Abstract
Extra chromosome copies markedly alter the physiology of eukaryotic cells, but the underlying reasons are not well understood. We created human trisomic and tetrasomic cell lines and determined the quantitative changes in their transcriptome and proteome in comparison with their diploid counterparts. We found that whereas transcription levels reflect the chromosome copy number changes, the abundance of some proteins, such as subunits of protein complexes and protein kinases, is reduced toward diploid levels. Furthermore, using the quantitative data we investigated the changes of cellular pathways in response to aneuploidy. This analysis revealed specific and uniform alterations in pathway regulation in cells with extra chromosomes. For example, the DNA and RNA metabolism pathways were downregulated, whereas several pathways such as energy metabolism, membrane metabolism and lysosomal pathways were upregulated. In particular, we found that the p62-dependent selective autophagy is activated in the human trisomic and tetrasomic cells. Our data present the first broad proteomic analysis of human cells with abnormal karyotypes and suggest a uniform cellular response to the presence of an extra chromosome.
Keywords: aneuploidy, autophagy, pathway analysis, proteomics, p62
Introduction
Aneuploidy is detrimental to eukaryotic cells. Chromosome number changes are the main cause of spontaneous miscarriages and surviving embryos are born with severe disabilities, but the reasons for the incompatibility of the karyotype changes with normal development are not well understood (Hassold et al, 2007). Cells isolated from patients with Down syndrome (trisomy of chromosome 21) grow substantially slower (Segal and McCoy, 1974) and addition of even a single chromosome to mouse cells causes multiple defects, such as growth delay or increased sensitivity to certain drugs (Williams et al, 2008; Tang et al, 2011). Abnormal karyotypes were also shown to affect cell physiology in yeasts and plants. Addition of chromosomes leads to growth defects (Torres et al, 2007), phenotypic variability (Selmecki et al, 2006; Pavelka et al, 2010; Chen et al, 2012) and increased genome instability that likely contributes to evolution (Niwa et al, 2006; Selmecki et al, 2006; Huettel et al, 2008; Sheltzer et al, 2011). On the other hand, variable aneuploid karyotypes can be found in tumors without an obvious adverse effect on cell proliferation and it has been hypothesized that aneuploidy facilitates tumorigenesis (Storchova and Pellman, 2004). Despite the frequent occurrence of aneuploidy and its link to pathological states, molecular mechanisms underlying the observed phenotypes remain unclear.
One of the important questions is how the presence of an extra chromosome affects the mRNA and protein content of a cell, and what is the physiological response to these changes. Transcriptome profiling suggests that the mRNA levels from genes encoded on the extra chromosomes mostly scale up proportionally with the gene copy numbers in various organisms such as in vitro generated aneuploid yeast, mouse and human cells (Upender et al, 2004; Torres et al, 2007; Williams et al, 2008), pathogenic Candida strains (Selmecki et al, 2006) and aneuploid plants (Makarevitch et al, 2008). Other reports suggest a feedback control that buffers the mRNA levels of amplified or underrepresented chromosomal regions in naturally occurring aneuploid yeast strains (Kvitek et al, 2008), plants (Birchler et al, 2005) or in Drosophila with partial or whole chromosomal aneuploidy (Stenberg et al, 2009). Detailed analysis of Down syndrome patients suggests that the transcription levels of some of the genes on chromosome 21 are compensated as well (Aït Yahya-Graison et al, 2007). So far, only little is known about the changes in protein content and pathway regulation in aneuploid cells. Recent studies in aneuploid budding yeasts yielded partially contradictory results. On one hand, an artificial introduction of a single chromosome into haploid cells led to a growth delay, general stress response and proteotoxic stress (Torres et al, 2007). Moreover, mRNAs and proteins coded on the disomes were expressed proportionally to the chromosome copy numbers with exception of the protein levels of subunits of multimolecular complexes that were partially compensated to maintain the stoichiometry (Torres et al, 2010). On the other hand, meiotically generated multi-chromosome aneuploidy resulted in a proportional scaling of protein expression with no significant compensation of the abundance of subunits of protein complexes, and no indications of proteotoxic stress or general stress response were identified (Pavelka et al, 2010). No similar analysis has been performed in human aneuploid cells so far.
To uncover the fate of the transcripts and proteins encoded on the extra chromosome and to investigate the global changes in human cells in response to supernumerary chromosomes, we created model tri- and tetrasomic cells derived from two different human chromosomally stable cell lines, HCT116 and RPE-1. We quantified the changes of genome, transcriptome and proteome and determined specific pathways whose regulation is altered in these cell lines. This analysis suggests a specific cellular response to the presence of extra chromosomes in human cells. In particular, we show that p62-dependent autophagy is activated in cell lines with extra chromosomes, where it may contribute to the maintenance of normal protein levels.
Results
Generation and characterization of human trisomic and tetrasomic cell lines
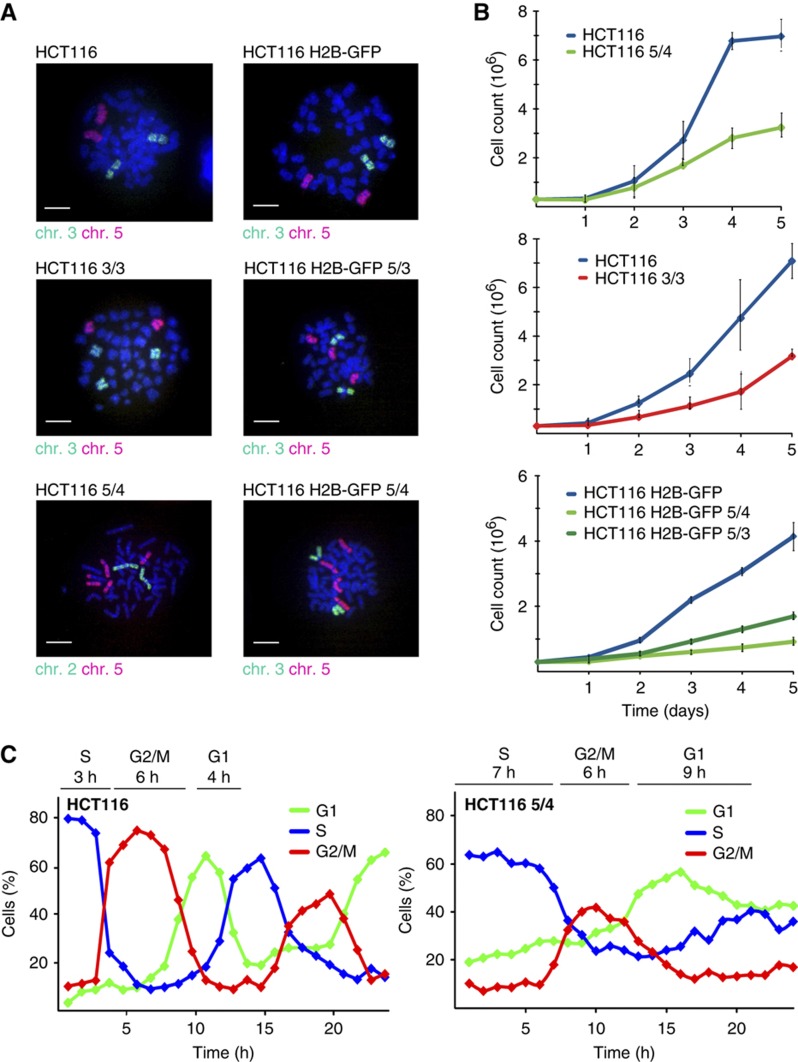
A detailed analysis of aneuploidy in human cells is hampered by the lack of an appropriate model with matching diploid and aneuploid cells. To circumvent this limitation, chromosome transfer via micronuclei was used to add an extra chromosome into HCT116 (Haugen et al, 2008) or HCT116 stably expressing H2B-GFP (Supplementary Figure S1A). This approach generated cognate trisomic and tetrasomic derivatives that carry additional copies of chromosome 3 (labeled HCT116 3/3) or chromosome 5 (trisomy: HCT116 H2B-GFP 5/3, tetrasomy: HCT116 5/4 and HCT116 H2B-GFP 5/4). HCT116 is a transformed cell line with several previously identified chromosomal changes such as the chromosome Y loss and amplified regions of chromosomes 8, 10 and 17 (Masramon et al, 2000). These aberrancies are mostly present in the new aneuploid cell lines (Supplementary Figure 1B) and thus likely do not affect the results. Nevertheless, to strengthen our analysis and to overcome this possible drawback, we generated cell lines trisomic for chromosomes 5 and 12, and another cell line trisomic for chromosome 21, both derived from the diploid primary epithelial cell line RPE-1 that was immortalized by the expression of hTert and that lacks substantial chromosomal aberrancies. The successful chromosome transfer was verified by chromosome paints (Figure 1A), comparative genomic hybridization (CGH) and multicolor fluorescence in situ hybridization (Supplementary Figure S1B and C). The analysis confirmed that original cell lines and their derivatives differ only by copy number of a specific chromosome.
Figure 1.
Characterization of HCT116 and its tri- and tetrasomic derivatives. (A) Chromosome paints of used tri- and tetrasomic cell lines. Bar—10 μm. (B) Growth curves of tri- and tetrasomic cell lines in comparison with their diploid counterparts. Each point represents the mean with standard deviation of three independent experiments. (C) Cell-cycle progression of HCT116 (left panel) and HCT116 5/4 (right panel) after release from thymidine block, analyzed by flow cytometry. The major delay occurs in the G1 and the S phase. The length of each cell-cycle phase is indicated above the graph. Population was considered to enter a specific phase of cell cycle if at least 50% of cells showed corresponding DNA content. See also Supplementary Figure S1. Source data is available for this figure in the Supplementary Information.
All trisomic and tetrasomic cell lines display marked growth impairment as previously observed (Segal and McCoy, 1974; Guo and Birchler, 1994; Torres et al, 2007; Williams et al, 2008), with tetrasomic cell lines growing significantly slower than the trisomic ones (Figure 1B). We found that the aneuploid cells progress slowly and less synchronously through G1 and S phase, with a delay of 5 and 4 h, respectively (Figure 1C). On the contrary, the progression through G2 phase and mitosis is not noticeably affected (Figure 1C; Supplementary Figure S1D and E) and there is no marked accumulation of dead and non-proliferating cells (Supplementary Figure S1F and G). Similar delay in cell-cycle progression was observed in the RPE-1 5/3 12/3 (Supplementary Figure S1H). Thus, the presence of an extra chromosome impairs the growth of human cells.
Comparison of genome, transcriptome and proteome of cells containing extra chromosomes
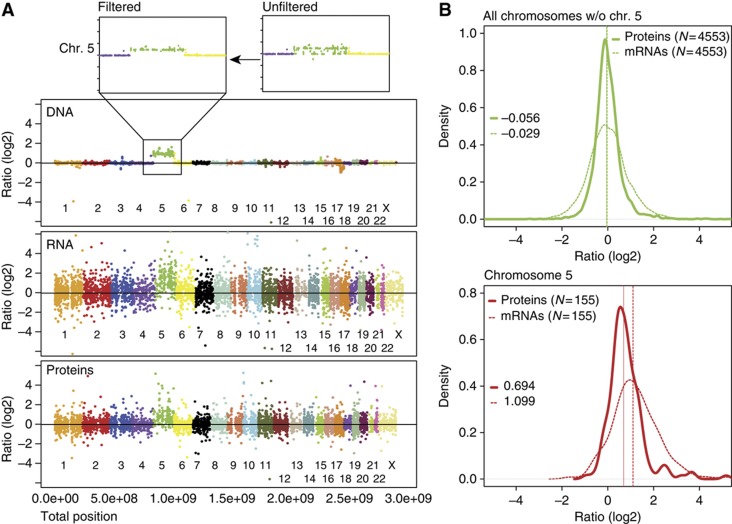
Using the series of isogenic diploid and aneuploid cell lines we asked how the mRNA and protein levels changed in response to the chromosome copy number changes. We examined the tetrasomic cell line HCT116 5/4, which guaranteed larger dynamic range of measured changes than in trisomic cells. To quantify the DNA levels, we used high-resolution CGH (Supplementary Table S1). To measure the corresponding mRNA, we determined the medians from microarray analysis of three independent samples (Supplementary Table S1). To compare the protein levels, we used stable isotope labeling with amino acids in cell culture (SILAC) followed by high-resolution mass spectrometry (Ong et al, 2002) and quantified the proteome to a depth of ∼6000 proteins (Supplementary Table S1). In total, we performed three biological replicates with six measurements for HCT116 5/4; all other cell lines were analyzed less extensively (up to three biological replicates, see Supplementary Table S1). The analysis of variability in technical and biological replicates demonstrated high quantification accuracy and reproducible measurements for all experiments including the reverse labeling, with Pearson correlation factors between 0.64 and 0.90 (Supplementary Figure S2A). We then determined the medians of quantitative protein changes. The log2 ratios of aneuploid-to-diploid abundance changes of DNA, the corresponding mRNAs and the proteins were matched directly using the annotated chromosomal positions (Figure 2A; Supplementary Figure S2B). In the case of equal levels in aneuploid and diploid cells, we expect the median aneuploid-to-diploid ratio (log2) to be 0 and this is observed for all DNA, mRNA and proteins coded on the disomes (DNA: 0.005, median mRNA: −0.03; proteins: −0.06; Figure 2B). The median ratio of mRNA encoded on the tetrasomic chromosome 5 is 1.09—very close to the expected value of 1.0—but in contrast, the median ratio of protein level changes is only 0.69 (Figure 2B). Indeed, 53 out of 197 proteins (27%) coded on the tetrasomic chromosome 5 are present at levels expected for disomes (median of diploid levels±twice the standard deviation). The calculated median of protein abundance of these 53 proteins is 0.26, whereas the median of the corresponding mRNAs is 1.16, further strengthening the idea that their expression is adjusted at the protein level. Similarly, 25% of proteins coded on chromosome 3 are present at diploid levels in HCT116 3/3 (Supplementary Table S1). We determined that the proteins coded on the multisomic chromosome are present at a lower level than expected in all analyzed cell lines (Supplementary Figure S3). In conclusion, whereas the mRNA expression corresponds to the increased chromosome copy numbers, approximately a quarter of the proteins are present at levels lower than expected and more similar to the disomic levels.
Figure 2.
Quantification of DNA, mRNA and protein abundance. (A) Ratios (log2) of DNA, mRNA and protein abundance changes between HCT116 5/4 and HCT116 aligned with respect to their chromosome position. Each dot represents abundance changes for one gene, the corresponding mRNA and the corresponding protein, respectively. CGH analysis revealed some deleted regions in chromosome 5 (upper panel); the disomic entries were omitted in all analyses (threshold 0.65 [log2]). (B) Overlays of mRNA and protein density histograms (HCT116 5/4 versus HCT116). The full line represents median of protein abundance changes, the dashed line median of mRNA abundance changes. Values of respective medians are plotted in the graph. The difference between distribution of proteins and mRNAs coded on chromosome 5 is statistically significant (Wilcoxon rank sum test). See also Supplementary Figure S2.
Specific proteins are maintained at stoichiometric levels
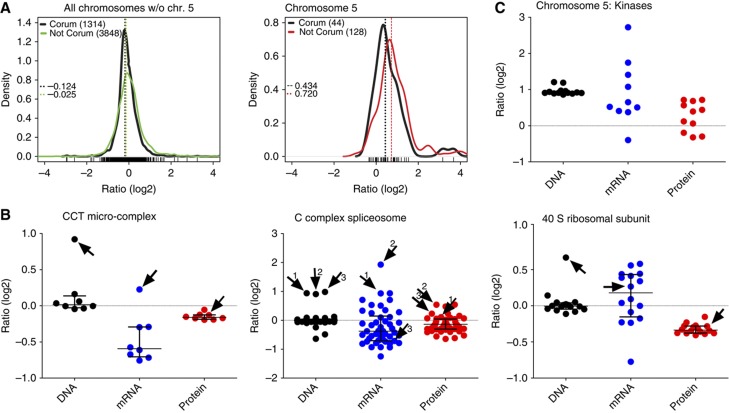
The fact that most proteins coded on the extra chromosomes are more abundant than proteins from diploid chromosomes indicates that there is no general efficient mechanism for ‘gene dosage compensation’ of the analyzed tri- or tetrasomies. Nevertheless, a remarkable proportion of these proteins are present at levels lower than expected according to the gene and mRNA copy numbers, suggesting that some proteins or protein categories might be adjusted to normal abundance. A long standing hypothesis posits that free subunits of multimolecular complexes may be degraded in cells (see e.g., Goldberg and Dice, 1974 and Guialis et al, 1979). Analysis of our data confirmed that the abundances of subunits of protein complexes (as annotated in the CORUM database; Ruepp et al, 2010) that are encoded on the tetrasomic chromosome are lower than expected based on the gene copy number, and more similar to the protein abundances observed in the parental cell line HCT116; the median of proteins of CORUM complexes coded on chromosome 5 is shifted down to 0.43 (Figure 3A). A similar shift toward the diploid levels for the CORUM-annotated proteins can be detected in all analyzed cell lines (Supplementary Figure S4A). We examined 14 different macromolecular complexes with at least one subunit coded on the tetrasomic chromosome and found that 8 of them (57%) maintain stoichiometry by decreasing the protein abundance close to the normal, disomic levels, while the mRNA levels vary (Figure 3B; Supplementary Figure S4B; Supplementary Table S3).
Figure 3.
Abundance of subunits of protein complexes and kinases in the tetrasomic cell line. (A) Density plots of subunits of protein complexes (as defined in the CORUM database) encoded on all disomic chromosomes compared with non-CORUM proteins (left panel); the same for proteins encoded on the tetrasomic chromosome 5 (right panel). The differences between CORUM and non-CORUM populations of chromosome 5 are statistically significant (Wilcoxon rank sum test). Dashed lines indicate medians of the populations. (B) Examples of protein complexes with at least one subunit coded on the tetrasomic chromosome 5. Each dot represents abundance changes of one gene (black), its corresponding mRNA (blue) and protein (red). Subunits coded on chromosome 5 are indicated with an arrow and number. (C) The abundance of DNA, mRNA and proteins of kinases coded on chromosome 5. See also Supplementary Figure S3. Source data is available for this figure in the Supplementary Information.
Moreover, we found that protein kinases coded on the supernumerary chromosomes shift toward near-diploid expression levels in HCT116 5/4 as well (Figure 3C). Interestingly, the abundance distribution shows a bi-modal pattern. Remarkably, the uncompensated kinases are preferentially associated with pathways that we determined to be globally upregulated in aneuploid cells (e.g., JNK2, see below); other kinases coded on chromosome 5 were present at near-diploid levels. A similar trend was observed in other cell lines, but the limited number of proteins did not allow evaluation of the statistical significance of these changes. In conclusion, trisomic and tetrasomic human cells maintain normal levels of proteins coded on chromosomes present in extra copies, but the levels are adjusted only for specific classes of proteins.
General cellular response to the presence of extra chromosomes in human cells
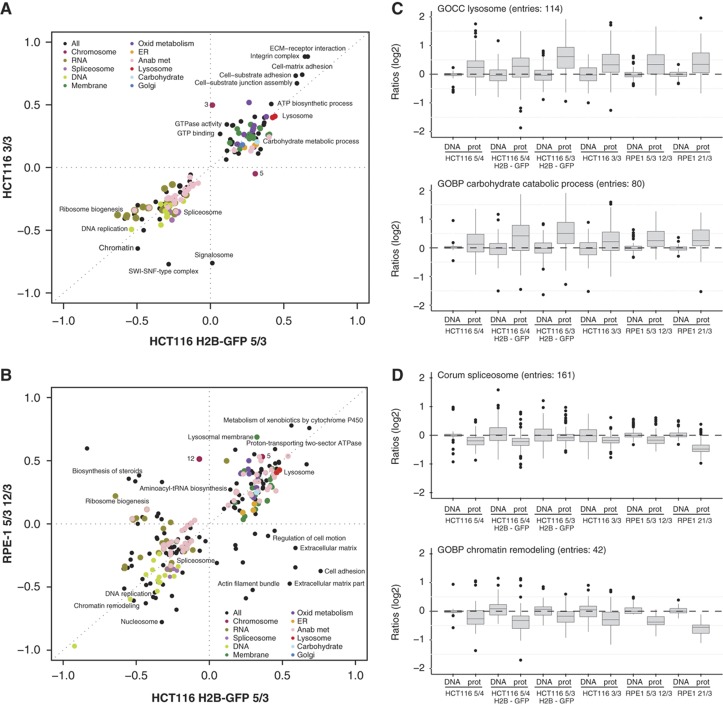
Next, we asked how aneuploidy affects global pathway regulation. To this end, we used a recently developed software called ‘2-D annotation enrichment analysis’ (see Supplementary Information). For each clone, we identified all significantly altered pathways (as defined by Gene Ontology categories, KEGG pathways and CORUM database) and ranked the relative abundance changes of proteins within the category compared with the complete measured data set. The resulting score is on the scale from −1 to +1, where the pathways close to −1 are most downregulated and pathways close to +1 are most upregulated (see Supplementary Information for further details). Plotting these calculated scores revealed remarkable similarities in pathway regulation among the analyzed aneuploid cell lines (Figure 4A and B). We found that pathways involved in DNA and RNA metabolism, such as replication, DNA repair, transcription and mRNA processing, were significantly downregulated (Figure 4A, B and D; Supplementary Figure S5A), which is consistent with the observed growth delay in the G1 and S phase (Figure 1C; Supplementary Figure S1D, E and H). Upregulated categories identified in all analyzed cell lines included pathways required for lipid and membrane biogenesis, endoplasmic reticulum, Golgi vesicles and lysosome functions as well as energy metabolic pathways such as mitochondrial respiratory metabolism and carbohydrate metabolism (Figure 4A–C; Supplementary Figure S5A). Remarkably, the 2-D enrichment analysis of the transcriptome data determined similar changes in the pathway regulation, suggesting that the pathway response cannot be caused by a bias in protein detection (Supplementary Figure S5B and C). The observed alterations in pathway regulation were not an artifact of the chromosome number changes, since excluding the proteins coded on chromosome 5 from the analysis did not affect the identified pathways (Supplementary Figure S5D). Additionally, we found a remarkable overlap in pathway alterations between all RPE-1- and HCT116-derived trisomic and tetrasomic cell lines (Figure 4B; Supplementary Figure S5A). Taken together, the uniformity of the response in different human trisomic and tetrasomic cells demonstrates that the presence of an extra chromosome itself, and not individual chromosome or cell types, is an important determinant of the general cellular response.
Figure 4.
Uniform global response to presence of an extra chromosome. (A) Two-dimensional annotation enrichment analysis. Pathways altered in the cell line with chromosome 3 trisomy in comparison with the cell line with chromosome 5 trisomy are plotted (Benjamini-Hochberg FDR threshold 0.02). Each dot represents one category as defined in the KEGG and GO database, the colors mark groups of related pathways as described in the inset. Axis—position scores of the pathways; negative values indicate downregulation, positive values indicate upregulation. See Supplementary Information for further details. (B) Pathway alterations identified in HCT116 and RPE-1 cells show similar trends. (C, D) Box-plots of all proteins within representative categories identified as significantly altered by the two-dimensional annotation enrichment analysis. Examples of upregulated (lysosome, carbohydrate catabolic process) and downregulated (spliceosome, chromatin remodeling) pathways show uniform response among all cell lines with extra chromosomes. All differences between DNA and protein levels are statistically significant (unpaired T-test with Welch’s correction, P<0.05). For more information, see also Supplementary Figure S4 and Supplementary Table S2.
Autophagy is activated in aneuploid cells
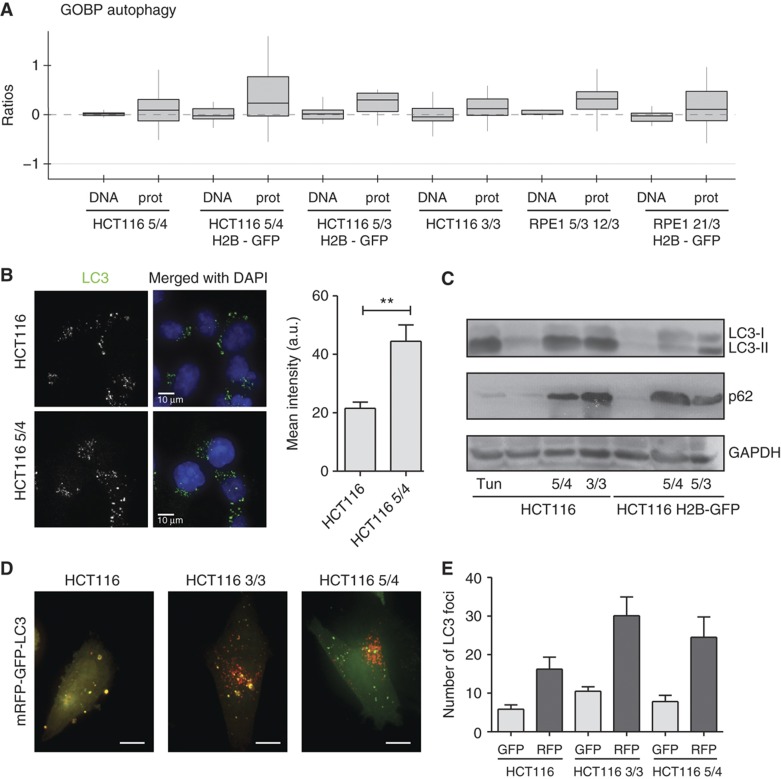
The analysis of the altered pathways indicates that lysosome proteins are upregulated in all aneuploid cell lines (Figure 4C; Supplementary Figure S5). The lysosome is essential for autophagy, a pathway involved in removal of damaged or superfluous proteins and organelles (He and Klionsky, 2009). A detailed analysis of the proteins involved in autophagic processes suggested that autophagy might be more active in aneuploid cell lines (Figure 5A). To substantiate this finding, we performed functional analysis of autophagy in aneuploid cells in comparison with parental diploid cell lines. By immunofluorescence, we observed an increased number of LC3 foci in HCT116 5/4 in comparison with HCT116 (Figure 5B). Similarly, immunoblot analysis of protein lysates from aneuploid clones showed accumulation of the autophagy marker LC3-II (Figure 5C), which is the lipidated form of LC3 that is conjugated to phosphatidylethanolamine (PE) when integrated into the membrane destined for autophagosomes (Kabeya et al, 2000). By monitoring the doubly tagged mRFP-GFP-LC3 (Kimura et al, 2007) we found that the turnover of LC3 in aneuploid clones is similar to that of the diploid control (Figure 5D and E) and can be blocked by inhibition of autophagy with Bafilomycin A1 (Supplementary Figure S6), a drug inhibiting the acidification of lysosomes and fusion of autophagosomes to lysosomes (Klionsky et al, 2008). This confirms that the elevated numbers of autophagosomes are not due to a defect in autophagosome-to-lysosome fusion, but indeed due to autophagy activation. In conclusion, our data suggest that autophagy is activated in trisomic and tetrasomic human cells.
Figure 5.
Activation of autophagy in trisomic and tetrasomic cell lines. (A) Abundance changes of the sub-category autophagy (GOBP) in all analyzed cell lines are depicted. CGH and proteome data are shown. (B) Fluorescence intensity of LC3-positive foci in HCT116 and HCT116 5/4. Right panel: quantification of the fluorescence intensity (non-parametric T-test, **P<0.01). (C) Western blot of LC3-II shows an increase in all analyzed aneuploids. Similarly, levels of p62/SQSTM1 are increased in aneuploids. Note that p62 is coded on chromosome 5, but its levels are increased in all aneuploids. Tun—diploid HCT116 treated with tunicamycin that activates unfolded protein response and hence autophagy. (D) Representative images of cells after transfection with the double-tagged mRFP-GFP-LC3. Yellow foci represent phagosomes (both GFP and mRFP signals visible), red foci represent lysosomes (only mRFP signal is insensitive to the acidic pH in lysosomes). Bar 10 μm. (E) Total number of LC3 foci within a defined area of each cell (2500 voxels). There are significantly more foci in the HCT116 3/3 cell line (non-parametric T-test, P<0.01), the levels in HCT116 5/4 are higher, but the difference from HCT116 is not statistically significant. See also Supplementary Figure S5. Source data is available for this figure in the Supplementary Information.
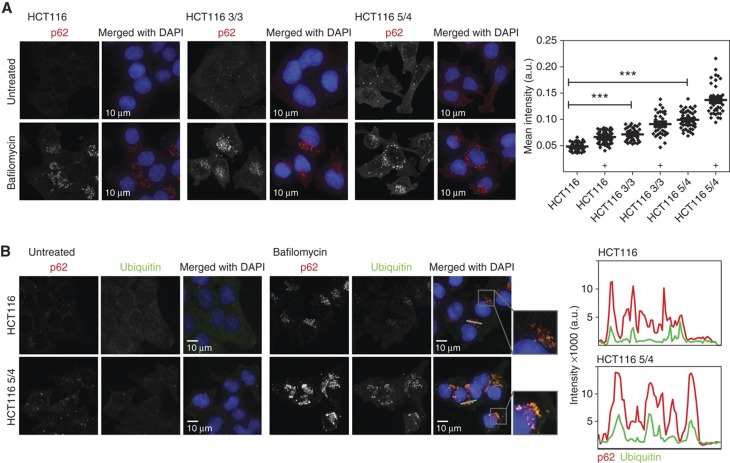
Markedly, the expression of p62/sequestosome (SQSTM1) was enhanced in all analyzed trisomic and tetrasomic cells (Figure 5C). p62 is a cytoplasmic stress response receptor that is activated by various cellular stresses such as oxidative stress (for review, see Lamark and Johansen, 2009). Misfolded or damaged ubiquitylated proteins are sequestered by p62 into aggregates, and targeted to autophagy via a direct interaction with LC3 (Pankiv et al, 2007). Immunoblotting (Figure 5C) as well as immunofluorescence staining showed a significant increase in p62 levels, which could be further enhanced by treatment with Bafilomycin A1 (Figure 6A). Moreover, indirect immunofluorescence revealed an increased co-localization of p62- and ubiquitin-positive foci in both the HCT116- and the RPE-1-derived cell lines, which is also enhanced by Bafilomycin (Figure 6B; Supplementary Figure S7A and B). Thus, our results suggest that cells with supernumerary chromosomes accumulate ubiquitylated proteins in the cytoplasm, likely as a consequence of the protein imbalance. The increase of p62-dependent autophagy could provide a new understanding of the pathways that allow aneuploid cells to maintain protein homeostasis despite chronic elevated expression of multiple genes.
Figure 6.
Analysis of autophagy in trisomic and tetrasomic cell lines. (A) Tri- and tetrasomic cells accumulate p62-positive foci. This effect was further increased by treatment with Bafilomycin A1. Right panel: Quantification of the fluorescence intensity changes; significance was evaluated by non-parametric T-test (***P<0.001), ‘+’ designates addition of Bafilomycin A1. (B) p62-positive foci co-localize with ubiquitin-positive foci and their fluorescence intensity is increased in aneuploid cell lines. Plots represent the signal intensity along the indicated gray line. Red line—p62, green line—ubiquitin. Source data is available for this figure in the Supplementary Information.
Discussion
To investigate the response of human cells to the presence of extra chromosomes, we generated human tri- and tetrasomic cell lines derived from diploid chromosomally stable cell lines HCT116 and RPE-1. The presence of additional chromosomes resulted in significant growth defect in particular in the G1 and S phase, further confirming the adverse effects of extra chromosomes on cellular growth that was observed in cells derived from Down syndrome patients (Segal and McCoy, 1974) or yeast and mouse cells with extra chromosomes (Torres et al, 2007; Williams et al, 2008). The cause of the growth delay remains unclear, but it has been proposed that it might result from a lack of energy, as these cells produce and degrade more proteins (Torres et al, 2007). Alternatively, the progression through the cell cycle might be delayed due to abnormal levels of cell-cycle regulators. Interestingly, highly aneuploid cancer cells do not show significant growth defects that would correlate with the extent of chromosomal alterations. This suggests that cells can adapt to the abnormal karyotype, and indeed, disomy tolerating mutations were recently identified in budding yeast (Torres et al, 2010). Our model tri- and tetrasomy cell lines provide a useful tool to identify the adaptations to abnormal karyotype in human cells and compare them with mutations frequently found in highly aneuploid cancers.
The comparison of DNA, mRNA and protein levels revealed that whereas mRNA abundance increases accordingly to the chromosome copy number changes, the abundance of ∼25% of the proteins coded on the extra chromosomes is lower than expected. This adjustment of the protein abundance is found specifically for protein kinases and subunits of protein complexes. Our results show that the previous notion that multimolecular complexes often maintain stoichiometric levels despite excess of one of the subunits can be generalized to most protein complexes. This question has been so far addressed only in budding yeast. On one hand, analysis of macromolecular complexes in disomic budding yeast revealed partial compensation of subunits of macromolecular complexes (Torres et al, 2010), whereas another study found no significant dosage changes in the protein abundance in budding yeast with complex aneuploid karyotype (Pavelka et al, 2010). Although some of these differences might be due to the technical issues as the changes in gene expression in aneuploids are subtle, another possible explanation is that in cells with multiple aneusomies more than one subunit of a multimolecular complex may be expressed at increased level. This would make the stoichiometry maintenance more complex and difficult to detect. Our results suggest that in human trisomic and tetrasomic cells the stoichiometry of protein subunits is often maintained despite the changes in gene copy numbers. Further analysis will be required to determine the underlying mechanisms.
Next, we asked whether there are global changes in pathway regulation in response to aneuploidy. Several pathways linked to RNA and DNA metabolism as well as cell-cycle regulators were strongly downregulated. This is in accordance with the growth defect of the model aneuploid cell lines. On the other hand, we observed upregulation of carbohydrate and oxidative metabolic processes, membrane metabolism, vesicle transport and lysosome-related pathways. Neither general stress response pathways nor heat-shock response was activated, supporting the idea that chronic protein overexpression does not trigger the same stress response as an acute proteotoxic stress (Gidalevitz et al, 2006). Remarkably, the changes in pathway regulation were very similar in all HCT116- and RPE-1-derived cell lines. Thus, this first pathway analysis in human trisomic and tetrasomic cells showed that there is a general cellular response to abnormal karyotype regardless of the type of chromosome or origin of the cells. As the trisomy of chromosome 21 elicits the same signature response, one can hypothesize that some of these alterations might contribute to the pathology of the Down syndrome. It should be noted that some of the observed changes in pathway regulation might be an indirect consequence of the altered phenotypes of aneuploid cells, such as the growth defect. We propose that the effect is rather modest, because the changes in pathway regulation are not proportional to the growth defects. Previously, a transcriptional analysis of yeast disomic strains revealed general environmental stress response (Torres et al, 2007), whereas no aneuploidy-specific signature was identified in budding yeast strains with complex whole chromosome aneuploidies (Pavelka et al, 2010). Thus, it will be important to compare the identified signature response with pathway regulation changes in human cells with more complex aneuploid karyotypes.
The existence of specific response to aneuploidy suggests that aneuploidy itself might be a useful target in cancer therapy. Markedly, the pathways identified by our approach indeed point to the growth requirements of aneuploid cells. We found upregulation of energy metabolism pathways and increased activation of autophagy in human trisomic cell lines, which corresponds with the recent finding that the energy stress inducer 5-aminoimidazole-4-carboxamide riboside (AICAR) and the autophagy inhibitor chloroquine impair the growth of trisomic mouse cells (Tang et al, 2011). Autophagy inhibiting drugs are considered for cancer therapy, and human tri- and tetrasomic cell lines might provide a useful model for elucidating the molecular mechanisms underlying their effect.
What is the function of autophagy activation in human tri- and tetrasomic cell lines? Autophagy increases in response to nutrient deprivation, which might exist in human cells with extra chromosomes since they activate energy metabolism pathways. However, starvation-induced autophagy usually results in decreased levels of p62 due to its increased turn-over, whereas we identified elevated amounts of p62 in all analyzed tri-and tetrasomic human cell lines. We propose that the p62-mediated selective autophagy is specifically activated to maintain protein homeostasis in cells with extra chromosomes. Future research should address how autophagy is activated in aneuploid cells and whether it contributes to the dosage compensation of proteins that are coded on the extra chromosomes.
In summary, model human tri- and tetrasomic cell lines presented in this study provide a novel system to analyze the cellular response to the presence of an extra chromosome. Identification of pathways altered in the cell lines with supernumerary chromosome may help to elucidate the pathological changes associated with aneuploidies and identify possible treatments for related pathologies such as trisomy syndromes or cancer.
Materials and methods
Cell lines
The cell lines HCT116, HCT116 5/4 and HCT116 3/3 were kindly provided by Minoru Koi, Baylor University Medical Centre, Dallas, TX, USA. The cell line RPE-1 hTERT (hereafter RPE-1) and RPE-1 hTERT H2B-GFP were a kind gift of Stephen Taylor (University of Manchester, UK). HCT116 H2B-GFP as well as RPE-1-derived cell lines were created in our laboratory. HCT116 H2B-GFP was generated by lipofection (FugeneHD, Roche) of HCT116 (ATCC No. CCL-247) with pBOS-H2B-GFP (BD Pharmingen) according to manufacturer’s protocols. All tri- and tetrasomic cell lines were generated by microcell fusion as described below. The donor mouse cell lines A9(Neo5) were purchased from the Health Science Research Resources Bank (HSRRB), Japan; the donor mouse cell line for a transfer of chromosome 21 was a kind gift of Professor Oshimura, Tottori University, Japan. All cell lines were maintained at 37°C with 5% CO2 atmosphere in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS), 100 U penicillin and 100 U streptomycin. The cell lines HCT116 3/3, HCT116 H2B-GFP 5/3, HCT116 H2B-GFP 5/4, RPE-1 5/3 12/3, RPE-1 3/3, RPE-1 H2B-GFP 21/3 and A9(Neo5) were grown in media supplemented with 400 μg/ml G418. The cell line HCT116 5/4 as well as the cell lines stably transfected with H2B-GFP were grown in media supplemented with 6 μg/ml blasticidin S. Before each experiment, tri- and tetrasomic cells were grown for two passages in medium lacking the antibiotics to avoid any effect on protein levels.
Antibodies and reagents
As primary antibody anti-LC3 (115-3) from Medical & Biological Laboratories, anti-p62 from BD Transduction Laboratories (610832, for western blot), anti-p62 from Progen (GP62-C, for Immunofluorescence), anti-Ubiquitin (P4D1) from Santa Cruz, anti-β-Tubulin (ab21057) from Abcam, anti-Cyclin B (05-373) from Millipore and anti-GAPDH (ab9483) from Abcam were used. As secondary antibodies for western blotting, we applied goat anti-mouse IgG HRP affinity purified PAb (HAF007) and donkey anti-goat IgG HRP affinity purified PAb (HAF109) from R&D systems, Minneapolis, USA. For immunofluorescence analysis, we used anti-mouse FITC (Abcam), anti-guinea pig dylight649 and anti-mouse dylight649 (Jackson ImmunoResearch). 5 μg/ml tunicamycin (Sigma) in dimethylsulfoxide (DMSO) was used to induce autophagy in HCT116. Bafilomycin (Sigma) was applied for 16–18 h to a final concentration of 50 nM.
Microcell fusion
To generate cell lines containing an additional chromosome, microcell fusion (Supplementary Figure S1A) was performed as described previously (Fournier, 1981). In brief, mouse A9(Neo5) donor cells containing an additional human chromosome with an antibiotic resistance gene were treated for 48 h with colchicine (final concentration 60 ng/ml). Donor cells were trypsinized and seeded on plastic bullets. After the cells attached to the surface, bullets were centrifuged at 15 000 r.p.m. for 30 min at 30–34°C in DMEM supplemented with 10 μg/ml cytochalasin B. Cell pellets were resuspended in serum-free DMEM and filtered (Whatman, pore size 8 and 5 μm) to clear suspension from mouse cells. Filtered microcells were mixed with phytohemagglutinin (PHA-P) and added to the recipient cell line HCT116 H2B-GFP, RPE-1 or RPE-1 H2B-GFP. Fusion of microcells with the recipient cells was facilitated by polyethylene glycol 1500 (PEG 1500).
Genomic analysis
Genomic DNA for aCGH analysis was extracted using the Qiagen Gentra Puregene Kit following manufacturer’s instructions. The aCGH analysis was performed by IMGM laboratories, Martinsried, Germany. Multicolor FISH (mFISH) was performed by the Chrombios GmbH, Raubling, Germany. For further details on the procedures, see Supplementary Experimental Procedures.
Microarrays
mRNA was purified using the Qiagen mRNeasy mini kit. mRNA array analysis was conducted by IMGM laboratories, Martinsried, Germany; see Supplementary Experimental Procedures. The data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al, 2002) and are accessible through GEO Series accession number GSE39768 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39768).
Cell-cycle analysis
Growth of the cells with extra chromosomes, cell-cycle progression after thymidine block release, cell death and number of senescent cells were analyzed according to previously established protocols. For details, see Supplementary Experimental Procedures.
Live-cell imaging
Freshly cultured cell were seeded sparsely in a 6 channel μ-slide (ibidi, Martinsried, Germany) 24 h before the experiment. Time laps movies were taken by imaging asynchronous cells in a 10 min or 4 min interval for 72 or 48 h, respectively. The slide was placed onto a sample stage with an incubator chamber (EMBLEM, Heidelberg, Germany) maintained at a 37°C, 40% humidity, in an atmosphere of 5% CO2. Imaging was performed using a Zeiss Axio Observer Z1 microscope equipped with a Plan Neofluar 20 × air objective. Metamorph 7.1 software (Molecular Devices) was used to control the microscope. Movies were evaluated using the ImageJ 1.42I software.
Indirect immunofluorescence
Cells were seeded either on glass slides or in an ibidi 8-well slide, grown to 50–70% confluency. The cells were fixed 2 days after seeding by 3.7% paraformaldehyde. To inhibit autophagy, cells were treated 18 h with bafilomycin at a final concentration of 50 nM and then fixed. Cells were labeled with anti-LC3, anti-p62 or anti-ubiquitin using the described antibodies. For p62 immunofluorescence intensity analysis, the cell cytoplasm was additionally stained by HCS cell mask red dye (Invitrogen) before fixation to allow automated cell segmentation. Mean intensity of p62/cell was analyzed using Cell Profiler software. Details on the quantitative analysis are in Supplementary Experimental Procedures.
Autophagy flux assay
Cells were seeded 1 day before transfection to achieve 40% confluency at the day of transfection. The ptfLC3 plasmid (mRFG-GFP-LC3, Addgene) was transfected using Lipofectamin LTX™ and PLUS™ reagent according to manufacturer’s protocol. Images were taken 2 days later. We quantified the number of GFP-positive and mRFP-positive LC3 foci within a fixed area (2500 voxels) and the co-localization of GFP- and RFP-positive foci. Details on the quantitative analysis are in Supplementary Experimental Procedures.
Fluorescence microscopy
The images were taken by a fully automated Zeiss inverted microscope (AxioObserver Z1) equipped with a MS-2000 stage (Applied Scientific Instrumentation, Eugene, OR), the CSU-X1 spinning disk confocal head (Yokogawa), LaserStack Launch with selectable laser lines (Intelligent Imaging Innovations, Denver, CO) and an X-CITE Fluorescent Illumination System. Images were captured using a CoolSnap HQ camera (Roper Scientific) under the control of the Slidebook software (Intelligent Imaging Innovations, Denver, CO). Signals were imaged with a 100 × oil objective by using a 561 nm (mRFP) and 473 nm (GFP) laser and a UV light.
SILAC labeling
Cells were cultured in DMEM (high glucose) devoid of arginine and lysine supplemented with 10% dialyzed FBS (10 kDa cutoff; Invitrogen), 1 × penicillin/streptomycin. Arginine and lysine (Sigma-Aldrich) were added in either light (Arg0; Lys0) or heavy (Arg10; Lys8) form to a final concentration of 33.6 μg/ml for arginine and 73 μg/ml for lysine. The cells were tested for full incorporation after the labeling. All experiments were performed with double labeling.
Protein sample preparation and analysis
Samples were prepared following the filter-aided sample preparation (FASP) protocol. In brief, the cells were lysed with 4% sodium dodecyl sulfate (SDS), 0.1 M dithiothreitol (DTT) in 100 mM Tris/HCl pH 7.6 followed by an incubation for 5 min at 95°C and subsequent sonication for 15 min. Heavy and light samples from one experiment were mixed with equal protein amounts. In all, 200–300 μg protein mix was alkylated and digested overnight with trypsin on Microcon YM-30 filter tubes (Millipore). Peptides were eluted from the membrane using 0.5 M NaCl or ddH2O and separated either using the OFFGEL fractionator (HCT116-derived samples) or a strong anion exchange fractionation (RPE-1-derived samples). Fractions were desalted with StageTips (Empore disk, C18 Reversed phase) before liquid-chromatography mass spectrometry (LC-MS)/MS analysis using CID fragmentation.
Liquid-chromatography mass spectrometry
Peptide mixtures were analyzed using nanoflow liquid chromatography (LC-MS/MS) on an EASY-nLC™ system (Proxeon Biosystems, Odense, Denmark) online connected to the LTQ Orbitrap XL or LTQ Orbitrap Velos instrument (Thermo Fisher Scientific, Bremen, Germany) through a Proxeon nanoelectrospray ion source. Peptide samples were directly autosampled onto a 15-cm in-house packed column (75 μl inner diameter; Proxeon Biosystems) with 3 μm reversed phase beads (ReproSil-Pur C18-AQ, Dr Maisch). Using a 170-min gradient (2–30% ACN), peptides were directly electrosprayed (2.2 kV) into the mass spectrometer. Mass spectrometer was operated in data-dependent mode switching automatically between one full scan MS and 7–15 MS/MS acquisitions. Instrument control was through Tune 2.6.0 and Xcalibur 2.1.0. Full scan MS spectra (m/z 300–1650) were acquired in the Orbitrap analyzer after accumulation to a target value of 106 in the linear ion trap (resolution of 60 000 at 400 m/z). Fragmentation was performed in CID mode applying 35% normalized collision energy after accumulation of the parent ions to a target value of 5000.
Data analysis
All double labeling SILAC experiments were analyzed together per cell line using the in-house developed software MaxQuant (version 1.0.14.10 and 1.2.0.25) with standardized workflow. Data were searched against the human International Protein Index protein sequence database (ipi.HUMAN.v3.62.dec) supplemented with frequently observed contaminants and concatenated with reversed copies of all sequences (target-decoy database). False discovery rates on peptide and protein level were fixed to 1%, including automatic filtering on peptide length, mass error precision estimates, and peptide scores of all forward and reversed peptide identifications. Reported protein groups had to be identified by at least one unique peptide to be accepted. Quantitation was based on unique and razor peptides only and a minimum of two ratio counts was required. Complete protein and peptide lists as well as the underlying *.RAW files are available on the TRANCHE database (https://proteomecommons.org/tranche/). For details, see Supplementary Experimental Procedures.
Statistics
All statistically evaluated experiments were performed in at least three independent biological replicates. The statistical evaluation was performed using GraphPad Prism 5 software, or R. All details on statistical analyses are in Supplementary Experimental Procedures.
Supplementary Material
A merged file of Supplementary Information Guide, Supplementary Figures 1 to 7 in high resolution and Supplementary Experimental Procedures
A complete summary of the quantitative genome, transcriptome and proteome data
Data used for the 2-D annotation enrichment analysis
A summary of the quantitative genome, transcriptome and proteome data of subunits of macromolecular complexes
Acknowledgments
HCT116 3/3 and HCT116 5/4 were kindly provided by Minoru Koi, Baylor School of Medicine, Dallas, TX, USA. We thank Christian Kuffer for the cell line HCT116 H2B-GFP, Sarah Schunter and Susanne Gutmann for technical assistance and Christoph Schaab and Silke Hauf for helpful discussions. We thank Sabine Langer and Doris Sollacher for help with the chromosome preparations and Stefan Jentsch and Tim Ammon for providing the anti-LC3 antibody and scientific advice. This research was supported by the Max-Planck Society and by the Center for Integrated Protein Science, Munich and by a grant from Deutsche Forschungsgemeinschaft to ZS. GS is supported by the Deutsche Krebshilfe (ON2 grant).
Author contributions: ZS initiated the study; ZS, SS and GS designed the experiments; SS and GS performed the experiments; KP established the chromosome transfer technique and created the RPE-1 derivatives; JC created the software; GS, SS and ZS analyzed the data; ZS wrote the manuscript, all authors discussed the results and commented on the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aït Yahya-Graison E, Aubert J, Dauphinot L, Rivals I, Prieur M, Golfier G, Rossier J, Personnaz L, Créau N, Bléhaut H, Robin S, Delabar JM, Potier MC (2007) Classification of human chromosome 21 gene-expression variations in Down syndrome: impact on disease phenotypes. Am J Hum Genet 81: 475–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Riddle NC, Auger DL, Veitia RA (2005) Dosage balance in gene regulation: biological implications. Trends Genet 21: 219–226 [DOI] [PubMed] [Google Scholar]
- Chen G, Bradford WD, Seidel CW, Li R (2012) Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature 482: 246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier RE (1981) A general high-efficiency procedure for production of microcell hybrids. Proc Natl Acad Sci USA 78: 6349–6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI (2006) Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311: 1471–1474 [DOI] [PubMed] [Google Scholar]
- Goldberg AL, Dice JF (1974) Intracellular protein degradation in mammalian and bacterial cells. Annu Rev Biochem 43: 835–869 [DOI] [PubMed] [Google Scholar]
- Guialis A, Morrison KE, Ingles CJ (1979) Regulated synthesis of RNA polymerase II polypeptides in Chinese hamster ovary cell lines. J Biol Chem 254: 4171–4176 [PubMed] [Google Scholar]
- Guo M, Birchler JA (1994) Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science 266: 1999–2002 [DOI] [PubMed] [Google Scholar]
- Hassold T, Hall H, Hunt P (2007) The origin of human aneuploidy: where we have been, where we are going. Hum Mol Genet 16: R203–R208 [DOI] [PubMed] [Google Scholar]
- Haugen AC, Goel A, Yamada K, Marra G, Nguyen TP, Nagasaka T, Kanazawa S, Koike J, Kikuchi Y, Zhong X, Arita M, Shibuya K, Oshimura M, Hemmi H, Boland CR, Koi M (2008) Genetic instability caused by loss of MutS homologue 3 in human colorectal cancer. Cancer Res 68: 8465–8472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Klionsky DJ (2009) Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43: 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel B, Kreil DP, Matzke M, Matzke AJ (2008) Effects of aneuploidy on genome structure, expression, and interphase organization in Arabidopsis thaliana. PLoS Genet 4: e1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19: 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T (2007) Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3: 452–460 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT et al. (2008) Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4: 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitek DJ, Will JL, Gasch AP (2008) Variations in stress sensitivity and genomic expression in diverse S. cerevisiae isolates. PLoS Genet 4: e1000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamark T, Johansen T (2009) Autophagy: links with the proteasome. Curr Opin Cell Biol 22: 192–198 [DOI] [PubMed] [Google Scholar]
- Makarevitch I, Phillips RL, Springer NM (2008) Profiling expression changes caused by a segmental aneuploid in maize. BMC Genomics 9: 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masramon L, Ribas M, Cifuentes P, Arribas R, García F, Egozcue J, Peinado MA, Miró R (2000) Cytogenetic characterization of two colon cell lines by using conventional G-banding, comparative genomic hybridization, and whole chromosome painting. Cancer Genet Cytogenet 121: 17–21 [DOI] [PubMed] [Google Scholar]
- Niwa O, Tange Y, Kurabayashi A (2006) Growth arrest and chromosome instability in aneuploid yeast. Yeast 23: 937–950 [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1: 376–386 [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145 [DOI] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R (2010) Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468: 321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A, Waegele B, Lechner M, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Mewes HW (2010) CORUM: the comprehensive resource of mammalian protein complexes--2009. Nucleic Acids Res 38: D497–D501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal DJ, McCoy EE (1974) Studies on Down's syndrome in tissue culture. I. Growth rates and protein contents of fibroblast cultures. J Cell Physiol 83: 85–90 [DOI] [PubMed] [Google Scholar]
- Selmecki A, Forche A, Berman J (2006) Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313: 367–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, Amon A (2011) Aneuploidy drives genomic instability in yeast. Science 333: 1026–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg P, Lundberg LE, Johansson A-M, Rydén P, Svensson MJ, Larsson J (2009) Buffering of Segmental and Chromosomal Aneuploidies in Drosophila melanogaster. PLoS Genet 5: e1000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchova Z, Pellman D (2004) From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol 5: 45–54 [DOI] [PubMed] [Google Scholar]
- Tang YC, Williams BR, Siegel JJ, Amon A (2011) Identification of aneuploidy-selective antiproliferation compounds. Cell 144: 499–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A (2010) Identification of aneuploidy-tolerating mutations. Cell 143: 71–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A (2007) Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317: 916–924 [DOI] [PubMed] [Google Scholar]
- Upender MB, Habermann JK, McShane LM, Korn EL, Barrett JC, Difilippantonio MJ, Ried T (2004) Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res 64: 6941–6949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A (2008) Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322: 703–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A merged file of Supplementary Information Guide, Supplementary Figures 1 to 7 in high resolution and Supplementary Experimental Procedures
A complete summary of the quantitative genome, transcriptome and proteome data
Data used for the 2-D annotation enrichment analysis
A summary of the quantitative genome, transcriptome and proteome data of subunits of macromolecular complexes