Over the period of just one decade, systems biology has emerged from obscurity and moved center stage. The novelty and rapidly growing notoriety have, unsurprisingly, piqued the interest of students in biology, and also in engineering, physics, computing, and a number of other fields. This growing interest has led to a demand for courses and even complete educational programs in systems biology. The creation of such courses is challenging due to the students' different backgrounds and also because systems biology has roots in an unusually wide variety of parent disciplines, which no student can fully master. Faced with these complications, one appears to be forced to teach a selection of representative topics and techniques. We believe that this default solution is not always optimal and that it is much more important instead to instill in students a ‘feel' for biological systems and for models that can be used to explore them. ‘Instilling a feel' may not sound scientifically rigorous, but the ability to gauge the complexity of a system, without executing a formal analysis, is arguably more beneficial to next-generation molecular biologists than mastery of selected techniques of systems analysis. Likewise, it will become increasingly more critical for biologists to assess how a computational model could be set up, how it might add genuine value, and where its limitations are.
Like others, we had previously taught the typical stepwise design, diagnosis, analysis, and utilization of models in the classroom. We discussed with the students that one must decide early whether to use deterministic or stochastic, continuous or discrete, static or dynamic models, partial differential equations, or agent-based methods. These initial dichotomies were followed by presentations on key topics of systems biology, such as dynamics and steady states, stability, sensitivity, and robustness, bifurcations toward limit cycles, hysteresis, chaos, and other emerging behaviors. All topics were illustrated with carefully selected examples demonstrating that even small systems have the capacity of exhibiting surprising responses.
Attempting to implement this structure in a new introductory graduate-level course, we soon realized that students would not receive real hands-on experience until the last few weeks of the semester. By this time, we feared, non-majors might have lost interest. We would also miss our goal of ‘instilling a feel' for systems and models and instead possibly create the impression that biological systems are simply too difficult for our intuition to handle. This impression could turn into unease and helplessness, possibly followed by a strong, long-lasting resistance to realizing and appreciating the genuine added value of systems biological models.
We therefore designed a radically different, hands-on problem-solving course, intended to scaffold the cognitive processes required to gauge systems, diagrams, and models. Our overarching goal was to afford students the ability to: (1) interpret a typical systems diagram of boxes, fluxes, and signals; (2) gauge whether the system might exhibit complex behaviors; (3) understand useful approaches for converting a diagram into a computable structure; and (4) discuss these approaches with biologists and modelers alike. To achieve these goals, we decided on a strategy opposite to the traditional paradigm: Right from the very beginning, we immersed the students in the tension between realistic biological complexity and a very simplified mesoscopic model. We selected cystic fibrosis (CF) of the lung as the overarching, clinically pertinent theme, and used this theme as an anchor for the entire semester.
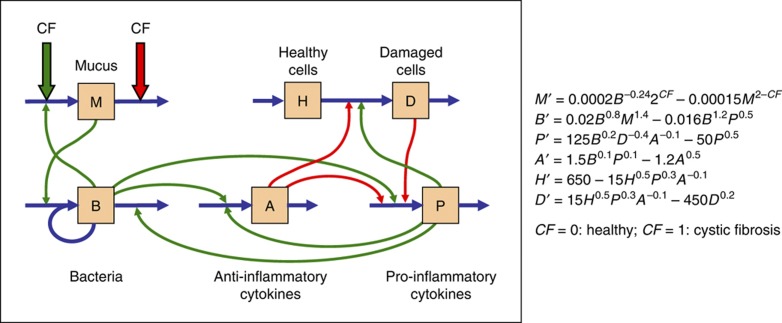
We started with a general introduction, followed by a class on CF, taught by an expert biologist. Directly afterwards, we introduced a simple dynamic model of CF (Figure 1), without offering details on ordinary differential equations (ODEs), except for their functionality. We suggested using the freeware PLAS ( http://enzymology.fc.ul.pt/software/plas), which permits ODE simulations and analyses with minimal overhead. The students learned immediately that the prepared model, like all initial models of complex systems, was greatly abstracted and simplified, that it captured some aspects of CF but was certain to fail in others, and that modeling is an iterative process that starts simple and becomes more realistic over time. Importantly, the students experienced, from the first week on, and through targeted, open-ended homework assignments, how to implement changes in the model, such as killing bacteria or treating patients with anti-inflammatory cytokines. After setting the stage, we taught classes on model diagnostics, features of ODE models, parameter estimation, and other aspects of dynamical systems. All topics were reinforced with homework assignments, which were formulated within the context of CF wherever feasible. We also discussed literature studies on CF and taught strategies for reviewing experimental and modeling articles. Throughout the semester, the students presented their findings to the class.
Figure 1.
Simple model of cystic fibrosis (CF) of the lung. Left panel: interaction diagram, accounting for direct effects of CF on mucus dynamics. Right panel: model in the format of Biochemical Systems Theory (Savageau, 1976; Voit, 2000). For CF=0, the system has a steady state at about M=1.3, B=2.1, P=1.700, A=2.0, H=1600, D=6.3. Typical simulations, which start at the steady state or the corresponding steady state for CF=1, might reset the number of bacteria at certain time points, modeling responses to infections, or treatments like the removal of mucus or the infusion of antibiotics or anti-inflammatory cytokines (see Supplementary information for further details).
The students soon realized that ODE models are terrific defaults, but also genuinely limited in scope. Because CF is strongly affected by random events like infections, we introduced concepts of stochasticity, progressively weaned the students off PLAS, and offered Matlab instruction on a need-to-know basis, for instance, with pre-formulated code for parameter estimation. Subsequently, we introduced approaches toward spatial aspects, including agent-based models. After about 10 weeks, we had introduced all theoretical topics we considered as important. For the remaining third of the semester, the students were charged with developing either extensions of the original model or new models of particular aspects of CF. They self-assembled into groups of between one and four students that worked together outside class and presented advances, challenges, and roadblocks in class for discussion and suggestions. Ultimately, the different groups: expanded the original model to account for the role of neutrophils; addressed scar tissue formation; developed a model for the synthesis and dynamics of mucus; and developed agent-based models of biofilms and the dynamics between bacteria and neutrophils. The final presentations were judged jointly by the class, the course instructors, and several CF experts. Throughout the semester, the students felt empowered by the autonomy and flexibility afforded to them. They were proud of their accomplishments in a highly relevant area and especially of the fact that they had created, de novo, a computational structure within a complex disease system. The course was evaluated with the usual institutional surveys and also through specific interviews by cognitive scientists. According to all metrics, the students were overwhelmingly positive about the course, except that they had missed a textbook as resource; in the future, we will use Voit (2012). Moreover, the experimentalists were eager to have students from the class work with them.
With respect to instilling a ‘feel for systems and models,' nothing is special about CF, and future courses will focus on other diseases or fundamental processes in biology. The syllabus for the completed course and further details can be found in Supplementary information.
Supplementary Material
The Supplements contain a file that can be copied into the software PLAS and used to explore the system described in the text. Second, it contains a few selected modeling results. Finally, this section contains the syllabus of the course that is described in the text, and which we taught in the Fall of 2011.
Acknowledgments
We are grateful for NSF (MCB-0958172 and DRL-REESE-097394084) support of this work.
Footnotes
The authors declare that they have no conflict of interest.
References
- Savageau MA (1976) Biochemical Systems Analysis: A Study of Function and Design in Molecular Biology Reading, MA: Addison-Wesley Pub. Co. Advanced Book Program, [Google Scholar]
- Voit EO (2000) Computational Analysis of Biochemical Systems. A Practical Guide for Biochemists and Molecular Biologists Cambridge, UK: Cambridge University Press, [Google Scholar]
- Voit EO (2012) A First Course in Systems Biology New York: Garland Science, [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Supplements contain a file that can be copied into the software PLAS and used to explore the system described in the text. Second, it contains a few selected modeling results. Finally, this section contains the syllabus of the course that is described in the text, and which we taught in the Fall of 2011.