Erythropoietin (EPO) is a glycoprotein hormone that plays important roles in regulating the production of red blood cells (erythrocytes).[1] Since human EPO was first isolated and purified from urine in 1977,[2] the structure and physiological properties of EPO have been thoroughly studied.[3] Mature human EPO found in nature consists of a polypeptide chain of 165 amino acids with four covalently attached oligosaccharides,[4] one of which is an O-linked oligosaccharide at Ser126, and the other three are N-linked oligosaccharides at residues Asn24, 38, 83.[ 5 ] EPO is used as a therapeutic agent to treat anemia caused by chronic kidney disease.[6] Commercial EPO is prepared using recombinant DNA technology. The carbohydrate moieties of both native and recombinant EPO[7] are heterogenerous and composed of multiple glycans with different lengths and composition at each glycosylation site.[5a,8] This heterogeneity makes it difficult to evaluate the effects of carbohydrate on EPO’s pharmacokinetic properties and, more importantly, complicates the understanding of mechanism of EPO’s action at the molecular level. For these reasons, it is important to develop an alternative strategy to prepare homogeneous EPO.
The production of homogeneous, glycosylated EPO by total synthesis is a major current objective of the organic synthesis community.[ 9 ] Recent developments in chemical synthesis of proteins[10] and glycoproteins[11] have made it possible in principle to prepare homogeneous EPO by total synthesis, thus offering the prospect of complete control of the covalent structure in order to perform systematic structure-activity studies. An early synthetic achievement was the preparation of a series of homogeneous polymer-modified EPO analogues of defined covalent structure and full biological activity.[12] These glycoprotein mimetics contained the full length 166 residue polypeptide chain encoded by the EPO gene, and were prepared by sequentially assembly of four synthetic peptide segments using thioester-mediated native chemical ligation.[13] This linear strategy of sequentially assembling multiple peptide segments involves repetitive ligation and purification, which results in low yields of final products.[ 14 ] More recently, a semisynthesis of an EPO containing thioether-linked complex glycan moieties has been reported.[15] Finally, the Danishefsky lab has set as a target the total synthesis of both the complex glycan moieties and the full length polypeptide chain of EPO; progress in this endeavour has been recently summarized.[ 16 ] Ultimately, a practical total synthesis of EPO will necessitate the fully convergent synthesis of the 165 residue mature polypeptide chain. None of the synthetic studies reported to date have successfully addressed this objective.
A primary function of the attached oligosaccharides is to extend EPO’s circulation time in vivo. Although the carbohydrate moiety accounts for about 35% of EPO molecule in weight, [1,4] it is not required for activity in vitro. The folded 165 amino acid polypeptide, with two native disulfide bonds, is responsible for binding to and activating the EPO receptor.[1b,3] Therefore, non-glycosylated EPO analogues are expected to be important for the study of structure-activity relationships. [Lys24,38,83]EPO produced by recombinant means has been previously used to determine the NMR structure of the EPO protein molecule[17] and the X-ray structure of EPO bound to its receptor;[18] In that work, lysine residues were placed at the N-glycosylation sties in order to increase the protein’s isoelectric point, and to thus prevent aggregation of non-glycosylated EPO at neutral or acidic pH conditions. This previous research[17, 18] showed that replacement of the three N-glycosylation sites with lysine residues does not affect the structure of EPO and its receptor binding activity.
We report here a convergent total chemical synthesis of [Lys24,38,83]EPO, containing two disulfide bonds which is composed of 165 amino acid residues with the three native N-glycosylation residues Asn24, 38, 83 mutated to Lys24, 38, 83. The synthetic protein is biologically active. This work is an important guide to the effective total synthesis of natural EPO and will enable the systematic preparation of neoglycoprotein analogues of EPO for the systematic dissection of the effect of glycan structure on the properties of the EPO molecule.
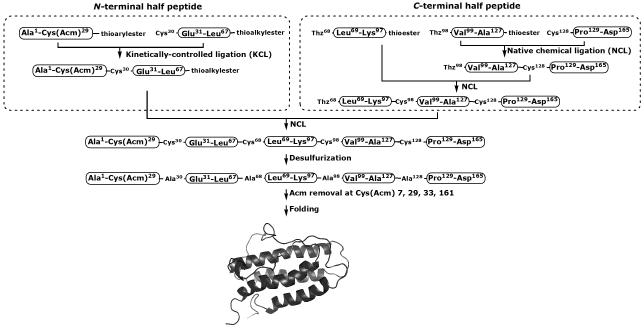
Our convergent synthetic strategy for the preparation of [Lys24,38,83]humanEPO is shown in Scheme 1. The combination of native chemical ligation[13] with kinetically controlled ligation strategy[19] makes it possible to efficiently assemble multiple peptide segments in a convergent fashion. Kinetically controlled ligation is based on the reaction of a peptide1-αthioarylester with a second Cys-peptide2-αthioalkylester to give a unique peptide1-peptide2-αthioalkylester product. Both native chemical ligation and kinetically controlled ligation use Xaa-Cys ligation sites. One challenge of EPO chemical synthesis is the limited number and uneven distribution of the natural cysteine residues (at amino acid positions 7, 29, 33 and 161) in the target polypeptide chain. To extend the application of native chemical ligation in the synthesis of cysteine-free proteins, Dawson and co-workers introduced the combination of native chemical ligation with subsequent desulfurization of cysteine residues to alanine residues.[20] Recently, two different protocols for selective desulfurization of cysteine residues in the presence of acetylamidomethyl (Acm)-protected cysteine and methionine residues have been reported: one uses Raney Nickel as catalyst;[21] the other is a phosphine-induced radical reduction.[22] Thus, Xaa-Ala sequences can be used as potential ligation sites in EPO synthesis, in the presence of suitably protected native Cys residues.
Scheme 1.
Convergent strategy for the total chemical synthesis of [Lys24,38,83] human EPO.
Based on the distribution of potential Xaa-Ala ligation sites in the EPO polypeptide chain, five synthetic peptide segments, Ala1-Cys29(29aa), Ala30-Leu67(38aa), Ala68-Lys97(30aa), Ala98-Ala127(30aa), Ala128-Asp165(38aa), of comparable lengths were selected as building blocks for assembling the full length EPO polypeptide chain. Full details of the preparation of these synthetic peptide segments are given in the Supporting Information. As shown in Scheme 1, the 165 amino acid polypeptide chain can be furnished by a final native chemical ligation of two large peptide segments: Ala1-Leu67-αthioalkylester and Cys68-Asp165. The N-terminal segment Ala1-Leu67-αthioalkylester can be prepared using a kinetically controlled ligation of peptide segments Ala1-Cys29-αthioarylester and Cys30-Leu67-αthioalkylester. The C-terminal segment Cys68-Asp165 can be prepared by native chemical ligation of Thz98-Ala127-αthioester and Cys128-Asp165, followed by a second native chemical ligation with peptide segment Thz68-Lys97-αthioester.
(R)-1, 3-Thiazolidine-4-carboxylic acid (Thz) was employed as a protected form of N-terminal L-cysteine, in order to avoid cyclization/oligomerization of Cys-peptide-α thioester segments during native chemical ligation.[23] The side chain thiols of the four native cysteine residues (at amino acid positions 7, 29, 33 and 161) were protected with the Acm group during the solid-phase synthesis of peptide segments Ala1-Cys29, Cys30-Leu67 and Cys128-Asp165. Four cysteine residues (at amino acid positions 30, 68, 98 and 128) were introduced as ligation sites and can subsequently be reduced to alanine residues using the selective desulfurization protocol[21,22] after the full length polypeptide chain is assembled. After removal of the Acm groups from native Cys7, 29, 33, 161 residues, the 165 residue polypeptide chain is folded with concomitant formation of two disulfide bonds to give synthetic [Lys24,38,83]EPO.
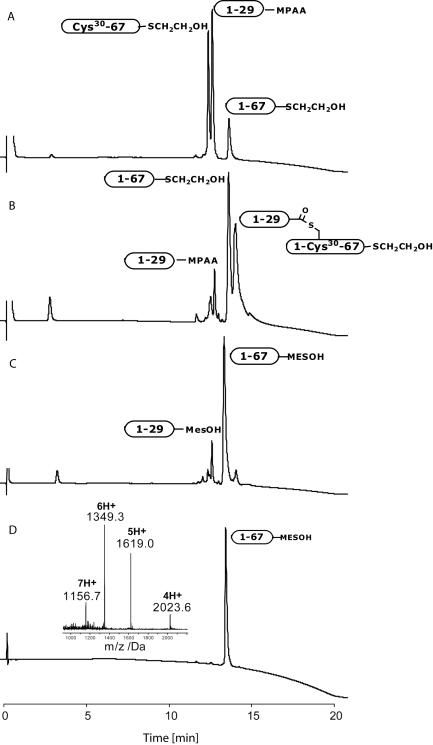
Kinetically controlled ligation of peptide segments Ala1-Cys(Acm)29-COSC6H4CH2COOH and Cys30-Leu67-αthioalkylester was performed at pH 6.8 without the use of added thiol catalyst (Figure 1). The starting peptide segments were essentially consumed after 1 hour. In addition to the formation of ligation product Ala1-Cys30-Leu67-αthioalkylester, a significant amount of branched thioester byproduct Ala1-Cys30(Ala1-Cys(Acm)29)-Leu67-αthioalkylester (Figure 1, B) was generated. Addition of an excess of sodium 2-mercaptoethanesulfonate (MESNa) regenerated the desired Ala1-Leu67-αthioalkylester (Figure 1, C), which was purified by reverse phase HPLC (Figure 1, D).
Figure 1.
Kinetically controlled ligation of peptide segments Ala1-Cys(Acm)29-αCOSC6H4CH2COOH and Cys30-Leu67-α COSCH2CH2OH. A) t < 1 min. B) t = 1 h. C) After adding MESNa for 2 h. D) After HPLC purification. ESI-MS (inset) gave an observed mass of 8090.0 ± 0.2 Da (calcd. 8089.1 Da, average isotopes). Analytical HPLC traces (λ = 214 nm) are shown.
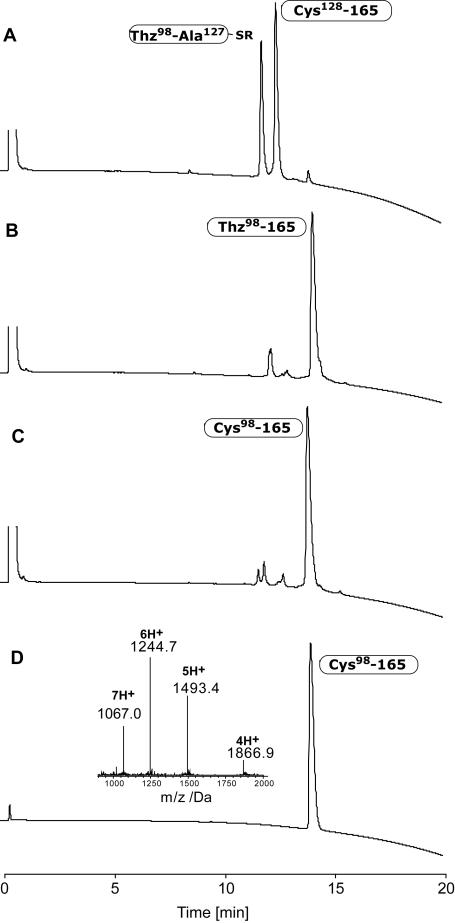
The native chemical ligation of peptide segments Thz98-Ala127-αthioalkylester and Cys128-Asp165 was performed at pH 6.8 with the use of the aryl thiol HSC6H4CH2COOH (MPAA) as catalyst (Figure 2). Reaction was essentially complete after 5 hours and gave the desired ligation product Thz98-Asp165, which was converted to Cys98-Asp165 by overnight reaction with methoxyamine hydrochloride at pH 4.0 (Figure 2, C).
Figure 2.
Native chemical ligation of peptide segments Thz98-Ala127-αCOSCH2CH2COLeu and Cys128-Asp165. A) t < 1 min. B) t = 5 h. C) After adding MeONH2·HCl for 14 h at pH = 4.0, to convert Thz- to Cys-peptide. D) After HPLC purification. ESI-MS (inset) gave an observed mass of 7462.5 ± 0.7 Da (calcd. 7462.7 Da, average isotopes).
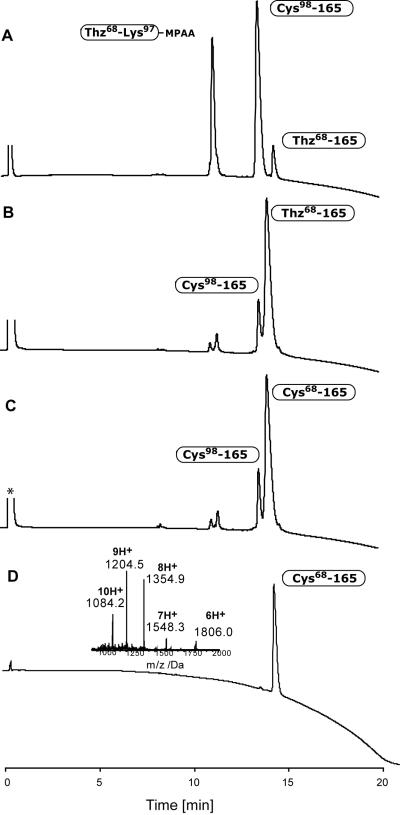
The dinitrophenyl (Dnp) group was removed from residue His94 by treating peptide segment Thz68-His(Dnp)94-Lys97-αthioalkylester with an excess of MPAA at pH 6.8, prior to the native chemical ligation of peptide segments Thz68-Lys97-αthioester and Cys98-Asp165. The aryl thiol reagent MPAA not only cleaved Dnp from histidine residues by thiolysis, it also generated a more reactive αthioarylester Thz68-His94-Lys97-αCOSC6H4CH2COOH through transthioesterification (see Supporting Information).[24] The native chemical ligation of segments Thz68-Lys97-αCOSC6H4CH2COOH and Cys98-Asp165 was performed at pH 6.8 for 8 hours (Figure 3, A & B). The reaction mixture was then treated with methoxyamine hydrochloride at pH 4.0 (Figure 3, C) to furnish the desired C-terminal half peptide Cys68-Asp165 (Figure 3, D) to be used in the final ligation to form the full length EPO polypeptide chain.
Figure 3.
Native chemical ligation of peptide segments Thz68-Lys97-αCOSC6H CH2COOH and Cys98-Asp165. A) t < 1 min. B) Ligation after 4 h, followed by another 4 h treatment with MPAA. C) After adding MeONH2·HCl for 15 h to convert Thz- to Cys-peptide. D) After HPLC purification. ESI-MS (inset) gave an observed mass of 10831.2 ± 0.7 Da (calcd. 10831.6 Da, average isotopes).
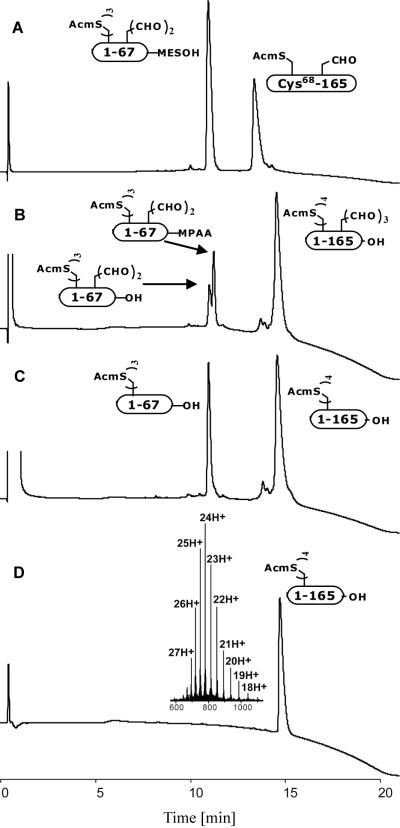
The final ligation of peptide Ala1-Leu67-SCH2CH2SO3 and peptide Cys68-Asp165 was performed at pH 6.8 with the use of MPAA as catalyst. This ligation, at a less reactive Leu-Cys site,[25] took about 15 hours to complete (Figure 4, B). An unusually large amount of hydrolyzed Ala1-Leu67-αthioester byproduct, Ala1-Leu67-OH (Figure 4, B), was formed during the reaction. The origin of this side reaction is uncertain; adjusting the pH of the reaction with more diluted NaOH solution did not reduce the amount of hydrolyzed byproduct. After ligation was complete, the three NIn-formyl protecting groups were removed from tryptophan residues at amino acid positions 51, 64 and 88 by adding 2-mercaptoethanol and piperidine into reaction mixture to a final concentration of 37 % (v/v) and 25 % (v/v), respectively.[19c] Formyl groups were quantitatively cleaved from the full length EPO polypeptide after 0.5 hours (Figure 4, C).
Figure 4.
Native chemical ligation of peptide segments Ala1-Leu67-αCOSCH2CH2SO3H and Cys68-Asp165. A) t < 1 min. B) t = 15 h. C) After adding 2-mercaptoethanol and piperidine for 0.5 h, to remove the Trp(In(formyl)) groups. D) After HPLC purification. ESI-MS (inset) gave an observed mass of 18696.5 ± 2.2 Da (calcd. 18694.7 Da, average isotopes).
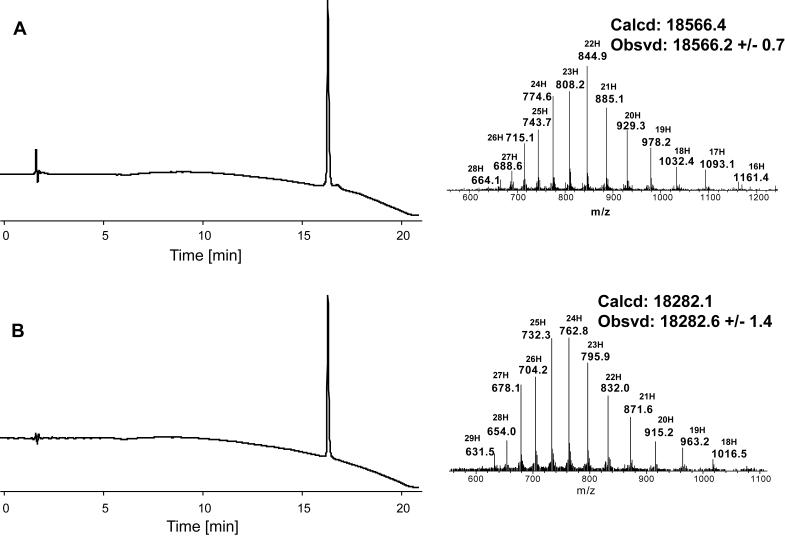
The full length synthetic EPO polypeptide chain was subject to selective desulfurization to reduce Cys30,68,98,128 to Ala30,68,98,128, while keeping four Acm-protected cysteine residues (at amino acid positions 7, 29, 33, 161) intact. Initial trials of selective desulfurization were performed using Raney nickel catalyst as described in previously published procedures.[21, 26 ] Although success has been shown in the selective desulfurization of the hormone amylin and a small model protein EETI-II in the presence of Acm-protected cysteine residues,[21] we found that under these conditions However, the differentiation in desulfurization rates between free cysteine residues and Acm-protected cysteine residues in the full length EPO polypeptide was not sufficient for selective desulfurization. A recently reported free radical-based desulfurization protocol[22] has been proven to be a mild and highly selective method. In this protocol, a water-soluble radical initiator 2-2′-azobis[2-(2-imidazolin-2-yl)propane]dihydrochloride (VA-044) and tris(2-carboxyethyl)phosphine (TCEP) were used for reducing cysteine residues. Full length EPO peptide Ala1-Cys(Acm)7, 29, 33, 161-Cys30, 68, 98, 128-Asp165 was exposed to a mixture of TCEP, EtSH, tBuSH and VA-044 in guanidine hydrochloride and phosphate buffer. EtSH and tBuSH are used as additional thiol radical source. A guanidine buffer was used in order to improve the solubility of full length EPO peptide. The desulfurization was performed at 37°C, and monitored by LCMS. After 5 hours, ESI-MS analysis showed near-quantitative conversion of free cysteine residues to alanine residues, although no obvious change in retention time was observed between starting peptide and desulfurized product. The analytical HPLC trace and ESI-MS spectrum of the purified desulfurization product Ala1-Cys(Acm)7, 29, 33, 161-Asp165 are shown in Figure 5A (mass: obsvd 18566.2 ± 0.7 Da; calcd. 18566.4 Da, average isotopes).
Figure 5.
Purified selective desulfurization product Ala1-Cys(Acm)7, 29, 33, 161-Ala30, 68, 98, 128-Asp165 (A); and, full-length polypeptide product Ala1-Cys7, 29, 33, 161-Ala30, 68, 98, 128-Asp165 (B) after Acm removal. Analytical HPLC traces (λ = 214 nm) and ESI-MS spectra are shown.
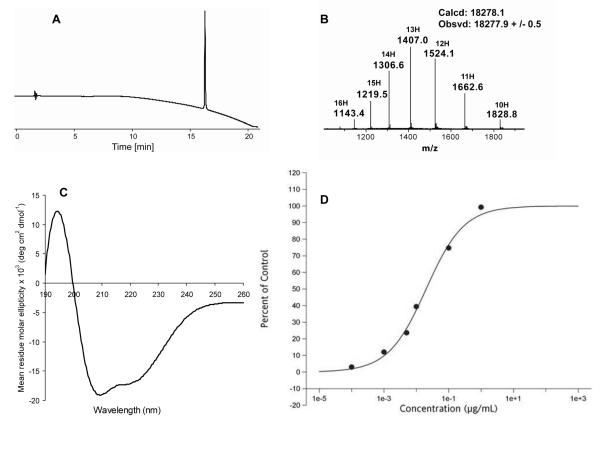
The four Acm protecting groups were removed by treating EPO polypeptide with AgOAc in 1:1 AcOH/H2O solution. Reaction progress was monitored by LCMS. Removal of the Acm groups from the EPO polypeptide chain gave no change in retention time on HPLC analysis, but on-line LCMS showed that reaction was completed after 6 hours. Then, a solution of 0.2 M 1, 4-dithiothreitol (DTT) in 6 M guanidine hydrochloride was added to quench excess AgOAc, and to convert peptide with silver thiolates to free thiols. The analytical HPLC and ESI-MS spectrum of purified Acm removal product Ala1-Cys7, 29, 33, 161-Ala30, 68, 98, 128-Asp165 are shown in Figure 5B (mass: obsvd 18282.6 ± 1.4 Da; calcd. 18282.1 Da, average isotopes). Folding of the synthetic 165 residue EPO polypeptide chain was performed according to the previously published procedure.[12] The reduced form of the polypeptide was first dissolved in 6 M guanidine hydrochloride solution. Then, it underwent a 3-step dialysis process sequentially against 3 M guanidine hydrochloride with 4 mM L-cysteine and 0.5 mM L-cystine (pH = 8.5), 1 M guanidine hydrochloride (pH = 8.0), and finally 10 mM Tris (pH = 7.0) solutions, to give folded protein containing two disulfide bonds. The synthetic protein was purified by HPLC. The analytical HPLC chromatogram of the synthetic [Lys24,38,83]EPO is shown in Figure 6 (A); the ESI-MS spectrum was obtained by direct infusion,[ 27 ] giving a mass of 18277.9 ± 0.5 Da (Figure 6, B), which is in excellent agreement with the calculated mass of 18278.1 Da. (average isotopes). The loss of 4 Daltons (4.7 ± 1 Da) in mass was consistent with the formation of two intramolecular disulfide bonds after folding. The CD spectrum of synthetic [Lys24,38,83]EPO was measured in 10 mM phosphate buffer (pH = 6.2) at a concentration of 5.9 μM. As shown in Figure 6C, the spectrum is in good agreement with the reported highly helical secondary structure of this protein molecule. The CD spectrum, with the characteristic shape of a minimum around 208 nm and a shoulder around 222 nm, is essentially identical to that reported for recombinant [Lys24,38,83]EPO[17] (see Supporting Information for comparison of CD spectra), and is closely similar to the CD spectra reported for recombinant EPO[28] and polymer-modified EPO analogues.[12a]
Figure 6.
Characterization of folded, lysine-mutated human EPO (synthetic [Lys24,38,83]EPO). A) Analytical HPLC trace (λ = 214 nm). The chromatographic separation was performed on an analytical C3 column (Agilent ZORBAX 300SB, 4.6 × 150 mm) using a linear gradient of 5-65% buffer B over 15 min (buffer A = 0.1% TFA in H2O; buffer B = 0.08% TFA in acetonitrile). B) ESI-MS spectrum of [Lys24,38,83]EPO by direct infusion. C) Circular dichroism spectrum of 5.9 μM lysine-mutated human EPO in 10 mM phosphate buffer (pH = 6.2). D) Cell proliferation assay.[29] The growth responses of TF-1 cells (Human erythroblast) to the addition of synthetic [Lys24,38,83]EPO and recombinant EPO (data not shown) were measured after 72 h incubation. Synthetic [Lys24,38,83]EPO had an EC50 value of 0.02 μg/mL.
The in vitro biological activity of synthetic [Lys24,38,83]EPO was evaluated in a cell proliferation assay.[ 29 ] TF-1 cells (human erythroblast) were incubated with different concentrations of synthetic [Lys24,38,83]EPO and native (recombinant) EPO for 72 hours before quantifying the uptake of [3H]Thymidine.[29,30 ] As shown in Figure 6D, lysine-mutated human EPO had an EC50 value of 0.02 μg/mL, which indicates that it is fully active for EPO receptor binding.
In summary, we have developed and prototyped a fully convergent chemical synthesis of a non-glycosylated human EPO. A key feature of the successful synthesis was native chemical ligation at Xaa-Ala sites enabled by the use of metal-free desulfurization (dethiylation)22. The combination of characterizations using HPLC, mass spectrometry, CD spectroscopy and cell proliferation assay showed that the synthetic [Lys24,38,83] human EPO has a well-defined covalent structure, was folded correctly, and was biologically active. This convergent chemical synthesis of EPO complements other synthetic and semi-synthetic approaches to the EPO glycoprotein, and will provide a flexible approach to the preparation of EPO analogues with any desired modification on the polypeptide backbone. Our convergent synthetic route also provides a versatile platform for the preparation of neoglycan analogues of EPO to explore the effects of carbohydrate structure on biological function.
Supplementary Material
Footnotes
[]This research was supported by the Office of Science (BER), U.S. Department of Energy, Grant No. DE-FG02 07ER64501 to S.B.H.K. and by the National Institutes of Health, Grant No. R01 GM075993 to S.B.H.K.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
References
- [1] a).Krantz SB. Blood. 1991;77:419–434. [PubMed] [Google Scholar]; b) Wognum AW. StemCell Technologies. Available online: http://www.stemcell.com/technical/EPO%20MiniReview.pdf. [Google Scholar]; c) Graber SE, Krantz SB. Annu. Rev. Med. 1978;29:51–66. doi: 10.1146/annurev.me.29.020178.000411. [DOI] [PubMed] [Google Scholar]
- [2].Miyake T, Kung CK-H, Goldwasser E. J. Biol. Chem. 1977;252:5558–5564. [PubMed] [Google Scholar]
- [3].Ribatti D. Leukemia Res. 2008;32:1169–1172. doi: 10.1016/j.leukres.2008.01.018. [DOI] [PubMed] [Google Scholar]
- [4].Lappin T. The Oncologist. 2003;8(Suppl. 1):15–18. doi: 10.1634/theoncologist.8-suppl_1-15. [DOI] [PubMed] [Google Scholar]
- [5] a).Sasaki H, Bothner B, Dell A, Fukuda M. J. Biol. Chem. 1987;262:12069–12076. [PubMed] [Google Scholar]; b) Takeuchi M, Kobata A. Glycobiology. 1991;1:337–346. doi: 10.1093/glycob/1.4.337. [DOI] [PubMed] [Google Scholar]; c) Lai P-H, Edverett R, Wang FF, Arakawa T, Goldwas E. J. Biol. Chem. 1986;261:3116–3121. [PubMed] [Google Scholar]
- [6].Lappin TR, Maxwell AP, Johnston PG. Stem Cells. 2002;20:485–492. doi: 10.1634/stemcells.20-6-485. [DOI] [PubMed] [Google Scholar]
- [7].Gillis S, Williams DE. Curr. Opin. Immunol. 1998;10:501–503. doi: 10.1016/s0952-7915(98)80213-6. [DOI] [PubMed] [Google Scholar]
- [8] a).Spivak JL, Hogans BB. Blood. 1989;73:90–99. [PubMed] [Google Scholar]; b) Misaizu T, Matsuki S, Strickland TW, Takeuchi M, Kobata A, Takasaki S. Blood. 1995;86:4097–4104. [PubMed] [Google Scholar]; c) Fukuda MN, Sasaki H, Lopez L, Fukuda M. Blood. 1989;73:84–89. [PubMed] [Google Scholar]
- [9].Kajihara Y, Yamamoto N, Okamoto R, Hirano K, Murase T. Chem. Rec. 2010;10:80–100. doi: 10.1002/tcr.200900024. [DOI] [PubMed] [Google Scholar]
- [10].Kent SBH. Chem. Soc. Rev. 2009;38:338–351. doi: 10.1039/b700141j. [DOI] [PubMed] [Google Scholar]
- [11].Liu L, Bennett CS, Wong C-H. Chem. Commun. 2006:21–33. doi: 10.1039/b513165k. [DOI] [PubMed] [Google Scholar]
- [12] a).Kochendoerfer GG, et al. Science. 2003;299:884–887. doi: 10.1126/science.1079085. [DOI] [PubMed] [Google Scholar]; b) Chen S-Y. Chem. & Biol. 2005;12:371–383. doi: 10.1016/j.chembiol.2005.01.017. [DOI] [PubMed] [Google Scholar]
- [13].Dawson PE, Muir TW, Lewis IC, Kent SBH. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- [14].Hendrickson JB. J. Am. Chem. Soc. 1977;99:5439–5450. [Google Scholar]
- [15].Hirano K, Macmillan D, Tezuka K, Tsuji T, Kajihara Y. Angew. Chem. Int. Ed. 2009;48:9557–9560. doi: 10.1002/anie.200904376. [DOI] [PubMed] [Google Scholar]
- [16].Kan C, Danishefsky SJ. Tetrahedron. 2009;65:9047–9065. doi: 10.1016/j.tet.2009.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Narhi LO, Arakawa T, Aoki K, Wen J, Elliott S, Boone T, Cheetham J. Protein Eng. 2001;14:135–140. doi: 10.1093/protein/14.2.135. [DOI] [PubMed] [Google Scholar]
- [18].Syed R, Reid S, Li C, Cheetham J, Aoki KH, Liu B, Zhan H, Osslund TD, Chirino AJ, Zhang J, Finer-Moore J, Elliott S, Sitney K, Katz BA, Matthews DJ, Wendoloski JJ, Egrie J, Stroud RM. Nature. 1998;395:511–516. doi: 10.1038/26773. [DOI] [PubMed] [Google Scholar]
- [19] a).Bang D, Pentelute BL, Kent SBH. Angew. Chem. Int. Ed. 2006;45:1–5. doi: 10.1002/anie.200600702. [DOI] [PubMed] [Google Scholar]; b) Torbeev VY, Kent SBH. Angew. Chem. Int. Ed. 2007;46:1667–1670. doi: 10.1002/anie.200604087. [DOI] [PubMed] [Google Scholar]; c) Durek T, Yu, Torbeev VY, Kent SBH. Proc. Natl. Acad. Sci. USA. 2007;104:4846–4851. doi: 10.1073/pnas.0610630104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yan LZ, Dawson PE. J. Am. Chem. Soc. 2001;123:526–533. doi: 10.1021/ja003265m. [DOI] [PubMed] [Google Scholar]
- [21].Pentelute BL, Kent SBH. Org. Lett. 2007;9:687–690. doi: 10.1021/ol0630144. [DOI] [PubMed] [Google Scholar]
- [22].Wan Q, Danishefsky SJ. Angew. Chem. Int. Ed. 2007;46:9248–9252. doi: 10.1002/anie.200704195. [DOI] [PubMed] [Google Scholar]
- [23] a).Villain M, Vizzavona J, Rose K. Chem. & Biol. 2001;8:673–679. doi: 10.1016/s1074-5521(01)00044-8. [DOI] [PubMed] [Google Scholar]; b) Bang D, Kent SBH. Angew. Chem. Int. Ed. 2004;43:2534–2538. doi: 10.1002/anie.200353540. [DOI] [PubMed] [Google Scholar]
- [24].Reaction details are listed in supporting information.
- [25].Hackeng TM, Griffin JH, Dawson PE. Proc. Natl. Acad. Sci. USA. 1999;96:10068–10073. doi: 10.1073/pnas.96.18.10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bang D, Makhatadze GI, Tereshko V, Kossiakoff AA, Kent SBH. Angew. Chem. Int. Ed. 2005;44:3852–3856. doi: 10.1002/anie.200463040. [DOI] [PubMed] [Google Scholar]
- [27].Mandal K, Kent SBH. Angew. Chem. Int. Ed. 2011;50:8029–8033. doi: 10.1002/anie.201103237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Davis JM, Arakawa T, Strickland TW, Yphantis DA. Biochemistry. 1987;26:2633–2638. doi: 10.1021/bi00383a034. [DOI] [PubMed] [Google Scholar]
- [29].The cell proliferation assay was performed by MDS Pharma Services (www.mdsps.com) using assay# 307850.
- [30].Kitamura T, et al. Blood. 1989;73:375–380. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.