Abstract
3-Methylcrotonyl-coenzyme A carboxylase (MCCase) is a mitochondrial biotin-containing enzyme whose metabolic function is not well understood in plants. In soybean (Glycine max) seedlings the organ-specific and developmentally induced changes in MCCase expression are regulated by mechanisms that control the accumulation of MCCase mRNA and the activity of the enzyme. During soybean cotyledon development, when seed-storage proteins are degraded, leucine (Leu) accumulation peaks transiently at 8 d after planting. The coincidence between peak MCCase expression and the decline in Leu content provides correlative evidence that MCCase is involved in the mitochondrial catabolism of Leu. Direct evidence for this conclusion was obtained from radiotracer metabolic studies using extracts from isolated mitochondria. These experiments traced the metabolic fate of [U-14C]Leu and NaH14CO3, the latter of which was incorporated into methylglutaconyl-coenzyme A (CoA) via MCCase. These studies directly demonstrate that plant mitochondria can catabolize Leu via the following scheme: Leu → α-ketoisocaproate → isovaleryl-CoA → 3-methylcrotonyl-CoA → 3-methylglutaconyl-CoA → 3-hydroxy-3-methylglutaryl-CoA → acetoacetate + acetyl-CoA. These findings demonstrate for the first time, to our knowledge, that the enzymes responsible for Leu catabolism are present in plant mitochondria. We conclude that a primary metabolic role of MCCase in plants is the catabolism of Leu.
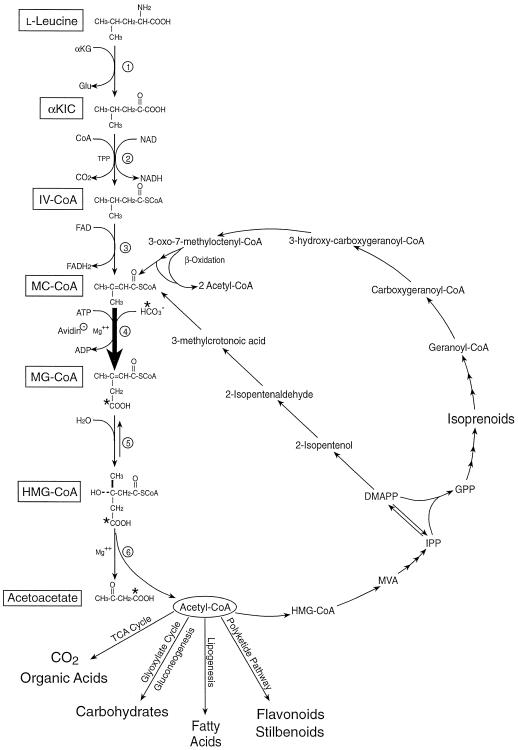
MCCase (EC 6.4.1.4) is a biotin-containing enzyme that catalyzes the Mg-ATP-dependent carboxylation of MC-CoA to form MG-CoA. MCCase was discovered nearly 40 years ago in bacteria and mammals (for review, see Moss and Lane, 1971) but its presence in plants was reported more recently (Wurtele and Nikolau, 1990). The reaction catalyzed by MCCase is required in several interconnected metabolic pathways (Fig. 1): Leu catabolism, isoprenoid catabolism, and the mevalonate shunt. The catabolism of Leu in animals and some bacteria requires the sequential action of six enzymes, which convert Leu to acetoacetate and acetyl-CoA. MCCase is the fourth enzyme in this catabolic pathway. In some species of bacteria, noncyclic isoprenoids such as geranoyl-CoA are catabolized to acetyl-CoA, probably via a set of reactions analogous to β-oxidation; one of the reactions in this pathway is the carboxylation of MC-CoA (Seubert and Remberger, 1963). The mevalonate shunt converts mevalonate to acetoacetate and acetyl-CoA in animals (Popják, 1971; Edmond and Popják, 1974) and in plants (Nes and Bach, 1985). The net result of this shunt is to reassimilate carbon that has been converted to mevalonate into primary metabolites rather than into isoprenoids.
Figure 1.
Potential metabolic functions of MCCase. MCCase catalyzes the ATP-dependent carboxylation of MC-CoA to form MG-CoA. This reaction may be required in the catabolism of Leu to acetoacetate and acetyl-CoA (reactions 1–6). MCCase may also function to convert mevalonate (MVA) to acetoacetate and acetyl-CoA (via isopentenyl pyrophosphate [IPP] and 3-methylcrotonoic acid) by the “mevalonate shunt.” A third function of MCCase may be as part of an isoprenoid catabolic pathway (via geranoyl-CoA). Reactions 4 to 6 are common to all three processes. The products of these processes, acetoacetate and acetyl-CoA, can be further metabolized to isoprenoids, polyketide derivatives (e.g. flavonoids, stilbenoids), and fatty acids, to Glc in tissues engaging the glyoxylate cycle, or respired to CO2 in the tricarboxylic acid cycle. The enzymes of Leu catabolism are: (a) branched-chain amino acid aminotransferase, (b) BCKDH complex, (c) branched-chain acyl-CoA dehydrogenase, (d) MCCase, (e) MG-CoA hydratase, and (f) HMG-CoA lyase. The asterisks denote the carbon atom expected to be radioactively labeled when NaH14CO3 is supplied for the MCCase reaction.
Despite the purification of MCCase from plants (Alban et al., 1993; Chen et al., 1993; Diez et al., 1994) and the isolation of cDNA and genomic clones for the biotin-containing subunit of MCCase (Song et al., 1994; Wang et al., 1994; Weaver et al., 1995), the physiological significance of this enzyme in plant metabolism has not been defined. Evidence for the catabolism of Leu has been provided by the observations that Leu-derived carbon is incorporated into sugars and organic acids (Stewart and Beevers, 1967; Sodek and Wilson, 1973) or isoprenoids (Overton, 1985; Koops et al., 1991), although there was no evidence implicating MCCase in the process. In vitro studies of Leu catabolism in plants indicate that this process occurs in peroxisomes by a mechanism that does not require MCCase (Gerbling and Gerhardt, 1988, 1989; Gerbling, 1993), a mitochondrial enzyme in plants (Baldet et al., 1992). To better understand the physiological role of MCCase in plants, we investigated the expression of MCCase during plant development, focusing on cotyledon development, when massive hydrolysis of seed-storage proteins and the remobilization and catabolism of the amino acids occurs. We also examined the metabolism of Leu and MC-CoA in protein extracts from isolated mitochondria. Together, these studies demonstrate the involvement of MCCase in the mitochondrial catabolism of Leu.
MATERIALS AND METHODS
Plant Material
Soybean (Glycine max cv Corsoy 79) seeds were germinated in a sterile mixture of 30% (v/v) black soil, 30% (v/v) peat moss, and 40% (v/v) perlite in a greenhouse at 22°C to 25°C, under a cycle of 15 h of illumination and 9 h of darkness, with a maximum daily irradiance of no less than 1200 μmol photons m−2 s−1. Plants were watered daily and fertilized twice a week with a solution of 20:10:20 (N:P:K). Plants were grown to maturity in 25-cm pots. Plants that were to be sampled before 20 DAP were grown in 50- × 30- × 6-cm flats. All samples were harvested between 4 and 6 h after the beginning of the illumination period. Organs of plants at different stages of development were harvested and frozen in liquid nitrogen. For long-term storage, plant materials for RNA isolation were stored at −70°C, and plant materials for protein isolation were stored in liquid nitrogen. Mitochondria were isolated from shoots of pea (Pisum sativum cv Progress no. 9) 14 to 16 DAP and grown in flats under conditions similar to those described for soybean.
Isolation and Analysis of RNA
RNA was isolated as described previously (Logemann et al., 1987). The concentration of RNA was determined by A260. RNA samples were denatured with formaldehyde and formamide and were fractionated by electrophoresis using 1.0% agarose gels buffered with 0.02 m Mops, 8 mm sodium acetate, 1 mm EDTA, and 2.2 m formaldehyde, pH 7.0 (Sambrook et al., 1989). After electrophoresis, the gel was washed twice with diethyl pyrocarbonate-treated water, soaked in 20× SSC (1× SSC is 150 mm NaCl and 15 mm sodium citrate, pH 7.0) for 10 min, and the RNA was transferred by capillary action with 20× SSC to a nitrocellulose filter. The filter was subsequently baked for 2 h at 80°C in a vacuum oven, prehybridized for 2 h at 68°C in a solution containing 6× SSC, 5× Denhardt's solution (1× Denhardt's solution is 0.02% [w/v] Ficoll, 0.02% [w/v] PVP-360, and 0.025% [w/v] BSA), 50 mm Tris-HCl, pH 8.0, 0.2% (w/v) SDS, 10 mm EDTA, and 100 μg/mL sheared salmon-sperm DNA, and hybridized overnight at 68°C in a similar solution containing 10% (w/v) dextran sulfate and 32P-labeled DNA probes. The DNA probes were radioactively labeled with [α-32P]dCTP (10 mCi/mmol; Amersham) using the random-primer labeling method (Feinberg and Vogelstein, 1983). After hybridization, the membrane was washed twice with 0.2× SSC, 0.5% SDS for 10 min at room temperature, followed by a third wash for 30 min at 68°C.
Preparation of Cell-Free Extracts
All of the procedures were performed at 4°C. Soybean organs (1–2 g fresh weight) were pulverized using a mortar and pestle in the presence of liquid nitrogen. The frozen, pulverized tissue was homogenized with 3 to 4 volumes of a buffer containing 0.1 m Hepes-KOH, pH 7.0, 1 mm EDTA, 0.1% (v/v) Triton X-100, 20% (v/v) glycerol, 20 mm β-mercaptoethanol, 100 μg/mL PMSF, and 10 μm trans-epoxysuccinyl-l-leucylamido(4-guanidone)-butane. The extract was filtered through four layers of cheesecloth, and the filtrate was centrifuged at 22,100g for 15 min. The extract was desalted in 200-μL portions using Sephadex G-25 columns (1-mL bed volume), and the pooled eluate was used for MCCase assays and western-blot analyses.
Enzyme Assays
MCCase activity was assayed by the incorporation of radioactivity from NaH14CO3 into an acid-stable product (Wurtele and Nikolau, 1990). The reaction mixture contained 0.1 m Tricine-KOH, pH 8.0, 5 mm MgCl2, 2.5 mm DTT, 5 mm KHCO3, 5 μCi NaH14CO3 (58 mCi/mmol; Amersham), 1 mm ATP, and 0.2 mm MC-CoA. The assay was initiated by the addition of the protein extract and incubated in a final volume of 200 μL at 37°C for 10 min. The reaction was terminated by the addition of 50 μL of 6 n HCl. A sample (100 μL) was applied to a strip of 3MM paper (Whatman), dried, and the acid-stable radioactivity was determined by liquid scintillation. Assays were performed in duplicate. Control assays lacking the MC-CoA substrate were carried out for each protein extract. Cyt c oxidase activity was assayed in 1-mL reactions containing 0.1 m KH2PO4, pH 7.0, 0.1% (v/v) Nonidet P-40, mitochondria (10–25 μg of protein), and 0.4 mg of reduced Cyt c (ε550 = 21 mm−1 cm−1). The oxidation state of Cyt c was adjusted to a standard level (A550 = 0.3 for a 0.16 mg/mL Cyt c solution) using sodium hydrosulfite as the reducing agent. Reactions were started with Cyt c and oxidation was monitored by the change in A550. Hydroxypyruvate reductase was assayed according to the method of Schwitzguebel and Siegenthaler (1984), and NADP-glyceraldehyde-3-phosphate dehydrogenase was assayed according to the method of Heber et al. (1963) as modified by Kaethner and ap Rees (1985).
Electrophoresis and Western-Blot Analysis
SDS-PAGE was performed in 7.5% acrylamide gels as described previously (Laemmli, 1970). After electrophoresis proteins were electrophoretically transferred to a nitrocellulose membrane (Kyhse-Andersen, 1984), and the biotin-containing proteins were detected by using 125I-streptavidin (Nikolau et al., 1985), or the biotin-containing polypeptide of MCCase was immunologically detected with specific antiserum (Song et al., 1994), diluted 1:200, and incubated with 125I-protein A.
Quantitation of Chlorophyll, Leu, and Protein
The chlorophyll content of developing soybean cotyledons was measured using the method of Arnon (1949). Free Leu content of developing soybean cotyledons was determined after conversion of Leu to its dansyl derivative and separation by HPLC. Cotyledons (0.5 g) were homogenized with 5 mL of 10% TCA and the insoluble material was removed by centrifugation at 10,000g for 10 min. The supernatant was extracted three times with cold diethyl ether to remove the TCA. The dansylation procedure was modified from that of Tapuhi et al. (1981). Equal volumes (200 μL) of TCA extracts, 0.2 m NaHCO3, and 1.5 mg/mL dansyl chloride were mixed and incubated for 60 min at 21°C in darkness. The reaction was terminated by addition of 20 μL of 2% (w/v) methylamine. The dansyl-Leu concentrations were determined from A254 after HPLC fractionation. The protein content of developing soybean cotyledons was determined from the protein concentration of the TCA precipitate of the cotyledon extracts after an overnight incubation in 0.1 m NaOH to resolubilize the protein (Bradford [1976] as modified by Jones et al. [1989]).
Isolation of Mitochondria
Mitochondria were isolated from pea shoots by a method modified from that of Sandalio et al. (1987). Reagents and equipment were maintained at 0°C to 4°C. Chilled plant material (480 g) was homogenized with a mortar and pestle in 80-g batches with 240 mL of 0.35 m mannitol, 30 mm Mops-NaOH, 1 mm EDTA, 0.2% (w/v) BSA, and 4 mm Cys, pH 7.5, and the homogenate was filtered through four layers of cheesecloth. The filtrate was centrifuged at 1,500g for 10 min to yield a pellet containing predominantly intact chloroplasts and other dense material. The supernatant was centrifuged at 12,000g for 20 min to yield a pellet enriched in mitochondria and peroxisomes. Pellets were resuspended in wash medium (0.3 m mannitol, 20 mm Mops-NaOH, 1 mm EDTA, and 0.2% [w/v] BSA, pH 7.2) and repelleted at 12,000g for 20 min. Washed pellets were resuspended in approximately 24 mL of wash medium and distributed among eight 40-mL polycarbonate tubes containing discontinuous Percoll gradients. Each gradient consisted of 6 mL of 60% (v/v) Percoll, 16 mL of 38% (v/v) Percoll, and 10 mL of 26% (v/v) Percoll in a buffer of 0.25 m Suc, 10 mm Mops-NaOH, and 0.2% (w/v) BSA, pH 7.2. These step gradients separated mitochondria (which form a band at the 26%/38% interface) from peroxisomes (which form a band at the 38%/60% interface), and to some extent from thylakoid membranes (atop and within the 26% Percoll layer). After centrifugation at 15,000g for 45 min using a JA-20 rotor (Beckman), mitochondria were removed, diluted at least 5-fold with wash medium, centrifuged at 12,000g for 20 min, and resuspended in a final volume of approximately 16 mL. This suspension was then distributed over four 40-mL polycarbonate tubes containing continuous Percoll gradients (self-generating gradients, initially 31% [v/v] Percoll). Gradients were subjected to centrifugation at 41,000g for 30 min. The highly purified mitochondria (which form a band near the bottom of the continuous gradient) were collected and washed twice by dilution (at least 5-fold) with BSA-free wash medium and centrifuged at 12,000g for 20 min. Pooled mitochondrial pellets (generally 15–25 mg of protein) were resuspended in a small volume of BSA-free wash medium (final volume of 1.5–2.5 mL). Mitochondrial purity was assessed by assaying Cyt c oxidase activity (mitochondrial marker), hydroxypyruvate reductase activity (peroxisomal marker), and NADP-glyceraldehyde-3-phosphate dehydrogenase activity (chloroplastic marker) in samples taken from the crude filtrate and from the final mitochondrial preparation. In mitochondrial purifications 12% to 21% of the Cyt c oxidase activity was retained, resulting in a 47- to 68-fold increase in specific activity. The purification procedure resulted in the removal of more than 99% of hydroxypyruvate reductase activity and more than 99.9% of NADP-glyceraldehyde-3-phosphate dehydrogenase activity.
In Vitro Mitochondrial Assays
Suspensions of freshly isolated mitochondria were adjusted to 10 mm DTT and 0.01% (v/v) Triton X-100 and sonicated (15 s/mL, 60% output power). Lysed mitochondria were immediately desalted on Sephadex G-25 columns (200 μL extract/mL bed volume) using 0.1 m KH2PO4, pH 7.2, 1 mm DTT as the elution buffer, and eluents were immediately frozen in liquid nitrogen and stored until later use.
The components of the Leu catabolic pathway were assessed as follows: Mitochondrial extracts were incubated with either 1 μCi of l-[U-14C]Leu (346 mCi/mmol; ICN) or with various intermediates of the Leu catabolic pathway (300 μm MC-CoA, 1 mm IV-CoA, 1 mm α-KIC, or 1 mm l-Leu) in the presence of 20 μCi of NaH14CO3 (54 mCi/mmol; Amersham). The basal reaction components of these incubations were: 0.3 m mannitol, 5 mm MgCl2, 10 mm KCl, 10 mm KH2PO4, 0.1% (w/v) BSA, 300 μm thiamine pyrophosphate, 300 μm NAD+, 300 μm FAD+, 1 mm ATP, 500 μm CoA, and 300 μg of mitochondrial protein, pH 7.2, in a total volume of 600 μL. In reactions containing either l-[U-14C]Leu or nonradioactive Leu, 1 mm α-ketoglutarate was added to support the transamination of Leu, and in reactions with l-[U-14C]Leu, 300 μm NaHCO3 was added to support the MCCase reaction. Mitochondrial extract was added immediately before the start of the reactions, which were initiated by the addition of the appropriate radioactively labeled compound. Reactions were incubated at 21°C for the appropriate duration on a nutator and terminated by adjusting the solution to 5% TCA. Precipitated protein was pelleted in a microcentrifuge, and 10 μL of the supernatant was transferred to a 0.75- × 0.75-cm section of 3MM paper (Whatman), allowed to dry overnight to remove 14CO2, and total acid-stable products were measured by liquid scintillation. The remainder of the supernatant was filtered through a 0.2-nm-pore nylon syringe filter for HPLC analysis.
HPLC Analysis
For the quantitation of Leu, dansylated amino acids were fractionated by HPLC using an octadecylsilane column (Supelcosil LC-18, 25-cm × 4.6-mm i.d., 5-μm particle size) with a 20-μL injection volume and a flow rate of 1.2 mL/min. The solvent program was isocratic at 7% solvent B for 1 min, linear gradients of 7% solvent B to 48% solvent B for 15 min, 48% solvent B to 65% solvent B for 2 min, 65% solvent B to 7% solvent B for 7 min, and isocratic at 7% solvent B for 3 min (solvent A was 10% acetonitrile and solvent B was 30% acetonitrile, both in 25 mm trifluoroacetic acid, pH 7.6). Eluted peaks were identified by A254 and quantification was performed with reference to standard dansyl-Leu.
Separation of radioactively labeled metabolites was performed using octadecylsilane columns (Supelcosil LC-18, 25-cm × 4.6-mm i.d., 5-μm particle size; Alltech [Deerfield, IL] Adsorbosphere, 25 cm × 4.6 mm i.d., 5-μm particle size). The solvents and gradient profiles were the same as those reported by King and Reiss (1985). Radioactively labeled metabolites were detected using a radioisotope detector (model 171, Beckman) with a scintillation-cocktail (ScintiSafe Plus 50%, Fisher) flow rate of 2 mL/min. Radioactive peaks were identified by co-elution with standards, measured at the maximal absorbance wavelength of the specific compounds (254 nm for acyl-CoA compounds and 210 nm for organic acids). The identity of acyl-CoA metabolites was confirmed by observing the disappearance of specific peaks after alkaline hydrolysis of the thioester bond (2.5 m NaOH, 3 h, 60°C). This treatment also affects α-KIC, which undergoes decarboxylation at alkaline pH.
Statistical Analyses
MCCase activity and Leu, protein, and chlorophyll data from Figures 2 and 3 are reported with se values for the individual treatments. For all other data se values were determined from the mean square of the error term from each respective analysis of variance. MCCase activity in Table I was analyzed as a factorial design with organ, developmental stage, and replication as the independent variables. The remaining data were analyzed as a factorial design with substrate, avidin, incubation duration, replication (Fig. 5; Table II) or reaction, incubation duration, and replication (Table III) as the independent variables. Incubation duration was treated as a linear variable within the model. Interactions that did not significantly reduce the error or were not meaningful were removed from the model, so only treatments giving a measurable response were included in the analysis. For the data in Tables II and III and Figure 5, the variance was proportional to the mean; consequently, a log transformation was used.
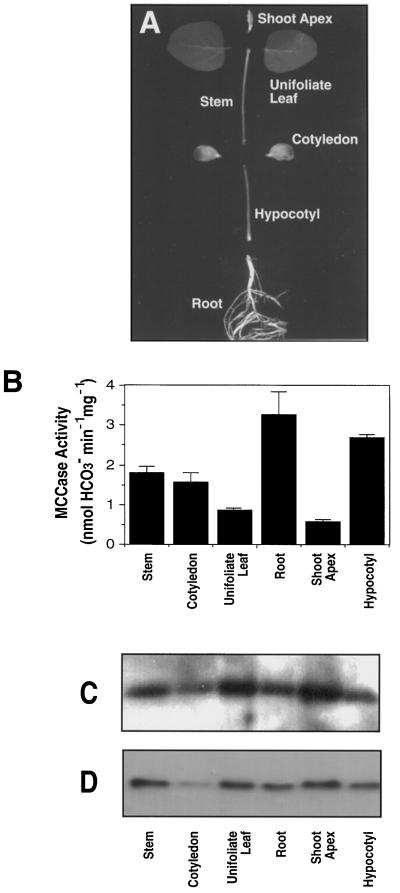
Figure 2.
MCCase activity and the accumulation of the biotin-containing subunit of MCCase in soybean seedlings at 13 DAP. A, Organs of a soybean seedling. B, MCCase activity (mean of five experiments ± se). C, Western blot probed with antiserum to detect the biotin-containing subunit of MCCase. D, Western blot identical to that shown in C, but instead probed with 125I-streptavidin to detect the biotin prosthetic group on the biotin-containing subunit of MCCase. In C and D protein samples were loaded on the basis of equal MCCase activity (0.1 nmol/min) to detect differences in the catalytic efficiency of the enzyme among organs. The data presented in C and D were gathered from a single experiment; five replicates of this experiment gave similar results.
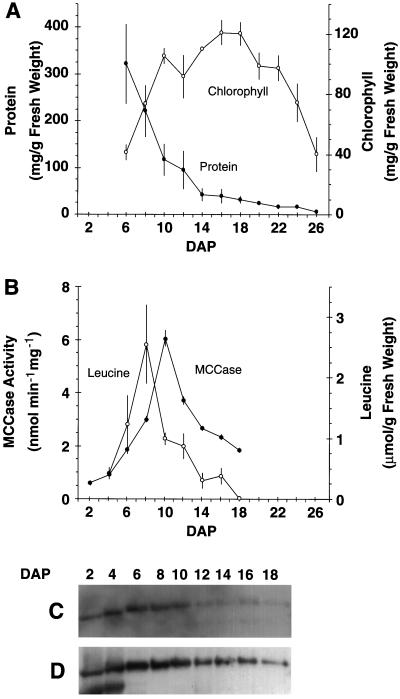
Figure 3.
Effect of cotyledon development on MCCase activity and Leu, protein, and chlorophyll accumulation. A, Chlorophyll and total protein contents. B, MCCase activity and free Leu content. C, Western blot probed with antiserum to detect the biotin-containing subunit of MCCase. D, Western blot identical to that shown in C, but instead probed with 125I-streptavidin to detect the biotin prosthetic group on the biotin-containing subunit of MCCase. In C and D protein samples were loaded on the basis of equal MCCase activity (0.1 nmol/min) to detect differences in the catalytic efficiency of the enzyme during cotyledon development. The data presented in A and B are means ± se from three replicates. The data presented in C and D were gathered from a single experiment; three replicates of this experiment gave similar results.
Table I.
MCCase activity in developing soybean organs
| Organ | Developmental Stage | MCCase Activitya |
|---|---|---|
| DAFb | nmol min−1mg−1 protein | |
| Developing seeds | 20 | 6.9 |
| 30 | 9.9 | |
| 40 | 5.6 | |
| 50 | 3.3 | |
| Dry seeds | 4.3 | |
| Seed pod coat | 10 | 2.5 |
| 20 | 5.0 | |
| 30 | 6.0 | |
| 40 | 3.3 | |
| 50 | 2.3 | |
| 60 | 0.8 | |
| Reproductive leafc | 10 | 1.2 |
| 20 | 1.3 | |
| 30 | 1.3 | |
| 40 | 1.3 | |
| 50 | 0.02 | |
| Young leafd | 3.4 | |
| Mature leafe | 2.7 |
Average of three replicates. se = 0.7 nmol min−1mg−1 protein.
Days after flowering.
Reproductive leaf is the leaf next to a seed pod and is staged relative to flowering.
Young leaf is the first true leaf of 14-d-old plants.
Mature leaf is the first true leaf of 30-d-old plants, before flowering.
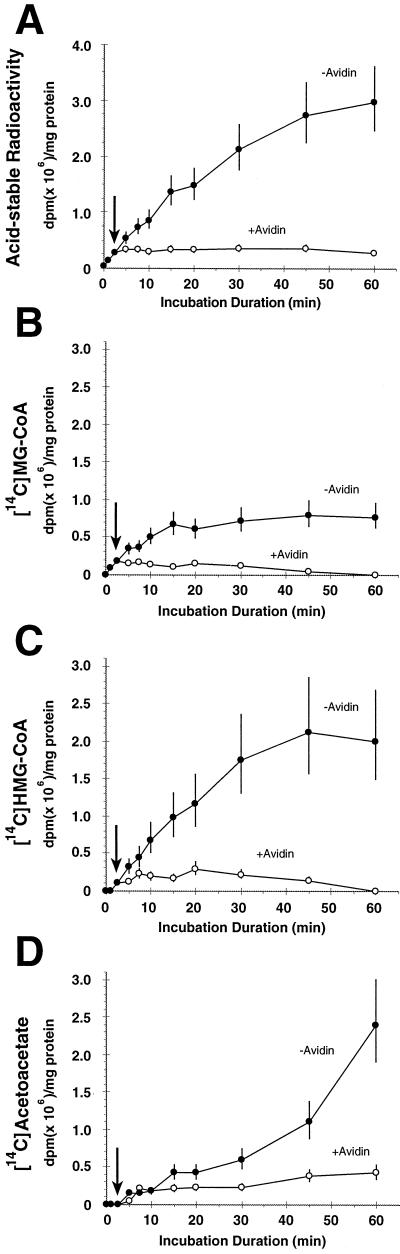
Figure 5.
Kinetics of the incorporation of radioactively labeled bicarbonate into acid-stable metabolites by mitochondrial extracts in the presence of MC-CoA and cofactors. Time course of the incorporation of radioactivity into acid-stable products (A), MG-CoA (B), HMG-CoA (C), or acetoacetate (D). Incubations were carried out either without or with 10 μg of avidin (to inhibit MCCase activity), which was added 2.5 min after the start of the reaction (arrows). Each time point is an independent sample and represents two replicates. Because of the rapid incorporation of NaH14CO3, the background radioactivity was impossible to obtain. Therefore, the 0-min time point was derived from samples incubated for 1 min in the presence of 10 μg of avidin.
Table II.
Mitochondrial metabolism of α-KIC and IV-CoA
| Substrate | Incubation Duration | Acid-Stable
Metabolites
|
MG-CoA
|
HMG-CoA
|
|||
|---|---|---|---|---|---|---|---|
| −Avidin | +Avidin | −Avidin | +Avidin | −Avidin | +Avidin | ||
| min | dpm (×103)/mg protein | % | |||||
| IV-CoA | 1 | 47.8 (+10.3) | – | NDa | – | ND | – |
| 5 | 80.3 (+17.3) | 64.6 (+13.9) | ND | ND | ND | ND | |
| 20 | 106.2 (+22.8) | 64.9 (+13.9) | ND | ND | 100 | ND | |
| 60 | 166.4 (+35.8) | 77.2 (+16.6) | 17 | ND | 83 | ND | |
| α-KIC | 1 | 49.9 (+10.7) | – | ND | – | ND | – |
| 5 | 63.7 (+13.7) | 64.5 (+13.9) | ND | ND | ND | ND | |
| 20 | 88.5 (+19.0) | 67.1 (+14.4) | ND | ND | 100 | ND | |
| 60 | 146.3 (+31.4) | 100.8 (+21.5) | ND | ND | 100 | ND | |
Mitochondrial extracts were incubated with α-KIC or IV-CoA, and their conversion to MC-CoA was monitored by the MCCase-catalyzed (avidin-sensitive) incorporation of radioactivity from NaH14CO3 into acid-stable products, which were identified as [14C]MG-CoA or [14C]HMG-CoA by HPLC analysis. The amount of radioactivity present in each metabolite is expressed as a percentage of the total radioactivity recovered during HPLC fractionation, discounting the residual [14C]bicarbonate peak. For acid-stable metabolites, the variance was dependent on the mean and a log transformation was used. Consequently, 1 se above the mean (shown) was greater than 1 se below the mean (not shown).
ND, Not detected.
Table III.
Catabolism of Leu by mitochondrial extracts
| Additions/Omissions | Incubation Duration | α-KIC | IV-CoA | MC-CoA |
|---|---|---|---|---|
| h | dpm (×103)/mg protein | |||
| None | 0.33 | 256.4 (+45.1) | NDa | ND |
| 1 | 585.3 (+102.9) | 19.7 (+2.3) | ND | |
| 2 | 1303.2 (+229.1) | 110.5 (+13.0) | ND | |
| 4 | 697.6 (+122.7) | 426.5 (+50.2) | 8.6 (+4.2) | |
| 6 | 362.1 (+63.7) | 508.6 (+59.8) | 29.9 (+14.4) | |
| −α-KG | 2 | 89.4 (+15.7) | ND | ND |
| −CoA | 2 | 1214.7 (+213.6) | ND | ND |
| +1 mm α-KIC | 2 | 1099.0 (+193.2) | ND | ND |
| 4 | 1835.0 (+322.6) | ND | ND | |
| 6 | 2270.3 (+399.1) | ND | ND | |
| +Avidin | 2 | 936.8 (+164.7) | 154.3 (+18.2) | ND |
| 4 | 800.7 (+139.9) | 420.6 (+49.5) | 51.4 (+24.7) | |
| 6 | 602.4 (+105.9) | 436.8 (+51.4) | 71.7 (+34.5) | |
Mitochondrial extracts were incubated with [U-14C]Leu and cofactors for various lengths of time and the resulting radioactive products were analyzed by HPLC. Additional reactions were also conducted without added α-ketoglutarate (α-KG) or CoA, or with the addition of 1 mm nonradioactive α-KIC or 10 μg of avidin. α-KG is required for the transamination of Leu; CoA is required for the decarboxylation and activation of α-KIC in the BCKDH reaction; nonradioactive α-KIC inhibits further metabolism of α-[14C]KIC by competition; and avidin inhibits MCCase. se values are reported as in Table II.
ND, Not detected.
RESULTS
MCCase Activity during Soybean Fruit Development
MCCase activity was determined in extracts from the developing soybean fruit, its subtending leaf, and in vegetative leaves (Table I). In developing seeds MCCase activity peaked at the onset of storage lipid and protein deposition at about 30 d after flowering, and subsequently declined to one-third of the peak activity as the seed matured. MCCase activity was also detectable in dry seeds. During seed set MCCase activity was somewhat lower in the pod (pericarp) than in the developing seed, and mirrored the developmentally induced changes that occurred in the seed until desiccation. In contrast to the seed, the pod did not retain MCCase activity upon desiccation. In the leaves subtending the fruit, MCCase activity changed little during the period when seeds were developing until late in seed development, when the leaves began to senesce and MCCase activity declined. MCCase activity in vegetative leaves was higher than in the leaves subtending the fruit and was somewhat higher in the young leaves than in the mature leaves.
Organ-Specific Expression of MCCase
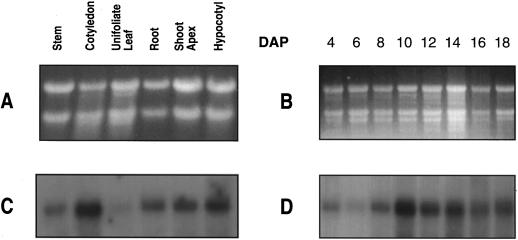
The distribution of MCCase was examined among organs of soybean seedlings at 13 DAP. At this stage of maturity the unifoliate leaves of the seedlings had fully unrolled (VC stage; Fehr and Caviness, 1977) and the cotyledons were beginning to senesce (Fig. 2A). MCCase activity was detectable in the shoot apex and in all organs of the seedling (Fig. 2B). The specific activity of MCCase was highest in roots and hypocotyls and lowest in unifoliate leaves and shoot apices. There was up to a 7-fold difference in the specific activities of MCCase among these organs.
To determine whether these differences in MCCase activity were caused by the differences in the accumulation of the biotin-containing subunit of MCCase, samples of extracts from each organ containing equal MCCase activity were subjected to SDS-PAGE, followed by western-blot analysis with antiserum against the biotin-containing subunit of MCCase (Fig. 2C). We would expect to observe equal intensities of the 85-kD MCCase subunit if the difference in activity among the organs were caused by differences in the accumulation of the biotin-containing subunit. This was not the case, however, so we conclude that the level of the biotin-containing subunit alone does not determine the activity of MCCase. One possible specific explanation that we tested is that MCCase activity is modified by the degree of biotinylation of the apoprotein, as was observed previously in tomato (Wang et al., 1995). However, in soybean the differences in the intensity of the MCCase biotin-containing subunit band, revealed by western-blot analysis with 125I-streptavidin, mirrored the changes detected by the antiserum to that subunit (Fig. 2, compare C and D), indicating that the relative state of biotinylation of the MCCase subunit was constant among the organs examined.
MCCase Expression in Developing Cotyledons
One possible metabolic function of MCCase in plants is in the catabolism of Leu. Examination of MCCase in organs in which free Leu is expected to be rapidly metabolized may provide insight into the role of MCCase in Leu catabolism. Developing soybean cotyledons were chosen as the model system. Using chlorophyll content as the criterion, under the growth conditions used in these experiments, development of the photosynthetic apparatus occurred between 6 and 10 DAP, after which the cotyledons maintained photosynthetic competence until 18 DAP, when senescence became evident. Protein content declined dramatically from 6 to 14 DAP, reflecting the degradation of seed-storage proteins (Fig. 3A). During this period of protein mobilization, free Leu accumulated, peaking at 8 DAP, and then sharply declined (Fig. 3B). During soybean cotyledon development, MCCase activity increased, reaching a peak at 10 DAP during the period of rapid protein degradation and coinciding with the decline in Leu concentration (Fig. 3B). The decrease in MCCase activity lagged just behind the decline in Leu content, which is consistent with a role of MCCase in Leu catabolism.
To ascertain whether these developmentally associated changes in MCCase activity were ascribable to changes in MCCase enzyme accumulation or to changes in enzyme efficiency, we determined the accumulation of the biotin-containing subunit of MCCase. Western blots were analyzed with either anti-MCCase serum to detect the subunit polypeptide (Fig. 3C) or 125I-streptavidin to detect the biotin prosthetic group of this polypeptide (Fig. 3D). In these analyses gels were loaded with samples containing equal amounts of MCCase activity. Hence, if the changes in activity during cotyledon development were caused by changes in the steady-state levels of the MCCase biotin-containing subunit, we would expect to observe protein bands of equal intensity. However, the MCCase biotin-containing subunit exhibited a modulation in intensity. Therefore, as we concluded with the organs of soybean seedlings, the level of the biotin-containing subunit alone does not determine the activity of MCCase. Furthermore, the close correlation between the levels of the biotin-containing subunit (Fig. 3C) and the biotin prosthetic group on this subunit (Fig. 3D) indicates that the biotinylation status of the MCCase biotin-containing subunit is unaltered during cotyledon development.
Abundance of MCCase mRNA in Soybeans
To ascertain whether the developmentally associated changes in MCCase expression may also be regulated by the accumulation of the mRNA coding for the biotin-containing subunit of MCCase, we determined the abundance of this mRNA in organs of soybean plants at 13 DAP and in cotyledons from seedlings of various ages (Fig. 4). In plants at 13 DAP, the steady-state level of the 2.4-kb MCCase mRNA was highest in the cotyledon, followed by, in decreasing order, the hypocotyl, shoot apex, root, and stem (Fig. 4C). Very little MCCase mRNA was detected in the leaf. In developing cotyledons the steady-state levels of the MCCase mRNA changed slightly from 4 to 8 DAP, increased by at least 10-fold between 8 and 10 DAP, and subsequently declined to about one-fifth of the level present at peak accumulation (Fig. 4D).
Figure 4.
Steady-state levels of the mRNA coding for the biotin-containing subunit of MCCase in organs of soybean seedlings at 13 DAP and during cotyledon development. Ethidium bromide-stained agarose gel containing approximately equal amounts (20 μg) of RNA, isolated from various organs (A) or from cotyledons at different stages of development (B). Northern blot depicting the accumulation of the mRNA coding for the biotin-containing subunit of MCCase in various organs (C) or in cotyledons at different stages of development (D). The data presented in this figure were gathered from a single experiment. Five replicates (A and C) and three replicates (B and D) of these experiments gave similar results.
Mitochondrial Catabolism of MC-CoA
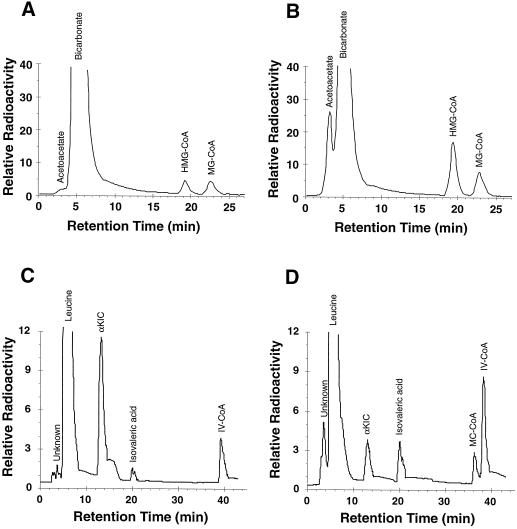
Because MCCase is located in the mitochondria (Baldet et al., 1992), we investigated MCCase-related metabolism in this organelle. In the presence of MC-CoA and cofactors (see Methods), mitochondrial extracts catalyzed the incorporation of radioactivity from NaH14CO3 into acid-stable metabolites in a time-dependent manner (Fig. 5A). Incorporation of label was precluded by withholding MC-CoA or ATP (data not shown), or by preincubating mitochondrial extracts with avidin (Wurtele and Nikolau, 1990; Fig. 5A), consistent with a requirement for MCCase. Analysis of the products by HPLC revealed four radioactively labeled peaks eluting at 3.3, 5, 19.2, and 22.6 min (Fig. 6, A and B). These were identified as acetoacetate, bicarbonate, HMG-CoA, and MG-CoA, respectively. Acetoacetate, HMG-CoA, and MG-CoA are predicted to be labeled if MCCase, MG-CoA hydratase, and HMG-CoA lyase are present and active in mitochondria (Fig. 1). The kinetics of metabolite accumulation (Fig. 5, B–D) support the sequential action of these enzymes. MG-CoA, the immediate product of MCCase, is labeled initially, followed sequentially by accumulation of [14C]HMG-CoA and [14C]acetoacetate. Interestingly, [14C]HMG-CoA accumulated to higher levels than [14C]MG-CoA, indicating that the hydratase reaction occurs at a faster rate than the lyase reaction, at least under the conditions we used.
Figure 6.
Separation of radioactive metabolites by HPLC. Representative chromatograms depicting metabolites arising from the catabolism of MC-CoA by mitochondrial extracts in the presence of NaH14CO3 and cofactors after 5-min (A) or 45-min (B) incubations. After 5 min [14C]MG-CoA and [14C]HMG-CoA were at similar levels and [14C]acetoacetate was barely detectable. After 45 min [14C]HMG-CoA had exceeded [14C]MG-CoA and [14C]acetoacetate had increased. Representative chromatograms depicting metabolites arising from the catabolism of [U-14C]Leu by mitochondrial extracts in the presence of 10 μg of avidin after a 2-h (C) or 6-h (D) incubation. After 2 h α-[14C]-KIC accumulated to a substantial level and [14C]IV-CoA accumulation was in progress. After 6 h the level of α-[14C]-KIC diminished, indicating that the rate of its disappearance had exceeded its formation. [14C]IV-CoA continued to accumulate and [14C]MC-CoA appeared. [14C]Isovaleric acid was likely a decarboxylation product of α-[14C]-KIC, possibly arising nonenzymatically.
Additional evidence for the sequential conversion of MC-CoA to MG-CoA, HMG-CoA, and acetoacetate was obtained in experiments in which avidin was used to inhibit MCCase activity after an initial incubation without avidin. In these experiments we essentially carried out a pulse-chase experiment in which the incubation without avidin (for 2.5 min) represented the pulse phase and the incubation after the addition of avidin represented the chase phase. As avidin inhibited further incorporation of radioactivity from NaH14CO3, [14C]MG-CoA and [14C]HMG-CoA sequentially declined as [14C]acetoacetate accumulated. The accumulation of [14C]acetoacetate indicates that further metabolism of acetoacetate in mitochondrial extracts is slower than the previous reaction catalyzed by HMG-CoA lyase, or that the catabolic mechanism (if it exists in mitochondria) had been disrupted in our mitochondrial extracts. These results clearly indicate the existence of MCCase, MG-CoA hydratase, and HMG-CoA lyase in plant mitochondria, enzymes that catalyze reactions common to Leu catabolism, isoprenoid catabolism, and the mevalonate shunt.
Mitochondrial Catabolism of Leu
If MCCase is involved in Leu catabolism, IV-CoA, α-KIC, and Leu would be precursors of MC-CoA (Fig. 1). We addressed this hypothesis by testing whether IV-CoA, α-KIC, and Leu could support the incorporation of radioactivity from NaH14CO3 into MG-CoA, HMG-CoA, and acetoacetate. The results presented in Table II indicate that both IV-CoA and α-KIC support the incorporation of radioactivity from NaH14CO3 into acid-stable products in a time-dependent manner. As expected, this incorporation was avidin sensitive. Both IV-CoA and α-KIC support the incorporation of radioactivity into HMG-CoA, which is detectable after a 20-min incubation. In the case of the incubation with IV-CoA, [14C]MG-CoA was detected after a 1-h incubation. These data indicate that both BCKDH and the branched-chain acyl-CoA dehydrogenase are present in mitochondria (Fig. 1). Leu was unable to support detectable incorporation of radioactivity from NaH14CO3 into MG-CoA or HMG-CoA (data not shown). A likely explanation is that Leu is too far upstream in the metabolic pathway from the MCCase reaction to be an effective source of MC-CoA.
To demonstrate that mitochondria possess the entire complement of Leu catabolic enzymes, the metabolic products of [U-14C]Leu were examined by HPLC. Upon incubation of mitochondrial extracts with [U-14C]Leu, six primary peaks of radioactive metabolites were observed, eluting at 3.6, 5.1, 13, 20, 36.5, and 38.5 min. The peak at 20 min co-elutes with isovaleric acid, possibly arising from decarboxylation of α-KIC. Other peaks were identified as Leu (5.1 min), α-KIC (13 min), MC-CoA (36.5 min), and IV-CoA (38.5 min). The peak at 3.6 min remains unidentified. α-[14C]-KIC was first detected after 20 min and continued to accumulate to a maximum at 2 h (Table III). The conversion of [U-14C]Leu to α-[14C]-KIC as well as subsequent catabolism was dependent on the addition of α-ketoglutarate, suggesting the presence of the branched-chain amino acid aminotransferase in mitochondria. Accumulation of [14C]IV-CoA was detected between 1 and 2 h, demonstrating the sequential action of the aminotransferase and BCKDH. This appearance of [14C]IV-CoA was prevented by competition with nonradioactive α-KIC or by withholding free CoA, which is needed by BCKDH, from the incubation mix. [14C]MC-CoA was detectable after 4 h of incubation and its presence was enhanced by the addition of avidin, which inhibits MCCase and prevents further catabolism of MC-CoA.
DISCUSSION
Developmental Regulation of MCCase
As a first step in elucidating the physiological functions of MCCase in plants, we determined the accumulation of MCCase mRNA and protein in different organs of soybean and during cotyledon development. MCCase is expressed in all organs of the soybean plant, although there is considerable variation in the level of expression, presumably reflecting the relative importance of MCCase in these organs. These studies indicated that MCCase expression can be regulated by at least two different mechanisms. Changes in the accumulation of the mRNA for the biotin-containing subunit of MCCase probably reflect developmentally regulated changes in its gene transcription or mRNA stability. For example, during the development of the cotyledons, the accumulation of this mRNA increased with increasing age of the organ. In addition, MCCase activity did not always correlate with the level of accumulation of the MCCase biotin-containing subunit, indicating that MCCase activity is controlled by a second mechanism. This lack of correlation could be explained by changes in the accumulation of the nonbiotinylated subunit (which we could not detect) or by changes in the catalytic efficiency of the MCCase enzyme. The catalytic efficiency of the MCCase enzyme might have been altered during development as a result of a posttranslational mechanism (e.g. covalent modification of the enzyme) or the differential expression of isozymes of MCCase that differ in their catalytic efficiency. One posttranslational modification that is essential for the activity of MCCase is biotinylation; MCCase activity is modulated by biotinylation in tomato (Wang et al., 1995). However, in seedling organs and developing cotyledons of soybean, differences in the abundance of the biotin prosthetic group closely paralleled differences in the abundance of the biotinylated subunit. Therefore, MCCase did not appear to be regulated by biotinylation in soybean. Clearly, isolation and characterization of the gene for the nonbiotinylated subunit of MCCase will facilitate studies of the regulation of the activity of this enzyme.
Cotyledons initially function as a source tissue, supporting the growth of the entire seedling. At this stage of development, seed storage proteins are hydrolyzed and the released amino acids are incorporated into new proteins, catabolized during respiration, or used in the formation of new compounds. In soybean cotyledons we observed that the maximal rate of protein degradation occurred between 4 and 6 DAP, which correlated with the peak accumulation of free Leu content. Subsequently, between 8 and 10 DAP, the free Leu concentration declined and this correlated with the peak in MCCase activity. These data indicate that MCCase may have a role in Leu catabolism in cotyledons. This conclusion is consistent with the finding that in carbohydrate-starved sycamore cells there is a correlation between a transient accumulation of Leu and the accumulation of the biotin-containing subunit of MCCase (Aubert et al., 1996). It is possible that Leu accumulation itself, or some consequence of Leu accumulation, is responsible for the induction of MCCase activity. Indeed, exogenously added Leu has been observed to induce MCCase activity in carrot cells (E.S. Wurtele and B.J. Nikolau, unpublished data). However, in sycamore cells exogenous Leu did not induce accumulation of the biotin-containing subunit of MCCase (Aubert et al., 1996), suggesting that Leu itself was not the signaling molecule in this system.
Role of MCCase in Leu Catabolism
To provide more direct evidence for the metabolic function of MCCase in plants, we analyzed the fate of the MCCase product MG-CoA in extracts from isolated mitochondria. Incubation of mitochondrial extracts with MC-CoA and NaH14CO3 resulted in the time-dependent incorporation of label into three compounds: MG-CoA, HMG-CoA, and acetoacetate. These three metabolites are predicted to arise from MC-CoA catabolism via the pathway depicted in Figure 1. Furthermore, the time course of accumulation of radioactivity in these products relative to each other, and the depletion of [14C]MG-CoA and [14C]HMG-CoA and the simultaneous accumulation of [14C]acetoacetate after the addition of avidin to the mitochondrial extracts, are consistent with the interdependence of these three metabolites within a single pathway. Additional radiotracer experiments showed that mitochondrial extracts can metabolize α-KIC and IV-CoA to MC-CoA, which could then be carboxylated by MCCase. Finally, mitochondrial extracts are able to catabolize [U-14C]Leu sequentially to α-[14C]-KIC, [14C]IV-CoA, and [14C]MC-CoA. In combination, these radiotracer metabolic studies have shown that mitochondria have the capacity to catabolize Leu to acetoacetate and, presumably, acetyl-CoA, as illustrated in Figure 1. These findings suggest that the six enzymes involved in Leu catabolism are housed in the mitochondria.
In animals, the mitochondrial location of all of the enzymes of Leu catabolism is well established, whereas in plants, these enzymes have received little attention. MG-CoA hydratase and HMG-CoA lyase have been studied in the context of their role in providing HMG-CoA for isoprenoid biosynthesis (Alam et al., 1991; Weber and Bach, 1993; van der Heijden et al., 1994; van der Heijden and Verpoorte, 1995). Studies of the branched-chain amino acid aminotransferase have concentrated on its role as the terminal enzyme in the biosynthesis of branched-chain amino acids (Aarnes, 1981; Pathre et al., 1987, 1989). Until our studies, the subcellular locations of these enzymes were unknown in plants.
Apart from MCCase, the only enzyme involved in mitochondrial Leu catabolism for which a cDNA or gene has been cloned is BCKDH (Fujiki et al., 1997; Luethy et al., 1997; Mooney et al., 1998). Interestingly, Fujiki et al. (1997) observed enhanced accumulation of BCKDH mRNA in response to dark-induced starvation and senescence, conditions that induce MCCase accumulation (Aubert et al., 1996). A BCKDH has been shown to be present in peroxisomes, and subsequent studies on the metabolism of α-KIC by isolated peroxisomes have led to the prediction of a parallel but MCCase-independent pathway for Leu catabolism (Gerbling and Gerhardt, 1988, 1989; Gerbling, 1993).
Our findings that mitochondria have the capacity to catabolize Leu by an MCCase-dependent mechanism, in combination with the studies of Gerbling and Gerhardt (1989) and Gerbling (1993), indicate that plants can catabolize Leu via two pathways that are separated into different subcellular compartments, mitochondria and peroxisomes. Studies are still required to determine the fate of carbon catabolized via these two pathways and the mechanisms that regulate the commitment of Leu to each catabolic pathway. The conditions under which such partitioning is altered should reveal much about the mechanisms that control the utilization of carbon reserves during plant metabolism.
In addition to its role in the mitochondrial catabolism of Leu, MCCase has been suggested to participate in two important elements of isoprenoid metabolism in plants: the mevalonate shunt (Nes and Bach, 1985) and the catabolism of isoprenoids via geranoyl-CoA (X. Guan, T.A. Diez, B.J. Nikolau, and E.S. Wurtele, unpublished data). The ultimate product of these pathways, acetyl-CoA, has a wide array of potential fates, including respiration, the glyoxylate cycle, lipogenesis, and the biogenesis of polyketides (Fig. 1). By virtue of its central position in Leu and isoprenoid metabolism, and the numerous potential fates of acetyl-CoA, MCCase might play a pivotal role in regulating the carbon flux among these pathways.
ACKNOWLEDGMENTS
We thank Dr. I.C. Anderson, Iowa State University, for providing soybean seeds. We are grateful to Beth L. Fatland for helpful discussion.
Abbreviations:
- BCKDH
branched-chain keto-acid dehydrogenase
- DAP
days after planting
- HMG-CoA
3-hydroxy-3-methylglutaryl-CoA
- IV-CoA
isovaleryl-CoA
- α-KIC
α-ketoisocaproate
- MCCase
3-methylcrotonyl-CoA carboxylase
- MC-CoA
3-methylcrotonyl-CoA
- MG-CoA
3-methylglutaconyl-CoA
Footnotes
This work was supported by National Science Foundation grant no. IBN-9507549 to E.S.W. and B.J.N. This is journal paper no. J-15558 of the Iowa Agriculture and Home Economics Experiment Station, Ames, IA; project nos. 2997 and 2913.
LITERATURE CITED
- Aarnes H. Branched-chain amino acid aminotransferase in barley seedlings. Z Pflanzenphysiol. 1981;102:81–89. [Google Scholar]
- Alam A, Britton G, Powls R, Goad J. Aspects related to 3-hydroxy-3-methylglutaryl-CoA synthesis in higher plants. Biochem Soc Trans. 1991;19:164S. doi: 10.1042/bst019164s. [DOI] [PubMed] [Google Scholar]
- Alban C, Baldet P, Axiotis S, Douce R. Purification and characterization of 3-methylcrotonyl-CoA carboxylase from higher plant mitochondria. Plant Physiol. 1993;102:957–965. doi: 10.1104/pp.102.3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. Copper enzymes in chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert S, Alban C, Bligny R, Douce R. Induction of beta-methylcrotonyl-coenzyme A carboxylase in higher plant cells during carbohydrate starvation: evidence for a role of MCCase in leucine catabolism. FEBS Lett. 1996;383:175–180. doi: 10.1016/0014-5793(96)00244-x. [DOI] [PubMed] [Google Scholar]
- Baldet P, Alban C, Axiotis S, Douce R. Characterization of biotin and 3-methylcrotonyl-coenzyme A carboxylase from higher plant mitochondria. Plant Physiol. 1992;99:450–455. doi: 10.1104/pp.99.2.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chen Y, Wurtele ES, Wang X, Nikolau BJ. Purification and characterization of 3-methylcrotonyl-CoA carboxylase from somatic embryos of Daucus carota. Arch Biochem Biophys. 1993;305:103–109. doi: 10.1006/abbi.1993.1398. [DOI] [PubMed] [Google Scholar]
- Diez TA, Wurtele ES, Nikolau BJ. Purification and characterization of 3-methylcrotonyl-coenzyme-A carboxylase from leaves of Zea mays. Arch Biochem Biophys. 1994;310:64–75. doi: 10.1006/abbi.1994.1141. [DOI] [PubMed] [Google Scholar]
- Edmund J, Popják G. Transfer of carbon atoms from mevalonate to n-fatty acids. J Biol Chem. 1974;249:66–71. [PubMed] [Google Scholar]
- Fehr WR, Caviness CE. Stages of soybean development. Iowa State University Cooperative Extension Service. 1977;80:1–12. [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Sato T, Yoshikawa Y, Ito M, Watanabe A. Dark induced expression of genes for branched-chain alpha-keto acid dehydrogenase complex in Arabidopsisleaves (abstract no. 400) Plant Physiol. 1997;114:S-95. [Google Scholar]
- Gerbling H. Peroxisomal degradation of 2-oxoisocaproate: evidence for free acid intermediates. Bot Acta. 1993;106:380–387. [Google Scholar]
- Gerbling H, Gerhardt B. Oxidative decarboxylation of branched-chain 2-oxo fatty acids by higher plant peroxisomes. Plant Physiol. 1988;88:13–15. doi: 10.1104/pp.88.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbling H, Gerhardt B. Peroxisomal degradation of branched-chain 2-oxo acids. Plant Physiol. 1989;91:1387–1392. doi: 10.1104/pp.91.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber U, Pon NG, Heber M. Localization of carboxydismutase and triosephosphate dehydrogenases in chloroplasts. Plant Physiol. 1963;38:355–360. doi: 10.1104/pp.38.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CG, Hare JD, Compton SJ. Measuring plant protein with the Bradford assay. J Chem Ecol. 1989;15:979–993. doi: 10.1007/BF01015193. [DOI] [PubMed] [Google Scholar]
- Kaethner TM, ap Rees T. Intracellular location of ATP citrate lyase in leaves of Pisum sativumL. Planta. 1985;163:290–294. doi: 10.1007/BF00393520. [DOI] [PubMed] [Google Scholar]
- King MT, Reiss PD. Separation and measurement of short-chain coenzyme-A compounds in rat liver by reversed-phase high-performance liquid chromatography. Anal Biochem. 1985;146:173–179. doi: 10.1016/0003-2697(85)90412-9. [DOI] [PubMed] [Google Scholar]
- Koops AJ, Italiaander E, Groeneveld HW. Triterpenoid biosynthesis in the seedling of Euphorbia lathyrisL. from sucrose and amino acids. Plant Sci. 1991;74:193–201. [Google Scholar]
- Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Luethy MH, Miernyk JA, Randall DD. Molecular analysis of a branched-chain keto-acid dehydrogenase from Arabidopsis thaliana(abstract no. 696) Plant Physiol. 1997;114:S-147. [Google Scholar]
- Mooney BP, Miernyk JA, Randall DD. Nucleotide sequence of a cDNA encoding the dihydrolipoylacyltransferase (E2) subunit of the branched-chain alpha-keto acid dehydrogenase complex from Arabidopsis thaliana (accession no. AF038505) (PGR 98-071) Plant Physiol. 1998;117:331. [Google Scholar]
- Moss J, Lane MD. The biotin-dependent enzymes. Adv Enzymol. 1971;35:321–442. doi: 10.1002/9780470122808.ch7. [DOI] [PubMed] [Google Scholar]
- Nes WD, Bach TJ. Evidence for a mevalonate shunt in a tracheophyte. Proc R Soc Lond B Biol Sci. 1985;225:425–444. [Google Scholar]
- Nikolau BJ, Wurtele ES, Stumpf PK. Use of streptavidin to detect biotin-containing proteins in plants. Anal Biochem. 1985;149:448–453. doi: 10.1016/0003-2697(85)90596-2. [DOI] [PubMed] [Google Scholar]
- Overton KH. The role of leucine in terpenoid metabolism: incorporation of leucine into sesquiterpenoids and phytosterols by andrographic tissue culture. In: Neumann KH, Barz W, Reinhard E, editors. Primary and Secondary Metabolism of Plant Cell Cultures. Berlin: Springer-Verlag; 1985. pp. 225–234. [Google Scholar]
- Pathre U, Singh AK, Sane PV. Inactivation of leucine aminotransferase with diethylpyrocarbonate and rose bengal: evidence for an active site histidine residue. Indian J Biochem Biophys. 1989;26:136–139. [PubMed] [Google Scholar]
- Pathre U, Singh AK, Viswanathan PN, Sane PV. Purification and properties of leucine aminotransferase from soybean seedlings. Phytochemistry. 1987;26:2913–2917. [Google Scholar]
- Popják G. Specificity of enzymes in sterol biosynthesis. Harvey Lect. 1971;65:127–156. [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sandalio LM, Palma JM, Del Rio LA. Localization of manganese superoxide dismutase in plant peroxisomes isolated from Pisum sativumL. Plant Sci. 1987;51:1–8. [Google Scholar]
- Schwitzguebel J-P, Siegenthaler P-A. Purification of peroxisomes and mitochondria from spinach leaf by Percoll gradient centrifugation. Plant Physiol. 1984;75:670–674. doi: 10.1104/pp.75.3.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seubert W, Remberger U. Untersuchungen uber den bakteriellen abbau von isoprenoiden. II. Die rolle der kohlensaure. Biochem Z. 1963;338:245–264. [PubMed] [Google Scholar]
- Sodek L, Wilson CM. Metabolism of lysine and leucine derived from storage protein during the germination of maize. Biochim Biophys Acta. 1973;304:353–362. doi: 10.1016/0304-4165(73)90253-5. [DOI] [PubMed] [Google Scholar]
- Song J, Wurtele ES, Nikolau BJ. Molecular cloning and characterization of the cDNA coding for the biotin-containing subunit of 3-methylcrotonyl-CoA carboxylase: identification of the biotin carboxylase and biotin-carrier domains. Proc Natl Acad Sci USA. 1994;91:5779–5783. doi: 10.1073/pnas.91.13.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CR, Beevers H. Gluconeogenesis from amino acids in germinating castor bean endosperm and its role in transport to the embryo. Plant Physiol. 1967;42:1587–1595. doi: 10.1104/pp.42.11.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapuhi YT, Schmidt DE, Lindner W, Karger BL. Dansylation of amino acids for high-performance liquid chromatography analysis. Anal Chem. 1981;48:1106–1109. doi: 10.1016/0003-2697(81)90534-0. [DOI] [PubMed] [Google Scholar]
- van der Heijden R, de Boer-Hlupa V, Verpoorte R, Duine JA. Enzymes involved in the metabolism of 3-hydroxy-3-methylglutaryl-coenzyme A in Catharanthus roseus. Plant Cell Tissue Organ Cult. 1994;38:345–349. [Google Scholar]
- van der Heijden R, Verpoorte R. Metabolic enzymes of 3-hydroxy-3-methylglutaryl-coenzyme A in Catharanthus roseus. Plant Cell Tissue Organ Cult. 1995;43:85–88. [Google Scholar]
- Wang X, Wurtele ES, Keller G, McKean AL, Nikolau BJ. Molecular cloning of cDNAs and genes coding for βmethylcrotonyl-CoA carboxylase of tomato. J Biol Chem. 1994;269:11760–11769. [PubMed] [Google Scholar]
- Wang X, Wurtele ES, Nikolau BJ. Regulation of βmethylcrotonyl-coenzyme A carboxylase activity by biotinylation of the apoenzyme. Plant Physiol. 1995;108:1133–1139. doi: 10.1104/pp.108.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Lebrun L, Franklin A, Huang L, Hoffman N, Wurtele ES, Nikolau BJ. Molecular cloning of the biotinylated subunit of 3-methylcrotonyl-coenzyme A carboxylase of Arabidopsis thaliana. Plant Physiol. 1995;107:1013–1014. doi: 10.1104/pp.107.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, Bach TJ. Partial purification and characterization of membrane-associated 3-hydroxy-3-methylglutaryl-coenzyme A lyase from radish seedlings. Z Naturforsch. 1993;48c:444–450. [Google Scholar]
- Wurtele ES, Nikolau BJ. Plants contain multiple biotin enzymes: discovery of 3-methylcrotonyl-CoA carboxylase, propionyl-CoA carboxylase and pyruvate carboxylase in the plant kingdom. Arch Biochem Biophys. 1990;278:179–186. doi: 10.1016/0003-9861(90)90246-u. [DOI] [PubMed] [Google Scholar]