Abstract
Affinity-matured antibodies can exhibit increased biological efficacy. Regardless of whether an antibody is isolated from a hybridoma or a human Fv phage library, the antibody affinity for its target may need improvement for therapeutic applications. An increased affinity may allow for a reduced dosage of a therapeutic antibody; toxic side effects may also be reduced. In the immune system, affinity maturation is a process involving somatic hypermutations in B cells. Therefore, germline hotspot residues are most likely to have a major impact on antibody affinity. Here, we describe procedures for germline hotspot mutagenesis with an emphasis on strategies for randomizing hotspots with PCR and phage display, using as an example the anti-CD22 monoclonal antibody.
Keywords: in vitro antibody affinity maturation, somatic hypermutation, single chain Fv or scFv, phage display, molecular evolution, antibody fragments, antibody engineering, CD22
1. Introduction
The hybridoma fusion technology, developed by Köhler and Milstein in 1975 (1), gave for the first time an access to monoclonal antibodies (mAb). Monoclonal antibodies that specifically target cancer cells while sparing normal tissues have become the most promising candidates for ‘magic bullets’ against cancer. Up to date, twenty one monoclonal antibodies have been approved by the U.S. Food and Drug Administration. Among them, nine target cancer cells. But unmet challenges in antibody engineering have still limited the number of therapeutic antibodies that can move from the bench to the clinic. Such challenges concern several serious issues such as affinity, specificity (or cross-reactivity), immunogenicity and half-life. Antibody affinity plays an important role in biological efficacy. Regardless of whether an antibody is isolated from hybridoma or a nonimmune human Fv phage library, the antibody affinity for its target may need improvement. An increased affinity may also allow for a reduced dosage of a therapeutic antibody and toxic side effects may be reduced.
Efforts have been made to improve the affinity of antibodies by random or semi-random mutations (2, 3). These approaches are often time-consuming and require an Fv library so large that it theoretically exceeds the limitations of all current physical display systems such as phage display and yeast display. Focused mutagenesis has the advantage that only very small libraries need to be screened. However, it is not always possible to identify the key residues for focused mutagnesis due to the fact that a crystral structure of an antibody-antigen complex is often not available. We have found that germline hotspot residues are most likely to have a major impact on affinity based on statistical analysis of independent monoclonal antibodies derived from the same germline genes recognizing the same epitopes (4). The number of germline hotspots tends to decrease in antibodies with high affinities in B cells, suggesting an overall correlation between affinity and number of hotspot-based somatic mutations that is characteristic of affinity maturation in vivo. The existence of germline hotspots in high affinity antibodies obtained in vivo provides the possibility that in vitro affinity maturation targeting these hotspots could further increase the binding affinity. We have developed an approach, called “hotspot mutagenesis”, using PCR and phage display to randomize germline hotspots to increase antibody affinity specific for various tumor antigens (4-7). Germline hotspots in the antibody complementarity determining regions (CDR) are naturally prone to hypermutations (8). We introduced random mutations into these sites by PCR and made phage-display libraries with a minimum size of 103 and 104 independent clones. Panning of these small hotspot libraries has yielded mutant antibodies with increased affinity.
We have also found an advantage of in vitro hotspot-based antibody evolution as compared to in vivo somatic hypermutation. Using our PCR-based method, we have found that several mutated hotspot residues in the evolved antibodies arise from double or even triple mutations at the first, second and third positions in each codon. It is improbable that such dramatic mutations can occur in vivo (9). It is well documented that hotspot mutagenesis in the somatic hypermutation process often involves only one nucleotide point mutation in each codon (9). This may explain why some hotspot mutations favorable for higher binding affinity fail to occur in vivo. In contrast to the in vivo scenario, a hotspot residue can be easily randomized in vitro by PCR. In addition to constraints arising from the mechanism of somatic hypermuation, the B cells response exhibits an apparent affinity ceiling (KD > 0.1nM) because of fast internalization rates of B cell receptors (10) (see Note 1).
Here, we describe procedures for germline hotspot mutagenesis with an emphasis on strategies for randomizing hotspot with PCR and phage display, using as an example the anti-CD22 monoclonal antibody (RFB4). Potential applications of this germline hotspot strategy extend beyond antibody affinity maturation and improvement of mAb or antibody fragments, to the modification of T-cell receptors, MHC molecules, and other biologically important proteins because somatic hypermutation may be a global phenomenon in the whole genome (11).
2. Materials
2.1. Sequence Analysis of Fv and Identification of Germline Hotspots
PC computer with Window operation system XP and Internet access.
Online programs for the Fv sequence analysis IMGT V-Quest (http://imgt.cines.fr/textes/vquest/) and for molecular modeling WAM (http://antibody.bath.ac.uk/index.html). Deep View-SwissPdb Viewer (www.expasy.org/spdbv/) for analysis of molecular models.
2.2. Construction of a Hotspot Phage Library
Phagemid used for phage display: pCANTAB-5E (GE Healthcare, formerly Amersham-Pharmacia Biotech, Piscataway, NJ) (see Note 2).
DNA gel eletrophoresis: 10% (w/w) ethidium bromide (Invitrogen, Carlsbad, CA), UltraPure Agarose (Invitrogen).
Cloning: Restriction endonucleases (all from New England Biolabs, Ipswich, MA): SfiI, NotI; T4 DNA ligase (New England Biolabs).
PCR: PCR dNTP mix (solution containing 25 mM each of dATP, dCTP, dGTP and dTTP) (Invitrogen); puReTaq Ready-To-Go PCR Beads (GE Healthcare, formerly Amersham Biosciences); QIAquick Gel Extraction Kit (Qiagen, Valencia, CA); Roche Expand High Fidelity PCR System (Roche, Indianapolis, IN) for 2nd PCR reaction in the 2-step extension PCR; UltraPure DNase/RNase-free distilled water (Invitrogen).
TG1 electroporation: Bacterial strains TG1 electroporation-competent cells (Stratagene); Electroporator (Bio-Rad, Hercules, CA); 0.1-cm gap electroporation cuvet (Bio-Rad).
Helper phage M13K07 (New England Biolabs).
E. coli cell culture and harvest: Shaker incubator; Centrifuge bottles; Benchtop centrifuge.
2YT broth (Invitrogen): suspend 31 g in 1 L of demineralized water. Autoclave for 15 min at 121°C.
2YT/Amp medium 2YT, 100 μg/mL ampicillin (Sigma, St. Louis, MO). 10. 2YT/Amp/Kan medium 2YT, 100 μg/mL ampicillin, 25 μg/mL kanamycin (Sigma).
Blocking buffer PBS (phosphate buffered saline), 1% (w/v) BSA (Bovine Serum Albumin Fraction V, heat shock, fatty acid ultra-free; Roche). Filter-sterilize.
Solution for microplate coating for ELISA: BupH Carbonate-Bicarbonate Buffer Packs (each pack yields 0.2M Carbonate-Bicarbonate Buffer, pH 9.4 when dissolved in 500 mL distilled water; Pierce, Rockford, IL).
LB medium, LB agar (Invitrogen), LB/Amp plates LB agar, 100 μg/mL ampicillin (Invitrogen).
PBS (Invitrogen).
PBST buffer PBS, 0.05% Tween 20 (Sigma), 0.5% BSA. Filter-sterilize.
PEG/NaCl 20% PEG-8000 (w/v) (USB), 2.5 M NaCl. Mix and autoclave.
SOC medium (Invitrogen).
Superbroth medium (Invitrogen).
10 × TAE buffer (Invitrogen).
2.3. Phage ELISA
3,3',5,5'-tetramethylbenzidine (TMB) (Kirkegaard & Perry Laboratories Inc., Gaithersburg, MD).
H2O2 peroxidase substrate (Kirkegaard & Perry Laboratories Inc.).
HRP/anti-M13 antibody conjugate (Abcam, Cambridge, MA).
96-well Maxisorp immunoplates (Nunc, Rochester, NY).
2.4. Cell Culture
Complete growth medium: Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% Fetal calf serum (Sigma), 1% L-Glutamine solution (Sigma), 1% Nonessential amino acids solution (Sigma) and Penicillin-streptomycin (Sigma).
Tissue culture flasks (Nunc).
Cells used for cell panning: Daudi (ATCC Catalog # CCL-213), MCF7 (ATCC Catalog # HTB-22) (see Note 3).
2.5. Flow Cytometry
FACS Solution: 5% (w/v) BSA, Bovine Serum Albumin Fraction V, heat shock, fatty acid ultra-free (Roche); 0.1% sodium azide (Sigma) in PBS.
Anti-M13 mAb (Abcam); anti-E-Tag antibody (GE Healthcare, Piscataway, NJ).
R-phycoerythrin labeled goat anti-mouse (Invitrogen/Biosource) to detect the phage binding on cells.
Target proteins (see Note 4).
Major equipment: Flor cytometer, BD FACSCalibur Flow Cytometer (Becton Dickinson, Franklin Lakes, NJ) (see Note 5).
3. Methods
High affinity antibodies are generated in vivo by somatic hypermutation. Somatic hypermutation does not occur randomly within immunoglobulin V genes but is preferentially targeted to certain nucleotide positions (hotspots) and away from others (cold spots) (14). Cold spots often coincide with residues essential for V gene folding. Hotspots, which appear to be strategically located to favor affinity maturation, are most frequently located in the CDRs. This process mainly results in the introduction of mutations that are located adjacent to A/G-G-C/T-A/T (RGYW) and AG-C/T (AGY) sequences. A total of 50–60% of all somatic hypermutations is found within or adjacent to RGYW motifs (9). To improve the affinity of a mAb, we have found that hotspots are the preferred regions to introduce random mutations. The first step in this process is to identify the DNA hotspot sequences in all the CDRs of the antibody under investigation.
3.1. Sequence Analysis and Identification of Germline Hotspots
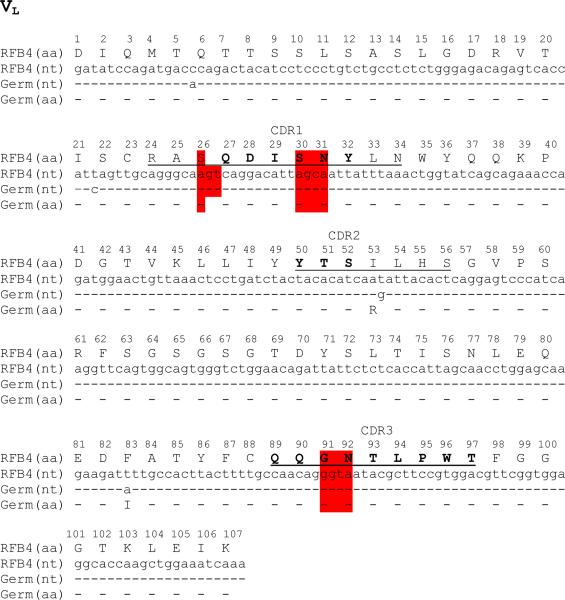
Five germline hotspot clusters can be identified in the CDR domains of RFB4. Among them, four contain RGYW in their nucleotide sequences. They are located in light chain CDR1 (L1) and CDR3 (L3) (Fig. 1, highlighted), and heavy chain CDR2 (H2) and CDR3 (H3) (data not shown). The VL of anti-CD22 antibody RFB4 is of the V10 class and has 96% identity at the amino acid level with the germline light chain IGKV10-96*01. The VL originates from recombination of the V gene (IGKV10-96*01) and the J1 gene IGKJ1*01. As shown in Fig. 1, the DNA sequence of the germline V and J gene fragments were manually assembled and translated into an amino acid sequence (4).
Fig. 1.
Sequence alignment of the VL domains of RFB4. The CDR regions are defined according to Katat et al. (15) (underline) and IMGT (16) (bold). A dash indicates an exact match between residues; a dot indicates a gap added to the sequence. In case of Germ (germline) genes, V gene (IGKV10-96*01) and J (IGKJ1*01) gene fragments were manually assembled for VL. Germline hotspot residues within CDR domains of RFB4 are highlighted. All residues are numbered according to the Kabat numbering scheme. (This figure reproduced in part from J. Biol. Chem. 280(1):607-617, January 7, 2005).
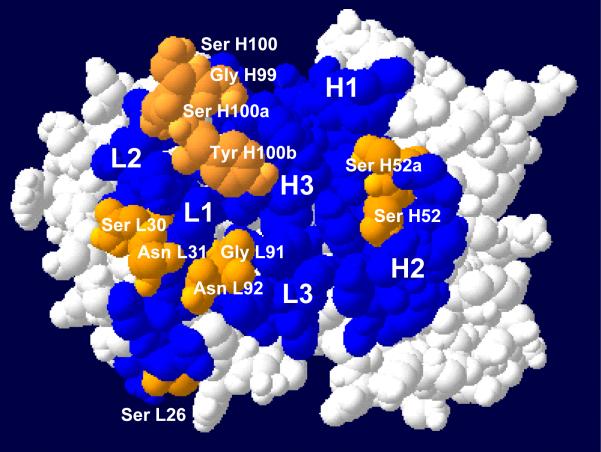
The structural models of the variable region and its germline counterpart can be made by molecular modeling using web-based antibody modeling software WAM (17) (http://antibody.bath.ac.uk/index.html). The modeling program uses a modified form of the algorithm used in antibody modeling software AbM of Oxford Molecular (Accelrys, San Diego, CA). A surface view of the RFB4 V region reveals that all the germline CDR hotspot residues except Ser-26 in light chain are exposed (Fig. 2, gray), suggesting that they may be located at the interface between the antibody and antigen.
Convert the heavy chain (VH) or light chain variable region (VL) DNA sequence into the FASTA format.
Perform a DNA sequence homology search using V-Quest at IMGT (http://imgt.cines.fr/textes/vquest/) (16) (see Note 6).
Identify germline hotspots in the nucleotide sequences of the CDR domains. Germline hotspot residues have the same nucleotide sequence as the germline gene, whereas in non-germline hotspot residues the nucleotide sequence at or near the RGYW motifs differs from the germline gene.
Make an antibody model by using web-based antibody modeling software WAM (http://antibody.bath.ac.uk/index.html) following the instruction. Use Deep View-SwissPdb Viewer (www.expasy.org/spdbv/) to view the molecular model.
Fig. 2.
A space-filling model of RFB4 Fv. The view is looking down on the antigen-binding surface. CDR 1, 2 and 3 for both chains of RFB4 are in black or gray. Germline hotspot residues within CDR domains are in gray. (This figure reproduced in part from J. Biol. Chem. 280(1):607-617, January 7, 2005).
3.2. Construction of a Phage-display Library Targeting a Germline Hotspot
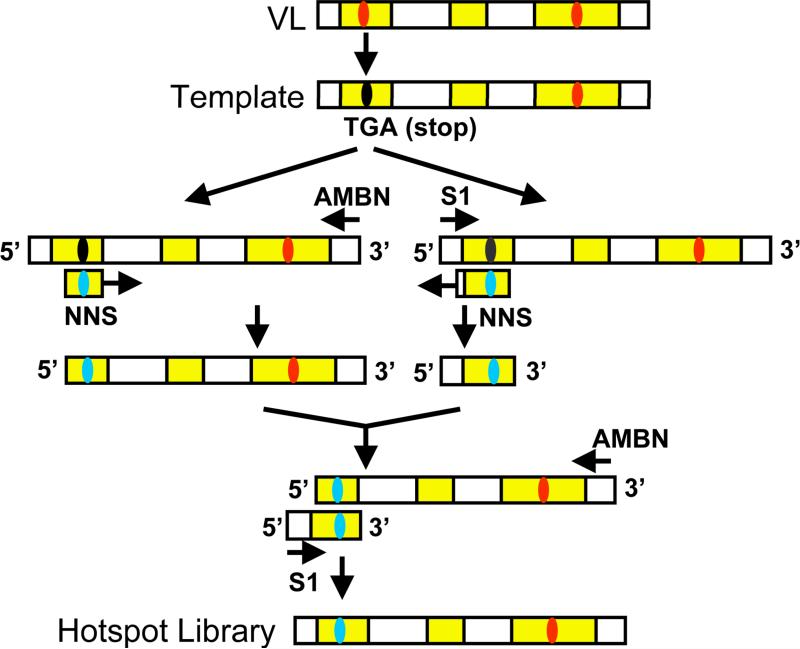
Here we use a germline hotspot (Ser30-Asn31) located in the light chain CDR1 of RFB4 as an example. We designed DNA oligomers to generate a library randomizing 6 nucleotides (2 consecutive amino acids). We used degenerate oligomers with the sequence NNS (N randomizing with all four nucleotides and S introducing only C or G).
Subclone scFv into the SfiI and NotI sites of phagemid pCANTAB5E to display single chain Fv (scFv) RFB4 on the surface of bacteriophage M13 according to the manufacturer's instruction (see Note 7).
Modify the scFv sequence by inserting a BspHI site (TCA TGA) containing a stop codon (underlined) at the hotspot positions using site-directed mutagenesis (QuickChange site-directed mutagenesis kit). Use the resulting phagemids pCANTAB5E-RFB4 VL30/31BspHI as a template to randomize two amino acid residues in a two-step overlap extension PCR (Fig. 3) (see Note 8).
The following oligonucleotides were used: S1, 5'-CAACGTGAAAAAATTATTATTCGC-3'; RFB4VL30/31F, 5'-AGTCAGGACATTNNSNNSTATTTAAACTGG-3'; RFB4VL30/31R, 5'-CCAGTTTAAATASNNSNNAATGTCCTGACT-3' and AMBN, 5'-GCTAAACAACTTTCAACAGTCTATGCGGCAC-3'.
In the first PCR, add 50 pg of the phagemid pCANTAB5E-RFB4 BspHI as the template and put it into two separate PCR tubes. In Tube One, add 20 pmol of DNA oligomers AMBN along with 20 pmol of the DNA oligomers RFB4VL30/31F. In Tube Two, add 20 pmol of DNA oligomers S1 along with 20 pmol of the DNA oligomers RFB4VL30/31R. Mix the template and oligonucleotides with two Ready-To-Go PCR beads in a 50-μL volume and then cycle using the following profile: 1 cycle at 95°C for 5 min, followed by 30 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min.
The PCR products in Tube One and Tube Two contain the mutations. Purify each PCR product using a Qiagen Quick Spin column and then quantify by visualization on a 1% agarose gel.
Add 25 pg of PCR product from Tube One and 25 pg of PCR product from Tube Two. Use the combined 1st PCR products (50 pg) as a template in a second PCR. In this reaction, use 20 pmol of the DNA oligomer S1 and AMBN. Mix the primers and PCR products with two PCR beads in a 50-μL volume and cycle using the profile described above. The reaction generates an 884-bp insert library.
Digest the PCR product with SfiI and NotI, and purify by using a QIAquick column.
Ligate the purified PCR product (150 ng) with 250 ng of pre-cut phage-display vector pCANTAB 5E and desalt using a Qiagen Quick Spin column.
Fig. 3.
Randomizing a hotspot in Fv by two-step overlap extension PCR. A germline hotspot (Ser30-Asn31) (highlighted) located in the light chain CDR1 of RFB4 (Fig. 1) is used as an example to show the procedure of randomizing a hotpsot by PCR. The scFv sequence was modified by inserting a BspHI site (TCA TGA) containing a stop codon at the hotspot positions using site-directed mutagenesis. Use the resulting phagemids as a template to randomize two amino acid residues in a two-step overlap extension PCR with degenerate oligomers with the sequence NNS (N randomizing with all four nucleotides and S introducing only C or G).
3.3. E. coli Electroporation and Sequence Confirmation of Library Phage
The final phase of library construction is the introduction of scFv-dsDNA into an E. coli host containing an F’ episome such as TG1 cells. M13 bacteriophage infects cells via the F-pilus. Therefore it is essential that host strains carry the F-pilus, which is encoded by the F’ episome. When cells carrying phagemids are infected with the appropriate filamentous helper bacteriophage such as M13K07, the library is packaged into phage particles. A major limitation to the production of highly diverse phage-displayed libraries has been the transformation efficiency into TG1 cells. Electroporation has been widely used. Transformation is directly related to ligation efficiency and the purity of the DNA to be transformed. We have used the TG1 E. coli strain for high-efficiency DNA transformation.
Use 50 ng of the ligation to transform TG1 electroporation-competent E. coli cells. Each transformation should produce a phage-display library containing more than 2 × 104 independent clones by counting the individual colonies grown on the plates with 0, 1:10, 1:100 or 1:1000 dilutions of cells (see Note 9).
The scFv expressed as a fusion with the minor phage coat protein pIII after rescue with a helper phage.
Sequence the scFv inserts of 10 individual clones using dideoxy method with primers S1 and AMBN. Perform DNA sequencing using PE Applied Biosystems Big Dye Terminator Cycle Sequencing kit.
Run and analyze the samples on a PE Applied Biosystem Model 310 automated sequencer.
Confirm the successful randomization of the hotspot in VL CDR1 by comparing all the sequences of randomly selected clones.
3.4. Phage Panning and Analysis
The phage panning can be done on purified proteins such as Fc fusion proteins or cells. The panning strategy has to be determined by each investigator. In this example, we decided to use cells expressing native CD22 molecules on the cell surface for phage panning (called “biopanning”). By doing that, we should not miss mutant antibodies recognizing conformational or carbohydrate epitopes. It may be especially important in this case because the precise epitope of the RFB4 antibody on CD22 has not been elucidated.
To mimic somatic hypermutation in the immune response, which optimizes the differential binding affinity for an antigen relative to other molecules, we used a strategy called “subtractive biopanning”. We screened for high affinity phage that could bind to CD22-positive Daudi but not CD22-negative MCF7 cells. To reduce the number of phage that bound nonspecifically, subtractive biopanning was performed first on CD22-negative MCF7 cells, and then enrichment was performed on CD22-positive Daudi cells as described here. This process is reiterated over several rounds to enrich for a pool of specific binders. A successful selection will enrich increasingly more specific phage clones to the target protein with each round. The panning will continue until desired clones with increased affinity are enriched.
Collect CD22-negative MCF7 cells (>5 × 107) and resuspend in 10 mL of cold panning blocking buffer (DPBS + 5% BSA + 0.1% NaN3).
Add phage (~ 1 × 1012 cfu) to the cell suspension and rotate the mixture slowly at 4°C for 60 min.
Pellet the cells and incubate the supernatant (10 mL) containing unbound phage with CD22-positive Daudi cells (1 × 106) and rotated slowly for 60 min at 4°C.
Pellet the Daudi cells and resuspend the cells in 10 mL of cold panning blocking buffer.
Wash the Daudi cells 6 times with 1 mL of cold panning blocking buffer for first and second round of panning.
At the third panning, wash the cells 16 times.
Elute bound phage by resuspending the washed cells in 0.5 mL of ice-cold 100 mM Glycine-HCL (pH2.5) and incubate on ice for 10 min.
Pellet Daudi cells and transfer the supernatant containing the eluted phage to a tube containing 100 μL of 1 M Tris (pH 8).
Titer the eluted phages to determine the number of phage captured.
Amplify the eluted phage (~0.5 mL) by reinfecting E. coli TG1 for use in the next round of panning (see Note 10).
3.5. Enzyme-linked Immunosorbent Assay (ELISA)
The progress of an affinity selection is conveniently monitored by phage ELISA calculating the enrichment ratio: the number of phage bound to a well coated with target protein divided by the number of phage bound to a well coated with an irrelevant protein. The affinity panning is generally carried through three or four rounds.
We describe a phage ELISA protocol that is used to identify phage-displayed scFv specific to the target protein (Fig. 4). The protein target (CD22-Fc) and negative controls (CD30-Fc or CD25-Fc) are immobilized on an immunoplate. Phage particles secreted by single phage clones are added to the target protein and control proteins under conditions appropriate for binding. Following a brief incubation period, the immunoplate is washed and detected by M13-specific antibodies conjugated to horseradish peroxidase (HRP). Upon addition of the HRP substrate, positive clones are confirmed by a spectrophotometric readout. Antibody-phage that generate an ELISA signal 10-fold higher for the CD22-Fc relative to CD30-Fc or CD25-Fc are considered as positive clones. The phagemid DNA of the positive phage clones is then purified and sequenced. We recommend sequencing 20 or more unique clones to generate a complete profile.
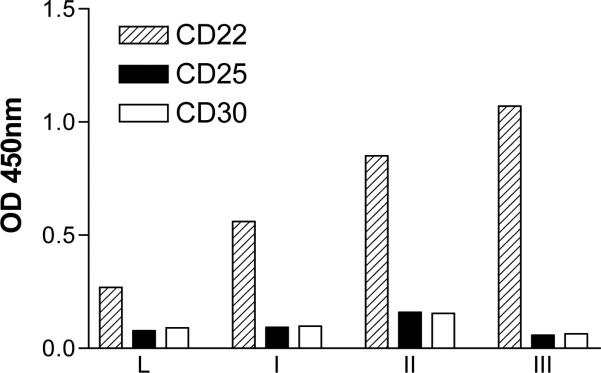
Fig. 4.
Pooled phage ELISA on CD22-Fc fusion proteins. Plates were coated with the indicated CD22-Fc fusion proteins. CD25- and CD30-Fc fusion proteins were used as background controls. The binding of the pooled phage population from the initial library (L) or phages eluted after each round of subtractive biopanning (I-III) is shown. (This figure reproduced in part from J. Biol. Chem. 280(1):607-617, January 7, 2005).
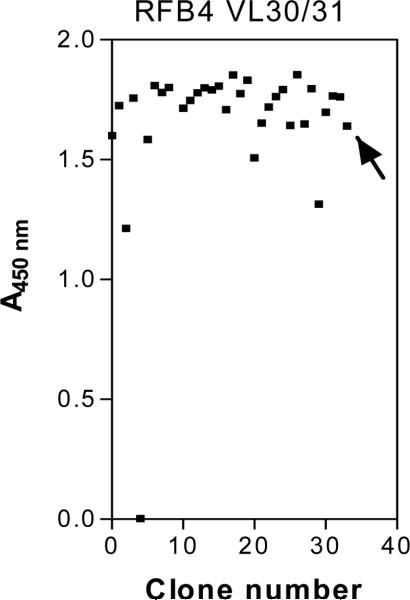
Before the addition of helper phage M13K07 at the end of each round of panning rescue, an aliquot of the phage-infected E. coli culture is taken and plated for single colonies on selection medium to titer the number of captured phage and monitor phage enrichment. Each single colony represents progeny from a single E. coli cell that harbors a phagemid expressing a unique scFv. Representative transformants from each round are subsequently tested for specific binding to the target protein by a phage ELISA. The ELISA signals on target protein are increased after each round of panning but the non-specific background signals on control proteins are not. Individual phage clones are prepared and tested for their ability to bind to CD22-Fc by ELISA (Fig. 5). In our case, we tested 33 clones and grouped them into four types: 1) 23 clones had high ELISA signals; 2) 4 clones had ELISA signals comparable to the original scFv phage (marked by an arrow); 3) 4 clones had ELISA signals lower than the starting parental phage; and 4) 2 clones had no signal. The most frequent residues were Gly30-Arg31 and Arg30-Gly31. Each of these appeared 6 times. More than one codon was used in the clones containing Gly30-Arg31 or Arg30-Gly31, indicating that there was a strong selection for these amino acid residues.
Prepare phages from single colonies of E. coli TG1 containing phagemids selected in biopanning.
Coat 96-well ELISA plates with goat anti-human IgG (0.5 μg/50 μL/well) in 50 mM bicarbonate buffer (pH9.5) at 4°C overnight.
Block the plate with 1% of BSA in PBS at room temperature for 1 hr.
Wash the wells with PBS containing 0.05% Tween 20 (PBST).
Add CD22-Fc fusion proteins and other Fc fusion proteins (e.g., 5 μg/mL in PBST containing 1% BSA, at 50 μL/precoated well (see Note 11).
After 1 hr at room temperature, wash the plates four times with PBST.
Incubate each well for 1 hr with 50 μL of a 1:10 dilution of culture supernatant containing phage clones (~109 phages/well).
After washing four times with PBST, incubate each well with HRP-conjugated anti-M13 antibody (1:10,000) at room temperature.
Develop assays with TMB/H2O2 (tetramethyl benzidine) substrate.
After 15 min incubation in the dark, stop the reaction with 100 μL of 1N sulfuric acid and read the absorbance spectro-photometrically at 450 nm.
Fig. 5.
Monoclonal phage scFv were tested for their binding to immobilized CD22-Fc fusion protein by ELISA. The wild-type phage scFv (marked by an arrow) was used as an internal standard. (This figure reproduced in part from J. Biol. Chem. 280(1):607-617, January 7, 2005).
3.6. Affinity Measurement by Flow Cytometry
Affinity (KD) can be measured by BIAcore on purified antigen proteins or by flow cytometry on cells. We describe measurement on Daudi cells using scFv by flow cytometry.
Incubate 5 × 105 Daudi cells in 0.5 mL of FACS blocking buffer containing scFv in different concentrations (e.g., 0.05nM, 0.1nM, 0.5nM, 1nM, 5nM, 10nM, 50nM) on ice for 60 min.
Wash cells two times with FACS blocking buffer containing 5% BSA plus 0.1% NaN3 in PBS.
Add 5 μg of anti E-Tag antibody to each sample.
Incubate the mixture on ice for 60 min and then wash twice with FACS blocking buffer.
Add a goat-anti-mouse-PE-labeled antibody (1:200), and incubate cells for 30 min on ice.
Wash cells two times, and perform analysis in a FACSCalibur machine.
Acquire data using Cell Quest software and analyze the data using FlowJo software.
Determine equilibrium constants and Scatchard plots by using the Marquardt-Levenberg algorithm for non-linear regression with the Prism sofware (see Note 12).
Table I.
Phage selection in subtractive biopanning
| Library for panning | Panning cycle | Input | Output | Enrichment over previous round | Total enrichment |
|---|---|---|---|---|---|
| RFB4LibVL30/31 | 1 | 1 × 1012 | 1.4 × 104 | - | |
| 2 | 1 × 1012 | 7 × 105 | 50 | ||
| 3 | 1 × 1012 | 1.5 × 108 | 214 | 10,700 |
(This table reproduced in part from J. Biol. Chem. 280(1):607-617, January 7, 2005).
Acknowledgements
This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
We have found that a directed evolution approach targeting a different hotspot in each generation allows us to sequentially increase the activity of an antibody. This uphill-climbing strategy has the major advantage that in each generation the library size is small and easy to make whereas it is technically very difficult to produce large phage libraries where many nucleotides and amino acids in the Fv are randomly mutated.
pCANTAB-5E is currently discontinued at GE Healthcare. We could provide the vector upon request. Other comparable phagemids such as pHEN1 for scFv (12) and pComb3 for Fab (13) can be used as well.
The panning strategy should be determined empirically. If cells are not used for phage panning or analysis, then no mammalian cell culture work is needed.
We prefer a Fc fusion protein as it uses a well-established expression system in HEK-293T cells and the one-step purification of Fc fusion proteins with a Protein A column is straightforward (4, 7). We can provide a detailed protocol for expression and purification of Fc fusion proteins upon request. Other choice of target proteins may suffice.
Most FACS core facilities may FACScalibur. However, other brands of flow cytometers may suffice.
Antibody variable regions of human and mouse origins as well as other species can be compared to their corresponding germline sequences.
As construction of scFv phage display libraries and other technical details about phage display were also well described in Phage display: a laboratory manual (18) and other literature. We do not provide such details here.
We have found that adding a stop codon and using the mutant containing a stop codon as a template for the following 2-step overlap extension PCR is very important to avoid over representation of wild type scFv in the library (4, 5, 7).
We typically have 104 – 106 independent clones per transformation. Because it is only necessary to produce a library of 1024 (32 × 32) independent clones to randomly mutate a hotspot sequence (e.g., AGCAAT) to NNSNNS, the size of the phage display library is much larger than the one required.
The overall enrichment for library at the end of the third round panning is often very large (see Table I).
We prefer a Fc fusion protein. However, other choice of antigen proteins may suffice if directly coated on ELISA plates.
We use Geometrical Mean values of flow cytometry to calculate KD. If the concentration of scFv in the culture supernatant is not high enough for affinity measurement by flow cytometry, we convert a scFv into an immunotoxin or whole IgG molecule before measuring its binding affinity.
References
- 1.Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 2.Presta LG. Engineering of therapeutic antibodies to minimize immunogenicity and optimize function. Adv. Drug Deliv. Rev. 2006;58:640–656. doi: 10.1016/j.addr.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 3.Sergeeva A, Kolonin MG, Molldrem JJ, Pasqualini R, Arap W. Display technologies: application for the discovery of drug and gene delivery agents. Adv. Drug Deliv. Rev. 2006;58:1622–1654. doi: 10.1016/j.addr.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho M, Kreitman RJ, Onda M, Pastan I. In vitro antibody evolution targeting germline hot spots to increase activity of an anti-CD22 immunotoxin. J. Biol. Chem. 2005;280:607–617. doi: 10.1074/jbc.M409783200. [DOI] [PubMed] [Google Scholar]
- 5.Chowdhury PS, Pastan I. Improving antibody affinity by mimicking somatic hypermutation in vitro. Nat. Biotechnol. 1999;17:568–572. doi: 10.1038/9872. [DOI] [PubMed] [Google Scholar]
- 6.Beers R, Chowdhury P, Bigner D, Pastan I. Immunotoxins with increased activity against epidermal growth factor receptor vIII-expressing cells produced by antibody phage display. Clin. Cancer Res. 2000;6:2835–2843. [PubMed] [Google Scholar]
- 7.Salvatore G, Beers R, Margulies I, Kreitman RJ, Pastan I. Improved cytotoxic activity toward cell lines and fresh leukemia cells of a mutant anti-CD22 immunotoxin obtained by antibody phage display. Clin. Cancer Res. 2002;8:995–1002. [PubMed] [Google Scholar]
- 8.Neuberger MS, Milstein C. Somatic hypermutation. Curr. Opin. Immunol. 1995;7:248–254. doi: 10.1016/0952-7915(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 9.Neuberger MS, Ehrenstein MR, Klix N, Jolly CJ, Yelamos J, Rada C, Milstein C. Monitoring and interpreting the intrinsic features of somatic hypermutation. Immunol. Rev. 1998;162:107–116. doi: 10.1111/j.1600-065x.1998.tb01434.x. [DOI] [PubMed] [Google Scholar]
- 10.Foote J, Eisen HN. Kinetic and affinity limits on antibodies produced during immune responses. Proc. Natl. Acad. Sci. USA. 1995;92:1254–1256. doi: 10.1073/pnas.92.5.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang CL, Harper RA, Wabl M. Genome-wide somatic hypermutation. Proc. Natl. Acad. Sci. USA. 2004;101:7352–7356. doi: 10.1073/pnas.0402009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoogenboom HR, Griffiths AD, Johnson KS, Chiswell DJ, Hudson P, Winter G. Multi-subunit proteins on the surface of filamentous phage: methologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 1991;19:4133–4137. doi: 10.1093/nar/19.15.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbas CF, Kang AK, Lerner RA, Benkovic SJ. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc. Natl. Acad. Sci. USA. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jolly CJ, Wagner SD, Rada C, Klix N, Milstein C, Neuberger MS. The targeting of somatic hypermutation. Semin. Immunol. 1996;8:159–168. doi: 10.1006/smim.1996.0020. [DOI] [PubMed] [Google Scholar]
- 15.Kabat EA, Wu TT, Reid-Miller M, Perry HM, Gottesman KS, Foeller C. Sequences of Proteins of Immunological Interest. 5th ed. NIH; Bethesda, MD: 1991. [Google Scholar]
- 16.Lefranc MP. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 2001;29:207–209. doi: 10.1093/nar/29.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitelegg NR, Rees AR. WAM: An improved algorithm for modelling antibodies on the WEB. Protein Eng. 2000;13:819–824. doi: 10.1093/protein/13.12.819. [DOI] [PubMed] [Google Scholar]
- 18.Barbas CF, Burton DR, Scott JK, Silverman GJ. Phage Display: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2000. [Google Scholar]