Summary
Infections with the azole-refractory yeast Candida glabrata are now commonly treated with the echinocandins caspofungin (CSF) or micafungin (MCF). True resistance (>32-fold decreased susceptibility) to these lipopeptide inhibitors of cell wall synthesis is rare and strictly associated with mutations in integral membrane proteins Fks1 or Fks2. In contrast, mutants exhibiting 4 to 32-fold CSF reduced susceptibility (CRS) were readily selected in vitro, and surprisingly demonstrated 4 to 32-fold MCF increased susceptibility (MIS). Sequencing and gene deletion demonstrated that CRS-MIS is Fks-independent. To explore alternative mechanisms, we initially employed Saccharomyces cerevisiae, and observed that CRS was conferred by multiple mutations (fen1Δ, sur4Δ, cka2Δ, and tsc10-ts) disrupting sphingolipid biosynthesis. Following this lead, C. glabrata fen1Δ and cka2Δ deletants were constructed, and shown to exhibit CRS-MIS. Sphingolipid analysis of CRS-MIS laboratory mutants and clinical isolates demonstrated elevated dihydrosphingosine (DHS) and phytosphingosine (PHS) levels, and consistent with this sequencing revealed fen1, sur4, ifa38, and sur2 mutations. Moreover, exogenous DHS or PHS conferred a CRS-MIS phenotype on wild-type C. glabrata. Exogenous PHS failed, however, to suppress CRS-MIS in a sur2 mutant blocked in conversion of DHS to PHS, implying that accumulation of these intermediates confers CRS-MIS. We conclude that membrane sphingolipids modulate echinocandin-Fks interaction.
Keywords: echinocandin, caspofungin, micafungin, Candida glabrata, sphingolipid, Fks1
Introduction
Opportunistic fungal infections have increased in recent years due to increased numbers of individuals who are immunocompromised. The antifungal agents available to combat these infections are limited, not only in number but also in terms of their activity, spectrum, and tolerability. Specifically, amphotericin B is fungicidal and broad spectrum, but toxicity limits its use to life-threatening infections. Conversely, azoles such as fluconazole are well tolerated but fungistatic, and resistance has been widely documented in both yeasts and molds. One such yeast is Candida glabrata, second only to Candida albicans as a cause of mucosal and invasive mycosis. C. glabrata exhibits intrinsically low azole susceptibility, and treatment is frequently complicated by efflux pump-mediated resistance (Morschhauser, 2010; Sanglard and Odds, 2002). The lipopeptide echinocandins, the most recent addition to the antifungal arsenal, combine low toxicity with high activity versus most Candida species as well as Aspergillus fumigatus and related molds (Cleary, 2009; Kauffman and Carver, 2008). Indeed, echinocandins have been elevated to first line agents for treatment of invasive infection with C. glabrata, or with C. albicans if azole resistance is suspected (Pappas et al., 2009). Clinically introduced echinocandins include caspofungin (CSF; FDA-approved in 2001), micafungin (MCF; 2005), and anidulafungin (ANF; 2006). All three are semisynthetic derivatives of natural products, with a common cyclic hexapeptide core but differing in their modifications to this core and to their lipid tails: alkyl in CSF, aryl-akyl in MCF and ANF (Denning, 2003).
Although extensively studied since their discovery in the 1970’s, important gaps remain in our understanding of echinocandin mechanism of action (Douglas, 2001; Perlin, 2007). It is well established that they are non-competitive inhibitors of UDP-glucose polymerization into the essential cell wall polysaccharide β-1,3-glucan (Sawistowska-Schroder et al., 1984; Taft et al., 1988). Further biochemical and structural characterization of the responsible enzyme has been hindered by its plasma membrane localization. As an alternative, genetic analysis of Saccharomyces cerevisiae echinocandin-resistant mutants was pursued, which initially identified the gene GNS1 (el-Sherbeini and Clemas, 1995). However, this putative glucan synthase gene was later revealed to be allelic to the fatty acid elongase ELO2, also known as FEN1 since mutation confers resistance to the ergosterol biosynthesis inhibitor fenpropimorph (Ladeveze et al., 1993). Subsequent genetic analysis in S. cerevisiae, as well as in Candida species, identified resistance-conferring mutations in FKS1 (or its paralog FKS2), encoding a large integral membrane protein (Douglas et al., 1997; Douglas et al., 1994). Partial purification by product entrapment, along with extensive kinetic analysis of Fks1 mutants, strongly suggest that Fks1 is indeed the catalytic component of β-1,3-glucan synthase (Garcia-Effron et al., 2009; Inoue et al., 1995; Okada et al., 2010). Nevertheless, attempts to directly confirm Fks1 as the echinocandin target by crosslinking failed (Radding et al., 1998), instead identifying Pil1 (Edlind and Katiyar, 2004), an eisosome component and putative mediator of sphingolipid signaling.
Echinocandin resistance in C. glabrata, while remaining rare, appears to be increasing in response to increasing clinical use (Garcia-Effron et al., 2010; Pfaller et al., 2011; Pfaller et al., 2010; Zimbeck et al., 2010). Resistant isolates demonstrate minimum inhibitory concentrations (MICs) ≥ 2 μg ml−1 (i.e., > 32-fold increase over susceptible isolates), are strictly associated with mutations in β-1,3-glucan synthase genes FKS1 and FKS2, and typically exhibit CSF, MCF, and ANF cross-resistance (Castanheira et al., 2010; Cleary et al., 2008; Garcia-Effron et al., 2009; Katiyar et al., 2006; Pfeiffer et al., 2010; Zimbeck et al., 2010). C. glabrata mutants with equivalent properties can be isolated in the laboratory, albeit at low frequency (ca. 10−7) (Healey et al., 2011).
To shed further light on mechanisms of echinocandin action and resistance, we recently described the isolation at relatively high frequency (ca. 10−5) of C. glabrata mutants exhibiting low-level CSF reduced susceptibility (CRS), with MICs = 0.12 to 0.5 μg ml−1 (Healey et al., 2011). Surprisingly, most of these mutants paradoxically exhibited MCF increased susceptibility (MIS), with MICs = 0.00025 to 0.004 μg ml−1. Sequencing and gene deletion demonstrated that this CRS-MIS phenotype is FKS-independent (Healey et al., 2011). Here we explore the CRS-MIS mechanism, initially employing S. cerevisiae as genetic model and subsequently by biochemical and genetic studies of C. glabrata. Intriguingly, these studies identified sphingolipid biosynthesis pathway mutations, and specifically the accumulation of long chain bases (LCBs) dihydrosphingosine or phytosphingosine as the basis for CRS-MIS. Furthermore, we show that CRS-MIS can be reproduced by exogenous LCB, suggesting that echinocandin therapy can be enhanced through addition of sphingolipid pathway inhibitors.
Results
Characterization of S. cerevisiae CRS-conferring deletants
Pooled homozygous deletants representing 4741 non-essential S. cerevisiae genes were selected on low-level (0.1 to 0.3 μg ml−1) CSF-containing YPD plates. Deletants exhibiting CRS were identified by amplification and sequencing of their gene-specific tags. Of the 34 characterized CRS deletants, 17 were lem3Δ/lem3Δ, 9 were cka2Δ/cka2Δ, and 8 were fen1Δ/fen1Δ. The selection was subsequently repeated with pooled heterozygous deletants representing 5996 essential and non-essential genes: 10 of the 13 identified disruptants were sur4Δ/SUR4 and the remainder lem3Δ/LEM3. Multiple isolations of each of these 4 different deletants imply that spontaneous mutations were not responsible. Nevertheless, the corresponding deletants unexposed to CSF were recovered from the arrayed collection and tested by broth microdilution assay, which confirmed their modestly reduced (2 to 8-fold) CSF susceptibility.
Characterization of an S. cerevisiae temperature-sensitive CRS mutant
Although sur4Δ was implicated in CRS only in the heterozygous deletant selection, it is not an essential gene. Its absence in the homozygous deletant selection can be explained by the slow growth of the sur4Δ/sur4Δ deletant recovered from the arrayed collection. To identify a true CRS-related essential gene, we selected for CRS mutants of haploid strain BY4742 at 25°C, and then screened 50 of these for temperature sensitivity at 38°C on drug-free YPD. Mutant CRS-ts1 identified in this manner was transformed with a single copy plasmid library of S. cerevisiae wild-type genomic DNA, and complementing clones selected at the non-permissive temperature. Plasmids were rescued from 16 clones, and characterized by restriction digestion followed by partial sequencing of their inserts. The inserts shared a single essential gene, TSC10. Sequence analysis of this gene in CRS-ts1 and its wild-type parent identified the mutation Lys35 to Glu (K35E).
S. cerevisiae sphingolipid pathway
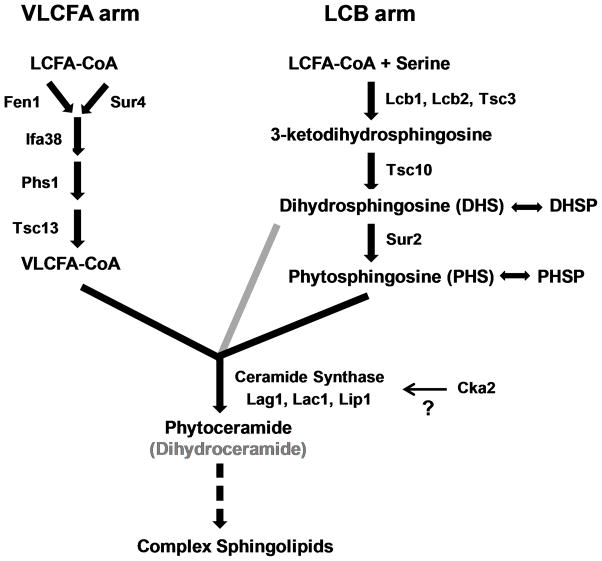
Three of the five S. cerevisiae CRS-related genes identified in these independent screens encode enzymes in the sphingolipid biosynthesis pathway (Fig. 1). This complex pathway has two arms which result in the production of very long chain fatty acids (VLCFA) > 20 carbons in length, and production of the LCBs dihydrosphingosine (DHS) and phytosphingosine (PHS) and their phosphorylated derivatives (e.g., PHS-1P). VLCFAs and LCBs are subsequently joined to form ceramide with carbon chains of varying length, and these ceramides are further modified with mannose and inositol side chains. Specifically, FEN1 and SUR4 encode partially redundant fatty acid elongases in the VLCFA arm, and TSC10 encodes ketodihydrosphingosine reductase in the LCB arm. Mutations in these genes result in reduced synthesis of ceramide and complex sphingolipids, and accumulation of LCBs (Beeler et al., 1998; Kobayashi and Nagiec, 2003; Oh et al., 1997). Additionally, CKA2 (alpha’ catalytic subunit of casein kinase 2) was previously implicated in the regulation of sphingolipid synthesis (potentially through phosphorylation of ceramide synthase; Fig. 1), since a cka2Δ strain demonstrated a sphingolipid profile very similar to that of a sur4Δ (elo3Δ) strain (Kobayashi and Nagiec, 2003). The fifth gene implicated in S. cerevisiae CRS, LEM3, plays a role in glycerophospholipid transport across the membrane (Saito et al., 2004) with no direct connection to sphingolipid biosynthesis. To determine the potential basis for CRS-MIS in C. glabrata, we pursued each of these leads. It should be noted, however, that the S. cerevisiae CRS deletants and mutants described above or selected by conventional methods (Healey et al., 2011) did not exhibit MIS since their MCF susceptibilities were minimally changed (data not shown).
Fig. 1. Sphingolipid biosynthesis pathway in yeast.
Synthesis of VLCFAs and LCBs are shown as separate arms, the products of which are linked by ceramide synthase. Relative to phytoceramides produced from PHS, dihydroceramides are typically produced from DHS at minor levels (grey). The mechanism by which Cka2 regulates sphingolipid biosynthesis is unclear (question mark), but has been suggested to involve ceramide synthase (Kobayashi & Nagiec, 2003). Complex sphingolipids include additional head groups such as mannose and inositol (not shown). Modified from Dickson (2010).
Characterization of C. glabrata fen1Δ, cka2Δ, and lem3Δ deletants
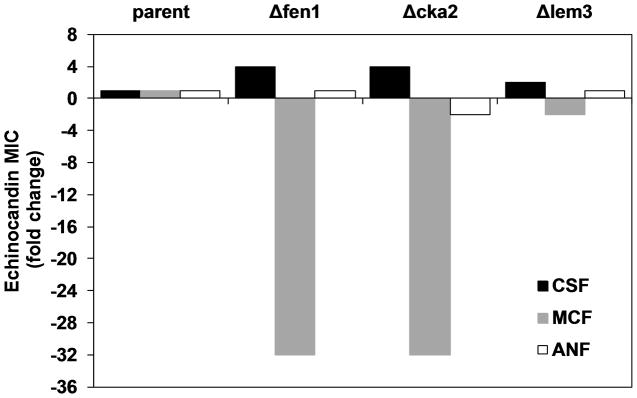
To test the role of the sphingolipid pathway in C. glabrata CRS-MIS, the syntenic FEN1 ortholog (CAGL0L08184g) was deleted and its echinocandin susceptibilities tested. Indeed, this fen1Δ strain exhibited a clear CRS-MIS phenotype (Fig. 2), with 4-fold reduced CSF and 32-fold increased MCF susceptibilities (128-fold differential). Moreover, deletion of the syntenic CKA2 ortholog (CAGL0G02035g) had an identical effect. In contrast to these two deletants relating to the sphingolipid pathway, a C. glabrata lem3Δ strain exhibited minimal change (≤ 2-fold) in both CSF and MCF susceptibilities. ANF susceptibilities were minimally changed for all three deletants; this was previously noted for C. glabrata CRS-MIS mutants (Healey et al., 2011).
Fig. 2. Deletion of C. glabrata genes involved in sphingolipid biosynthesis confers CRS-MIS.
Three genes implicated in S. cerevisiae CRS were deleted in C. glabrata and tested for echinocandin susceptibility by broth microdilution; results are expressed as fold change in MIC relative to parent. FEN1 encodes a fatty acid elongase (partially redundant with Sur4) in the VLCFA arm and CKA2 encodes a protein kinase catalytic subunit that regulates (directly or indirectly) ceramide synthase. LEM3 is involved in glycerophospholipid transport and has no known role in sphingolipid biosynthesis. Results are representative of three independent experiments.
Sphingolipid analysis of C. glabrata CRS-MIS mutants
To extend these promising results obtained with C. glabrata deletants, three genetically uncharacterized CRS-MIS mutants and their wild-type parent (Healey et al., 2011) were subjected to sphingolipid analysis by LC-MS/MS. Compared to its parent strain 66032u, mutant 66032u-C1 exhibited 7 and 6-fold increases in DHS and PHS, respectively (Table 1). Similarly, mutant 66032u-C3 exhibited 5 and 7-fold increases in these two LCBs. By analogy to S. cerevisiae (Kobayashi and Nagiec, 2003; Oh et al., 1997) these changes suggest a loss-of-function mutation in one of the multiple enzymes involved in VLCFA elongation (Fig. 1). The third mutant, 66032-C2, also exhibited an increase (25-fold) in DHS; however, PHS did not increase but rather decreased 60-fold in this mutant compared to its parent (Table 1). These changes suggest a loss-of-function mutation in the C4-hydroxylase responsible for conversion of DHS to PHS (Fig. 1). The sphingolipid analysis performed also detected phosphorylated LCBs and phytoceramides. PHSP was present at levels about 10-fold below those of PHS for each mutant, varying in an equivalent pattern (data not shown). DHSP and phytoceramide levels were near or below the limit of detection, although the expected decrease in the ratio of C26 to C24 phytoceramides in the mutants was apparent.
Table 1.
LCBs accumulate in CRS-MIS laboratory mutants and clinical isolates.
| Strain/mutant | Mutation | pmol/sample
|
|
|---|---|---|---|
| DHS | PHS | ||
| 66032u | WT | 110 | 180 |
| 66032u-C1 | Fen1-N156D | 820 | 1110 |
| 66032u-C2 | Sur2-M1I | 2800 | 3 |
| 66032u-C3 | Sur4-Y329stop | 590 | 1200 |
| Isolate 380 | WT | 110 | 190 |
| Isolate 4719 | Fen1-Y298stop | 950 | 4000 |
| Isolate 4743 | Ifa38-I339M | 400 | 360 |
| Isolate 4771 | Fen1-K175M | 31 | 630 |
Sequence analysis of candidate sphingolipid pathway genes
Based on its sphingolipid profile, we initiated sequence analysis of the genes involved in VLCFA elongation in 66032u-C1 in search of the responsible mutation. This promptly identified a Fen1-N156D substitution (Table 2), confirmed as a mutation by sequencing the corresponding region from parent 66032u. This substitution is non-conservative; furthermore, BLASTP analysis indicates that N156 is highly conserved in Fen1 homologs of other fungi. Thus, this mutation is consistent with loss of Fen1 function leading to DHS/PHS accumulation. In mutant 66032u-C3, which similarly accumulates DHS/PHS, a premature stop codon mutation (Y329stop) was identified in the C. glabrata SUR4 ortholog (CAGL0G04851g), strongly implying loss-of-function in this second VLCFA elongase. With respect to mutant 66032u-C2, as predicted by its sphingolipid profile (increased DHS, decreased PHS) a substitution in the C. glabrata SUR2 ortholog (CAGL0H01375g) was identified. The mutation, Sur2-M1I (Table 2), is similarly consistent with loss-of-function since an alternative upstream start codon is absent (not shown).
Table 2.
CRS-MIS is mediated by mutation of the sphingolipid biosynthesis pathway.
| Strain/mutant | MIC (μg ml−1)
|
CRS-MIS fold differentiala | Mutation | |
|---|---|---|---|---|
| CSF | MCF | |||
| 66032u | 0.03 | 0.008 | - | - |
| 66032u-C1 | 0.5 | 0.00025 | 512 | Fen1-N156D |
| 66032u-C2 | 0.5 | 0.0005 | 256 | Sur2-M1I |
| 66032u-C3 | 0.5 | 0.001 | 128 | Sur4-Y329stop |
| 66032u-C4 | 1 | 0.002 | 128 | Sur2-E299stop |
| 66032 | 0.03 | 0.008 | - | - |
| 66032-C1 | 0.5 | 0.00025 | 512 | Sur2-M197I |
| 945 | 0.008 | 0.008 | - | - |
| 945-C1 | 0.25 | 0.001 | 256 | Fen1-W145stop |
| 945-C2 | 0.12 | 0.0005 | 256 | Sur4-S141F |
| 945-C3 | 0.06 | 0.0005 | 128 | Fen1-P206L |
| 945-C4 | 0.06 | 0.0005 | 128 | Sur4-S141F |
| CE14 | 0.016 | 0.008 | - | - |
| CE14-C1 | 0.25 | 0.0005 | 256 | Fen1-W235stop |
| CE14-C2 | 0.25 | 0.001 | 128 | Fen1-V255E |
| CE14-C3 | 0.25 | 0.001 | 128 | Fen1-V255E |
| CE14-C4 | 0.12 | 0.0005 | 128 | Sur4-K201E |
| CE08 | 0.016 | 0.008 | - | - |
| CE08-C1 | 0.25 | 0.00025 | 512 | Sur4-L144R |
| DSY753 | 0.016 | 0.008 | - | - |
| DSY753-C1 | 0.5 | 0.0005 | 512 | Fen1-E17stop |
| DSY753-C2 | 0.12 | 0.001 | 64 | Sur2-N86K |
| 2805615 | 0.008 | 0.008 | - | - |
| 2805615-C1 | 0.25 | 0.0005 | 512 | Fen1-D166Y |
| 2805615-C2 | 0.25 | 0.002 | 128 | Ifa38-G214D |
| 2807990 | 0.03 | 0.008 | - | - |
| 2807990-C1 | 0.5 | 0.001 | 128 | Fen1-ΔI115-A138 |
| Isolate 4771 | 0.25 | 0.001 | - | Fen1-K175M |
| Isolate 4719 | 0.25 | 0.0005 | - | Fen1-Y298stop |
| Isolate 4743 | 0.12 | 0.0005 | - | Ifa38-I339M |
Relative to wild-type parent, fold increase in CSF MIC x fold decrease in MCF MIC.
Sequence analysis of sphingolipid pathway genes in additional CRS-MIS mutants
Based on these initial results, we sequenced FEN1, SUR4, and SUR2 from 16 additional CRS-MIS mutants derived from 7 total C. glabrata strain backgrounds. Fen1 mutations were identified in 8 of these, including 4 mutants with premature stop codons and one with a 24 amino acid deletion, all consistent with full loss of function (Table 2). Four mutants exhibited Sur4 mutations, and 3 mutants exhibited Sur2 mutations, including a premature stop codon (Table 2). In the remaining mutant, it was necessary to sequence the remaining VLCFA-pathway genes, which identified a mutation in the C. glabrata ortholog of IFA38 (CAGL0H07513g). In all cases, the implicated gene was sequenced in the parent strain to confirm mutation.
Characterization of CRS-MIS clinical isolates by sphingolipid and sequence analysis
Our previous identification of C. glabrata bloodstream isolates 4719, 4743, and 4771 exhibiting a CRS-MIS-like phenotype provided initial support for the clinical relevance of CRS-MIS (Healey et al., 2011). For all three, the FKS1 (CAGL0G01034g) and FKS2 (CAGL0K04037g) coding regions were sequenced in their entirety and no differences at the amino acid level with the corresponding sequences from type strain CBS138 were detected (Healey et al., 2011, and data not shown). Nevertheless, since the wild-type parents of these clinical isolates are unavailable, confirmation of these isolates as bonafide CRS-MIS mutants required elucidation of the CRS-MIS mechanism. With this now in hand, we first subjected these clinical isolates to sphingolipid analysis, and subsequently sequenced candidate genes. As shown in Table 1, isolate 4719 exhibited 9 and 21-fold elevated levels of DHS and PHS, respectively, relative to representative wild-type isolate 380 analyzed in parallel. Consistent with this, sequence analysis identified a Fen1 premature stop codon mutation likely to yield loss of function. The sphingolipid profiles of the remaining isolates revealed more modest increases, with 2 to 4-fold elevated DHS and PHS in isolate 4743 and 3-fold elevated PHS in isolate 4771, but again both carry substitutions in VLCFA elongation enzymes. Specifically, the Ifa38-I339M substitution in isolate 4743 involves a residue that is otherwise conserved in 7 genotypically diverse C. glabrata isolates, although it is polymorphic in other yeast species (data not shown). The Fen1-K175M substitution in isolate 4771 involves a residue that is similarly conserved in C. glabrata as well as 16 other yeast species for which genome sequences are available. Note that the DHS level in this isolate was unexpectedly low rather than elevated (Table 1); the basis for this is unknown, although it could reflect a compensating mutation.
We confirmed that the Fen1-Y298stop and Fen1-K175M mutations of clinical isolates 4719 and 4771 conferred loss-of-function by introducing these alleles (with flanking regulatory sequences) on a single-copy plasmid into the fen1Δ deletion strain described above. As with the Fen1-N156D allele from mutant 66032u-C1 tested in the same manner, the 128-fold CRS-MIS differential of fen1Δ was unchanged or only minimally reduced to 64-fold by expression of the 4719 or 4771 alleles, respectively. In contrast, plasmid-based expression of a wild-type Fen1 allele reduced the CRS-MIS differential to 8-fold. (Lack of full reversal may be a consequence of lower plasmid-based expression due, for example, to competition for regulatory factors from the chromosomal FEN1 promoter.)
Effects of exogenous long chain bases and myriocin
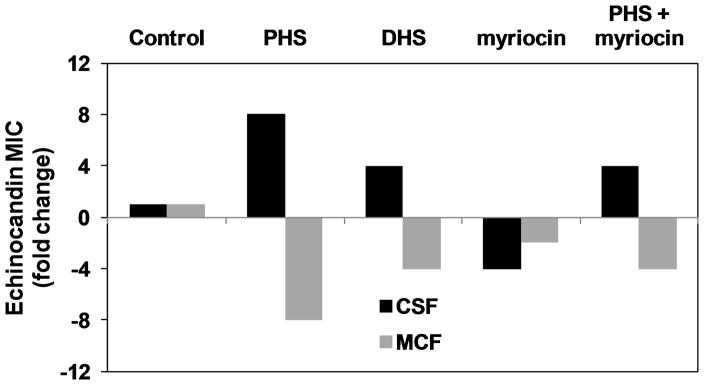
Analogous to the effects of the sphingolipid pathway mutations described above, exogenous DHS or PHS (1.25 μg ml−1) conferred a CRS-MIS phenotype on all four wild-type C. glabrata strains tested (Fig. 3A and data not shown). Specifically, exogenous PHS increased the CSF MIC 8-fold and decreased the MCF MIC 8-fold (64-fold differential), while DHS led to a 4-fold increase and a 4-fold decrease in CSF and MCF MICs, respectively.
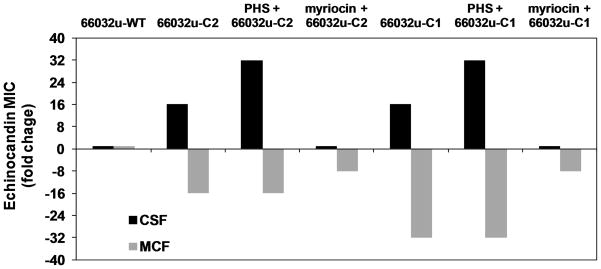
Fig. 3. LCB accumulation is responsible for C. glabrata CRS-MIS.
A. C. glabrata strain 66032u was pretreated with PHS, DHS, myriocin, or PHS + myriocin (all at 1.25 μg ml−1) for 1 h and then assayed for echinocandin susceptibility. Results are expressed as fold-change in MIC relative to non-pretreated controls, and are representative of three independent experiments. B. CRS-MIS mutants 66032u-C2 (Sur2-M1I) and 66032u-C1 (Fen1-N156D) were pretreated with PHS or myriocin (1.25 μg ml−1) and then assayed as in A above.
For comparison, we tested myriocin, a specific inhibitor of the LCB1/LCB2-encoded serine palmitoyltransferase representing the first, essential enzymatic step in the sphingolipid pathway (Fig. 1). In contrast to DHS/PHS, myriocin at a subinhibitory concentration (1.25 μg ml−1) did not confer CRS-MIS, but rather conferred increased susceptibility to both CSF and MCF (Fig. 3A). This enhancement of echinocandin susceptibility by inhibition of serine palmitoyltransferase was independently confirmed in S. cerevisiae by testing a tetO promoter strain in which doxycyline addition downregulates LCB1 (data not shown). As expected, PHS addition reversed the effects of myriocin (Fig. 3A).
In S. cerevisiae, it has been shown that exogenous LCBs are incorporated into complex sphingolipids and rescue serine palmitoyltransferase mutants (Nagiec et al., 1994; Wells and Lester, 1983). As noted above, loss of Sur2 function in C. glabrata mutant 66032u-C2 resulted in DHS accumulation, PHS depletion, and a CRS-MIS phenotype. We reasoned that exogenous PHS might reduce the CRS-MIS phenotype of this mutant by permitting synthesis of complex sphingolipids. Intriguingly, PHS did not have this effect (Fig. 3B). On the other hand, myriocin addition reduced CRS-MIS in both the sur2 mutant 66032u-C2 and fen1 mutant 66032u-C1 (Fig. 3B). Together these data argue that LCB accumulation, as opposed to complex sphingolipid depletion, is responsible for the CRS-MIS phenotype in C. glabrata.
Discussion
We were motivated to study the mechanism behind the paradoxical CRS-MIS phenotype for two reasons. First, it may suggest novel approaches to enhancing echinocandin therapy or, relatedly, reducing the likelihood of resistance. Second, it is likely to shed much needed light on the mechanism of echinocandin action. Initial studies in S. cerevisiae provided several apparently unrelated leads, since CRS in this model yeast was conferred by mutations in the LEM3 gene involved in glycerophospholipid transport, the protein kinase gene CKA2, the fatty acid elongase genes FEN1 and SUR4, and the ketodihydrosphingosine reductase gene TSC10. In fact, 4 of these 5 genes relate to sphingolipid biosynthesis (Fig. 1). Following these leads, the C. glabrata orthologs of FEN1 and CKA2 were disrupted and shown to confer CRS-MIS. Returning to the spontaneous laboratory mutants described in our initial report (Healey et al., 2011), sphingolipid analysis revealed elevated levels of long chain bases DHS or DHS plus PHS. Finally, gene sequencing confirmed the presence of mutations in FEN1, SUR4, and IFA38 of the VLCFA arm and SUR2 of the LCB arm of the sphingolipid biosynthesis pathway. In addition to defining its mechanism, these results explain the relatively high frequency (ca. 10−5) at which CRS-MIS mutants arise (Healey et al., 2011), since loss of function in any one of four different genes will suffice.
In retrospect, FEN1 (a.k.a. GNS1) was one of the first S. cerevisiae genes implicated in reduced susceptibility to echinocandins, or more specifically the pneumocandin B0 derivative L-733,560 (el-Sherbeini and Clemas, 1995). More recently, screens of the S. cerevisiae genome-wide deletion library for clones exhibiting reduced CSF susceptibility identified lem3Δ (Lesage et al., 2004) and cka2Δ (Markovich et al., 2004), as we did here. However, these screens failed to identify fen1Δ, sur4Δ, or any other sphingolipid pathway deletant. A potential explanation for this discrepancy is that both previous screens employed deletants in a haploid background while a diploid background was used here. Since the diploid deletants were generated by mating independently generated haploid deletants, they are less susceptible to aneuploidy artifacts associated with the latter (Hughes et al., 2000). Validation of our S. cerevisiae screens was ultimately provided by their accurate prediction of the basis for CRS-MIS in C. glabrata.
Confirmation and extension of our genetic analysis was provided by studies with exogenously added DHS and PHS, both of which conferred the CRS-MIS phenotype on wild-type C. glabrata strains. Contrary to expectation, exogenous PHS did not suppress the CRS-MIS phenotype of Sur2 mutant 66032u-C2 which is blocked in the conversion of DHS to PHS. This implies that CRS-MIS is mediated not by reduced synthesis of complex sphingolipids but rather by the accumulation of LCB intermediates. Myriocin, an inhibitor of the first step in sphingolipid synthesis and hence LCB accumulation, did reverse CRS-MIS. Indeed, myriocin enhanced susceptibility to all three echinocandin derivatives in wild-type strains, most likely a consequence of decreased Fks1 function due to altered membrane environment.
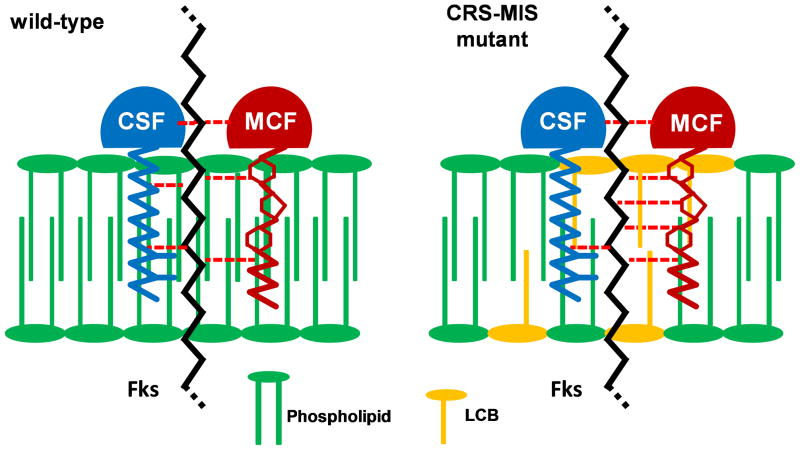
The mechanism by which LCB accumulation results in reduced susceptibility to one echinocandin but increased susceptibility to another remains to be elucidated. While sphingolipids have regulatory roles which include cell wall maintenance (Dickson, 2010) such regulation should affect susceptibility to all echinocandins equally. It is more likely that CRS-MIS reflects a direct interaction between sphingolipids, echinocandins, and Fks within the context of the plasma membrane. A model illustrating this potential relationship is presented in Fig. 4. It is based on a key structural difference among echinocandins: the lipid tail, which is essential to echinocandin activity (Boeck et al., 1989), is purely alkyl in CSF but mixed aryl-alkyl in MCF. Furthermore, it draws upon our recent topological analysis of S. cerevisiae Fks1 indicating that all three hotspots for resistance-conferring mutations are located externally, with hotspot 1 adjacent to and hotspot 3 embedded within the outer leaflet of the plasma membrane (Johnson and Edlind, 2012). Thus, in this model we speculate that increased LCB levels (e.g., from Fen1 mutation) within the membrane weaken the interaction between the membrane-spanning hotspot regions of Fks1 (or Fks2) and the lipid tail of CSF, but conversely strengthen the interaction with the lipid tail of MCF (Fig. 4). Testing this model may be challenging; among other requirements there is a need for echinocandin derivatives that differ only in their lipid tails (CSF and MCF differ as well in modifications to their hexapeptide cores).
Fig. 4.
Model illustrating potential mechanism by which LCB accumulation confers CRS-MIS. Elevated LCB levels within the plasma membrane are hypothesized to weaken the interaction between Fks (most likely at hotspot 3 which is membrane embedded) and CSF but strengthen the interaction with MCF. (Not drawn to scale.)
The mechanism by which cka2Δ confers CRS-MIS also awaits further study. Cka2-mediated phosphorylation may be required for activity of one or more sphingolipid pathway enzymes; e.g., Fen1 of the VLCFA arm, Sur2 of the LCB arm, or the Lag1/Lac1 ceramide synthase that joins them. Indeed, Kobayashi and Nagiec (2003) reported impaired ceramide synthase activity in their S. cerevisiae cka2Δ mutant, and we note that the C-termini of Lag1 and Lac1 are enriched in potential Cka2 phosphorylation sites.
Importantly, the three clinical isolates exhibiting a CRS-MIS phenotype described in our initial report (Healey et al., 2011) were confirmed here as bonafide CRS-MIS mutants based on sphingolipid analysis and gene sequencing. It remains to be seen, however, whether or not C. glabrata CRS-MIS mutants routinely accumulate during CSF therapy. One argument against this is that some sphingolipid pathway mutations are deleterious with respect to growth in vitro and hence likely to confer reduced virulence in vivo. On the other hand, there are numerous examples of deleterious mutations in diverse pathogens that are tolerated due to the strong selective pressure exerted by antimicrobial therapy, especially in immunocompromised patients. Echinocandin resistance-conferring Fks mutations provide a particularly pertinent example (Ben-Ami et al., 2011; Garcia-Effron et al., 2009). Another argument is that serum, which significantly affects echinocandin susceptibility (Paderu et al., 2007), could significantly alter CRS-MIS; however, in assays with 15% human serum (higher concentrations inhibit C. glabrata growth) this was not observed (data not shown). Ultimately, it will be necessary to survey isolates from patients before and after CSF therapy to establish the clinical relevance of CRS-MIS mutants. If indeed they do accumulate, a switch to MCF would be preferable to the usual response of switching antifungal classes. Alternatively, our results suggest novel ways in which echinocandin therapy could be enhanced; e.g., by combining MCF with a sphingolipid pathway intermediate or inhibitor that mimics the effects of CRS-MIS mutation.
Experimental Procedures
Strains
C. glabrata strain 66032u (ura3) and its CRS-MIS mutants, clinical isolate 945 and its CRS-MIS mutants, and CRS-MIS clinical isolates 4719, 4771, and 4743 have been described (Healey et al., 2011). Strains 66032 and 200989 (ura3, trp1Δ, his3Δ) are from the American Type Culture Collection (Manassas, Va.). Additional CRS-MIS mutants were selected on CSF-containing medium as previously described (Healey et al., 2011). Pooled and arrayed S. cerevisiae homozygous and heterozygous deletion libraries, their diploid parent BY4743, and haploid BY4742 were obtained from Research Genetics (Invitrogen, Carlsbad, CA). The S. cerevisiae genomic DNA library in shuttle vector YCp50 was generously provided by J. Broach (Princeton).
Media, drugs, and sphingolipid intermediates
SOB medium with or without 50 μg ml−1 ampicillin was used to culture E. coli DH5a. The yeast media used were YPD (1% yeast extract, 2% peptone, 2% dextrose) or, where indicated, SD-ura or SD-trp (DOB plus CSM-ura or CSM-trp; Sunrise Science Products, San Diego, CA). CSF (Merck, Rahway, N.J.), MCF (Astellas, Deerfield, IL), ANF (Pfizer, New York, NY), myriocin, dihydrosphingosine, and phytosphingosine (Biomol/Enzo Life Sciences, Farmingdale, N.Y.) were dissolved or diluted in dimethyl sulfoxide; final solvent concentration was ≤ 0.5% in all experiments.
Isolation and characterization of S. cerevisiae temperature-sensitive CRS mutant
Haploid BY4742 cells (1 × 107) were plated on 0.25–0.5 μg ml−1 CSF-containing YPD, and CRS mutants selected following incubation at 25°C for 3 days. Mutants (50) were screened for no growth at 38°C on drug-free YPD, which identified temperature-sensitive mutant CRS-ts1. This mutant was transformed with a YCp50 single copy plasmid library of Sau3A partially digested S. cerevisiae genomic DNA with selection on SD-ura plates at 25°C. The 17,000 transformants were pooled and 1 × 106 cells plated on YPD with incubation at 38°C. From 16 of the 800 colonies obtained, plasmids complementing the temperature-sensitive mutation were rescued by transformation into E. coli DH5α and purified as described below. Inserts were characterized by restriction digestion, and sequences flanking the inserts were determined with primers YCp50F and YCp50R (Table 3). This identified one essential gene, which was amplified from CRS-ts1 and its wild-type parent and sequenced to identify the mutation.
Table 3.
DNA primers used in this study and their application.
| Primera | Application | Sequence (5′ – 3′)b |
|---|---|---|
| KanMXuF | amplify uptag | ATGGATGTCCACGAGGTCTCT |
| KanMXuR | amplify uptag | CGCACCTGATTGCCCGACAT |
| KanMXuseq | sequence uptag | GCGCACGTCAAGACTGTCAAG |
| KanMXdF | amplify downtag | CTCACCGGATTCAGTCGTC |
| KanMXdR | amplify downtag | ACGGTGTCGGTCTCGTAG |
| KanMXdseq | sequence downtag | TCATCTGCCCAGATGCGAAG |
| YCp50F | sequence | CGCTTCGCTACTTGGAGCCACT |
| YCp50R | sequence | GCGCCAGCAACCGCACCTGT |
| ScTSC10u74F | amplify, sequence | CTCTGCATTTACTGGAAGCGA |
| ScTSC10d46R | amplify, sequence | ATAAAGAGAGTCACGGGGAAT |
| CgFEN1-TRP1F | fen1Δ | ATATCCTTTGCTGTAATTTCACTGATCAATAAGAACTGATAATCACTTATAAGTACTATGTCTGTTATTAATTTCACAGG |
| CgFEN1-TRP1R | fen1Δ | TTAGATGCAAACTCCGAGTATAAGAGAAAAAGAGGTTATCACTTGACCTCAACGAGCCTATTTCTTAGCATTTTTGACGA |
| CgFEN1u248F | fen1Δ screen | CACCAACGTATAAGGACTGTA |
| CgFEN1c201R | fen1Δ screen | AGGAAGTTCACCAGCGACAA |
| ScTRP1c345R | fen1Δ screen | TGGCAAACCGAGGAACTCTT |
| CgCKA2-URA3F | cka2Δ | AAGCTGTGAGGGATAGCCAGGAGGGGGAGCAGCAAGAGAGACCAAGCCAAGCGATGTCGAAAGCTACATATAAG |
| CgCKA2-URA3R | cka2Δ | GCAAATCCTTTCTGATAATTGCGTGCTTGGAGATAATTCAGAATGTACATATATGTGTTTAGTTTTGCTGGCCGCATC |
| CgCKA2u418F | cka2Δ screen | CCTGGTTATCAACTTCGACC |
| CgCKA2c507R | cka2Δ screen | GCTTCACGTCCCTGTGCAT |
| ScURA3c349R | cka2Δ screen | CTGCCCATTCTGCTATTCTG |
| CgFEN1u481F | amplify | CCCTTGCTCAATGACACATC |
| CgFEN1d192R | amplify | TCCCAACATGTGAATAGGTCT |
| CgFEN1u361F | sequence | CCTCATCGCAAAAGGCAAAC |
| CgFEN1c378F | sequence | TGACTCCAATGATCTACCATC |
| CgSUR4u340F | amplify | GATTTGTCCTTGTCGTCGTC |
| CgSUR4d91R | amplify | AAAGCCACATCTAACTGTCAG |
| CgSUR4c495R | sequence | GATAGCCCAGAACAAACCGT |
| CgSUR4c355F | sequence | ATCAACGCCTCTCCAATGCA |
| CgIFA38u280F | amplify | ACTGTAGAACCTGCATTGGC |
| CgIFA38d137R | amplify | GACATAGGGTAAGGACCACT |
| CgIFA38u134F | sequence | TCAAGTATCGGCTGCTAGCT |
| CgIFA38c400F | sequence | TCCCTTTGTGATGACCTTCC |
| CgPHS1u178F | amplify | ACATAACCGAGCACGATTGTA |
| CgPHS1d185R | amplify | CTTAGCCAGCAAAGTCCGTA |
| CgPHS1u136F | sequence | CTCTAGCAAGAGACTGTGAG |
| CgTSC13u276F | amplify | CGAAGCTCATCTCATCGCAA |
| CgTSC13d111R | amplify | GGTAGAACACATGATAAGGGA |
| CgTSC13u158F | sequence | GGGATATCCAATTTCCTGGG |
| CgTSC13c359F | sequence | TCCACAGAGACCCATTCTTG |
| CgSUR2u483F | amplify | GGATATGGGAACTTCTCAATG |
| CgSUR2d72R | amplify | CGCTCAATAATTATCGGAGCA |
| CgSUR2c241F | sequence | GAGATCGAGAAGCGTAACAG |
| CgSUR2c436R | sequence | CGTAGTAGACAGCAGCATCT |
| pGRB-CgFEN1u587F | gap-repair | CGAGGTCGACGGTATCGATAAGCTTGATATCGAATTCCTGACAAGTCTTCTGTCGAAACC |
| pGRB-CgFEN1d193R | gap-repair | GGAGCTCCACCGCGGTGGCGGCCGCTCTAGAACTAGTGATCCCAACATGTGAATAGGTC |
| CgFEN1c141R | screen | AGGAAGTTCACCAGCGACAA |
| T7 | screen | TAATACGACTCACTATAGGG |
Numbers in primer names correspond to location upstream (u) or within the coding region (c) relative to the start codon, or downstream (d) relative to the stop codon.
Underlined regions of deletion primers correspond to S. cerevisiae TRP1 or URA3 coding sequences; underlined regions of gap-repair cloning primers correspond to C. glabrata FEN1 upstream or downstream sequences.
Isolation and characterization of CRS clones within the S. cerevisiae pooled deletion library
Pooled homozygous or heterozygous deletion libraries (2 × 105 cells) were plated on 0.1 to 0.3 μg ml−1 CSF-containing YPD agar, and following incubation at 30°C for 3 days, CRS clones were streaked for isolation and their DNA purified as described below. Deletant-specific tags flanking the KanMX4 cassette were amplified as described below with primer pairs KanUF/KanUR (uptag) and KanDF/KanDR (downtag) and sequenced with primers KanUseq or KanDseq, respectively (Table 3). The deleted gene was identified by comparison to the tag database (www-sequence.stanford.edu/group/yeast_deletion_project/ResGenftp.html). The corresponding unexposed deletant from the arrayed collection was subsequently tested by broth microdilution as described below to confirm the CRS phenotype.
C. glabrata gene deletion
Deletant fen1Δ was generated in C. glabrata 200989 by transformation with a PCR-amplified TRP1 PRODIGE cassette (for primers, see Table 3) and selection on SD-trp as previously described (Edlind et al., 2005). Deletant cka2Δ was similarly generated in strains 66032u and BG14 with a URA3 PRODIGE cassette and selection on SD-ura. Transformants were screened by PCR with the indicated primers (Table 3). Deletant lem3Δ was described previously (Edlind et al., 2005).
DNA purification, amplification, and sequencing
Plasmids were purified from lysozyme-treated E. coli or glass bead-disrupted S. cerevisiae by the STET buffer/boiling procedure (Holmes and Quigley, 1981). Genomic DNAs were isolated by phenol extraction of glass bead-disrupted cells followed by two cycles of ethanol precipitation (Edlind et al., 2005). PCR with the indicated template and primers (Table 3) employed Taq polymerase and the conditions recommended by the supplier (New England Biolabs, Ipswich, Mass.). Plasmids, or PCR products treated with exonuclease I plus shrimp alkaline phosphatase (New England Biolabs) were sequenced (Genewiz, South Plainfield, N.J.) with gene-specific primers (Table 3).
Gap-repair cloning
Plasmid pGRB2.0 (pRS416 with URA3 marker and C. glabrata CEN/ARS; gift of B. Cormack, Johns Hopkins) was linearized with SmaI and treated with alkaline phosphatase as recommended by the supplier (New England Biolabs). FEN1 plus flanking sequence was amplified using DNA from strains 66032u, 66032u-C1, 4719, or 4771 as template and the pGRB-CgFEN1 primers (Table 3) which include 38 or 39 bases at their 5′ ends homologous to pGRB2.0 sequences flanking the SmaI site. Competant fen1Δ cells were cotransformed with linearized plasmid and PCR product with selection on SD-ura, and transformants screened by PCR and sequencing. For an empty vector control, fen1Δ was also transformed with untreated pGRB2.0.
Sphingolipid analysis
Approximately 2 × 108 cells from log phase cultures in YPD at 35°C were pelleted, washed in sterile water three times, and frozen on dry ice. After thawing, LCBs were extracted and analyzed by liquid chromatography-mass spectrometry (LC-MS/MS) (Bielawski et al., 2010) by the Lipidomics Shared Resource of the Medical University of South Carolina (www.musc.edu/BCMB/lipidomics).
Broth microdilution assays
Echinocandin MICs (concentration inhibiting growth ≥ 80% relative to drug-free control) were determined by broth microdilution in YPD as described (Vermitsky et al., 2006). For combination studies, PHS, DHS, or myriocin at the indicated concentrations were added to diluted cells 1 h prior to the broth microdilution assay. All assays were repeated at least twice on separate days, and comparable results obtained.
Acknowledgments
We thank J. Broach for the YCp50 library, and M. Pfaller, D. Diekema, M. Castanheira, and D. Sanglard for C. glabrata strains. These studies were supported by National Institutes of Health grant AI075272.
References
- Beeler T, Bacikova D, Gable K, Hopkins L, Johnson C, Slife H, Dunn T. The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3-ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of the Ca2+-sensitive csg2Delta mutant. J Biol Chem. 1998;273:30688–30694. doi: 10.1074/jbc.273.46.30688. [DOI] [PubMed] [Google Scholar]
- Ben-Ami R, Garcia-Effron G, Lewis RE, Gamarra S, Leventakos K, Perlin DS, Kontoyiannis DP. Fitness and virulence costs of Candida albicans FKS1 hot spot mutations associated with echinocandin resistance. J Infect Dis. 2011;204:626–635. doi: 10.1093/infdis/jir351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielawski J, Pierce JS, Snider J, Rembiesa B, Szulc ZM, Bielawska A. Sphingolipid analysis by high performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) Adv Exp Med Biol. 2010;688:46–59. doi: 10.1007/978-1-4419-6741-1_3. [DOI] [PubMed] [Google Scholar]
- Boeck LD, Fukuda DS, Abbott BJ, Debono M. Deacylation of echinocandin B by Actinoplanes utahensis. J Antibiot (Tokyo) 1989;42:382–388. doi: 10.7164/antibiotics.42.382. [DOI] [PubMed] [Google Scholar]
- Castanheira M, Woosley LN, Diekema DJ, Messer SA, Jones RN, Pfaller MA. Low prevalence of fks1 hot spot 1 mutations in a worldwide collection of Candida strains. Antimicrob Agents Chemother. 2010;54:2655–2659. doi: 10.1128/AAC.01711-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary JD. Echinocandins: pharmacokinetic and therapeutic issues. Curr Med Res Opin. 2009;25:1741–1750. [PubMed] [Google Scholar]
- Cleary JD, Garcia-Effron G, Chapman SW, Perlin DS. Reduced Candida glabrata susceptibility secondary to an FKS1 mutation developed during candidemia treatment. Antimicrob Agents Chemother. 2008;52:2263–2265. doi: 10.1128/AAC.01568-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning DW. Echinocandin antifungal drugs. Lancet. 2003;362:1142–1151. doi: 10.1016/S0140-6736(03)14472-8. [DOI] [PubMed] [Google Scholar]
- Dickson RC. Roles for sphingolipids in Saccharomyces cerevisiae. Adv Exp Med Biol. 2010;688:217–231. doi: 10.1007/978-1-4419-6741-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas CM. Fungal beta(1,3)-D-glucan synthesis. Med Mycol. 2001;39(Suppl 1):55–66. doi: 10.1080/mmy.39.1.55.66. [DOI] [PubMed] [Google Scholar]
- Douglas CM, D’Ippolito JA, Shei GJ, Meinz M, Onishi J, Marrinan JA, et al. Identification of the FKS1 gene of Candida albicans as the essential target of 1,3-beta-D-glucan synthase inhibitors. Antimicrob Agents Chemother. 1997;41:2471–2479. doi: 10.1128/aac.41.11.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas CM, Foor F, Marrinan JA, Morin N, Nielsen JB, Dahl AM, et al. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-beta-D-glucan synthase. Proc Natl Acad Sci U S A. 1994;91:12907–12911. doi: 10.1073/pnas.91.26.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlind TD, Henry KW, Vermitsky JP, Edlind MP, Raj S, Katiyar SK. Promoter-dependent disruption of genes: simple, rapid, and specific PCR-based method with application to three different yeast. Curr Genet. 2005;48:117–125. doi: 10.1007/s00294-005-0008-3. [DOI] [PubMed] [Google Scholar]
- Edlind TD, Katiyar SK. The echinocandin “target” identified by cross-linking is a homolog of Pil1 and Lsp1, sphingolipid-dependent regulators of cell wall integrity signaling. Antimicrob Agents Chemother. 2004;48:4491. doi: 10.1128/AAC.48.11.4491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Sherbeini M, Clemas JA. Cloning and characterization of GNS1: a Saccharomyces cerevisiae gene involved in synthesis of 1,3-beta-glucan in vitro. J Bacteriol. 1995;177:3227–3234. doi: 10.1128/jb.177.11.3227-3234.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Effron G, Chua DJ, Tomada JR, DiPersio J, Perlin DS, Ghannoum M, Bonilla H. Novel FKS mutations associated with echinocandin resistance in Candida species. Antimicrob Agents Chemother. 2010;54:2225–2227. doi: 10.1128/AAC.00998-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Effron G, Lee S, Park S, Cleary JD, Perlin DS. Effect of Candida glabrata FKS1 and FKS2 mutations on echinocandin sensitivity and kinetics of 1,3-beta-D-glucan synthase: implication for the existing susceptibility breakpoint. Antimicrob Agents Chemother. 2009;53:3690–3699. doi: 10.1128/AAC.00443-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healey KR, Katiyar SK, Castanheira M, Pfaller MA, Edlind TD. Candida glabrata mutants demonstrating paradoxical reduced caspofungin susceptibility but increased micafungin susceptibility. Antimicrob Agents Chemother. 2011;55:3947–3949. doi: 10.1128/AAC.00044-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes DS, Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Roberts CJ, Dai H, Jones AR, Meyer MR, Slade D, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- Inoue SB, Takewaki N, Takasuka T, Mio T, Adachi M, Fujii Y, et al. Characterization and gene cloning of 1,3-beta-D-glucan synthase from Saccharomyces cerevisiae. Eur J Biochem. 1995;231:845–854. doi: 10.1111/j.1432-1033.1995.tb20770.x. [DOI] [PubMed] [Google Scholar]
- Johnson ME, Edlind TD. Topological and mutational analysis of Saccharomyces cerevisiae Fks1. Euk. Cell. 2012 doi: 10.1128/EC.00082-12. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar S, Pfaller M, Edlind T. Candida albicans and Candida glabrata clinical isolates exhibiting reduced echinocandin susceptibility. Antimicrob Agents Chemother. 2006;50:2892–2894. doi: 10.1128/AAC.00349-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman CA, Carver PL. Update on echinocandin antifungals. Semin Respir Crit Care Med. 2008;29:211–219. doi: 10.1055/s-2008-1063859. [DOI] [PubMed] [Google Scholar]
- Kobayashi SD, Nagiec MM. Ceramide/long-chain base phosphate rheostat in Saccharomyces cerevisiae: regulation of ceramide synthesis by Elo3p and Cka2p. Eukaryot Cell. 2003;2:284–294. doi: 10.1128/EC.2.2.284-294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladeveze V, Marcireau C, Delourme D, Karst F. General resistance to sterol biosynthesis inhibitors in Saccharomyces cerevisiae. Lipids. 1993;28:907–912. doi: 10.1007/BF02537499. [DOI] [PubMed] [Google Scholar]
- Lesage G, Sdicu AM, Menard P, Shapiro J, Hussein S, Bussey H. Analysis of beta-1,3-glucan assembly in Saccharomyces cerevisiae using a synthetic interaction network and altered sensitivity to caspofungin. Genetics. 2004;167:35–49. doi: 10.1534/genetics.167.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovich S, Yekutiel A, Shalit I, Shadkchan Y, Osherov N. Genomic approach to identification of mutations affecting caspofungin susceptibility in Saccharomyces cerevisiae. Antimicrob Agents Chemother. 2004;48:3871–3876. doi: 10.1128/AAC.48.10.3871-3876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morschhauser J. Regulation of multidrug resistance in pathogenic fungi. Fungal Genet Biol. 2010;47:94–106. doi: 10.1016/j.fgb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Nagiec MM, Baltisberger JA, Wells GB, Lester RL, Dickson RC. The LCB2 gene of Saccharomyces and the related LCB1 gene encode subunits of serine palmitoyltransferase, the initial enzyme in sphingolipid synthesis. Proc Natl Acad Sci U S A. 1994;91:7899–7902. doi: 10.1073/pnas.91.17.7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh CS, Toke DA, Mandala S, Martin CE. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem. 1997;272:17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- Okada H, Abe M, Asakawa-Minemura M, Hirata A, Qadota H, Morishita K, et al. Multiple functional domains of the yeast l,3-beta-glucan synthase subunit Fks1p revealed by quantitative phenotypic analysis of temperature-sensitive mutants. Genetics. 2010;184:1013–1024. doi: 10.1534/genetics.109.109892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paderu P, Garcia-Effron G, Balashov S, Delmas G, Park S, Perlin DS. Serum differentially alters the antifungal properties of echinocandin drugs. Antimicrob Agents Chemother. 2007;51:2253–2256. doi: 10.1128/AAC.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas PG, Kauffman CA, Andes D, Benjamin DK, Jr, Calandra TF, Edwards JE, Jr, et al. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:503–535. doi: 10.1086/596757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin DS. Resistance to echinocandin-class antifungal drugs. Drug Resist Updat. 2007;10:121–130. doi: 10.1016/j.drup.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller M, Boyken L, Hollis R, Kroeger J, Messer S, Tendolkar S, Diekema D. Use of epidemiological cutoff values to examine 9-year trends in susceptibility of Candida species to anidulafungin, caspofungin, and micafungin. J Clin Microbiol. 2011;49:624–629. doi: 10.1128/JCM.02120-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Boyken L, Hollis RJ, Kroeger J, Messer SA, Tendolkar S, et al. Wild-type MIC distributions and epidemiological cutoff values for the echinocandins and Candida spp. J Clin Microbiol. 2010;48:52–56. doi: 10.1128/JCM.01590-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer CD, Garcia-Effron G, Zaas AK, Perfect JR, Perlin DS, Alexander BD. Breakthrough invasive candidiasis in patients on micafungin. J Clin Microbiol. 2010;48:2373–2380. doi: 10.1128/JCM.02390-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radding JA, Heidler SA, Turner WW. Photoaffinity analog of the semisynthetic echinocandin LY303366: identification of echinocandin targets in Candida albicans. Antimicrob Agents Chemother. 1998;42:1187–1194. doi: 10.1128/aac.42.5.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Fujimura-Kamada K, Furuta N, Kato U, Umeda M, Tanaka K. Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:3418–3432. doi: 10.1091/mbc.E03-11-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D, Odds FC. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect Dis. 2002;2:73–85. doi: 10.1016/s1473-3099(02)00181-0. [DOI] [PubMed] [Google Scholar]
- Sawistowska-Schroder ET, Kerridge D, Perry H. Echinocandin inhibition of 1,3-beta-D-glucan synthase from Candida albicans. FEBS Lett. 1984;173:134–138. doi: 10.1016/0014-5793(84)81032-7. [DOI] [PubMed] [Google Scholar]
- Taft CS, Stark T, Selitrennikoff CP. Cilofungin (LY121019) inhibits Candida albicans (1–3)-beta-D-glucan synthase activity. Antimicrob Agents Chemother. 1988;32:1901–1903. doi: 10.1128/aac.32.12.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermitsky JP, Earhart KD, Smith WL, Homayouni R, Edlind TD, Rogers PD. Pdr1 regulates multidrug resistance in Candida glabrata: gene disruption and genome-wide expression studies. Mol Microbiol. 2006;61:704–722. doi: 10.1111/j.1365-2958.2006.05235.x. [DOI] [PubMed] [Google Scholar]
- Wells GB, Lester RL. The isolation and characterization of a mutant strain of Saccharomyces cerevisiae that requires a long chain base for growth and for synthesis of phosphosphingolipids. J Biol Chem. 1983;258:10200–10203. [PubMed] [Google Scholar]
- Zimbeck AJ, Iqbal N, Ahlquist AM, Farley MM, Harrison LH, Chiller T, Lockhart SR. FKS mutations and elevated echinocandin MIC values among Candida glabrata isolates from U.S. population-based surveillance. Antimicrob Agents Chemother. 2010;54:5042–5047. doi: 10.1128/AAC.00836-10. [DOI] [PMC free article] [PubMed] [Google Scholar]