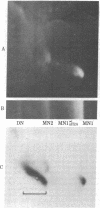
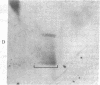
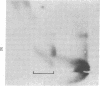
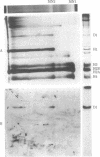
Abstract
Our previous work [Levinger, L. & Varshavsky, A. (1982) Cell 28, 375-385] has shown that D1, a 50-kilodalton chromosomal protein of Drosophila melanogaster, is specifically associated with isolated nucleosomes that contain a complex A + T-rich satellite DNA with buoyant density of 1.688 g/ml. We show here that D1 is also a component of nucleosomes containing a simple-sequence, pure A + T satellite DNA, buoyant density 1.672 g/ml. Furthermore, using a modification of a protein blotting technique in which proteins are not exposed to dodecyl sulfate denaturation, we have found that D1 preferentially binds to A + T-rich double-stranded DNA in vitro, and it is apparently the only abundant nuclear protein in cultured D. melanogaster cells that possesses this property. Synthetic poly[d(A-T)].poly[d(A-T)] and poly(dA).poly(dT) duplexes effectively compete in vitro with A + T-rich D. melanogaster satellite DNAs for binding to D1, whereas total Escherichia coli DNA is an extremely poor competitor. These findings strongly suggest that D1 is a specific component of A + T-rich, tandemly repeated, heterochromatic regions, which constitute up to 15-20% of the total D. melanogaster genome. Possible functions of D1 protein include compaction of A + T-rich heterochromatin and participation in microtubule-centromere interactions in mitosis. In addition, D1 may prevent nonspecific binding to A + T-rich satellite DNA of other nuclear proteins that have a preference for AT-DNA, such as RNA polymerase or regulatory proteins, and may also participate in the higher-order chromatin organization outside tandemly repetitive regions by binding to nonrandomly positioned stretches of A + T-rich DNA.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alfageme C. R., Rudkin G. T., Cohen L. H. Locations of chromosomal proteins in polytene chromosomes. Proc Natl Acad Sci U S A. 1976 Jun;73(6):2038–2042. doi: 10.1073/pnas.73.6.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett F. C., Rosenfeld B. I., Huang C. H., Yeoman L. C. Evidence for two conformational forms of nonhistone protein BA which differ in their affinity for DNA. Biochem Biophys Res Commun. 1982 Jan 29;104(2):649–656. doi: 10.1016/0006-291x(82)90686-6. [DOI] [PubMed] [Google Scholar]
- Bowen B., Steinberg J., Laemmli U. K., Weintraub H. The detection of DNA-binding proteins by protein blotting. Nucleic Acids Res. 1980 Jan 11;8(1):1–20. doi: 10.1093/nar/8.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutlag D. L. Molecular arrangement and evolution of heterochromatic DNA. Annu Rev Genet. 1980;14:121–144. doi: 10.1146/annurev.ge.14.120180.001005. [DOI] [PubMed] [Google Scholar]
- Brutlag D., Appels R., Dennis E. S., Peacock W. J. Highly repeated DNA in Drosophila melanogaster. J Mol Biol. 1977 May 5;112(1):31–47. doi: 10.1016/s0022-2836(77)80154-x. [DOI] [PubMed] [Google Scholar]
- Brutlag D., Fry K., Nelson T., Hung P. Synthesis of hybrid bacterial plasmids containing highly repeated satellite DNA. Cell. 1977 Mar;10(3):509–519. doi: 10.1016/0092-8674(77)90038-1. [DOI] [PubMed] [Google Scholar]
- Busch H., Goldknopf I. L. Ubiquitin - protein conjugates. Mol Cell Biochem. 1981 Nov 13;40(3):173–187. doi: 10.1007/BF00224611. [DOI] [PubMed] [Google Scholar]
- Carlson M., Brutlag D. Cloning and characterization of a complex satellite DNA from Drosophila melanogaster. Cell. 1977 Jun;11(2):371–381. doi: 10.1016/0092-8674(77)90054-x. [DOI] [PubMed] [Google Scholar]
- Catino J. J., Busch H., Daskal Y., Yeoman L. C. Subcellular localization of DNA-binding protein BA by immunofluorescence and immunoelectron microscopy. J Cell Biol. 1979 Nov;83(2 Pt 1):462–467. doi: 10.1083/jcb.83.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M., Clarke L., Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982 May;29(1):235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Hsieh T., Brutlag D. L. A protein that preferentially binds Drosophila satellite DNA. Proc Natl Acad Sci U S A. 1979 Feb;76(2):726–730. doi: 10.1073/pnas.76.2.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack R. S., Gehring W. J., Brack C. Protein component from Drosophila larval nuclei showing sequence specificity for a short region near a major heat-shock protein gene. Cell. 1981 May;24(2):321–331. doi: 10.1016/0092-8674(81)90322-6. [DOI] [PubMed] [Google Scholar]
- Karch F., Török I., Tissières A. Extensive regions of homology in front of the two hsp70 heat shock variant genes in Drosophila melanogaster. J Mol Biol. 1981 May 25;148(3):219–230. doi: 10.1016/0022-2836(81)90536-2. [DOI] [PubMed] [Google Scholar]
- Kornberg R. The location of nucleosomes in chromatin: specific or statistical. Nature. 1981 Aug 13;292(5824):579–580. doi: 10.1038/292579a0. [DOI] [PubMed] [Google Scholar]
- Lengyel J., Spradling A., Penman S. Methods with insect cells in suspension culture. II. Drosophila melanogaster. Methods Cell Biol. 1975;10:195–208. doi: 10.1016/s0091-679x(08)60738-4. [DOI] [PubMed] [Google Scholar]
- Levinger L., Barsoum J., Varshavsky A. Two-dimensional hybridization mapping of nucleosomes. comparison of DNA and protein patterns. J Mol Biol. 1981 Mar 5;146(3):287–304. doi: 10.1016/0022-2836(81)90389-2. [DOI] [PubMed] [Google Scholar]
- Levinger L., Varshavsky A. High-resolution fractionation of nucleosomes: minor particles, "whiskers," and separation of mononucleosomes containing and lacking A24 semihistone. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3244–3248. doi: 10.1073/pnas.77.6.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinger L., Varshavsky A. Selective arrangement of ubiquitinated and D1 protein-containing nucleosomes within the Drosophila genome. Cell. 1982 Feb;28(2):375–385. doi: 10.1016/0092-8674(82)90355-5. [DOI] [PubMed] [Google Scholar]
- Moreau J., Marcaud L., Maschat F., Kejzlarova-Lepesant J., Lepesant J. A., Scherrer K. A + T-rich linkers define functional domains in eukaryotic DNA. Nature. 1982 Jan 21;295(5846):260–262. doi: 10.1038/295260a0. [DOI] [PubMed] [Google Scholar]
- Nelson P. P., Albright S. C., Wiseman J. M., Garrard W. T. Reassociation of histone H1 with nucleosomes. J Biol Chem. 1979 Nov 25;254(22):11751–11760. [PubMed] [Google Scholar]
- Panyim S., Chalkley R. High resolution acrylamide gel electrophoresis of histones. Arch Biochem Biophys. 1969 Mar;130(1):337–346. doi: 10.1016/0003-9861(69)90042-3. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rodriguez Alfageme C., Rudkin G. T., Cohen L. H. Isolation, properties and cellular distribution of D1, a chromosomal protein of Drosophila. Chromosoma. 1980;78(1):1–31. doi: 10.1007/BF00291907. [DOI] [PubMed] [Google Scholar]
- Schaffhausen B. S., Benjamin T. L. Deficiency in histone acetylation in nontransforming host range mutants of polyoma virus. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1092–1096. doi: 10.1073/pnas.73.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A., Galas D. J. The interaction of RNA polymerase and lac repressor with the lac control region. Nucleic Acids Res. 1979 Jan;6(1):111–137. doi: 10.1093/nar/6.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed B. Diazotizable arylamine cellulose papers for the coupling and hybridization of nucleic acids. Nucleic Acids Res. 1982 Mar 11;10(5):1799–1810. doi: 10.1093/nar/10.5.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M. Drosophila genome organization: conserved and dynamic aspects. Annu Rev Genet. 1981;15:219–264. doi: 10.1146/annurev.ge.15.120181.001251. [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Alberts B. The interaction of estradiol-receptor protein with the genome: an argument for the existence of undetected specific sites. Cell. 1975 Apr;4(4):301–310. doi: 10.1016/0092-8674(75)90150-6. [DOI] [PubMed] [Google Scholar]