Abstract
We report a proteomic analysis of microdissected material from formalin-fixed and paraffin-embedded colorectal cancer, quantifying >7500 proteins between patient matched normal mucosa, primary carcinoma, and nodal metastases. Expression levels of 1808 proteins changed significantly between normal and cancer tissues, a much larger fraction than that reported in transcript-based studies. Tumor cells exhibit extensive alterations in the cell-surface and nuclear proteomes. Functionally similar changes in the proteome were observed comparing rapidly growing and differentiated CaCo-2 cells. In contrast, there was minimal proteomic remodeling between primary cancer and metastases, suggesting that no drastic proteome changes are necessary for the tumor to propagate in a different tissue context. Additionally, we introduce a new way to determine protein copy numbers per cell without protein standards. Copy numbers estimated in enterocytes and cancer cells are in good agreement with CaCo-2 and HeLa cells and with the literature data. Our proteomic data set furthermore allows mapping quantitative changes of functional protein classes, enabling novel insights into the biology of colon cancer.
Keywords: colon cancer, FASP, FFPE tissue, microdissection, quantitative proteomics
Introduction
In the past decades, colorectal cancer (CRC) has been comprehensively studied with ever more refined molecular biology methods (Fearon, 2011). A large number of expression and genomic profiling studies have contributed to our current understanding of the different cellular mechanisms underlying colorectal tumor formation, maintenance, and development of metastasis (Cardoso et al, 2007). However, these achievements did not provide direct information on the composition of the major active components of the cell, which are its proteins. To fill this gap in understanding of the disease, many proteomic studies have been performed. They investigated clinical material or cultured cells, typically providing extensive lists of protein changes in CRC. Usually, these observations were restricted to highly abundant molecules (Jimenez et al, 2010). The application of proteomics has been hampered by a lack of methods allowing exploration of tissues to a depth comparable to that provided by molecular biology methods, which analyze alterations in the cellular genome or transcript levels (Nambiar et al, 2010). At best proteomic studies were able to identify and quantify a few thousand proteins, but most studies focused only on changes in the levels of a few proteins. This is in stark contrast to microarray or deep sequencing-based transcriptomic studies that can cover large parts of the transcriptome. In particular, proteomics so far did not provide systematic insights into changes in signaling pathway, alterations in the cell-surface proteome, or variations in processing genetic information in CRC. Moreover, proteomic approaches have usually required large amounts of sample and therefore could not profit from microdissection techniques enabling isolation of defined populations of cells.
Recently, our group has reported advances in sample preparation methods that enabled studying membrane proteins in the same manner as soluble proteins (Wisniewski et al, 2009b; Wisniewski, 2011) and allowed identification and comparison of thousands of proteins from samples containing only few microgram of total protein (Wisniewski et al, 2011a). In addition, we have recently developed an efficient protocol that enables the analysis of proteomes and post-translational modifications in formalin-fixed and paraffin-embedded (FFPE) tissues (Ostasiewicz et al, 2010). Taking advantage of these technological developments in conjunction with ongoing refinements in high accuracy mass spectrometry (Nagaraj et al, 2011), we set out to analyze proteomes of normal mucosal tissue, adenocarcinoma, and its nodal metastasis from laser microdissected patient matched FFPE samples.
Using label-free quantitative analysis, we wanted to obtain insights into overall changes in subcellular composition, molecular function, and pathways in CRC. We report relative quantitation of 7576 proteins between the three stages and show that a large fraction of the proteome is changed between the normal mucosa and cancer cells, whereas the altered proteome profile is preserved largely unchanged in nodal metastases.
Results
Identification of 8000 proteins from laser microdissected colonic mucosa, primary cancer, and its nodal metastasis
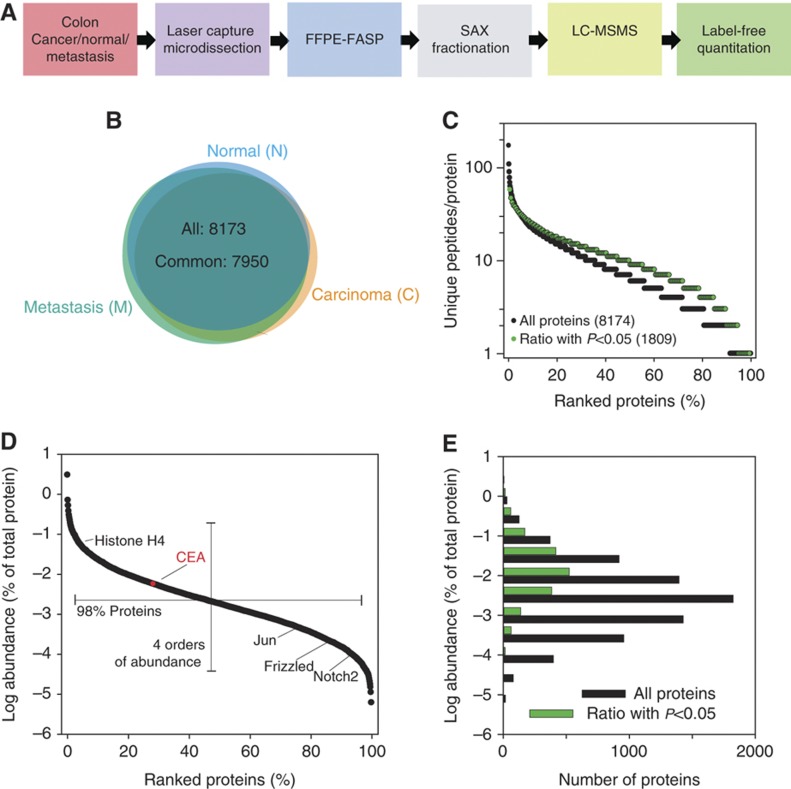
To survey and compare the proteomes of colonic mucosa (normal or N) and colon cancer (cancer or C), we analyzed archival FFPE clinical samples originating from eight patients (Supplementary Table 1; Materials and methods). In addition, for seven of the patients we analyzed the proteomes of nodal metastases (M). Laser capture microdissection was used to obtain enriched populations of enterocytes, primary cancer, and metastasizing cells (Supplementary Figure 1). We made an effort to reduce any contamination by stroma and therefore did not emphasize identification of secreted proteins. From each sample, a volume of about 175 nl of cells were collected and processed using the FFPE-FASP (filter aided sample preparation) procedure (Wisniewski et al, 2011a). Yields were 6.1±1.8 μg total peptide per sample. To maximize depth of proteome coverage, peptides were fractionated by anion exchange chromatography into six fractions before they were analyzed by LC-MS/MS on a linear ion trap Orbitrap mass spectrometer (Wisniewski et al, 2011a; Figure 1A). Fragment spectra were obtained by Higher Energy Collisional Dissociation (HCD) with high mass accuracy (Olsen et al, 2009). The complete data acquisition took 23 days. The obtained mass spectrometric raw data were analyzed in the MaxQuant environment (Cox and Mann, 2008) with the integrated Andromeda searching engine (Cox et al, 2011). Label-free quantitation (LFQ) algorithms in MaxQuant (Luber et al, 2010) allowed quantitative comparison of the individual samples. In our analysis, 72 000 unique peptides corresponding to 8173 proteins were identified at a false discovery rate (FDR) of 1% (Supplementary Tables 2 and 3). Comparison of the N, C, and M proteomes revealed that 99% of the identified proteins were common in all three stages (Figure 1B). Of these, 7576 proteins were identified at least four times in one of the states (N, C, or M) and only these were subjected to further analysis and statistical evaluation (Supplementary Table 4). In all, 90% of these proteins and 94% of the proteins that were found to be significantly changed between the normal and cancer cells were identified by at least two peptides (Figure 1C). The summed mass spectrometric peptide intensities—a proxy for the protein abundances of the identified proteins—span six orders of magnitude, but the levels of 98% of the proteins were within a 10 000-fold expression range (Figure 1D). Comparison of the abundances of all proteins and those identified as significantly changed showed that with descending abundance the frequency of detection of statistically significant protein changes decreases (Figure 1E). For highly abundant proteins, expression levels of half of the proteins changed significantly, whereas for low abundant proteins this proportion was much smaller (Figure 1E). This is likely due to technical factors and indicates that a substantial portion of quantitative differences remains undetected. Assuming that over the entire range of abundance the portion of proteins with altered levels is similar, we expect that about 50% of the proteome is changed between normal and cancer cells.
Figure 1.
Proteomic analysis of FFPE archival samples of colonic mucosa, cancer, and metastasis. (A) Proteomic workflow applied to microdissected samples of colonic mucosa (N), cancer (C), and metastasis (M). (B) Overlap of the proteins identified from colonic mucosa, cancer, and metastasis. (C) Peptide-based identification of proteins. (D) Distribution of protein abundances with selected examples. In red, relative abundance of CEA. Examples of lower abundant proteins identified in this study: Jun, proto-oncogene Jun; Frizzled, a G protein-coupled receptor protein that serves as receptor in the Wnt signaling pathway; and Notch 2. (E) Comparison of abundance distribution of all proteins and proteins that were significantly changed in adenocarcinomas. The protein abundances were calculated on the basis of total peptide intensities of all the quantified proteins.
Colon carcinoma is accompanied by extensive subcellular and functional changes
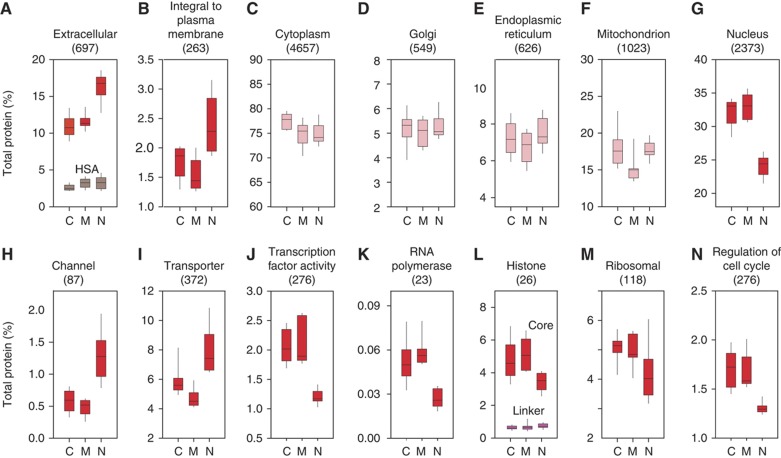
To describe the protein content constituting defined cellular compartments or molecular functions and to provide insight into differences between cellular states, we used the total signal intensities of the peptides identifying each protein as determined in the MaxQuant software (normalized values for LFQ intensities). These peptide intensities are a good proxy for the absolute abundance of the proteins, especially when a set of proteins is considered because inaccuracies for specific proteins are averaged out. Panels A–G in Figure 2 show representative examples of the protein content of selected subcellular compartments. Clear alterations were apparent between N and C, but not between C and M. In particular, the abundance of extracellular and integral to plasma membrane proteins in the N proteome was 20–30% higher than in the C proteome whereas the abundance of nuclear proteins was about 30% less (Figure 2A, B). We did not find clear differences between N and C in the overall abundance of proteins belonging to cytoplasm (Figure 2C) and its major components such as the Golgi body, mitochondrion, and endoplasmic reticulum (Figure 2D–F). We next investigated selected functional categories related to the altered subcellular compartments (Figure 2H–N). Partial loss of integral plasma membrane proteins in cancer was accompanied by a 50 and 30% decrease in the abundance of channel and transporter proteins, respectively (Figure 2H and I). Several individual examples of this global decrease are shown in Tables I and II. They included a >10-fold decrease of chloride and sodium channels and three- to five-fold reduction of FXYD ion transport regulators 1 and 3 (Table I). The latter proteins are also known as Phospholemnan and Mat-8 and are known modulators of Na,K-ATP-ases that affect their kinetic properties (Garty and Karlish, 2006). The increase of the total content of nuclear proteins correlates well with changes in the content of histones, subunits of the RNA polymerases, and transcription factors (Figure 2J, K). We found that general transcription factors and chromatin activators such as histone deacetylases and the high mobility group proteins were 2- to 10-fold upregulated in the cancer samples (Table I). A similar increase of protein abundance was found for a number of nuclear transporters (Table II).
Figure 2.
Abundance of proteins matching selected subcellular locations (A–G) and functions (H–N). The numbers of proteins belonging to each class are given in the parentheses. The protein abundances were calculated on the basis of total spectral intensities of all quantified proteins. Red boxes indicate that the changes between cancer and normal were significant with P<0.01. Note that serum albumin levels (gray boxes) are similar between the three types of samples, indicating absence of differential contamination with plasma proteins. In (L), summed abundance of core and linker histones are shown by separate boxes. The purple boxes are for linker histones whereas the core histones by red boxes.
Table 1. Examples of transporter and channel proteins with changed abundances in colon cancer.
| Protein | Gene | Uniprot | Abundancea | Ratio cancer/normal | P-value |
|---|---|---|---|---|---|
| Nuclear transporters | |||||
| Importin-4 | IPO4 | Q8TEX9-2 | MED | 3.9 | 0.0037 |
| Importin-5 | IPO5 | O00410 | HIGH | 11.3 | 0.0007 |
| Importin-7 | IPO7 | O95373 | HIGH | 6.0 | 0.0038 |
| Importin-8 | IPO8 | O15397 | HIGH | 2.2 | 0.048 |
| Importin-9 | IPO9 | Q96P70 | HIGH | 4.9 | 0.0022 |
| Importin-11 | IPO11 | B4DZ73 | HIGH | 3.4 | 0.020 |
| Importin subunit α-2 | KPNA2 | P52292 | MED | 5.8 | 0.0083 |
| Importin subunit α-3 | KPNA3 | O00505 | MED | 2.4 | 0.033 |
| Importin subunit β-1 | KPNB1 | Q14974 | MED | 2.2 | 0.013 |
| Exportin-2 | CSE1L | P55060-1 | MED | 8.8 | 0.0033 |
| Non-nuclear channels and transporters | |||||
| Chloride intracellular channel protein 2 | CLIC2 | O15247 | LOW | 0.026 | 0.0030 |
| Calcium-activated chloride channel regulator 1 | CLCA1 | A8K7I4 | HIGH | 0.031 | 0.0010 |
| Calcium-activated chloride channel regulator 4 | CLCA4 | A8MQC9 | MED | 0.052 | 0.0055 |
| Sodium channel protein type 7 subunit α | SCN7A | Q01118 | MED | 0.068 | 0.0186 |
| FXYD domain-containing ion transport regulator 3 | FXYD3 | Q14802-2 | MED | 0.17 | 0.0462 |
| FXYD domain-containing ion transport regulator 1 | FXYD1 | O00168 | LOW | 0.32 | 0.0171 |
| Chloride anion exchanger | SLC26A3 | P40879 | MED | 0.12 | 0.00708 |
| Fatty acid-binding protein, intestinal | FABP2 | P12104 | HIGH | 0.070 | 0.00533 |
| Fatty acid-binding protein, liver | FABP1 | P07148 | HIGH | 0.16 | 0.0461 |
aProtein abundances assigned using MS intensities. Values correspond to the 0–25th percentile for LOW, 25th–75th percentiles for MEDIUM, and 75th–100th percentile for HIGH.
Table 2. Examples of nuclear proteins with changed abundances in colon cancer.
| Protein | Gene | Uniprot | Abundancea | Ratio cancer/normal | P-value |
|---|---|---|---|---|---|
| General transcription factors | |||||
| General transcription factor IIE1 | GTF2E1 | P29083 | MED | 4.4 | 0.0123 |
| General transcription factor IIF2 | GTF2F2 | P13984 | MED | 2.3 | 0.0290 |
| General transcription factor IIH2 | GTF2H2 | Q13888 | MED | 2.4 | 0.0062 |
| General transcription factor IIH4 | GTF2H4 | Q92759 | LOW | 2.8 | 0.0101 |
| General transcription factor II-I | GTF2I | P78347-1 | HIGH | 4.4 | 0.0009 |
| Transcription elongation factor S-II | GTF2S | P23193-1 | HIGH | 4.5 | 0.0033 |
| General transcription factor 3C4 | GTF3C4 | Q9UKN8 | MED | 3.4 | 0.0476 |
| Nucleolar transcription factor 1 | UBF | P17480-1 | MED | 1.8 | 0.0114 |
| Deacetylases | |||||
| Histone deacetylase 1 | HDAC1 | Q13547 | HIGH | 2.4 | 0.0481 |
| Histone deacetylase 2 | HDAC2 | Q92769-2 | HIGH | 4.2 | 0.0262 |
| High mobility group proteins | |||||
| High mobility group protein 20A | HMG20A | Q9NP66-1 | MED | 6.6 | 0.0134 |
| High mobility group AT-hook protein 1 | HMGA1 | P17096-1 | MED | 9.4 | 0.0027 |
| High mobility group protein B1 | HMGB1 | P09429 | HIGH | 3.7 | 0.0009 |
| High mobility group protein B2 | HMGB2 | P26583 | MED | 2.6 | 0.0328 |
| High mobility group protein N1 | HMGN1 | P05114 | MED | 5.4 | 0.0047 |
| Nuclear ubiquitous casein and cyclin-dependent kinases substrate | NUCKS | Q9H1E3-1 | MED | 4.3 | 0.0028 |
aProtein abundances assigned using MS intensities. Values correspond to the 0–25th percentile for LOW, 25th–75th percentiles for MEDIUM, and 75th–100th percentile for HIGH.
According to the canonical model of chromatin one linker histone H1 molecule occurs per one core histone octamer. Since the molecular masses of the linker and the core histones are about 20 and 10 kDa, respectively, the total amount of H1 should be a quarter of the core histones (Figure 2E). Indeed, this is the ratio we observed in the N samples (0.24). In contrast, in the cancer cells the content of core histones was increased and the ratio of H1 to core histone decreased to only 0.14. This suggests a reduced compaction of chromatin accompanying elevated transcriptional activity.
Additionally, elevated expression of proteins involved in cell-cycle regulation in C and M reflects higher cell division rates of cancer cells in comparison with normal cells (Figure 2N). Similarly, elevated expression of ribosomal proteins in C and M reflects an upregulation of translation (Figure 2M). Interestingly, this change is not reflected in the content of endoplasmic reticulum (Figure 2E), suggesting that this functionality is not needed to a higher degree in cancer cells.
Identification of proteins changed in colon cancer
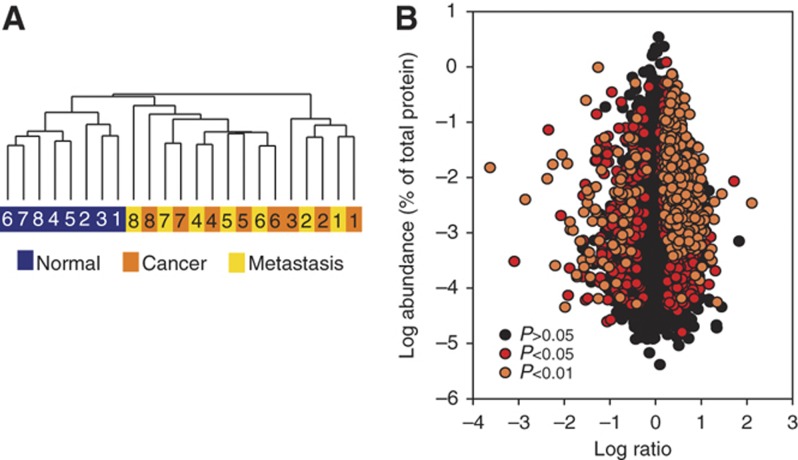
To assess the similarities of the analyzed proteomes in a global manner, we performed unsupervised clustering, in which samples are grouped on the basis of their expression patterns (Figure 3A). All eight samples of normal mucosa clustered together. In striking contrast, cancer and nodal metastases clustered into seven patient matched pairs. In these pairs, the primary tumor had a higher similarity to its metastatic counterpart than to other tumors. To identify potential driver proteins of cancer, we first compared the intensities of detected proteins between colonic mucosa, primary cancer, and its nodal metastasis. Paired t-test was used to determine the significance of protein differences. In total, 1808 proteins were significantly (P<0.05) downregulated or upregulated in tumor tissues when compared with normal mucosa (Figure 3B; Supplementary Table 5). Changes of 762 proteins were observed at a P-value of 0.01 (Figure 3B). The same statistical analysis of our proteomics data did not identify any significant changes between the primary tumor and the nodal metastasis.
Figure 3.
Quantitative comparison of proteins identified in colonic mucosa, cancer, and metastasis. (A) Unsupervised hierarchical clustering of intensities of 7576 proteins for patient matching primary adenocarcinomas and adjacent normal mucosa (8 pairs) and 7 nodal metastases. (B) Distribution of protein ratios versus protein abundance.
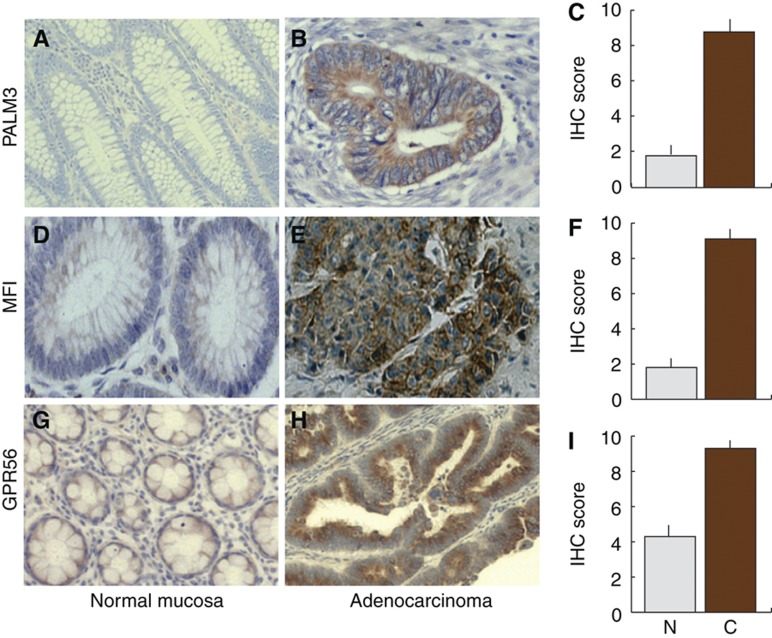
Proteins that are significantly upregulated in cancer samples can be considered as potential biomarkers. Among these, cell-surface proteins are particularly attractive because they may be targeted by antibodies. Table III lists 34 upregulated plasma membrane proteins. Two low abundant and one medium abundant protein were selected for further validation by immunohistochemistry. Antibodies against PALM3, MFI, and GPR56 strongly stained cancer cells whereas only weak staining was observed in normal tissue, thus validating upregulation of the proteins (Figure 4). Knowledge on proteins significantly changed between normal tissue and adenocarcinoma can be used for subsequent targeted analysis of larger number of samples, using either IHC or mass spectrometry-based methods. This may identify clinically relevant correlations between protein upregulation in cancer and the disease outcome. Another attractive follow-up to this study would be to search for the proteins identified as outliers as secreted proteins in plasma samples (Hanash et al, 2008). In this way, clinically useful biomarkers could be identified and potentially later monitored by conventional ELISA.
Table 3. In cancer upregulated plasma membrane proteins.
| Protein names | Gene names | Uniprot | Protein abundancea | Cancer/normal ratio (fold) | P-value |
|---|---|---|---|---|---|
| Retinoic acid-induced protein 3 | GPCR5A | Q8NFJ5 | Medium | 131.0 | 0.0017 |
| Dipeptidase 1 | DPEP1 | P16444 | High | 54.0 | 0.0184 |
| Paralemmin-3 | PALM3 | A6NDB9 | Low | 22.8 | 0.0049 |
| Solute carrier family 2 (facilitated glucose transporter) | SLC2A1 | P11166 | Medium | 20.0 | 0.0045 |
| Large neutral amino acids transporter small subunit 1 | SLC7A5 | Q01650 | Medium | 17.4 | 0.0062 |
| Melanocyte-stimulating hormone receptor | MC1R | Q01726 | High | 9.6 | 0.0084 |
| Calponin-2 | CNN2 | Q99439 | High | 8.9 | 0.0090 |
| 4F2 cell-surface antigen heavy chain | SLC3A2 | P08195 | High | 7.3 | 0.0123 |
| C-terminal tensin-like protein;Tensin-4 | TNS4 | Q8IZW8 | Low | 7.2 | 0.0464 |
| Melanotransferrin | MFI2 | P08582 | Medium | 6.7 | 0.0250 |
| G protein-coupled receptor 56 | GPR56 | Q9Y653 | Low | 6.1 | 0.0305 |
| 1,2-Dihydroxy-3-keto-5-methylthiopentene dioxygenase | ADI1 | Q9BV57 | Medium | 6.0 | 0.0067 |
| Epidermal growth factor receptor kinase substrate 8 | EPS8 | Q12929 | High | 5.3 | 0.0327 |
| von Willebrand factor | VWF | P04275 | High | 5.2 | 0.0055 |
| Tyrosine-protein kinase Lyn | LYN | P07948 | Medium | 4.9 | 0.0017 |
| Exocyst complex component 4 | EXOC4 | Q96A65 | Medium | 4.8 | 0.0066 |
| Serine/threonine-protein kinase N1 | PKN1 | Q16512 | Low | 4.7 | 0.0344 |
| Protocadherin fat 1 | FAT1 | Q14517 | Medium | 4.6 | 0.0176 |
| Focal adhesion kinase 1 | PTK2 | Q05397 | Medium | 4.5 | 0.0218 |
| Neutral amino-acid transporter B(0) | SLC1A5 | Q15758 | High | 4.5 | 0.0085 |
| Junctophilin-1 | JPH1 | Q9HDC5 | Medium | 4.4 | 0.0283 |
| Syntenin-1 | SDCBP | O00560 | Medium | 4.1 | 0.0060 |
| Protein kinase C iota type | PRKCI | P41743 | Medium | 4.1 | 0.0234 |
| Rho-related GTP-binding protein RhoB | RHOB | P62745 | Medium | 4.1 | 0.0061 |
| Early endosome antigen 1 | EEA1 | Q15075 | High | 3.7 | 0.0057 |
| Amyloid β A4 protein | A4;AD1;APP | P05067 | Medium | 3.6 | 0.0123 |
| Transferrin receptor protein 1 | TFRC | P02786 | High | 3.5 | 0.0152 |
| STE20-like serine/threonine-protein kinase | SLK | Q9H2G2 | Medium | 3.5 | 0.0478 |
| Protein diaphanous homolog 1 | DIAP1 | O60610 | High | 3.5 | 0.0031 |
| Gephyrin | GPH | Q9NQX3 | Medium | 3.4 | 0.0189 |
| Lymphocyte antigen 75 | LY75 | O60449 | High | 3.4 | 0.0212 |
| Protein kinase C and casein kinase substrate in neurons 3 | PACSIN3 | Q9UKS6 | Low | 3.2 | 0.0254 |
| Periplakin | PPL | O60437 | Medium | 3.2 | 0.0488 |
| CD166 antigen | ALCAM | Q13740 | Medium | 3.1 | 0.0194 |
| Interferon-induced guanylate-binding protein 1 | GBP1 | P32455 | Medium | 3.0 | 0.0039 |
aProtein abundances assigned using MS intensities. Values correspond to the 0–25th percentile for LOW, 25th–75th percentiles for MEDIUM, and 75th–100th percentile for HIGH.
Figure 4.
Immunohistochemical staining of adenocarcinoma (A, D, G) and normal mucosa (B, E, H) with antibodies against PALM3, MFI2, and GPR56. (C, F, I) Statistical evaluation of the staining of 20 sample pairs. All differences between cancer and normal were significant at P<0.0001.
Proteomic screen captures quantitative changes in regulatory pathways
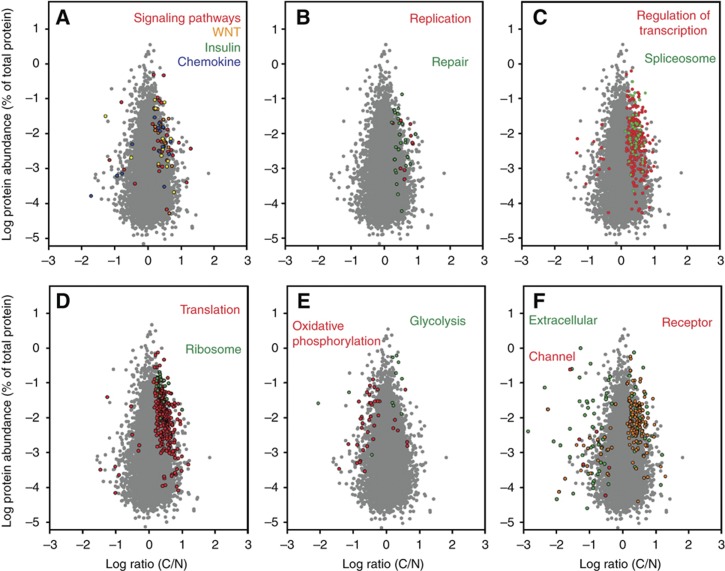
The high number of identified proteins allowed systematic investigation of functional alterations characteristic for colon cancer cells such as signaling pathways, metabolic processes, ion transport, replication, and transcriptional regulation, among many others. Alterations in signaling pathways due to gene mutations are recognized drivers of cancer development, but so far it has been difficult to measure the corresponding proteins directly by quantitative methods. We found that in CRC the WNT, p53, ERBB, or TGFβ pathways are most frequently affected. We identified 147 proteins involved in at least 1 of these pathways; 29 of them were significantly elevated in tumor and none significantly downregulated (Figure 5A). We observed a similar distribution of changes for insulin and chemokine pathways, with a majority of upregulated and only a few downregulated members (Figure 5A). The coverage of key members of these pathways and their quantitative changes is shown in Supplementary Figure 2.
Figure 5.
Distribution of proteins with significant C/N ratio (P<0.05) representing selected pathways and specific functions. (A) Cellular signaling; (B) DNA replication and repair; (C) regulation of transcription and the spliceosome; (D) translation and the ribosomal proteins; (E) oxidative phosphorylation and glycolysis/gluconeogenesis; (F) plasma membrane channel, extracellular and receptor proteins. Gray background dots show the distribution of all quantified proteins without significant change between cancer and normal tissue.
Accelerated growth is a common feature of cancer cells and is reflected in an increase of the abundance of proteins involved in processing the genetic information. A considerable number of proteins involved in these processes were quantified in our proteomic data. All significantly changed proteins matching the GO categories of DNA replication and repair occurred at higher levels in cancer (Figure 5B). Similarly, of the 234 and 300 significantly changed proteins involved in the regulation of transcription and translation, respectively, 95% were upregulated (Figure 5C and D). Likewise, all quantified ribosomal and spliceosomal proteins with significant changes appear in cancer at higher levels (Figure 5C and D).
One of the most common biochemical phenotype of cancer cells, including those from colon, is their elevated glycolysis (the Warburg effect, Warburg, 1956; Vander Heiden et al, 2009) and, under hypoxia, reduced oxidative phosphorylation. Our list of changed proteins includes 11 proteins involved in glycolysis/gluconeogenesis (Figure 5E). It contains upregulated glycolytic enzymes such as phosphofructokinase, pyruvate kinase, or enolase. In contrast, 80% of the changes in the level of proteins involved in oxidative phosphorylation showed downregulation (Figure 5E). Examples are NADH dehydrogenase subunits 3, 4, 5, and 6 with at least five-fold decrease and cytochrome c oxidase subunits 2, 3, 4, 5, and 6 with an average four-fold decrease in expression (Supplementary Table 5). Many of these changes were generally expected, providing a positive control for our experiments. However, knowledge of the magnitude of protein expression changes in a near comprehensive data set of this pathway in vivo should provide interesting information for the characterization of CRC.
In summary, in this study we observed extensive downregulation in the global abundance of proteins occurring in the extracellular space and those having transporter or channel activity (see above). All plasma membrane channels were significantly downregulated in cancer (Figure 5F). Interestingly, receptor proteins occurring in the plasma membrane were upregulated.
Differentiation-related changes in cultured colon cancer cells resemble the extensive subcellular and functional differences between cancer and normal cells
The unexpectedly large differences between cancer and normal cells prompted us to investigate another system where differentiated cancer cells (enterocyte-like cells) can be compared with undifferentiated ones. For this purpose, we selected the human cell line CaCo-2, which is a well-established model system to study cellular differentiation of enterocytes. These cells have the capability to differentiate spontaneously into polarized cells with the morphological and after reaching confluence exhibit the biochemical properties of enterocytes (Zweibaum et al, 1983). We compared the proteomes of cells that reached 100% confluence with cells that were harvested after 4, 11, and 18 days beyond this point. Proteomics identified 9712 proteins of which 7504 were quantified between the stages (Supplementary Table 6).
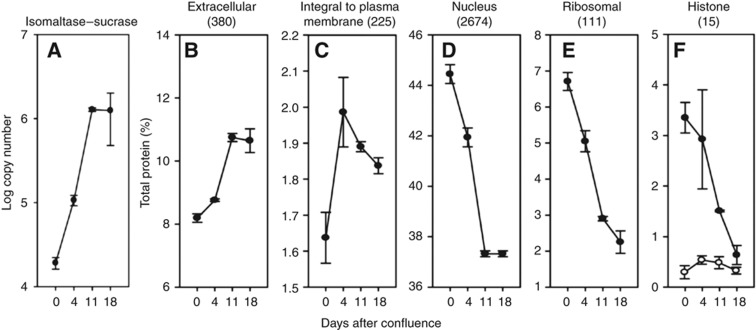
Sucrase-isomaltase is the standard marker of differentiation in CaCo-2 cells and its transcript levels increase 70 times during this process (Buhrke et al, 2011). Our data revealed a 63- to 68-fold increase of the abundance of this protein between day 0 and days 11 and 18 (Figure 6A), indicating that the cells were well differentiated.
Figure 6.
Abundance of proteins matching selected subcellular locations and functions in CaCo-2 cells. The numbers of proteins belonging to each class are given in the parentheses. The protein abundances were calculated on the basis of total spectral intensities of all quantified proteins. Each point represents data from four independent experiments for the 0, 11, and 18 days post confluence. The values for 4 days are for three experiments. Open circles are for linker histone H1 abundance.
Analysis of the temporary changes in the CaCo-2 cell proteome showed that already during the first 4 days of the differentiation process, 23% of all quantified proteins changed significantly (Supplementary Table 6). At days 11 and 18 after confluence expression levels of more than half of the quantified proteins had significantly changed (Supplementary Table 6). These extensive alterations in protein abundances are reflected in changes in the cellular content of organelles and protein functional categories (Figure 6). Differentiation of CaCo-2 cells is accompanied by an increase in extracellular and cell-surface proteins (Figure 6B and C) and a decrease of the abundance of nuclear proteins including the core histones (Figure 6D and F), and the ribosomal proteins (Figure 6E). Notably, the content of linker histones changed little between days 0 and 18, similarly to the finding reported above for colon cancer. The overall changes observed during CaCo-2 cell differentiation thus resemble the large-scale alterations between cancer and normal cells in vivo, including both the large number of individual proteins changed and the alteration in the cell architecture.
Estimation of absolute copy number of proteins
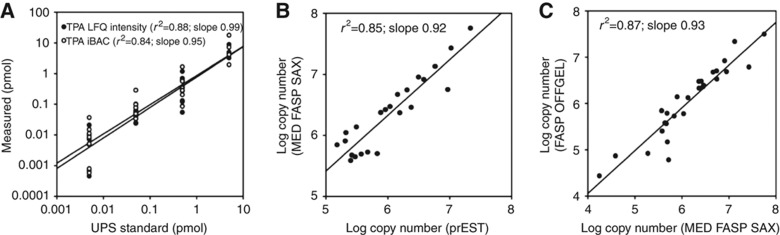
Recent large-scale proteomic analyses estimated protein copy numbers per cell by extrapolating from added standards (Beck et al, 2011; Schwanhausser et al, 2011). Here, we determine copy numbers per cell from our data simply on the basis of individual LFQ intensities compared with the total MS signal of the measured proteome. We named this method as total protein approach (TPA). By dividing these values by the molecular weight and multiplying by the Avogadro constant and by the protein content of a single cell, they can be converted into copy number values. To validate this approach, we first used a mixture of proteins with known protein concentrations. We found a linear response of the calculated protein amount with the amount of protein digested and measured by LC-MS/MS (Figure 7A). Calculations using summed peptide intensities or the iBAQ algorithm resulted in similar values. Next, we applied our calculation method to an LC-MS/MS analysis of a HeLa cell lysate. We compared the copy numbers of 23 proteins for which such values have recently been measured using stable isotope-labeled PrEST standards (Zeiler et al, 2012). We found that the values obtained by the TPA and SILAC-PrEST approaches were similar (r2=0.85; Figure 7B). This is even more remarkable considering that the SILAC-PrEST study used standard tryptic digestion and was performed on an Orbitrap Velos instrument whereas the TPA analysis employed two enzyme digestion and SAX fractionation (MED-FASP-SAX) followed by peptide analysis on the Q Exactive mass spectrometer. Next, we employed the TPA method to calculate protein copy numbers from a HeLa data set generated using FASP, OFFGEL peptide separation, and Orbitrap Velos analysis (Wisniewski et al, 2009b) and compared the values with the copy numbers obtained in this study. Figure 7C shows that the copy number values are similar in both experiments (r2=0.87). This evaluation demonstrates that the TPA-based copy number estimation is applicable to diverse large-scale proteomic data set generated in the past.
Figure 7.
Validation of total protein approach (TPA) for estimation of absolute copy numbers. (A) TPA calculation applied to analysis of a mixture of standard proteins (UPS2; Sigma). Protein standards were solubilized in SDS containing buffer and processed with the FASP method using trypsin. The digest was analyzed by LC-MS/MS using 4 h acetonitrile gradient. The protein concentrations were calculated either using directly protein intensities or iBAQ values. (B) Comparison of protein copy numbers in HeLa cells determined using isotope-labeled standards (SILAC-PrEST) with values obtained by label-free protein intensity-based TPA calculation. HeLa cell lysates were analyzed using the MED FASP-SAX method (Wisniewski and Mann, 2012). The SILAC-PrEST values are from Zeiler et al (2012). (C) Comparison of the protein copy numbers calculated using data from the current analysis (B) and a previously published analysis of HeLa cells (Wisniewski et al, 2009b).
To calculate copy numbers of proteins in the enterocytes, we assumed the volume of these cells to be about 1400 μm3 (Buschmann and Manke, 1981; MacLeod et al, 1991; Crowe and Marsh, 1993) and the protein content of mammalian cells to be about 20% (Ellis, 2001). The same volume and proteins content values were used for calculation of the copy numbers in carcinoma (C) (Materials and methods; Supplementary Table 7). For comparison, we also analyzed CaCo-2 and HeLa cells (Supplementary Table 7). To validate the calculated values, we furthermore compared them with the values obtained by biochemical methods in the past.
Core histones are among the most abundant proteins in the eukaryotic cells. Our copy numbers for these proteins were 1.1 × 108 in normal mucosa to 4.4 × 108 in undifferentiated CaCo-2 cells (Table IV). Assuming that all enterocytes are diploid, their DNA content is 2 × 2.85 billion base pairs (International Human Genome Sequencing Consortium, 2004) corresponding to 3.0 × 107 nucleosomes (assuming an average of 190 bp per nucleosome; van Holde, 1989). We determined a value of 3.5–14 core histone molecules per nucleosome, while the theoretical number is 8 molecules. For the adenocarcinoma and undifferentiated CaCo-2 cells, we found higher values than in normal mucosa and in the differentiated cells (Table IV), which likely reflects the pre-mitotic status of these cells in which the DNA was already duplicated. The numbers of linker histone were between 1.2 × 107 and 4.3 × 107 whereas the theoretical value is 3.3 × 107. This corresponds to 0.4–1.4 H1 molecule per nucleosome—similar to the average value of 0.95 observed across a variety of organisms (van Holde, 1989).
Table 4. Examples of estimated copy numbers per cella.
| Protein | Literature/theoretical | HeLa cells | Differentiatedb CaCo-2 cells | Undifferentiatedb CaCo-2 cells | Normal mucosa (N) | Carcinoma (A) |
|---|---|---|---|---|---|---|
| Linker histone H1 | 3.0 × 107 | 3.3 × 107 | 4.3 × 107 | 1.2 × 107 | 2.4 × 107 | 1.9 × 107 |
| Core histones | 2.4 × 108 | 1.6 × 108 | 1.7 × 108 | 4.4 × 108 | 1.1 × 108 | 1.5 × 108 |
| Linker histones/nucleosomec | 1 | 1.1 | 1.4 | 0.4 | 0.8 | 0.6 |
| Core histones/nucleosome | 8 | 5.2 | 5.7 | 14 | 3.5 | 4.9 |
| RNA Pol II largest subunitd | 3.2 × 105 | 2.2 × 105 | 1.2±0.1 × 105 | 1.3±0.2 × 105 | 1.6±1.5 × 104 | 2.6±1.4 × 104 |
| General transcription factor IIB | 2.2 × 105 | 1.7 × 105 | 6.7±1.1 × 104 | 5.8±0.9 × 104 | 1.9±0.7 × 105 | 2.5±1.0 × 105 |
| General transcription factor IIE1 | 1.3 × 105 | 1.7 × 105 | 4.0±1.1 × 104 | 1.2±0.3 × 105 | 1.1±0.4 × 104 | 3.0±1.8 × 104 |
| General transcription factor IIE2 | 3.6 × 105 | 3.5 × 105 | 1.3±0.4 × 105 | 2.5±0.4 × 105 | 1.3±1.8 × 105 | 6.4±5.2 × 104 |
| General transcription factor IIF1 | 1.6 × 105 | 5.3 × 105 | 1.7±0.3 × 105 | 3.1±0.3 × 105 | 1.5±1.8 × 105 | 1.9±0.6 × 105 |
| General transcription factor IIE2 | 3.2 × 105 | 9.6 × 105 | 2.0±0.3 × 105 | 3.8±0.9 × 105 | 6.1±3.0 × 104 | 1.2±0.5 × 105 |
| Total proteinse | 7.9 × 109 | 8.3 × 109 | 4.1±0.5 × 109 | 4.3±0.7 × 109 | 4.8±2.4 × 109 | 4.8±2.3 × 109 |
| 2.6 × 109 |
aCalculations were performed assuming an average HeLa cell volume of 2800 μm3 (calculated using an average diameter of 17.5 μm) the enterocyte volume of 1400 μm3 (average of values from references, Buschmann and Manke, 1981; MacLeod et al, 1991; Crowe and Marsh, 1993) and the average cellular protein content of 20% of the cell (Ellis, 2001).
bThe ‘Differentiated’ and ‘Undifferentiated’ CaCo-2 cells refer to cells harvested at confluence and 11 days after the confluence, respectively. Theoretical values for histones are given in italics.
cThe number of nucleosomes was calculated using the value 2 × 2.85 billion base pairs (International Human Genome Sequencing Consortium, 2004) for diploid human cell, and 190 bp DNA per nucleosome (Yanagawa et al, 2001).
dThe literature values for RNA II polymerase subunits are from reference (Kimura et al, 1999) the HeLa cell.
eThe values are from references Lodish et al (2000) and Princiotta et al (2003).
Western blot-based quantitative characterization of RNA polymerase II complex in HeLa cells by Kimura et al (1999) revealed that its subunits are present at 1.5–3.0 × 105 copies per cell (Table IV). Our TPA-based estimation in HeLa and CaCo-2 cells resulted in similar values of 1.2–2.2 × 105 copies per cell. In contrast, in the microdissected cells the abundance of polymerase II was about one order of magnitude lower. This was accompanied by a reduction of the copy numbers of TFIIE and TFIIF, suggesting a lower transcriptional activity in N and C compared with cultured cells. The copy numbers of the polymerase and TFIIB changed little between N and C. Similar changes were observed between non-differentiated and differentiated CaCo-2 cells. Finally, the TPA estimates of the total number of proteins per cell are in the range of 4.1–8.3 × 109 (Table IV) and these are generally within two-fold agreement with the literature values.
The absolute copy numbers calculated for cultured CaCo-2 cells were much more reproducible than those obtained for microdissected tissue (Table IV). The reason for this difference is the high variability of the abundances in different clinical samples in comparison with cultured cells (Supplementary Figure 3). Whereas the absolute values measured in N and C vary within an order of magnitude the variation in the cultured cells is usually much lower. A large variation in the absolute numbers in the human material has been reported in the past. For example, a recent MRM-based study of a large number of human hepatocyte proteins resulted in differences of up to one order of magnitude for some proteins between donors (Schaefer et al, 2012).
Comparison with previous transcriptomics and proteomics results
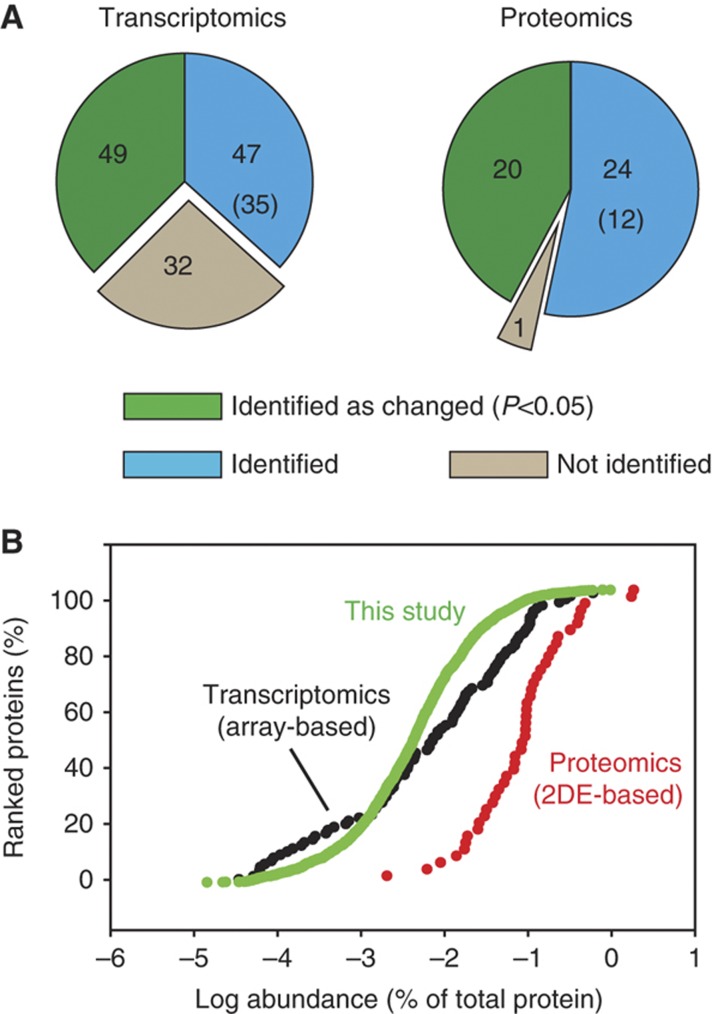
Over the past decade a large number of transcriptomic studies aimed at identification of genes that are upregulated or downregulated in CRC have been carried out. Cardoso et al (2007) summarized the results of 30 investigations using array platforms for identification of genes differentially expressed in colorectal carcinomas when compared with normal mucosa. Although in total close to 1000 changes were reported in these studies only 128 genes were identified in at least 3 of them. Strikingly, for the majority of these genes (96) we identified the corresponding proteins in our data set. At the protein level, we found 49 of these to be significantly changed between carcinoma and normal mucosa and in each case the direction of change was the same as that observed by transcriptomics analysis (Supplementary Table 8). This high overlap of the changed proteins found in a single proteomic study with a compiled data set from 30 transcriptomic studies demonstrates the potential and the robustness of proteomics for unveiling gene expression changes in cancer. Of note, the abundances of the 49 proteins whose abundance changed in both our proteomic and transcriptomic data sets are distributed over 4 orders of magnitude of transcript abundance (Figure 8A). This is similar to the abundance distribution of all changed proteins found in this study, suggesting that the sensitivity and dynamic range of proteomic analysis can be comparable to that of microarray-based approaches.
Figure 8.
Comparison of our data set with previous transcriptomic and proteomic studies. (A) Coverage of our data set of the most frequently observed upregulated or downregulated genes ‘Transcriptomics’ (Cardoso et al, 2007) and proteins ‘Proteomics’ (Jimenez et al, 2010) in CRC. The numbers of matching genes and proteins are indicated by the ‘green’ and ‘blue’ areas and numbers within them. The number of proteins changed in the same direction (up or down), but with P>0.05 is given in parentheses. Genes and proteins reported as changed in cancer, but not identified in our study are indicated as gray areas ‘not identified’. (B) Abundances of proteins corresponding to previously found differentially expressed genes ‘Transcriptomics’ (Cardoso et al, 2007) and changed protein levels ‘Proteomics’ (Jimenez et al, 2010) measured in this study.
The absence of significant changes between adenocarcinoma and nodal metastasis was an unexpected observation of this study. This could have been caused simply by the inadequate sensitivity of our analysis, which could have precluded identification of proteins whose changes specifically accompany metastasis. We tested this possibility by comparing the list of 41 proteins reported to be changed between normal mucosa and cancer and cancer and metastasis in a microarray study (Nambiar et al, 2010) with our data. We found 80% of these proteins in our data set (Supplementary Table 9), suggesting that our analysis had the potential to at least identify the proteins previously reported to be different between primary cancer and nodal metastases.
During recent years many proteomic studies of CRC have been published, each providing sets of proteins observed to change between cancer and normal mucosa. Comparison of these analyses showed that only 45 proteins were common in at least 3 of these studies (Jimenez et al, 2010). Out of these, we identified 44 proteins and found 19 of them to be significantly changed (in the same direction, Supplementary Table 10). Since the previous studies were mainly based on 2D gel separation, they were usually restricted to soluble and abundant proteins. Indeed, the 44 common proteins are on average two orders of magnitude more abundant than the average of proteins identified in this study (Figure 8B). This illustrates the advantages of proteomics workflows that are based on gel-free, high-resolution mass spectrometric methods.
In addition to 2D gel-based studies, there are also a few reports of CRC analysis using one-dimensional electrophoresis coupled to LC-MS/MS (Han et al, 2011; Van Houdt et al, 2011; de Wit et al, 2012). In these studies, the number of proteins identified and compared between different CRC stages was in a range of 2000–3000, considerably fewer than the number of transcripts probed in array-based studies and of the 7500 proteins identified here. This suggests that those studies were also biased toward the more abundant proteins in the cancer proteome. For example, proteins reported to be upregulated in cancer discussed as potential biomarkers in very recent studies—STOML2 (Han et al, 2011), BIRC6 (Van Houdt et al, 2011), OLFM4 (Conrotto et al, 2008; Besson et al, 2011), NGAL (Conrotto et al, 2008), and GLUT1 (de Wit et al, 2012)—have relatively high abundances between 1.6 × 10−2% and 1.9 × 10−3% of total protein mass, respectively (Supplementary Table 4). Our data confirm that STOML2, BIRC6, and GLUT1 are 2.4- to 20-fold upregulated in cancer samples (Supplementary Table 5). In contrast, the extent of upregulation of OLFM4 and NGAL was very variable, with cancer to normal ratios changing from 0.09 to 109 (Supplementary Table 4). The variability in the abundance of OLFM4 has already been observed by IHC analysis and likely reflects its transient rise during pre- and early cancer stages (Besson et al, 2011). The variability in C/N ratios of NGAL may be due to the fact that this protein appears abundantly in stroma (Conrotto et al, 2008), which can contaminate the microdissected material.
Since our large-scale analysis covers previously identified potential biomarkers very well, our set of upregulated proteins in cancer may be useful in future research aimed at the identification of novel biomarkers. In particular, our analysis contains almost all of the proteins identified in studies involving subcellular fractionation of clinical material (Conrotto et al, 2008; Albrethsen et al, 2010; de Wit et al, 2012). Such fractionation strategies typically require large amounts of starting material and are seldom of a purity that can substantially increase the depth of analysis. For example, nuclear and cell-surface proteomes contain albumin and abundant mitochondrial proteins (Conrotto et al, 2008; Albrethsen et al, 2010).
Discussion
The present proteomic study demonstrates unequivocally that proteomics is now capable of analyzing cancer proteomes in great depth. Using only minute amounts of archival, laser-microdissected FFPE tissues, we identified >8000 proteins and quantified 7576 of them between normal mucosa, cancer, and metastasis. To our knowledge, this is by far the greatest proteome coverage achieved in studies of clinical material. Nevertheless, this data set still contains many proteins for which changes were observed between normal and tumor cells but for which these changes were not statistically significant due to high variation. For instance, >600 proteins were identified in only a few of the analyzed samples and were therefore not used for quantitative analysis. Further developments in sample preparation and mass spectrometric analysis as well as larger numbers of patients are still required for extending our knowledge of the CRC proteome.
An important implication of this study is that proteomics does not require fresh or frozen material for studying human material for studying diseases. The archival FFPE material appears to be a valuable source of proteins, which can readily be compared between various stages of disorders and between different tissues. Furthermore, the label-free quantification approach used in this study, while not as accurate as stable isotope-based approaches (Geiger et al, 2010), offers a straightforward way to quantitatively analyze major proteomics features of clinical samples, and was also used in several recent proteomic studies on cancer (Gamez-Pozo et al, 2012; Meding et al, 2012; Pan et al, 2012). It requires neither specific reagents for labeling of the peptides nor standards. Clearly, these and other developments are now making proteomics readily applicable to the exploration of clinical samples.
Our study provides insights into the composition of three proteomes: normal colonic mucosa, CRC, and its proximal metastases. Most strikingly, we uncovered drastic proteomic remodeling between healthy and neoplastic epithelial cells, apparently involving half of the expressed proteins. This stands in a clear contrast to the results of individual gene expression profiling studies by microarrays, which typically report only a relatively small fraction of genes (1.8–2.5%) to be differentially expressed (Notterman et al, 2001; Birkenkamp-Demtroder et al, 2002; Cardoso et al, 2007). This difference between microarray technique and proteomics may reflect both biological and technical factors. For instance, differential turnover of RNA and protein is a major determinant of differences of the transcriptome and the proteome (Schwanhausser et al, 2011). The latter are not mirrored by transcriptomic analyses but are taken account of in proteomics analysis and may be especially important in tissue analysis, where many cells are slowly growing or post-mitotic. Furthermore, proteins are generally much more stable than RNA during sample preparation, making protein quantification potentially more robust and opening the possibility to readily analyze archival FFPE material as demonstrated in this study.
In striking contrast to the large-scale rearrangement of the proteome between normal mucosa and cancer, we found little if any significant changes between cancer and its nodal metastases. This suggests that the proteome of the cancer cells does not appear to need major further adaption at distal sites. Of course, limited quantitative accuracy as well as the relatively small number of tumor samples could also partially contribute to these conclusions. However, this finding is in accordance with systemic biology studies on breast cancer malignancy indicating no major differences between gene expression pattern in cells of primary cancer and its nodal metastases (Weigelt et al, 2003, 2005; Harrell et al, 2011).
CRC has been studied to a lesser extent than breast cancer and most gene expression studies have focused on differences in gene signatures of primary cancers either in localized or in generalized stages of that disease (Nannini et al, 2009), although there are some studies comparing expression profiles of primary and metastatic cells (Yanagawa et al, 2001; Koehler et al, 2004; Kwong et al, 2005; Kleivi et al, 2007; Lin et al, 2007, 2011). Most of these studies report only a few differentially expressed genes in metastases (Yanagawa et al, 2001; Ki et al, 2007; Kleivi et al, 2007; Lin et al, 2011) and only two such genes were in common in at least three papers (summarized in Lin et al, 2011). Therefore, the transcriptomic analyses for both CRC and breast cancer agree with our proteomic finding that gene expression changes between primary tumor and metastases are relatively minor. Interestingly, a recent 2DE-based proteomic study also did not report significant changes in about 1000 compared proteins between primary colon cancer and liver metastasis (Shi et al, 2011). Despite the fact that our analysis is the most comprehensive proteomic study on CRC so far, we cannot exclude the possibility that some proteins which are specifically upregulated in nodal metastases escaped detection due to their lower abundance or due to high variation between samples originating from different donors. It is also possible that proteins enabling the primary tumor to metastasize were already upregulated in the primary cancer and were therefore not detected in our differential analysis.
Here, we also introduced a simple method for estimation of absolute protein copy numbers without using external standards or isotope labels, the TPA. The method is based on the observation that the 3000 most abundant proteins of the cell already constitute >99% of the proteome mass. Thus, using intensity values for each protein, a fractional value of the MS signal (LFQ intensity) of a protein compared with the total MS signal is a good proxy of the percentage of its protein mass to total protein mass. This can then be converted into numbers of molecules per cell by measuring or estimating the volume and protein content of the analyzed cells. Potentially, the method can be applied to any protein mixture—as simple as several proteins, purified complexes, or organelles. Despite the simplicity of the approach we obtained good agreement with the theoretical values for the histones and found high consistency between our estimates and biochemical data in the abundance of RNA polymerase II and its general transcription factors in HeLa cells. Several of these proteins were about one order of magnitude less abundant in the enterocytes than in the adenocarcinoma cells and the cultured colon cancer cells. Thus, even the abundance of protein involved in basic cellular processes can be quite different between cells and these differences can be studied by the TPA method without using standards. Importantly, the TPA approach can be used on any previously generated large-scale data set.
The presented data comparing proteomes of normal mucosa and the primary cancer can be further studied at two distinct levels: First, they allow GO category-based analysis of proteome-wide changes in the cell architecture and functional classes. In a second step, the proteomic data for each of the members of the category can be examined to discover the extent of the quantitative change. Depending on the abundance of the protein, statistically significant upregulation can involve a percent change or it can be several-fold. We believe this is a particular strength of proteomics as opposed to transcriptomics. In conclusion, we here demonstrate proteomics analysis of colon cancer to a depth approaching standard microarray studies, yielding novel insights into proteome remodeling during cancer development.
Materials and methods
FFPE human tissue
Archival FFPE samples of grade 2 of CRC were obtained from the Department of Pathology of Wrocław Medical University. Analysis of the samples followed an informed consent approved by the local ethics committee.
Tissue microdissection and lysis
To obtain enriched populations of enterocytes, primary cancer, and metastasizing cells, tissue was dissected with the Laser Pressure Catapulting (LPC) PALM Instrument (Zeiss, Göttingen, Germany). The enterocytes (‘normal appearing’) were dissected from normal tissue adjacent to cancer. Collected tissues were lysed in a buffer consisting of 0.1 M Tris–HCl, pH 8.0, 0.1 M DTT, 0.5% (w/v) polyethylene glycol 20 000, and 4% SDS at 99°C as described previously (Ostasiewicz et al, 2010; Wisniewski et al, 2011a).
Protein digestion and peptide fractionation
Detergent was removed from the lysates and the proteins were digested with trypsin using the FASP protocol (Wisniewski et al, 2009b) using the 30 k filtration units (Cat No. MRCF0R030; Millipore) (Wisniewski et al, 2011b). The resulting peptides were fractionated according to the previously described pipette tip protocol (FASP-SAX) (Wisniewski et al, 2009a).
Proteomic analysis of HeLa and CaCo-2 cells
Frozen cells were lysed in 2% SDS, 0.1 M Tris–HCl, pH 8.0, and 0.1 M DTT. Aliquots of HeLa cells containing 20 μg of total protein were processed according to the MED-FASP protocol using consecutive two step digestion with LysC and trypsin (Wisniewski and Mann, 2012). Peptides released by LysC and trypsin were fractionated into four and two SAX fractions, respectively (Wisniewski and Mann, 2012).
LC-MS/MS analysis
The analysis of the microdissected cells was performed as described previously. Briefly, SAX-fractionated peptides were separated on a reverse phase (15 cm × 75 μm inner diameter) ReproSil-Pur C18-AQ, 3 μm resin (Dr Maisch GmbH, Ammerbuch-Entringen, Germany) column using a 230-min acetonitrile gradient and were analyzed with an LTQ-Orbitrap Velos mass spectrometer using a ‘high-high’ strategy with HCD (Olsen et al, 2007).
The analysis of the cultured cells and protein standards was performed using reverse phase column (20 cm × 75 μm inner diameter) packed with 1.8 μm C18 particles (Dr Maisch GmbH) using a 4-h acetonitrile gradient in 0.1% formic acid at a flow rate of 250 nl/min. The LC was coupled to a Q Exactive mass spectrometer (Michalski et al, 2011; Thermo Fisher Scientific, Germany) via a nanoelectrospray source (Proxeon Biosystems, now Thermo Fisher Scientific). The Q Exactive was operated in data-dependent mode with survey scans acquired at a resolution of 50 000 at m/z 400 (transient time 256 ms). Up to the top 10 most abundant isotope patterns with charge ≥2 from the survey scan were selected with an isolation window of 1.6 Th and fragmented by HCD with normalized collision energies of 25. The maximum ion injection times for the survey scan and the MS/MS scans were 20 and 60 ms, respectively. The ion target value for both scan modes was set to 106. The samples were analyzed in the order of 1 (C, N, M), 2 (C, N, M)…8 (C, N, M), but the order of N, C, and M in each group was random. The files are available at ‘Tranche’.
CRC Data: Hash key: 4J7wto4aNCmjeYCcihnNWakm2M94h8orULE0obgNeg2f/8f1mPMCuTtbGmIoeqRRvCltK4pGmvGWSdC3VFmON9e/ X90AAAAAAABiXw==
HeLa Data: Hash key: y9r+XWuGRc9pnH5LtJ/n0qarp4j KHWQpk87dFCVLIU4ZmtD9hqnR4TeBXvIWWKxyzP1HPFdo+1n8EzCezrYxH62kGrMAAAAAAAALGw==
Data analysis
The MS data were analyzed using the software environment MaxQuant (Cox and Mann, 2008) version 1.2.2.8. Proteins were identified by searching MS and MS/MS data of peptides against a decoy version of the International Protein Index (IPI) human database (v.3.68). Carbamidomethylation of cysteines was set as fixed modification. The maximum false peptide discovery rate was specified as 0.01. Label-free quantification was carried out in MaxQuant as previously described (Luber et al, 2010). Protein abundance was calculated on the basis of the normalized spectral protein intensity (LFQ intensity). Quantifiable proteins in the analysis of clinical samples were defined as those identified at least four times (50%) in at least one type of sample (N, C, M). In CaCo-2 experiment, the quantifiable proteins occurred at least three times in at least one time point.
Statistical analysis
Statistical analysis was handled in R ( http://www.R-project.org). Zero intensities were filled with intensities from the lower part of normal distribution (imputation width=0.3, shift=1.8). Unsupervised hierarchical clustering analysis was performed to identify groups that show similar characteristics. A paired t-test was applied for testing of differences in protein intensities in clinical samples. Analysis of CaCo-2 data was performed using t-test. Significance of outliers was calculated by multiple hypothesis testing (Benjamini and Hochberg, 1995) with the threshold value of 0.05.
Calculation of intensity-based total protein values and copy numbers per cell
The total protein content was defined as a sum of peptide intensities integrated over the elution profile of each peptide. The amount of individual proteins was calculated as the ratio of their LFQ intensity (LFQ MS intensity) to the sum of all LFQ intensities (total protein) in the measured sample. TPA calculations were performed assuming an average HeLa cell volume of 2800 μm3 (calculated using an average diameter of 17.5 μm) and the enterocyte volume of 1400 μm3 (average of values from references, Buschmann and Manke, 1981; MacLeod et al, 1991; Crowe and Marsh, 1993). The latter value was also used for calculation of protein copy numbers in adenocarcinoma cells and CaCo-2 cells. All calculation used an average cellular protein content of 20% of the cell (Ellis, 2001).
Immunohistochemistry
Five-micrometer paraffin sections were cut, mounted on slides, and rehydrated. For antigen retrieval, sections were microwaved in citrate buffer (pH 6.0) for 10 min at 750 W. Non-specific bindings were blocked with Protein Block Solution (Abcam) for 10 min. Then, the sections were washed with PBST solution and incubated for 1 h at room temperature with rabbit primary antibodies: anti-PALM3 (Abgent, 1:10 dilution), anti-MFI2 (Sigma-Aldrich, 1:50 dilution), and anti-GPR56 (Abcam, 1:100 dilution). After washing in PBST and subsequent 15-min incubation with secondary HRP-labeled antibodies (Abcam), sections were blocked with 0.3% hydrogen peroxide for 10 min and stained for 5 min with DAB. Finally, sections were counterstained with hemotoxyline. Intensity of staining was assessed by two independent observers using the IHC-score scale described elsewhere (Kok et al, 2010) with values ranging from 0 points (no staining) to 12 points (>75% of cells strongly stained).
Supplementary Material
Acknowledgments
We thank Piotr Ziołkowski (Wroclaw Medical University) for providing the samples and Korbinian Mayr for assistance in mass spectrometric analysis and Katharina Zettl for excellent technical assistance. This work was supported by the Max-Planck Society for the Advancement of Science, by the European Commission's 7th Framework Program (grant agreement HEALTH-F4-2008-201648/PROSPECTS), the Munich Center for Integrated Protein Science (CIPSM), and the Polish National Center of Science (DEC-2011/01/N/NZ5/04253).
Author contributions: JRW designed the experiments, performed the MS analysis, analyzed the data, supervised the work, and wrote the manuscript; PO designed the experiments, selected clinical samples, and performed microdissection; KD selected clinical samples and performed the IHC experiments; DZ and FG analyzed the data. MM supervised the work and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Albrethsen J, Knol JC, Piersma SR, Pham TV, de Wit M, Mongera S, Carvalho B, Verheul HM, Fijneman RJ, Meijer GA, Jimenez CR (2010) Subnuclear proteomics in colorectal cancer: identification of proteins enriched in the nuclear matrix fraction and regulation in adenoma to carcinoma progression. Mol Cell Proteomics 9: 988–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R (2011) The quantitative proteome of a human cell line. Mol Syst Biol 7: 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful aproach to multiple testing. J R Stat Soc 57: 289–300 [Google Scholar]
- Besson D, Pavageau AH, Valo I, Bourreau A, Belanger A, Eymerit-Morin C, Mouliere A, Chassevent A, Boisdron-Celle M, Morel A, Solassol J, Campone M, Gamelin E, Barré B, Coqueret O, Guette C (2011) A quantitative proteomic approach of the different stages of colorectal cancer establishes OLFM4 as a new nonmetastatic tumor marker. Mol Cell Proteomics 10: M111.009712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenkamp-Demtroder K, Christensen LL, Olesen SH, Frederiksen CM, Laiho P, Aaltonen LA, Laurberg S, Sorensen FB, Hagemann R, ØRntoft TF (2002) Gene expression in colorectal cancer. Cancer Res 62: 4352–4363 [PubMed] [Google Scholar]
- Buhrke T, Lengler I, Lampen A (2011) Analysis of proteomic changes induced upon cellular differentiation of the human intestinal cell line Caco-2. Dev Growth Differ 53: 411–426 [DOI] [PubMed] [Google Scholar]
- Buschmann RJ, Manke DJ (1981) Morphometric analysis of the membranes and organelles of small intestinal enterocytes. I. Fasted Hamster. J Ultrastruct Res 76: 1–14 [DOI] [PubMed] [Google Scholar]
- Cardoso J, Boer J, Morreau H, Fodde R (2007) Expression and genomic profiling of colorectal cancer. Biochim Biophys Acta 1775: 103–137 [DOI] [PubMed] [Google Scholar]
- Conrotto P, Roesli C, Rybak J, Kischel P, Waltregny D, Neri D, Castronovo V (2008) Identification of new accessible tumor antigens in human colon cancer by ex vivo protein biotinylation and comparative mass spectrometry analysis. Int J Cancer 123: 2856–2864 [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10: 1794–1805 [DOI] [PubMed] [Google Scholar]
- Crowe PT, Marsh MN (1993) Morphometric analysis of small intestinal mucosa. IV. Determining cell volumes. Virchows Arch A Pathol Anat Histopathol 422: 459–466 [DOI] [PubMed] [Google Scholar]
- de Wit M, Jimenez CR, Carvalho B, Belien JA, Delis-van Diemen PM, Mongera S, Piersma SR, Vikas M, Navani S, Ponten F, Meijer GA, Fijneman RJ (2012) Cell surface proteomics identifies glucose transporter type 1 and prion protein as candidate biomarkers for colorectal adenoma-to-carcinoma progression. Gut 61: 855–864 [DOI] [PubMed] [Google Scholar]
- Ellis RJ (2001) Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci 26: 597–604 [DOI] [PubMed] [Google Scholar]
- Fearon ER (2011) Molecular genetics of colorectal cancer. Annu Rev Pathol 6: 479–507 [DOI] [PubMed] [Google Scholar]
- Gamez-Pozo A, Sanchez-Navarro I, Calvo E, Agullo-Ortuno MT, Lopez-Vacas R, Diaz E, Camafeita E, Nistal M, Madero R, Espinosa E, López JA, Fresno Vara JÁ (2012) PTRF/cavin-1 and MIF proteins are identified as non-small cell lung cancer biomarkers by label-free proteomics. PLoS ONE 7: e33752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garty H, Karlish SJ (2006) Role of FXYD proteins in ion transport. Annu Rev Physiol 68: 431–459 [DOI] [PubMed] [Google Scholar]
- Geiger T, Cox J, Ostasiewicz P, Wisniewski JR, Mann M (2010) Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat Methods 7: 383–385 [DOI] [PubMed] [Google Scholar]
- Han CL, Chen JS, Chan EC, Wu CP, Yu KH, Chen KT, Tsou CC, Tsai CF, Chien CW, Kuo YB, Lin PY, Yu JS, Hsueh C, Chen MC, Chan CC, Chang YS, Chen YJ (2011) An informatics-assisted label-free approach for personalized tissue membrane proteomics: case study on colorectal cancer. Mol Cell Proteomics 10: M110.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanash SM, Pitteri SJ, Faca VM (2008) Mining the plasma proteome for cancer biomarkers. Nature 452: 571–579 [DOI] [PubMed] [Google Scholar]
- Harrell JC, Prat A, Parker JS, Fan C, He X, Carey L, Anders C, Ewend M, Perou CM (2011) Genomic analysis identifies unique signatures predictive of brain, lung, and liver relapse. Breast Cancer Res Treat 132: 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. (2004) Finishing the euchromatic sequence of the human genome. Nature 431: 931–945 [DOI] [PubMed] [Google Scholar]
- Jimenez CR, Knol JC, Meijer GA, Fijneman RJ (2010) Proteomics of colorectal cancer: overview of discovery studies and identification of commonly identified cancer-associated proteins and candidate CRC serum markers. J Proteomics 73: 1873–1895 [DOI] [PubMed] [Google Scholar]
- Ki DH, Jeung HC, Park CH, Kang SH, Lee GY, Lee WS, Kim NK, Chung HC, Rha SY (2007) Whole genome analysis for liver metastasis gene signatures in colorectal cancer. Int J Cancer 121: 2005–2012 [DOI] [PubMed] [Google Scholar]
- Kimura H, Tao Y, Roeder RG, Cook PR (1999) Quantitation of RNA polymerase II and its transcription factors in an HeLa cell: little soluble holoenzyme but significant amounts of polymerases attached to the nuclear substructure. Mol Cell Biol 19: 5383–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleivi K, Lind GE, Diep CB, Meling GI, Brandal LT, Nesland JM, Myklebost O, Rognum TO, Giercksky KE, Skotheim RI, Lothe RA (2007) Gene expression profiles of primary colorectal carcinomas, liver metastases, and carcinomatoses. Mol Cancer 6: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler A, Bataille F, Schmid C, Ruemmele P, Waldeck A, Blaszyk H, Hartmann A, Hofstaedter F, Dietmaier W (2004) Gene expression profiling of colorectal cancer and metastases divides tumours according to their clinicopathological stage. J Pathol 204: 65–74 [DOI] [PubMed] [Google Scholar]
- Kok LF, Lee MY, Tyan YS, Wu TS, Cheng YW, Kung MF, Wang PH, Han CP (2010) Comparing the scoring mechanisms of p16INK4a immunohistochemistry based on independent nucleic stains and independent cytoplasmic stains in distinguishing between endocervical and endometrial adenocarcinomas in a tissue microarray study. Arch Gynecol Obstet 281: 293–300 [DOI] [PubMed] [Google Scholar]
- Kwong KY, Bloom GC, Yang I, Boulware D, Coppola D, Haseman J, Chen E, McGrath A, Makusky AJ, Taylor J, Steiner S, Zhou J, Yeatman TJ, Quackenbush J (2005) Synchronous global assessment of gene and protein expression in colorectal cancer progression. Genomics 86: 142–158 [DOI] [PubMed] [Google Scholar]
- Lin AY, Chua MS, Choi YL, Yeh W, Kim YH, Azzi R, Adams GA, Sainani K, van de Rijn M, So SK, Pollack JR (2011) Comparative profiling of primary colorectal carcinomas and liver metastases identifies LEF1 as a prognostic biomarker. PLoS ONE 6: e16636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HM, Chatterjee A, Lin YH, Anjomshoaa A, Fukuzawa R, McCall JL, Reeve AE (2007) Genome wide expression profiling identifies genes associated with colorectal liver metastasis. Oncol Rep 17: 1541–1549 [DOI] [PubMed] [Google Scholar]
- Lodish H, Berk A, Zipursky S, Matsudaira P, Baltimore D, Darnell J (eds) (2000) Molecular Cell Biology 4th edn, New York: W.H. Freeman, [Google Scholar]
- Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, Tschopp J, Akira S, Wiegand M, Hochrein H, O‘Keeffe M, Mann M (2010) Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 32: 279–289 [DOI] [PubMed] [Google Scholar]
- MacLeod RJ, Hamilton JR, Bateman A, Belcourt D, Hu J, Bennett HP, Solomon S (1991) Corticostatic peptides cause nifedipine-sensitive volume reduction in jejunal villus enterocytes. Proc Natl Acad Sci USA 88: 552–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meding S, Balluff B, Elsner M, Schone C, Rauser S, Nitsche U, Maak M, Schafer A, Hauck SM, Ueffing M, Langer R, Höfler H, Friess H, Rosenberg R, Walch A (2012) Tissue based proteomics reveals FXYD3, S100A11 and GSTM3 as novel markers for regional lymph node metastasis in colon cancer. J Pathol advance online publication, 19 March 2012; doi:; DOI: 10.1002/path.4021 [DOI] [PubMed] [Google Scholar]
- Michalski A, Damoc E, Hauschild JP, Lange O, Wieghaus A, Makarov A, Nagaraj N, Cox J, Mann M, Horning S (2011) Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol Cell Proteomics 10: M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Paabo S, Mann M (2011) Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol 7: 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar PR, Gupta RR, Misra V (2010) An ‘Omics’ based survey of human colon cancer. Mutat Res 693: 3–18 [DOI] [PubMed] [Google Scholar]
- Nannini M, Pantaleo MA, Maleddu A, Astolfi A, Formica S, Biasco G (2009) Gene expression profiling in colorectal cancer using microarray technologies: results and perspectives. Cancer Treat Rev 35: 201–209 [DOI] [PubMed] [Google Scholar]
- Notterman DA, Alon U, Sierk AJ, Levine AJ (2001) Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res 61: 3124–3130 [PubMed] [Google Scholar]
- Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M (2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat Methods 4: 709–712 [DOI] [PubMed] [Google Scholar]
- Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, Lange O, Remes P, Taylor D, Splendore M, Wouters ER, Senko M, Makarov A, Mann M, Horning S (2009) A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol Cell Proteomics 8: 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostasiewicz P, Zielinska DF, Mann M, Wisniewski JR (2010) Proteome, phosphoproteome, and N-glycoproteome are quantitatively preserved in formalin-fixed paraffin-embedded tissue and analyzable by high-resolution mass spectrometry. J Proteome Res 9: 3688–3700 [DOI] [PubMed] [Google Scholar]
- Pan S, Chen R, Brand RE, Hawley S, Tamura Y, Gafken PR, Milless BP, Goodlett DR, Rush J, Brentnall TA (2012) Multiplex targeted proteomic assay for biomarker detection in plasma: a pancreatic cancer biomarker case study. J Proteome Res 11: 1937–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Princiotta MF, Finzi D, Qian SB, Gibbs J, Schuchmann S, Buttgereit F, Bennink JR, Yewdell JW (2003) Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity 18: 343–354 [DOI] [PubMed] [Google Scholar]
- Schaefer O, Ohtsuki S, Kawakami H, Inoue T, Liehner S, Saito A, Sakamoto A, Ishiguro N, Matsumaru T, Terasaki T, Ebner T (2012) Absolute quantification and differential expression of drug transporters, cytochrome P450 enzymes, and UDP-glucuronosyltransferases in cultured primary human hepatocytes. Drug Metab Dispos 40: 93–103 [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M (2011) Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Shi H, Hood KA, Hayes MT, Stubbs RS (2011) Proteomic analysis of advanced colorectal cancer by laser capture microdissection and two-dimensional difference gel electrophoresis. J Proteomics 75: 339–351 [DOI] [PubMed] [Google Scholar]
- van Holde KE (1989) Chromatin New York: Springer Verlag, [Google Scholar]
- Van Houdt W, Emmink B, Pham T, Piersma S, Verheem A, Vries R, Fratantoni S, Pronk A, Clevers H, Borel Rinkes I, Jimenez CR, Kranenburg O (2011) Comparative proteomics of colon cancer stem cells and differentiated tumor cells identifies birc6 as a potential therapeutic target. Mol Cell Proteomics 10: M111.011353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324: 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O (1956) On respiratory impairment in cancer cells. Science 124: 269–270 [PubMed] [Google Scholar]
- Weigelt B, Glas AM, Wessels LF, Witteveen AT, Peterse JL, van’t Veer LJ (2003) Gene expression profiles of primary breast tumors maintained in distant metastases. Proc Natl Acad Sci USA 100: 15901–15905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B, Wessels LF, Bosma AJ, Glas AM, Nuyten DS, He YD, Dai H, Peterse JL, van’t Veer LJ (2005) No common denominator for breast cancer lymph node metastasis. Br J Cancer 93: 924–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR (2011) Tools for phospho- and glycoproteomics of plasma membranes. Amino Acids 41: 223–233 [DOI] [PubMed] [Google Scholar]
- Wisniewski JR, Mann M (2012) Consecutive proteolytic digestion in an enzyme reactor increases depth of proteomic and phosphoproteomic analysis. Anal Chem 84: 2631–2637 [DOI] [PubMed] [Google Scholar]
- Wisniewski JR, Ostasiewicz P, Mann M (2011a) High recovery FASP applied to the proteomic analysis of microdissected formalin fixed paraffin embedded cancer tissues retrieves known colon cancer markers. J Proteome Res 10: 3040–3049 [DOI] [PubMed] [Google Scholar]
- Wisniewski JR, Zielinska DF, Mann M (2011b) Comparison of ultrafiltration units for proteomic and N-glycoproteomic analysis by the filter-aided sample preparation method. Anal Biochem 410: 307–309 [DOI] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Mann M (2009a) Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J Proteome Res 8: 5674–5678 [DOI] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M (2009b) Universal sample preparation method for proteome analysis. Nat Methods 6: 359–362 [DOI] [PubMed] [Google Scholar]
- Yanagawa R, Furukawa Y, Tsunoda T, Kitahara O, Kameyama M, Murata K, Ishikawa O, Nakamura Y (2001) Genome-wide screening of genes showing altered expression in liver metastases of human colorectal cancers by cDNA microarray. Neoplasia 3: 395–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiler M, Straube WL, Lundberg E, Uhlen M, Mann M (2012) A Protein Epitope Signature Tag (PrEST) library allows SILAC-based absolute quantification and multiplexed determination of protein copy numbers in cell lines. Mol Cell Proteomics 11: O111.009613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweibaum A, Triadou N, Kedinger M, Augeron C, Robine-Leon S, Pinto M, Rousset M, Haffen K (1983) Sucrase-isomaltase: a marker of foetal and malignant epithelial cells of the human colon. Int J Cancer 32: 407–412 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.