18:2), C2 |
|---|---|---|---|---|
| (A) Metabolite as continuous variable (n=589) | ||||
| Per s.d. | 0.75 (0.58–0.95) | 0.72 (0.54–0.93) | 0.92 (0.73–1.14) | 0.36 (0.20–0.67) |
| P | 0.02 | 0.02 | 0.50 | 0.001 |
| (B) Metabolite as categorical variable (n=589) | ||||
| First quartile | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) |
| Second quartile | 1.0 (0.80–1.46) | 0.96 (0.73–1.27) | 0.89 (0.66–1.23) | 0.54 (0.30–0.97) |
| Third quartile | 1.0 (0.74–1.34) | 0.71 (0.51–0.99) | 0.93 (0.69–1.26) | 0.66 (0.37–1.18) |
| Fourth quartile | 0.78 (0.55–1.06) | 0.78 (0.54–1.12) | 0.99 (0.73–1.35) | 0.36 (0.19–0.69) |
| P for trend | 0.06 | 0.05 | 0.79 | 0.0082 |
| (C) Metabolite as continuous variable (n=876) | ||||
| Per s.d. | 0.73 (0.55–0.97) | 0.70 (0.51–0.94) | 0.94 (0.74–1.18) | 0.39 (0.21–0.71) |
| P | 0.04 | 0.02 | 0.59 | 0.0002 |
| (D) Metabolite as categorical variable (n=876) | ||||
| 1st quartile | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) |
| 2nd quartile | 0.87 (0.71–1.07) | 0.95 (0.77–1.17) | 1.05 (0.85–1.31) | 0.50 (0.33–0.76) |
| 3rd quartile | 0.82 (0.67–1.01) | 0.70 (0.56–0.88) | 0.97 (0.78–1.19) | 0.57 (0.38–0.88) |
| 4th quartile | 0.67 (0.54–0.84) | 0.68 (0.54–0.88) | 1.21 (0.98–1.50) | 0.33 (0.21–0.52) |
| P for trend | 0.00061 | 0.00021 | 0.19 | 1.8E−05 |
| (E) Linear regression (n=843) | ||||
| β Estimatesa (95% CI) | −2.47 (−4.64, −0.29) | −4.57 (−6.90, −2.24) | 1.02 (−1.11, 3.15) | −4.23 (−6.52, −2.31) |
| P | 0.026 | 0.00013 | 0.59 | 8.8E−05 |
Odds ratios (ORs, 95% confidence intervals) and P-values of multivariate logistic regression results are shown in (A) and (B) for IGT and in (C) and (D) for T2D, respectively, whereas β estimates and P-values from linear regression analysis between metabolite concentration in baseline KORA S4 and 2-h glucose values in follow-up KORA F4 are shown in (E). All models were adjusted for age, sex, BMI, physical activity, alcohol intake, smoking, systolic BP and HDL cholesterol.
aβ Estimate indicates the future difference in the glucose tolerance corresponding to the one s.d. differences in the normalized baseline metabolite concentration.
Baseline metabolite concentrations correlate with future glucose tolerance
We next investigated the associations between baseline metabolite concentrations and follow-up 2-h glucose values after an oral glucose tolerance test. Consistent results were observed for the three metabolites: glycine and LPC (18:2), but not acetylcarnitine C2 levels, were found to be significantly associated, indicating that glycine and LPC (18:2) predict glucose tolerance. Moreover, the three metabolites (glycine, LPC (18:2) and C2) revealed high significance even in the fully adjusted model 2 in the cross-sectional KORA S4 cohort (Supplementary Table S8). As expected, a very significant association (P=1.5 × 10−22) was observed for hexose H1 in model 1, while no significance (P=0.12) was observed for it in the fully adjusted model 2 (Supplementary Table S8).
Prospective population-based versus nested case–control designs
To investigate the predict value of the five branched-chain and aromatic amino acids (isoleucine, leucine, valine, tyrosine and phenylalanine) (Wang et al, 2011) in our study, we correlated the baseline metabolite concentrations with follow-up 2-h glucose values. We found none of them to be associated significantly, indicating that the five amino acids cannot predict risk of IGT (β estimates and P-values are shown in Supplementary Table S9). Furthermore, none of these five amino acids showed associations with 2-h glucose values in the cross-sectional KORA S4 study (Supplementary Table S8).
To replicate the identified five branched-chain and aromatic amino acids (Wang et al, 2011), we matched our baseline samples to the 91 incident T2D using the same method described previously (Wang et al, 2011). We replicated four out of the five branched-chain and aromatic amino acids (characteristics of the case–control and non-T2D samples are shown in Supplementary Table S10; ORs and P-values are given in Supplementary Table S11). As expected, the three identified IGT-specific metabolites did not significantly differ between the matched case control samples, because the selected controls were enriched with individuals accompanied by high-risk features such as obesity and elevated fasting glucose as described by Wang et al (2011). In fact, the 91 matched controls include about 50% pre-diabetes individuals, which is significantly higher than the general population (about 15%).
Replication in the cross-sectional EPIC-Potsdam cohort
Metabolomics data from serum samples of a randomly drawn EPIC-Potsdam subcohort (n=2500) were used for replication. Glycine (OR=0.60 (0.47–0.77), P=7.4 × 10−5) and LPC (18:2) (OR=0.79 (0.63–0.98), P=0.037) were replicated when 133 T2D patients were compared with 1253 individuals with NGT at baseline (Supplementary Table S12). However, acetylcarnitine C2 (OR=0.98 (0.81–1.19), P=0.858) could not be replicated when T2D patients were compared with NGT individuals, since the IGT participants were not available in the data set. The absolute levels of these three metabolites were in a similar range, with only slight differences that were due probably to the differences of the two cohorts or to potential batch effects of metabolomics measurements (Supplementary Tables S12 and S15). Thus, these data therefore provide an independent validation of the metabolomics study.
Metabolite–protein interaction networks confirmed by transcription levels
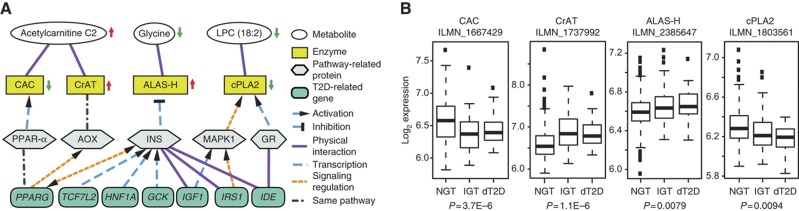
To investigate the underlying molecular mechanism for the three identified IGT metabolites, we studied their associations with T2D-related genes by analyzing protein-metabolite interaction networks (Wishart et al, 2009; Szklarczyk et al, 2011). In all, 7 out of the 46 known T2D-related genes (PPARG, TCF7L2, HNF1A, GCK, IGF1, IRS1 and IDE) were linked to these metabolites through related enzymes or proteins (Figure 3A; the list of 46 genes is shown in Supplementary Table S13). To validate the networks, the links between metabolites, enzymes, pathway-related proteins and T2D-related genes were manually checked for biochemical relevance and classified into four groups: signaling regulation, transcription, physical interaction and the same pathway (Supplementary Table S14).
Figure 3.
Three candidate metabolites for IGT associated with seven T2D-related genes. (A) Metabolites (white), enzymes (yellow), pathway-related proteins (gray) and T2D-related genes (blue) are represented with ellipses, rectangles, polygons and rounded rectangles, respectively. Arrows next to the ellipses and rectangles indicate altered metabolite concentrations in persons with IGT as compared with NGT, and enzyme activities in individuals with IGT. The 21 connections between metabolites, enzymes, pathway-related proteins and T2D-related genes were divided after visual inspections into four categories: physical interaction (purple solid line), transcription (blue dash line), signaling regulation (orange dash line) and same pathway (gray dot and dash line). The activation or inhibition is indicated. For further information, see Supplementary Table S12. (B) Log-transformed gene expression results of the probes of CAC, CrAT, ALAS-H and cPLA2 in 383 individuals with NGT, 104 with IGT and 26 patients with dT2D are shown from cross-sectional analysis of the KORA S4 survey. The P-values were adjusted for sex, age, BMI, physical activity, alcohol intake, smoking, systolic BP, HDL cholesterol, HbA1c and fasting glucose when IGT individuals were compared with NGT participants.
Gene expression analysis in whole-blood samples of participants from the KORA S4 revealed significant variations (P-values ranging from 9.4 × 10−3 to 1.1 × 10−6) of transcript levels of four enzymes, namely, carnitine/acylcarnitine translocase (CAC), carnitine acetyltransferase (CrAT), 5-aminolevulinate synthase 1 (ALAS-H) and cytosolic phospholipase A2 (cPLA2), which are known to be strongly associated with the levels of the three metabolites (Figure 3B). The clear relationship between changes in metabolites and transcription levels of associated enzymes strongly suggests that these metabolites are functionally associated with T2D genes in established pathways.
Discussion
Using a cross-sectional approach (KORA S4, F4), we analyzed 140 metabolites and identified three (glycine, LPC (18:2) and C2) which are IGT-specific metabolites with high statistical significance. Notably, these three metabolites are distinct from the currently known T2D risk indicators (e.g., age, BMI, systolic BP, HDL cholesterol, HbA1c, fasting glucose and fasting insulin). A prospective analysis (KORA S4→F4) shows that low levels of glycine and LPC at baseline predict the risks of developing IGT and/or T2D. Glycine and LPC especially were shown to be strong predictors of glucose tolerance, even 7 years before disease onset. Moreover, those two metabolites were independently replicated in the EPIC-Potsdam cross-sectional study. Finally, based on our analysis of interaction networks, and supported by gene expression profiles, we found that seven T2D-related genes are functionally associated with the three IGT candidate metabolites.
Different study designs reveal progression of IGT and T2D
From a methodological point of view, our study is unique with respect to the large sample sizes and the availability of metabolomics data from two time points. This allowed us to compare results generated with cross-sectional and prospective approaches directly, as well as with results from prospective population-based cohort and nested case–control designs. We found that individuals with IGT have elevated concentrations of the acetylcarnitine C2 as compared with NGT individuals only in the cross-sectional study, whereas C2 was unable to predict IGT and T2D 7 years before the disease onset. We speculate that the acetylcarnitine C2 might be an event with a quick effect.
Our analysis could replicate four out of the five branched-chain and aromatic amino acids recently reported to be predictors of T2D using nested/selected case–control samples (Wang et al, 2011). However, the population-based prospective study employed in our study revealed that these five amino acids are in fact not associated with future 2-h glucose values. It should be taken into account, however, that more pre-diabetes individuals (∼50%) were in the control group of that study design, and that these markers were unable to be extended to the general population (with only 0.4% improvement from the T2D risk indicators as reported in the Framingham Offspring Study) (Wang et al, 2011). Most likely, changes in these amino acids happen at a later stage in the development of T2D (e.g., from IGT to T2D); indeed, similar phenomenon was also observed in our study (Supplementary Figure S1D). In contrast, we found that combined glycine, LPC (18:2) and C2 have 2.6 and 1% increment in predicting IGT and T2D in addition to the common risk indicators of T2D. This suggests they are better candidate for early biomarkers, and specifically from NGT to IGT, than the five amino acids.
IFG and IGT should be considered as two different phenotypes
By definition (WHO, 1999; ADA, 2010), individuals with IFG or IGT or both are considered as pre-diabetics. Yet we observed different behaviors regarding the change of the metabolite panel from NGT to i-IFG or to IGT, indicating that i-IFG and IGT are two different phenotypes. For future studies, we therefore suggest separating IFG from IGT.
Glycine
The observed decrease in the serum concentration of glycine in individuals with IGT and dT2D may result from insulin resistance (Pontiroli et al, 2004). It was already reported that insulin represses ALAS-H expression (Phillips and Kushner, 2005). As insulin sensitivity progressively decreases during diabetes development (McGarry, 2002; Stumvoll et al, 2005; Faerch et al, 2009; Tabak et al, 2009), it is expected that the expression levels of the enzyme increase in individuals with IGT and dT2D, since ALAS-H catalyzes the condensation of glycine and succinyl-CoA into 5-aminolevulinic acid (Bishop, 1990). This may explain our observation that glycine was lower in both individuals with IGT and those with dT2D. However, the level of fasting insulin in IGT and T2D individuals was higher than in NGT participants in the KORA S4 study, suggesting that yet undetected pathways may also play roles here.
Acetylcarnitine C2
Acetylcarnitine is produced by the mitochondrial matrix enzyme, CrAT, from carnitine and acetyl-CoA, a molecule that is a product of both fatty acid β-oxidation and glucose oxidation and can be used by the citric acid cycle for energy generation. We observed higher transcriptional level of CrAT in individuals with IGT and T2D, most probably due to an activation of the peroxisome proliferator activated receptor alpha (PPAR-α) pathway in peroxisomes (Horie et al, 1981). Higher expression of CrAT would explain the elevated levels of acetylcarnitine C2 in IGT individuals. Although it is not clear if mitochondrial CrAT is overexpressed when there is increased fatty acid β-oxidation (e.g., in diabetes; Noland et al, 2009), it is expected that additional acetylcarnitine will be formed by CrAT due to increased substrate availability (acetyl-CoA), thereby releasing pyruvate dehydrogenase inhibition by acetyl-CoA and stimulating glucose uptake and oxidation. An increase of acylcarnitines, and in particular of acetylcarnitine C2, is a hallmark in diabetic people (Adams et al, 2009). Cellular lipid levels are increased in humans with IGT or overt T2D who also may have altered mitochondrial function (Morino et al, 2005; Szendroedi et al, 2007). Together, these findings reflect an important role of increased cellular lipid metabolites and impaired mitochondrial β-oxidation in the development of insulin resistance (McGarry, 2002; Szendroedi et al, 2007; Koves et al, 2008).
LPC (18:2)
In our study, individuals with IGT and dT2D had lower cPLA2 transcription levels, suggesting reduced cPLA2 activity. As a result, a concomitant decrease in the concentration of arachidonic acid (AA), a product of cPLA2 activity, is expected. AA has been shown to inhibit glucose uptake by adipocytes (Malipa et al, 2008) in a mechanism that is probably insulin independent and that involves the GLUT-1 transporter. Therefore, our findings may point to regulatory effects in individuals with IGT, since the inhibition of AA production would result in an increased glucose uptake.
Limitations
While our metabolite profiles provide a snapshot of human metabolism, more detailed metabolic profile follow-ups, with longer time spans and more time points, are necessary to further evaluate the development of the novel biomarkers. Moreover, the influence from long-term dietary habits should not be ignored, even though we used only serum from fasting individuals (Altmaier et al, 2011; Primrose et al, 2011). Furthermore, additional tissue samples (e.g., muscle and adipocytes) and experimental approaches are needed to characterize the causal pathways in detail.
Conclusions
Three novel metabolites, glycine, LPC (18:2) and C2, were identified as pre-diabetes-specific markers. Their changes might precede other branched-chain and aromatic amino acids markers in the progression of T2D. Combined levels of glycine, LPC (18:2) and C2 can predict risk not only for IGT but also for T2D. Targeting the pathways that involve these newly proposed potential biomarkers would help to take preventive steps against T2D at an earlier stage.
Materials and methods
Ethics statement
Written informed consent was obtained from each KORA and EPIC-Potsdam participant. The KORA and EPIC-Potsdam studies were approved by the ethics committee of the Bavarian Medical Association and the Medical Society of the State of Brandenburg, respectively.
Sample source and classification
The KORA surveys are population-based studies conducted in the city of Augsburg and the surrounding towns and villages (Holle et al, 2005; Wichmann et al, 2005). KORA is a research platform in the field of epidemiology, health economics and health-care research. Four surveys were conducted with 18 079 participants recruited from 1984 to 2001. The S4 consists of 4261 individuals (aged 25–74 years) examined from 1999 to 2001. From 2006 to 2008, 3080 participants (with an age range of 32–81) took part in an F4 survey. Ascertainments of anthropometric measurements and personal interviews, as well as laboratory measurements of persons, from the KORA S4/F4 have been described elsewhere (Rathmann et al, 2009; Meisinger et al, 2010; Jourdan et al, 2012).
Sampling
In the KORA cohort, blood was drawn into S-Monovette® serum tubes (SARSTEDT AG & Co., Nümbrecht, Germany) in the morning between 0800 and 1030, h after at least 8 h of fasting. Tubes were gently inverted twice, followed by 30 min resting at room temperature, to obtain complete coagulation. For serum collection, blood was centrifuged at 2750 g at 15°C for 10 min. Serum was filled into synthetic straws, which were stored in liquid nitrogen until the metabolic analyses were conducted.
Metabolite measurements and exclusion of metabolites
For the KORA S4 survey, the targeted metabolomics approach was based on measurements with the AbsoluteIDQ™ p180 kit (BIOCRATES Life Sciences AG, Innsbruck, Austria). This method allows simultaneous quantification of 188 metabolites using liquid chromatography and flow injection analysis–mass spectrometry. The assay procedures have been described previously in detail (Illig et al, 2010; Römisch-Margl et al, 2012). For each kit plate, five references (human plasma pooled material, Seralab) and three zero samples (PBS) were measured in addition to the KORA samples. To ensure data quality, each metabolite had to meet two criteria: (1) the coefficient of variance (CV) for the metabolite in the total 110 reference samples had to be smaller than 25%. In total, seven outliers were removed because their concentrations were larger than the mean plus 5 × s.d.; (2) 50% of all measured sample concentrations for the metabolite should be above the limit of detection (LOD), which is defined as 3 × median of the three zero samples. In total, 140 metabolites passed the quality controls (Supplementary Table S15): one hexose (H1), 21 acylcarnitines, 21 amino acids, 8 biogenic amines, 13 sphingomyelins (SMs), 33 diacyl (aa) phosphatidylcholines (PCs), 35 acyl-alkyl (ae) PCs and 8 lysoPCs. Concentrations of all analyzed metabolites are reported in μM.
Measurements of the 3080 KORA F4 samples and the involved cleaning procedure have already been described in detail (Mittelstrass et al, 2011; Yu et al, 2012).
Gene expression analysis
Peripheral blood was drawn under fasting conditions from 599 KORA S4 individuals at the same time as the serum samples used for metabolic profiling were prepared. Blood samples were collected directly in PAXgene (TM) Blood RNA tubes (PreAnalytiX). The RNA extraction was performed using the PAXgene Blood miRNA kit (PreAnalytiX). Purity and integrity of RNA was assessed on the Bioanalyzer (Agilent) with the 6000 Nano LabChip reagent set (Agilent). In all, 500 ng of RNA was reverse-transcribed into cRNA and biotin-UTP labeled, using the Illumina TotalPrep-96 RNA Amplification Kit (Ambion). In all, 3000, ng of cRNA was hybridized to the Illumina HumanHT-12 v3 Expression BeadChip. Chips were washed, detected and scanned according to manufacturer’s instructions. Raw data were exported from the Illumina ‘GenomeStudio’ Software to R. The data were converted into logarithmic scores and normalized using the quantile method (Bolstad et al, 2003). The sample sets comprised 383 individuals with NGT, 104 with IGT and 26 with dT2D. The known T2D individuals were removed as had been done for the metabolomics analysis.
Data availability
Metabolite concentrations of Glycine, LPC (18:2) and C2 with T2D status in the KORA S4 and F4 are provided (Supplementary Table S16). Additional data from the KORA S4 and F4 studies, including the metabolite concentrations and the gene expression with clinical phenotypes used in this study, are available upon request from KORA-gen (http://epi.helmholtz-muenchen.de/kora-gen). Requests should be sent to kora-gen@helmholtz-muenchen.de and are subject to approval by the KORA board to ensure that appropriate conditions are met to preserve patient privacy. Formal collaboration and co-authorship with members of the KORA study is not an automatic condition to obtain access to the data published in the present paper. More general information about KORA, including S4 and F4 study design and clinical variables, can be found at http://epi.helmholtz-muenchen.de/kora-gen/seiten/variablen_e.php and http:/helmholtz-muenchen.de/en/kora-en/information-for-scientists/current-kora-studies.
Statistical analysis
Calculations were performed under the R statistical environment (http://www.r-project.org/).
Multivariate logistic regression and linear regression
In multivariate logistic regression analysis, ORs for single metabolites were calculated between two groups. The concentration of each metabolite was scaled to have a mean of zero and an s.d. of one; thus, all reported OR values correspond to the change per s.d. of metabolite concentration. Various T2D risk factors were added to the logistic regression analysis as covariates. To handle false discovery rates from multiple comparisons, the cutoff point for significance was calculated according to the Bonferroni correction, at a level of 3.6 × 10−4 (for a total use of 140 metabolites at the 5% level). Because the metabolites were correlated within well-defined biological groups (e.g., 8 lysoPCs, 33 diacyl PCs, 35 acyl-alkyl PCs and 13 SMs), this correction was conservative.
Additionally, the categorized metabolite concentrations and combined scores (see below) were analyzed, and the ORs were calculated across quartiles. To test the trend across quartiles, we assigned all individuals either the median value of the concentrations or the combined scores, and obtained the P-values using the same regression model.
For linear regression analyses, β estimates were calculated from the concentration of each metabolite and the 2-h glucose value. The concentration of each metabolite was log-transformed and normalized to have a mean of zero and an s.d. of one. Various risk factors in the logistic regression were added as covariates, and the same significance level (3.6 × 10−4) was adopted.
Combination of metabolites
To obtain the combined scores of metabolites, the scaled metabolite concentrations (mean=0, s.d.=1) were first modeled with multivariate logistic regression containing all confounding variables. The coefficients of these metabolites from the model were then used to calculate a weighted sum for each individual. In accordance with the decreasing trend of glycine and LPC (18:2), we inverted these values as the combined scores.
Residuals of metabolite concentrations
To avoid the influence of other confounding factors when plotting the concentration of metabolites, we used the residuals from a linear regression model. Metabolite concentrations were log-transformed and scaled (mean=0, s.d.=1), and the residuals were then deduced from the linear regression that included the corresponding confounding factors.
Random forest, stepwise selection methods and candidate biomarker selection
To select candidate biomarkers, we applied two additional methods: the random forest selection (Breiman, 2001) and the stepwise selection, which assess the metabolites as a group.
Between two groups, the supervised classification method of random forest was first used to select the metabolites among the 30 highest ranking variables of importance score, allowing the best separation of the individuals from different groups. T2D risk indicators were also included in this method with all the metabolites.
We further selected the metabolites using stepwise selection on the logistic regression model. Metabolites with significantly different concentrations between the compared groups in logistic regression, and which were also selected using random forest, were used in this model along with all the risk indicators. Akaike’s Information Criterion (AIC) was used to evaluate the performance of these subsets of metabolites used in the models. The model with minimal AIC was chosen. The AUC was used to evaluate the models.
Network analysis
Metabolite–protein interactions from the Human Metabolome Database (HMDB; Wishart et al, 2009) and protein–protein interactions in the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; Szklarczyk et al, 2011) were used to construct a network containing relationships between metabolites, enzymes, other proteins and T2D-related genes. The candidate metabolites were assigned to HMDB IDs using the metaP-Server (Kastenmuller et al, 2011), and their associated enzymes were derived according to the annotations provided by HMDB. These enzymes were connected to the 46 T2D-related genes (considered at that point), allowing for 1 intermediate protein (other proteins) through STRING protein functional interaction and optimized by eliminating edges with a STRING score of <0.7 and undirected paths. The subnetworks were connected by the shortest path from metabolites to T2D-related genes.
Replication
The EPIC-Postdam is part of the multicenter EPIC study (Boeing et al, 1999; Riboli et al, 2002). It was drawn from the general adult population in Potsdam and surrounding areas and consists of 27 548 participants recruited from 1994 to 1998 (Boeing et al, 1999). At baseline, participants underwent anthropometric and BP measurements, completed an interview on prevalent diseases, a questionnaire on socioeconomic and lifestyle factors and submitted a validated food frequency questionnaire. Follow-up questionnaires were administered every 2–3 years (Bergmann et al, 1999).
From the EPIC-Potsdam population, a substudy of 2500 participants was randomly selected from all participants who had provided blood samples at baseline (n=26 444). The substudy had a limited number of fasting samples available. Therefore, non-fasting samples were also considered. Out of the substudy, 814 participants were excluded because of missing information on relevant covariates or missing fasting samples. Individuals with NGT and T2D were determined according to HbA1c categories defined by the American Diabetes Association in 2010 (ADA, 2010).
In the EPIC-Potsdam study, 30 ml of blood was drawn by qualified medical staff during the baseline examination, immediately fractionated into serum, plasma, buffy coat and erythrocytes and aliquoted into straws. The blood samples were stored in liquid nitrogen (at −196°C) until the metabolic analyses.
Metabolite measurements for the EPIC-Potsdam samples were performed using the same kit and the same method as for the KORA F4 samples (Floegel et al, 2011).
Calculations were performed using the Statistical Analysis System (SAS), Version 9.2 (SAS Institute, Inc., Cary, NC, USA).
Supplementary Material
Acknowledgments
We express our appreciation to all KORA and EPIC-Potsdam study participants for donating their blood and time. We thank the field staff in Augsburg who conducted the KORA studies. The KORA group consisted of HE Wichmann (speaker), A Peters, C Meisinger, T Illig, R Holle and J John, as well as their co-workers, and they were responsible for the design and conduction of the studies. We thank all the staff of the Institute of Epidemiology, Helmholtz Zentrum München, and the Genome Analysis Center, as well as the Metabolomic Platform, who helped in the sample logistics, the metabolite profiling assays and the genetic expression analyses, especially A Sabunchi, H Chavez, B Hochstrat, F Scharl, N Lindemann and J Scarpa. We thank M Sattler, W Mewes, VA Raker and J Mendes for comments and suggestions. This study was supported in part by a grant from the German Federal Ministry of Education and Research (BMBF) to the German Center for Diabetes Research (DZD e.V). In addition, this work was partly supported by the BMBF project ‘Metabolomics of ageing’ (FKZ: 01DO12030) and Project ‘SysMBo: Systems Biology of Metabotypes’ (FKZ: 0315494A). Further support for this study was obtained from the Federal Ministry of Health (Berlin, Germany), the Ministry of Innovation, Science, Research and Technology of the state North-Rhine Westphalia (Düsseldorf, Germany) and the Federal Ministry of Education, Science, Research and Technology (NGFN-Plus AtheroGenomics/01GS0423; Berlin, Germany). The KORA research platform and the KORA Augsburg studies are financed by the Helmholtz Zentrum München, German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education, Science, Research and Technology and by the State of Bavaria. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author contributions: RWS, ZY, CHe, KS, HP, AP, TM, HEW, TP, JA and TI designed the research. RWS, CHe, CP, WRM, MC, KH and HP performed the experiments. RWS, ZY, CHe, ACM, AF, YH, KH, MC, CHo, BT, HG, TX, EB, AD, KM, HYO, YL, LX, KS, AP, HP, TM, MR, HEW, TP, JA and TI analyzed the data. RWS, ZY, CHe, ACM, AF, YH, CHo, HP, TM, AP, MR, TP and JA wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- ADA. (2010) Executive summary: standards of medical care in diabetes--2010. Diabetes Care 33: Suppl 1S4–S10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams SH, Hoppel CL, Lok KH, Zhao L, Wong SW, Minkler PE, Hwang DH, Newman JW, Garvey WT (2009) Plasma acylcarnitine profiles suggest incomplete long-chain fatty acid beta-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic African-American women. J Nutr 139: 1073–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmaier E, Kastenmuller G, Romisch-Margl W, Thorand B, Weinberger KM, Illig T, Adamski J, Doring A, Suhre K (2011) Questionnaire-based self-reported nutrition habits associate with serum metabolism as revealed by quantitative targeted metabolomics. Eur J Epidemiol 26: 145–156 [DOI] [PubMed] [Google Scholar]
- Bergmann MM, Bussas U, Boeing H (1999) Follow-up procedures in EPIC-Germany--data quality aspects. European Prospective Investigation into Cancer and Nutrition. Ann Nutr Metab 43: 225–234 [DOI] [PubMed] [Google Scholar]
- Bishop DF (1990) Two different genes encode delta-aminolevulinate synthase in humans: nucleotide sequences of cDNAs for the housekeeping and erythroid genes. Nucleic Acids Res 18: 7187–7188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeing H, Wahrendorf J, Becker N (1999) EPIC-Germany--A source for studies into diet and risk of chronic diseases. European Investigation into Cancer and Nutrition. Ann Nutr Metab 43: 195–204 [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Breiman L (2001) Random Forests. Machine Learning 45: 5–32 [Google Scholar]
- Cheng S, Rhee EP, Larson MG, Lewis GD, McCabe EL, Shen D, Palma MJ, Roberts LD, Dejam A, Souza AL, Deik AA, Magnusson M, Fox CS, O'Donnell CJ, Vasan RS, Melander O, Clish CB, Gerszten RE, Wang TJ (2012) Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation 125: 2222–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faerch K, Vaag A, Holst JJ, Hansen T, Jorgensen T, Borch-Johnsen K (2009) Natural history of insulin sensitivity and insulin secretion in the progression from normal glucose tolerance to impaired fasting glycemia and impaired glucose tolerance: the Inter99 study. Diabetes Care 32: 439–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floegel A, Drogan D, Wang-Sattler R, Prehn C, Illig T, Adamski J, Joost HG, Boeing H, Pischon T (2011) Reliability of serum metabolite concentrations over a 4-month period using a targeted metabolomic approach. PLoS ONE 6: e21103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goek ON, Doring A, Gieger C, Heier M, Koenig W, Prehn C, Romisch-Margl W, Wang-Sattler R, Illig T, Suhre K, Sekula P, Zhai G, Adamski J, Kottgen A, Meisinger C (2012) Serum metabolite concentrations and decreased GFR in the general population. Am J Kidney Dis 60: 197–206 [DOI] [PubMed] [Google Scholar]
- Holle R, Happich M, Lowel H, Wichmann HE (2005) KORA--a research platform for population based health research. Gesundheitswesen 67: S19–S25 [DOI] [PubMed] [Google Scholar]
- Horie S, Ishii H, Suga T (1981) Changes in peroxisomal fatty acid oxidation in the diabetic rat liver. J Biochem 90: 1691–1696 [DOI] [PubMed] [Google Scholar]
- Illig T, Gieger C, Zhai G, Romisch-Margl W, Wang-Sattler R, Prehn C, Altmaier E, Kastenmuller G, Kato BS, Mewes HW, Meitinger T, de Angelis MH, Kronenberg F, Soranzo N, Wichmann HE, Spector TD, Adamski J, Suhre K (2010) A genome-wide perspective of genetic variation in human metabolism. Nat Genet 42: 137–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdan C, Petersen AK, Gieger C, Doring A, Illig T, Wang-Sattler R, Meisinger C, Peters A, Adamski J, Prehn C, Suhre K, Altmaier E, Kastenmuller G, Romisch-Margl W, Theis FJ, Krumsiek J, Wichmann HE, Linseisen J (2012) Body fat free mass is associated with the serum metabolite profile in a population-based study. PLoS ONE 7: e40009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenmuller G, Romisch-Margl W, Wagele B, Altmaier E, Suhre K (2011) metaP-server: a web-based metabolomics data analysis tool. J Biomed Biotechnol 2011: 1–7, pii: 839862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, Walker EA, Nathan DM (2002) Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 346: 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, Bain J, Stevens R, Dyck JR, Newgard CB, Lopaschuk GD, Muoio DM (2008) Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab 7: 45–56 [DOI] [PubMed] [Google Scholar]
- Krebs M, Krssak M, Bernroider E, Anderwald C, Brehm A, Meyerspeer M, Nowotny P, Roth E, Waldhausl W, Roden M (2002) Mechanism of amino acid-induced skeletal muscle insulin resistance in humans. Diabetes 51: 599–605 [DOI] [PubMed] [Google Scholar]
- Malipa AC, Meintjes RA, Haag M (2008) Arachidonic acid and glucose uptake by freshly isolated human adipocytes. Cell Biochem Funct 26: 221–227 [DOI] [PubMed] [Google Scholar]
- McGarry JD (2002) Banting lecture 2001: dysregulation of fatty acid metabolism in the etiology of type 2 diabetes. Diabetes 51: 7–18 [DOI] [PubMed] [Google Scholar]
- Meisinger C, Strassburger K, Heier M, Thorand B, Baumeister SE, Giani G, Rathmann W (2010) Prevalence of undiagnosed diabetes and impaired glucose regulation in 35-59-year-old individuals in Southern Germany: the KORA F4 Study. Diabet Med 27: 360–362 [DOI] [PubMed] [Google Scholar]
- Mittelstrass K, Ried JS, Yu Z, Krumsiek J, Gieger C, Prehn C, Roemisch-Margl W, Polonikov A, Peters A, Theis FJ, Meitinger T, Kronenberg F, Weidinger S, Wichmann HE, Suhre K, Wang-Sattler R, Adamski J, Illig T (2011) Discovery of sexual dimorphisms in metabolic and genetic biomarkers. PLoS Genet 7: e1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morino K, Petersen KF, Dufour S, Befroy D, Frattini J, Shatzkes N, Neschen S, White MF, Bilz S, Sono S, Pypaert M, Shulman GI (2005) Reduced mitochondrial density and increased IRS-1 serine phosphorylation in muscle of insulin-resistant offspring of type 2 diabetic parents. J Clin Invest 115: 3587–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muoio DM, Newgard CB (2008) Mechanisms of disease: molecular and metabolic mechanisms of insulin resistance and beta-cell failure in type 2 diabetes. Nat Rev Mol Cell Biol 9: 193–205 [DOI] [PubMed] [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, Rochon J, Gallup D, Ilkayeva O, Wenner BR, Yancy WS Jr, Eisenson H, Musante G, Surwit RS, Millington DS, Butler MD et al. (2009) A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 9: 311–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noland RC, Koves TR, Seiler SE, Lum H, Lust RM, Ilkayeva O, Stevens RD, Hegardt FG, Muoio DM (2009) Carnitine insufficiency caused by aging and overnutrition compromises mitochondrial performance and metabolic control. J Biol Chem 284: 22840–22852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JD, Kushner JP (2005) Fast track to the porphyrias. Nat Med 11: 1049–1050 [DOI] [PubMed] [Google Scholar]
- Pietilainen KH, Rog T, Seppanen-Laakso T, Virtue S, Gopalacharyulu P, Tang J, Rodriguez-Cuenca S, Maciejewski A, Naukkarinen J, Ruskeepaa AL, Niemela PS, Yetukuri L, Tan CY, Velagapudi V, Castillo S, Nygren H, Hyotylainen T, Rissanen A, Kaprio J, Yki-Jarvinen H et al. (2011) Association of lipidome remodeling in the adipocyte membrane with acquired obesity in humans. PLoS Biol 9: e1000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontiroli AE, Pizzocri P, Caumo A, Perseghin G, Luzi L (2004) Evaluation of insulin release and insulin sensitivity through oral glucose tolerance test: differences between NGT, IFG, IGT, and type 2 diabetes mellitus. A cross-sectional and follow-up study. Acta Diabetol 41: 70–76 [DOI] [PubMed] [Google Scholar]
- Primrose S, Draper J, Elsom R, Kirkpatrick V, Mathers JC, Seal C, Beckmann M, Haldar S, Beattie JH, Lodge JK, Jenab M, Keun H, Scalbert A (2011) Metabolomics and human nutrition. Br J Nutr 105: 1277–1283 [DOI] [PubMed] [Google Scholar]
- Rathmann W, Kowall B, Heier M, Herder C, Holle R, Thorand B, Strassburger K, Peters A, Wichmann HE, Giani G, Meisinger C (2010) Prediction models for incident type 2 diabetes mellitusin the older population: KORA S4/F4 cohort study. Diabet Med 27: 1116–1123 [DOI] [PubMed] [Google Scholar]
- Rathmann W, Strassburger K, Heier M, Holle R, Thorand B, Giani G, Meisinger C (2009) Incidence of Type 2 diabetes in the elderly German population and the effect of clinical and lifestyle risk factors: KORA S4/F4 cohort study. Diabet Med 26: 1212–1219 [DOI] [PubMed] [Google Scholar]
- Rhee EP, Cheng S, Larson MG, Walford GA, Lewis GD, McCabe E, Yang E, Farrell L, Fox CS, O'Donnell CJ, Carr SA, Vasan RS, Florez JC, Clish CB, Wang TJ, Gerszten RE (2011) Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest 121: 1402–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riboli E, Hunt KJ, Slimani N, Ferrari P, Norat T, Fahey M, Charrondiere UR, Hemon B, Casagrande C, Vignat J, Overvad K, Tjonneland A, Clavel-Chapelon F, Thiebaut A, Wahrendorf J, Boeing H, Trichopoulos D, Trichopoulou A, Vineis P, Palli D et al. (2002) European Prospective Investigation into Cancer and Nutrition (EPIC): study populations and data collection. Public Health Nutr 5: 1113–1124 [DOI] [PubMed] [Google Scholar]
- Römisch-Margl W, Prehn C, Bogumil R, Roehring C, Suhre KJA (2012) Procedure for tissue sample preparation and metabolite extraction for high-throughput targeted metabolomics. Metabolomics 8: 133–142 [Google Scholar]
- Shaham O, Wei R, Wang TJ, Ricciardi C, Lewis GD, Vasan RS, Carr SA, Thadhani R, Gerszten RE, Mootha VK (2008) Metabolic profiling of the human response to a glucose challenge reveals distinct axes of insulin sensitivity. Mol Syst Biol 4: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumvoll M, Goldstein BJ, van Haeften TW (2005) Type 2 diabetes: principles of pathogenesis and therapy. Lancet 365: 1333–1346 [DOI] [PubMed] [Google Scholar]
- Szendroedi J, Schmid AI, Chmelik M, Toth C, Brehm A, Krssak M, Nowotny P, Wolzt M, Waldhausl W, Roden M (2007) Muscle mitochondrial ATP synthesis and glucose transport/phosphorylation in type 2 diabetes. PLoS Med 4: e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39: D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak AG, Herder C, Rathmann W, Brunner EJ, Kivimaki M (2012) Prediabetes: a high-risk state for diabetes development. Lancet 379: 2279–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak AG, Jokela M, Akbaraly TN, Brunner EJ, Kivimaki M, Witte DR (2009) Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet 373: 2215–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomilehto J, Lindstrom J, Eriksson JG, Valle TT, Hamalainen H, Ilanne-Parikka P, Keinanen-Kiukaanniemi S, Laakso M, Louheranta A, Rastas M, Salminen V, Uusitupa M (2001) Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Engl J Med 344: 1343–1350 [DOI] [PubMed] [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, O'Donnell CJ, Carr SA, Mootha VK, Florez JC, Souza A, Melander O, Clish CB, Gerszten RE (2011) Metabolite profiles and the risk of developing diabetes. Nat Med 17: 448–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. (1999) Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus. Report of a WHO ConsultationGenevapp 59
- Wichmann HE, Gieger C, Illig T (2005) KORA-gen--resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen 67: Suppl 1S26–S30 [DOI] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P et al. (2009) HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res 37: D603–D610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wopereis S, Rubingh CM, van Erk MJ, Verheij ER, van Vliet T, Cnubben NH, Smilde AK, van der Greef J, van Ommen B, Hendriks HF (2009) Metabolic profiling of the response to an oral glucose tolerance test detects subtle metabolic changes. PLoS ONE 4: e4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Zhai G, Singmann P, He Y, Xu T, Prehn C, Romisch-Margl W, Lattka E, Gieger C, Soranzo N, Heinrich J, Standl M, Thiering E, Mittelstrass K, Wichmann HE, Peters A, Suhre K, Li Y, Adamski J, Spector TD et al. (2012) Human serum metabolic profiles are age dependent. Aging Cell (e-pub ahead of print 26 July 2012; doi:; DOI: 10.1111/j.1474-9726.2012.00865.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Fritsche J, Wang J, Chen J, Rittig K, Schmitt-Kopplin P, Fritsche A, Haring HU, Schleicher ED, Xu G, Lehmann R (2010) Metabonomic fingerprints of fasting plasma and spot urine reveal human pre-diabetic metabolic traits. Metabolomics 6: 362–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Metabolite concentrations of Glycine, LPC (18:2) and C2 with T2D status in the KORA S4 and F4 are provided (Supplementary Table S16). Additional data from the KORA S4 and F4 studies, including the metabolite concentrations and the gene expression with clinical phenotypes used in this study, are available upon request from KORA-gen (http://epi.helmholtz-muenchen.de/kora-gen). Requests should be sent to kora-gen@helmholtz-muenchen.de and are subject to approval by the KORA board to ensure that appropriate conditions are met to preserve patient privacy. Formal collaboration and co-authorship with members of the KORA study is not an automatic condition to obtain access to the data published in the present paper. More general information about KORA, including S4 and F4 study design and clinical variables, can be found at http://epi.helmholtz-muenchen.de/kora-gen/seiten/variablen_e.php and http:/helmholtz-muenchen.de/en/kora-en/information-for-scientists/current-kora-studies.