SUMMARY
Eukaryotic cells possess many transcriptionally regulated mechanisms to alleviate the nucleosome barrier. Dramatic changes to the chromatin structure of Drosophila melanogaster Hsp70 gene loci are dependent on the transcriptional activator, heat shock factor (HSF), and poly(ADP-ribose) polymerase (PARP). Here, we find that PARP is associated with the 5′ end of Hsp70, and its enzymatic activity is rapidly induced by heat shock. This activation causes PARP to redistribute throughout Hsp70 loci and Poly(ADP-ribose) to concurrently accumulate in the wake of PARP’s spread. HSF is necessary for both the activation of PARP’s enzymatic activity and its redistribution. Upon heat shock, HSF triggers these PARP activities mechanistically by directing Tip60 acetylation of histone H2A lysine 5 at the 5′ end of Hsp70, where inactive PARP resides before heat shock. This acetylation is critical for the activation and spread of PARP as well as for the rapid nucleosome loss over the Hsp70 loci.
INTRODUCTION
Packaging of eukaryotic DNA into the nucleus relies on compaction of DNA into nucleosomes and higher order chromatin structures. Consequently, critical DNA sequences can be occluded from proteins that function by accessing particular DNA elements. Transcription is one such nuclear process that can be significantly inhibited by the barriers created by nucleosomes. In vitro studies demonstrate that nucleosomes are sufficient to block transcription initiation and also impair efficient transcription elongation by RNA Polymerase II (Pol II) (Knezetic and Luse, 1986; Lorch et al., 1987). Genome-wide, in vivo studies demonstrate that nucleosome disruption and turnover is greatest at the most highly transcribed mRNA-encoding genes (Deal et al., 2010). These results indicate that eukaryotic cells possess mechanisms to alleviate the nucleosomal barrier to efficiently transcribe genes.
Addressing how Pol II traverses the nucleosome has resulted in the discovery of unique features of transcriptional regulation. Eukaryotes utilize five general mechanisms to change the occupancy, position, structure, or composition of nucleosomes. First, chromatin-remodeling complexes change the position and occupancy of nucleosomes through their ability to slide entire nucleosomes as well as remove or transfer individual histones (Saha et al., 2006). Second, histone chaperones facilitate transcription by the disassembly and reassembly of nucleosomes in the path of Pol II (Das et al., 2010). Third, the composition of individual histones are changed by the numerous posttranslational modifications including acetylation, methylation, phosphorylation, ubiquitination, and sumoylation, which can either directly affect chromatin structure or promote association of chromatin-binding proteins (Taverna et al., 2007). Fourth, canonical core histones can be replaced by histone variants, like H2A.Z, which can change both the composition and structure of chromatin (Talbert and Henikoff, 2010). Lastly, many other nonhistone proteins that bind to nucleosomes or naked DNA can synergize or compete with nucleosomes to affect position, occupancy, structure, and composition (Gilchrist et al., 2010; Saunders et al., 2006).
Model inducible gene systems continue to elaborate the many ways in which these five methods are utilized to coordinate transcription-coupled nucleosome changes and thereby regulate changes in gene expression (Weake and Workman, 2010). The D. melanogaster heat shock (HS)-inducible Hsp70 genes employ these methods (Saunders et al., 2006) as well as an additional method that leads to rapid loss of nucleosomes extending beyond the Hsp70 gene to the flanking scs and scs’ insulator elements (Petesch and Lis, 2008). Interestingly, the nucleosome loss that occurs within the first minute of HS can be uncoupled from active transcription of Hsp70, whereas the additional changes in nucleosome structure by 2 min of HS are dependent on transcription. A directed RNAi screen identified both heat shock factor (HSF) as well as poly(ADP-ribose) polymerase (PARP) as factors necessary for both transcription-independent and -dependent nucleosome loss during HS (Petesch and Lis, 2008). Although both HSF and PARP have been identified to bring about a rapid change in the entire HS locus independently of transcription, the mechanism through which these proteins accomplish this feat is still unknown.
HSF is the master regulatory protein of HS genes and acts as a traditional transcriptional activator of HS genes. Within seconds of HS, HSF trimerizes and binds to a specific HS DNA element (HSE) repeated multiple times within the promoter of Hsp70 (Boehm et al., 2003; Zobeck et al., 2010). Activated HSF is thought to increase transcription by recruiting transcriptional coactivators, such as the mediator complex, P-TEFb, and histone acetyltransferases (HATs) (Lis et al., 2000; Park et al., 2001; Smith et al., 2004). These coactivators are thought to facilitate transcription by regulating additional recruitment and initiation of Pol II, releasing paused Pol II, or altering the structure and composition of nucleosomes to enhance Pol II elongation. It is yet to be determined if HSF-mediated, transcription-independent nucleosome loss is also achieved through a coactivator, and what, if any, are the connections between HSF, coactivators, and PARP.
PARP is an enzyme well studied for its function in DNA repair, apoptosis, transcriptional regulation, as well as altering chromatin structure (Krishnakumar and Kraus, 2010a; Rouleau et al., 2010). PARP’s enzymatic activity uses NAD+ donor molecules to catalyze the polymerization of poly(ADP-ribose) (PAR) onto acceptor proteins as a posttranslational modification. The Zn fingers of PARP bind to many aberrant DNA structures, especially broken DNA, but also bind to nucleosomes (Kim et al., 2004; Krishnakumar and Kraus, 2010a). Damaged DNA, individual histones, and polynucleosomes can activate the enzymatic activity of PARP (Kim et al., 2004; Pinnola et al., 2007). In vivo, PARP primarily targets itself, although other proteins, including histones, are also known to be substrates of PARylation (D’Amours et al., 1999). Even though PARP is also able to affect transcription independently of its enzymatic activity at some genes (Hassa et al., 2001; Pavri et al., 2005), the catalytic activity of PARP is known to be able to affect transcription at many genes, including Hsp70 (Ju et al., 2006; Krishnakumar and Kraus, 2010b; Tulin and Spradling, 2003). PARP and its catalytic activity are necessary for decompaction of chromatin structure at HS loci as well as transcription-independent nucleosome loss at Hsp70 (Petesch and Lis, 2008; Tulin and Spradling, 2003). Although formation of PAR is necessary to bring about changes in chromatin structure at HS loci, little is known about how PARP is activated at Hsp70 and, more so, how this activation can lead to nucleosome loss over such a broad domain.
Here, we map the immediate kinetic changes that occur to PARP and its catalytic product, PAR, at Hsp70 loci immediately following HS. We observe that PARP colocalizes with the first 2 nucleosomes at the beginning of the Hsp70 transcription unit before HS. Upon HS, PARP spreads rapidly throughout the Hsp70 gene and beyond, to the scs’ insulator element, with kinetics similar to those previously measured to track changes in chromatin structure. The activation of PARP leads to accumulation of PAR throughout the locus in the wake of spreading PARP. Additionally, we find that HSF is necessary for both the activation and spread of PARP at Hsp70 following HS and triggers these changes by altering the level of histone acetylation at the site where PARP is found prior to HS. Finally, we identify Drosophila Tip60 (dTip60) as the HAT responsible for increasing acetylation of histone H2A lysine 5 at Hsp70 following HS. This increase in acetylation induces the activation and spread of PARP and a resulting nucleosome loss that is independent of transcription.
RESULTS
PARP Rapidly Redistributes along Hsp70 upon Heat Shock
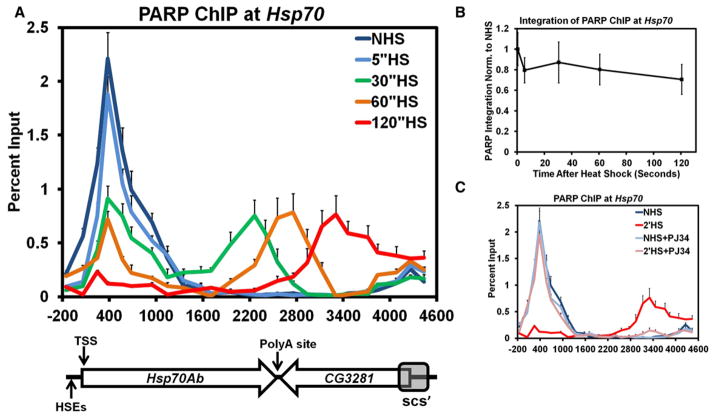
To investigate potential mechanisms by which PARP is able to bring about rapid and drastic changes in chromatin structure at the Hsp70 gene loci upon HS, we first examined PARP’s location on Hsp70 before and in the seconds following HS using chromatin immunoprecipitation (ChIP). In our assay, Drosophila S2 cells are instantaneously heat shocked, cooled, and immediately crosslinked with formaldehyde for durations previously used to characterize changes in nucleosomes (Petesch and Lis, 2008). The immunoprecipitated DNA was measured using quantitative PCR (qPCR) with 25 primer sets spaced, on average, 200 base pairs (bp) apart to canvas the Hsp70 HS loci from the Hsp70Ab promoter to the scs’ insulator element (Figure 1A).
Figure 1. PARP Rapidly Redistributes across theHsp70 Heat Shock Locus upon Heat Shock.
(A) Kinetic ChIP analysis of PARP at the Hsp70Ab HS locus. S2 cells are heat shocked for 0 (dark blue), 5 (light blue), 30 (green), 60 (orange), or 120 s (red). Twenty-five primer sets, spaced approximately 200 bp apart, provide high spatial resolution. The y axis represents the percent of input material immunoprecipitated. The x axis corresponds to base pair units with 0 being the TSS. Error bars represent the SEM of three independent experiments. The diagram below depicts the Hsp70Ab locus, noting the sites of the HSEs, its TSS, the Poly A site (+2343), the downstream gene CG3281, and the site of the scs’ insulator element. The diagram is on the same scale as the x axis. Each subsequent figure labeled as ChIP will be for the Hsp70Ab HS locus and contains the same error bars, y axis, and x axis that correspond to the diagram depicted.
(B) The amount of PARP detected by ChIP at the Hsp70Ab HS locus stays constant during the first 2 min of HS. The ChIP values for each of the 5 HS time points were integrated across the region depicted in (A) using the 25 percent input values as points to trapezoids. Each sum was normalized back to the NHS summation and plotted on the y axis. The x axis represents the elapsed time following HS in seconds. Error bars represent the propagated error associated with the summation of the SEM of each percent input.
(C) ChIP of PARP in the presence of PJ34, a catalytic inhibitor of PARP. Untreated NHS (dark blue) and 2′ HS (red) S2 cells are compared to those NHS (light blue) and 2′ HS (pink) cells pretreated with 300 nM PJ34 for 10 min. Error bars represent the SEM of three independent experiments.
Prior to HS, PARP is bound to the Hsp70 gene downstream of the transcription start site (TSS) (Figure 1A, dark blue), and overlaps with the previously mapped position of the first 2 well-protected nucleosomes at Hsp70 (Petesch and Lis, 2008). These results are consistent with those studies in Drosophila that detect PARP present at the Hsp70 HS loci (Kotova et al., 2011; Tulin and Spradling, 2003; Zobeck et al., 2010) and studies in humans that show PARP concentrates on regions near the TSS (Krishnakumar et al., 2008). Within 5 s of HS (Figure 1A, light blue) PARP begins to be lost from its site occupied prior to HS, and it decreases monotonically by 30, 60, and 120 s of HS (Figure 1A, green, orange, and red, respectively). Interestingly, the rate of PARP loss from this region mirrors the rate of nucleosome loss previously measured at this region. More surprising is the transient accumulation of detectable PARP ChIP signal progressively further downstream by 30, 60, and 120 s of HS, extending all the way to the scs’ insulator element. These results can be explained by two simple models: (1) local PARP molecules are lost and new PARP is recruited to downstream sites after HS, or (2) prebound PARP rapidly spreads across the Hsp70 gene loci upon HS.
Previous live cell imaging experiments of PARP at Hsp70 during HS show that the total amount of PARP at the loci, both unbound and bound to chromatin, remains unchanged from the non-heat-shock (NHS) state (Zobeck et al., 2010). To determine if the amount of PARP associated with the chromatin at the Hsp70Ab locus remains constant throughout the HS time course, we took the integral of each PARP ChIP time point curve (Figure 1B). The level of total PARP ChIP signals for 5, 30, 60, and 120 s of HS does not significantly change from the NHS time point. This is in contrast to most factors which progressively accumulate at Hsp70 loci following the first few minutes of HS (Boehm et al., 2003). Together these experiments are consistent with a model in which PARP is prebound to the Hsp70 loci before HS and this prebound PARP rapidly redistributes upon HS.
Since PARP’s catalytic activity is necessary for the changes in chromatin structure that occur at Hsp70 upon HS (Petesch and Lis, 2008), we tested whether PARP’s enzymatic activity was also required for rapid loss and redistribution of PARP upon HS. S2 cells treated with PJ34, a catalytic inhibitor of PARP, did not alter the position or level of PARP present at the Hsp70 gene before HS, indicating that PARP is bound to the Hsp70 promoter independently of its catalytic activity (Figure 1C). Treatment with PJ34 for 10 min prior to a 2 min HS prevented PARP redistribution. The loss of PARP from its NHS site upon HS is consistent with in vitro data showing that PARP binds to chromatin through its DNA binding domain and dissociates following activation of its catalytic domain (Kim et al., 2004; Poirier et al., 1982; Wacker et al., 2007).
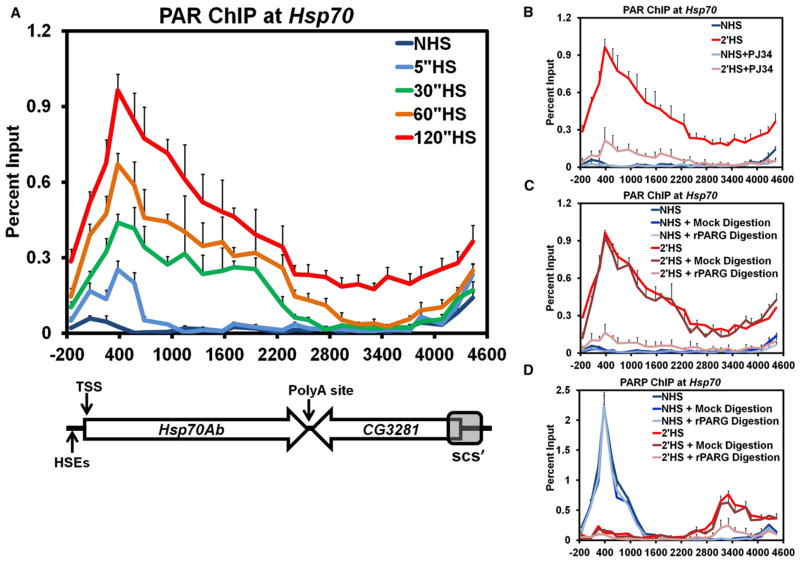
PAR Rapidly Accumulates in the Wake of PARP and Tethers PARP to the Locus following Heat Shock
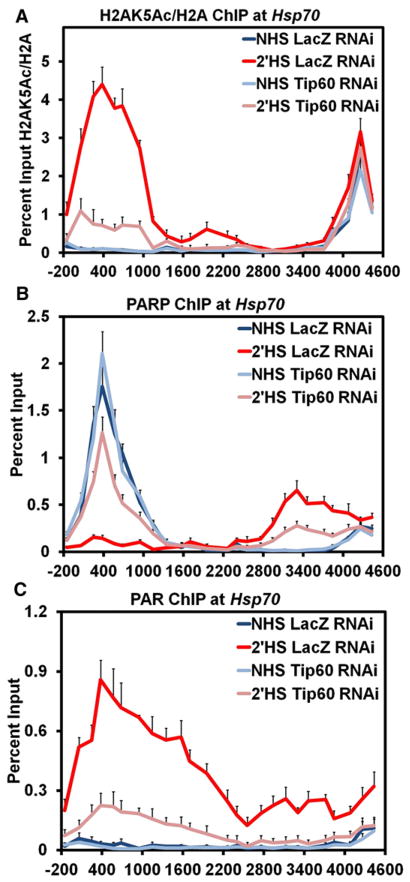
As the catalytic activation of PARP is necessary for the loss and redistribution of PARP along the Hsp70Ab locus, we employed the same kinetic ChIP assay outlined in Figure 1 to track the changes in PAR. Prior to HS, PAR is absent from Hsp70 (Figure 2A, dark blue); immediately following 5 s of HS, PAR begins to accumulate at the 5′ end of Hsp70 (Figure 2A, light blue). Interestingly, the initial site of PAR polymerization overlaps with PARP’s position before HS. By 30, 60, and 120 s of HS, PAR monotonically increases at the 5′ end of Hsp70 and, more surprisingly, accumulates further downstream until it reaches the scs’ insulator element by 120 s of HS (Figure 2A, green, orange, and red, respectively). As expected, the accumulation of PAR at Hsp70 after HS is dependent on both PARP (Figure S2A) and PARP’s catalytic activity (Figure 2B). The position of PARP before HS and its downstream position by 30, 60, and 120 s of HS bookends the region of PAR accumulation at the corresponding time point of HS. These kinetic results demonstrate that PAR accumulates in the wake of PARP spreading along the Hsp70Ab locus.
Figure 2. PAR Rapidly Accumulates across theHsp70 Heat Shock Locus upon Heat Shock.
(A) Kinetic ChIP analysis of PAR at the Hsp70Ab HS locus, as in Figure 1A. S2 cells are heat shocked for 0 (dark blue), 5 (light blue), 30 (green), 60 (orange), or 120 s (red). Error bars represent the SEM of three independent experiments for all four panels.
(B) ChIP of PAR in the presence of the catalytic inhibitor of PARP, PJ34. Untreated NHS (dark blue) and 2′ HS (red) S2 cells are compared to NHS (light blue) and 2′ HS (pink) cells pretreated with 300 nM PJ34 for 10 min.
(C) ChIP of PAR at the Hsp70Ab HS locus with PARG digestion. ChIP extracts prepared for PAR IP were treated with 1200 nM of purified recombinant rat PARG (rPARG) prior to IP with either NHS (light blue) or 2′ HS samples (pink). These values were compared to control NHS (medium blue) or 2′ HS (dark red) samples that were mock-treated at 37°C for 30 min. The values of the mock treatment were not significantly different from those obtained without any additional treatment for NHS (dark blue) or 2′ HS (red).
(D) ChIP for PARP with rPARG digestion as in (C).
Two nonmutually exclusive models can explain the results from the kinetic ChIP analysis of PARP and PAR. The first model asserts that PARP progressively modifies the underlying histones, or another component of chromatin, that fall withinits path and cross-linking during ChIP captures PARP directly interacting with chromatin. The second model asserts that activated PARP modifies itself and the polymerization of PAR results in continuously further reaching contacts with distal chromatin and crosslinking captures PARP bridged indirectly to chromatin through PAR.
To test these two models, we purified recombinant poly(ADP-ribose) glycohydrolase (PARG) and confirmed its ability to cleave PAR in ChIP extracts from crosslinked cells (Figure S2B). ChIP for PAR after PARG treatment of crosslinked material, in comparison to the mock treated samples, confirmed catabolism of PAR at Hsp70 (Figure 2C). ChIP for PARP with PARG treated extracts resulted in no change in the ChIP signal under NHS conditions (Figure 2D), supporting the conclusion that PARP bound to the 5′ end of Hsp70 before HS is inactive and directly bound to chromatin. However, when 2 min HS samples were treated with PARG the level of PARP ChIP decreased in comparison to the mock treated sample (Figure 2D, pink compared to dark red). These results indicate that PARP detected by ChIP downstream during HS arises from crosslinks that capture PAR bridging the interaction between PARP and chromatin and not from a direct interaction with the chromatin. Similar results are also found after 30 s of HS (Figure S2C). These results are additionally supported by the kinetics of PARP’s redistribution more closely correlating with a constant rate of expansion in 3 dimensions (Figure S2D) rather than a 1-dimensional movement along the chromatin fiber (Figure S2E).
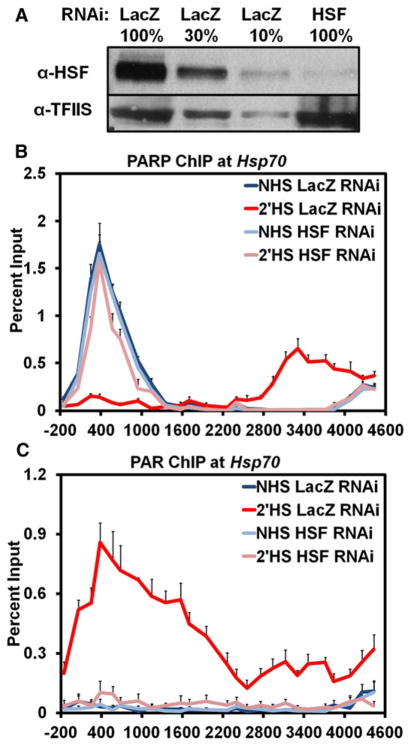
HSF Is Necessary for Activation and Spread of PARP Following Heat Shock
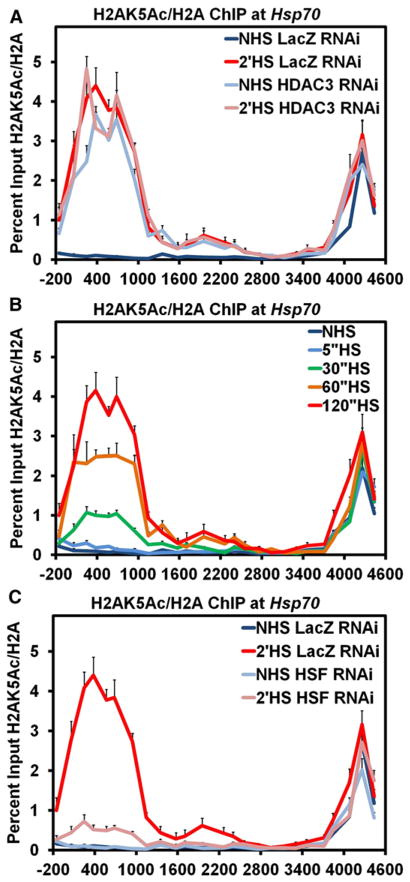
Our initial RNAi screen for factors that affected the loss of chromatin structure at Hsp70 identified both PARP and HSF as critical components in both the transcription-dependent and -independent changes that occurred following HS (Petesch and Lis, 2008). To determine if these two factors were working together in an ordered pathway, we RNAi-depleted either HSF or PARP and used ChIP to assay if the other factor was affected. When PARP was either depleted or inhibited, the recruitment of HSF to the promoter of Hsp70 following HS was not affected (Figures S1A–S1D). In contrast, HSF knockdown (Figures 3A and S3) prevented the redistribution (Figure 3B) and activation (Figure 3C) of PARP at Hsp70 following HS, but not PARP deposition prior to HS. These results order a mechanism whereby HSF recruitment to Hsp70 results in the rapid activation and spread of PARP.
Figure 3. HSF Is Necessary for the Activation and Redistribution of PARP at theHsp70 Heat Shock Locus following Heat Shock.
(A) Western blot confirming the RNAi knockdown of HSF. A serial dilution of the LacZ RNAi control cells were used to quantify the extent of HSF knockdown, shown in the last lane, with 100% equivalent to 5 × 106 cells. The top panel shows the immunoblot of HSF, and the bottom panel shows the immunoblot of TFIIS as a loading control.
(B) ChIP of PARP at the Hsp70Ab HS locus in HSF RNAi cells. S2 cells RNAi depleted of HSF are heat shocked for either 0 (light blue) or 2 min (pink) and compared to those LacZ RNAi NHS (dark blue) and 2′ HS (red) cells. Error bars represent the SEM of three independent experiments for (B) and (C).
(C) ChIP of PAR at the Hsp70Ab HS locus in HSF RNAi cells as in (B).
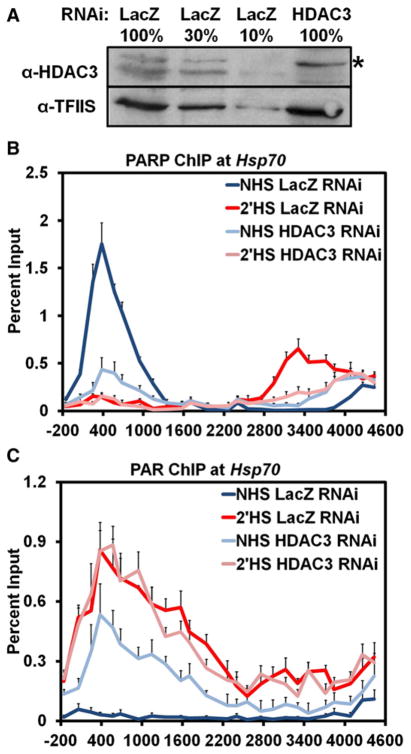
The Activity of HDAC3 Maintains PARP Inactivity at Hsp70 Prior to Heat Shock
Activated HSF binds to multiple HSEs at the promoter of Hsp70 upon HS and directs changes in transcription by signaling to downstream paused Pol II through multiple coactivators. We hypothesized that HSF recruitment to Hsp70 also triggers a signaling mechanism through its coactivators that leads to activation of downstream PARP and the resulting transcription-independent loss of nucleosomes.
To identify a potential coactivator responsible for activating PARP upon HS, we examined data from our previous RNAi screen looking for factors that affected the loss of chromatin structure at Hsp70 before and after HS. Of the 28 factors tested, only one factor, HDAC3, had a significant effect on the loss of chromatin structure prior to HS (Petesch and Lis, 2008). Interestingly, RNAi depletion of HDAC3 in NHS cells results in a loss in chromatin structure to the level seen by 1 min of HS when the initial, transcription-independent loss occurs. To determine if HDAC3 was responsible for the maintenance of PARP in an inactive state at Hsp70 prior to HS, we RNAi-depleted HDAC3 (Figure 4A) and looked by ChIP to determine if PARP or PAR was affected in NHS conditions. RNAi depletion of HDAC3 resulted in loss of PARP from its 5′ binding site prior to HS and redistribution further downstream (Figure 4B, light blue). Strikingly, HDAC3 knockdown under NHS conditions also resulted in the accumulation of PAR across the Hsp70Ab locus, indicating HDAC3 is responsible for the maintenance of inactive PARP bound to Hsp70 under NHS conditions (Figure 4C, light blue). HS and HDAC3 RNAi did not result in an additive effect as only a small increase in PAR accumulation was observed upon HS, comparable to normal HS levels. This increase in PAR upon HS is likely due to the activation and loss of the remaining PARP at the 5′ end in HDAC3 RNAi NHS cells. Similar results were obtained by treating S2 cells with trichostatin A (TSA), an inhibitor of class I and II histone deacetylases, which includes HDAC3 (Figures S4A and S4B). Interestingly, the inhibition of the enzymatic activity of HDAC3 is sufficient to bring about activation of PARP at Hsp70 independent of HS-induced activated binding of HSF (Figure S4C).
Figure 4. HDAC3 Knockdown Activates PARP at theHsp70 Heat Shock Locus under Non-Heat-Shock Conditions.
(A) Western blot confirming the RNAi knockdown of HDAC3 as in Figure 3A, with the top panel showing the immunoblot of HDAC3 and the bottom panel showing the immunoblot of TFIIS as a loading control. * Indicates a slower-migrating, cytoplasmic, and cross-reactive protein to the HDAC3 antibody that is not affected by HDAC3 RNAi.
(B) ChIP of PARP at the Hsp70Ab HS locus in HDAC3 RNAi cells. S2 cells RNAi depleted of HDAC3 were heat shocked for either 0 (light blue) or 2 min (pink) and compared to those LacZ RNAi NHS (dark blue) and 2′ HS (red) cells. Error bars represent the SEM of three independent experiments for (B) and (C).
(C) ChIP of PAR at the Hsp70Ab HS locus in HDAC3 RNAi cells, as in (B).
Heat Shock Factor Facilitates Rapid Acetylation of Histone H2A at Lysine 5 upon Heat Shock
HDAC3 has multiple in vivo targets of deacetylation (Karagianni and Wong, 2007), but perhaps its best known targets are the N-terminal tails of histones H2A at lysine 5 (H2AK5) and H4 at lysines 5 and 12 (H4K5 and H4K12) (Johnson et al., 2002). We depleted cells of HDAC3 by RNAi knockdown and used ChIP assays to examine if either H2AK5Ac or H4K5Ac and H4K12Ac were affected using an antibody that specifically detects H2AK5Ac or an antibody recognizing 4 acetylation marks on H4 (K5, K8, K12, and K16) (Egelhofer et al., 2011). Under NHS conditions, HDAC3 knockdown results in an increase of H2AK5Ac as compared to control NHS LacZ RNAi cells (Figure 5A) and also moderate increases in the acetylation of H4 (Figure S5A). Again, similar results were found using TSA inhibition (Figures S5B and S5C).
Figure 5. HSF Directs Acetylation of Histone H2A Lysine 5 upon Heat Shock at theHsp70 Heat Shock Locus.
(A) ChIP of H2AK5 acetylation at the Hsp70Ab HS locus in HDAC3 knockdown cells. S2 cells RNAi-depleted of HDAC3 and heat-shocked for either 0 (light blue) or 2 min (pink) are compared to LacZ RNAi cells that were heat shocked for 0 (dark blue) or 2 min (red). The level of H2AK5 acetylation is normalized to ChIP values for histone H2A and plotted on the y axis. Error bars represent the SEM of three independent experiments for all three panels.
(B) Kinetic ChIP analysis of H2AK5 acetylation normalized to histone H2A as in (A). S2 cells were heat shocked for 0 (dark blue), 5 (light blue), 30 (green), 60 (orange), or 120 s (red).
(C) ChIP of H2AK5 acetylation normalized to H2A as in (A) but with HSF RNAi-depleted cells.
To address if HS-induced loss of nucleosomes is accompanied by rapid histone acetylation, we performed a kinetic ChIP analysis. H2AK5Ac and H4Ac rapidly accumulate at the Hsp70 gene following HS (Figures 5B and S5D, respectively) with kinetics similar to the extremely rapid recruitment of HSF (Boehm et al., 2003). Interestingly, the buildup of histone acetylation overlaps with the site occupied by PARP prior to HS. The rate of histone acetylation is also consistent with both the rates of loss of PARP and nucleosomes from the 5′ end of Hsp70. Additionally, knockdown of HSF severely inhibits the acetylation of H2AK5 and H4 following HS (Figures 5C and S5E). However, inhibition or RNAi depletion of PARP did not affect acetylation following HS (Figures S5F and S5G). Together, these results demonstrate that HSF is necessary for acetylation of H2AK5 and H4 upon HS, and acetylation either under NHS conditions by HDAC3 knockdown or after HS by HSF recruitment both result in the activation of PARP and reduced nucleosome occupancy at Hsp70.
dTip60 Is Responsible for Acetylation of Histone H2A Lysine 5 and Activation of PARP upon Heat Shock
To demonstrate that acetylation of histones H2A or H4 was necessary for the activation and spread of PARP following HS at Hsp70, we sought to identify the responsible histone acetyltransferase (HAT). Previously, knockdown of CBP or Gcn5, two major HATs recruited to Drosophila Hsp70 following HS (Lebedeva et al., 2005; Smith, Petruk, et al., 2004), did not block nucleosomes loss upon HS (Petesch and Lis, 2008). Instead, we sought to target the dTip60 HAT, which was previously shown to acetylate H2AK5 as well as other sites on H4 (Kimura and Horikoshi, 1998; Kusch et al., 2004). Indeed, knockdown of dTip60 (Figure S6A) prevented the full acetylation of H2AK5Ac following a 2 min HS (Figure 6A), but only mildly affected acetylation of H4 (Figure S6B), indicating that dTip60 specifically targets H2AK5 for acetylation at Hsp70 following HS. Strikingly, depletion of dTip60 prevented the full loss of PARP from its 5′ site and its spread further downstream (Figure 6B). Correspondingly, dTip60 RNAi inhibited full activation of PARP at Hsp70 following HS (Figure 6C). Loss of dTip60, however, did not prevent recruitment of HSF to Hsp70 (Figure S6C). These results demonstrate that, upon HS, HSF stimulates dTip60 to specifically acetylate H2AK5 at Hsp70, which results in activation of PARP.
Figure 6. The dTip60 Histone Acetyltransferase Is Necessary for Acetylation of Histone H2A Lysine 5 and Activation of PARP upon Heat Shock.
(A) ChIP of H2AK5 acetylation at the Hsp70Ab HS locus in dTip60 knockdown cells. S2 cells RNAi-depleted of dTip60 and heat-shocked for either 0 (light blue) or 2 min (pink) are compared to LacZ RNAi cells heat-shocked for 0 (dark blue) or 2 min (red). The level of H2AK5 acetylation is normalized to ChIP values for histone H2A and plotted on the y axis. Error bars represent the SEM of three independent experiments for all three panels.
(B) ChIP of PARP in dTip60 knockdown cells. S2 cells RNAi depleted of dTip60 and heat-shocked for either 0 (light blue) or 2 min (pink) are compared to LacZ RNAi treated cells heat-shocked for 0 (dark blue) or 2 min (red).
(C) ChIP of PAR in dTip60 knockdown cells as in (B).
dTip60 Is Necessary for the Loss of Nucleosomes and Full Transcriptional Activation of Hsp70 upon Heat Shock
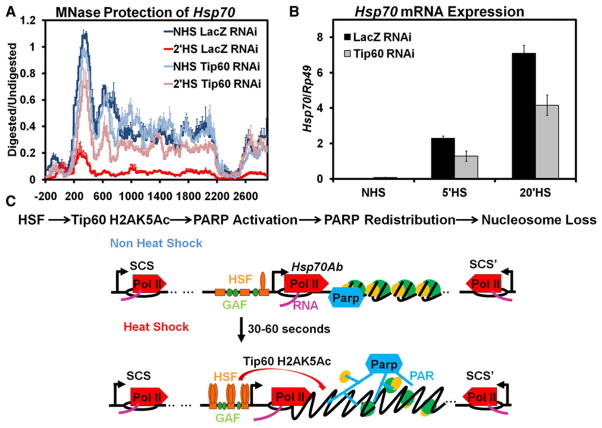
As dTip60 is necessary for the activation of PARP upon HS, changes in chromatin structure at Hsp70 should also be dependent on dTip60. We utilized our previously developed high-resolution MNase protection assay of mononucleosomes at Hsp70 both before and after HS (Petesch and Lis, 2008). Briefly, S2 cells from NHS and 2 min HS samples are crosslinked, and their chromatin is isolated and split into mock and MNase-treated samples. The DNAs from both matched samples are probed using qPCR to quantify the amount of protection for each of the 100 primer sets amplifying 100 bp regions of Hsp70Ab spaced 30 bp apart. The MNase protection assay of nucleosomes at Hsp70 before HS does not show significant differences with dTip60 depletion. However, following a 2 min HS, knockdown of dTip60 significantly inhibits the loss in chromatin structure of Hsp70 that occurs both initially from PARP’s activation as well as from Pol II’s movement through the gene by 2 min of HS (Figure 7A, compare pink to red). This pattern of nucleosome retention following HS in dTip60-depleted cells matches those found when PARP is inhibited or depleted (Petesch and Lis, 2008).
Figure 7. The dTip60 Histone Acetyltransferase Is Necessary for Nucleosomal Loss and Transcriptional Activation of theHsp70 Heat Shock Locus.
(A) dTip60 affects the rapid loss of nucleosomes at the Hsp70Ab HS locus. Control S2 cells treated with RNAi targeting LacZ or dTip60 were heat shocked for 0 (dark blue and light blue, respectively) or 2 min (red and pink, respectively), immediately cooled to room temperature and crosslinked, and their chromatin was isolated. Purified DNA products from samples were treated with 0 or 500 U of MNase and assayed by qPCR. The chromatin profile of Hsp70 was determined by using approximately 100 PCR amplicons with an average size of 100 ± 5 bp, spaced 30 ± 6 bp apart. To assess relative levels of protection, the amount of MNase-digested PCR product was normalized to the undigested product using the ΔC(t) method (y axis), which is plotted against the gene nucleotide location (x axis). Values from overlapping primer sets are averaged. The x axis represents base pair units with 0 being the TSS. Lines represent the average of three separate experiments with error bars representing the SEM of three independent experiments.
(B) Hsp70 mRNA levels following 0, 5, and 20 min of HS were measured for S2 cells RNAi targeting of LacZ (black) or dTip60 (gray). Hsp70 expression levels were measured by oligo dT primed reverse transcription followed by qPCR using Hsp70-specific primers and normalized to the Rp49 gene with error bars representing the SEM of three replicates.
(C) Model of HSF-mediated actions at Hsp70 to activate PARP and affect nucleosome loss. Top: Linear order of steps of the model depicted below. Bottom: Under NHS conditions, GAGA Factor (GAF) is bound to the promoter of Hsp70, Pol II is transcriptionally paused 20–40 base pairs downstream of the TSS, and inactive PARP is bound to the region occupied by the first well positioned nucleosome at Hsp70. Upon HS, by 30–60 s, HSF trimerizes and binds to multiple HSEs found at the promoter of Hsp70. The recruitment of HSF stimulates dTip60-mediated H2AK5 acetylation of nearby nucleosomes, which in turn activates PARP’s catalytic activity, PAR chain formation, and release from the nucleosome it was bound to prior to HS. The activation of PARP leads to the spread of active PARP and buildup of PAR across the HS locus. The progressive and rapid buildup of PAR within the region ultimately leads to nucleosome loss contained within the HS locus.
To test if retention of chromatin structure by 2 min of HS in dTip60-depleted cells has functional consequences on Hsp70 transcription, we measured mRNA levels before HS and after a 5 and 20 min HS. dTip60 depletion does not significantly affect Hsp70 NHS mRNA levels but significantly reduces mRNA levels of Hsp70 by 50% to that of control cells following both 5 and 20 min of HS (Figure 7B), mirroring those results obtained with PARP depletion (Petesch and Lis, 2008).
Our previous results demonstrated that nucleosome loss upon HS could occur independently of Hsp70 transcription. Treatment of S2 cells with sodium salicylate induces recruitment of HSF to the Hsp70 promoter under NHS conditions, but it does not result in Pol II movement into the gene (Petesch and Lis, 2008; Wine-garden et al., 1996). To both assess PARP and dTip60’s role in this transcription-independent nucleosome loss and solidify an ordered mechanism whereby HSF recruitment results in nucleosome loss, we performed ChIP in S2 cells treated with sodium salicylate. Under NHS conditions, sodium salicylate causes the activation, loss, and spread of PARP at Hsp70 (Figures S7A and S7B). Additionally, under NHS conditions, sodium salicylate also induces H2AK5Ac and H4Ac (Figures S7C and S7D). These results support a mechanism whereby HSF-induced histone acetylation triggers PARP activation and spread and nucleosome loss at Hsp70 that can be decoupled from active transcription.
DISCUSSION
This study establishes an ordered mechanism by which a transcription activator binding to a gene’s regulatory region leads to rapid removal of nucleosomes throughout the gene locus. Specifically, the transcriptional activator, HSF, stimulates dTip60 acetylation of H2AK5 that in turn activates PARP, causing its redistribution along Hsp70 and reduced nucleosome occupancy over the locus (Figure 7C). Moreover, all of these steps can be accomplished independently of transcription. This activation of PARP and its rapid spread throughout the Hsp70 HS loci demonstrate an interesting mechanism by which the nucleosome barrier can be alleviated to facilitate efficient transcription by Pol II.
HSF and many other transcriptional activators have been classically studied for their ability to recruit or release Pol II into transcriptional elongation. Our results speak to another function of HSF as an activator to direct changes in chromatin structure upon HS. HSF is able to achieve this function through physically interacting with the dTip60 complex and facilitating its recruitment to Hsp70 following HS (personal communication by Thomas Kusch). Just as the presence of paused Pol II in NHS conditions primes the Hsp70 gene for rapid transcriptional induction (Rougvie and Lis, 1988), inactive PARP bound in NHS conditions primes Hsp70 for rapid changes in chromatin structure. Interestingly, trimerization and binding of HSF to the promoter of Hsp70 precipitates the activation of both Pol II and PARP through distinct pathways that ultimately synergize to facilitate rapid and robust transcriptional activation. In vitro studies have shown that the DNA-binding and catalytic domains of PARP comprise the minimal structure sufficient for inactive PARP to bind and locally compact nucleosomes and, upon activation, release PARP from chromatin and decompact chromatin structure (Kim et al., 2004; Leduc et al., 1986; Poirier et al., 1982; Wacker et al., 2007). Activation of PARP is known to result in the formation of linear and branched anionic polymers with upwards of 200 units of ADP-ribose (D’Amours et al., 1999). Electron micrograph structures of branched PAR make it easy to visualize how creation of these voluminous, dendritic structures causes automodified PARP to expand 3-dimensionally throughout the Hsp70 loci following HS (de Murcia et al., 1983). Our results also indicate that PARP is crosslinked to Hsp70 after HS through a PAR linkage to chromatin. Although PARylation of another target, such as histones, cannot be ruled out, our results fit the simplest model where PARP is its own target. In agreement with the aforementioned in vitro studies, PARP automodification would result in its release from nucleosomes bound prior to HS, and the PAR created from this automodification could create a bridging interaction between PARP and chromatin formed during crosslinking, as seen in Figure 2D. This also is consistent with in vivo studies showing the major target of PARylation is PARP itself (D’Amours et al., 1999; Kim et al., 2004). Antibodies specifically recognizing ADP-ribosylated target proteins, such as PARP or histones, are needed to identify the target of PARP following HS at Hsp70.
The accumulation of PAR throughout the Hsp70 locus provides additional functional insight into how activation of PARP upon HS can affect chromatin structure and transcriptional activation. PAR has remarkable chemical similarity to other nucleic acids, such as DNA and RNA, but it has twice the charge per nucleic acid residue and the potential to form nonlinear, branched structures. As such, in vitro reconstitution assays have shown that PAR has the ability to locally compete with DNA to bind histones and potentially disrupt native chromatin structure (Althaus et al., 1994). The transient formation of PAR to alter chromatin structure followed by catabolism of PAR to return histones to its DNA template has been referred to as histone shuttling (Althaus et al., 1994). While initially investigated to explain PARP’s role in DNA damage repair, this phenomenon can be equally extended to PARP’s role in facilitating transcription. Indeed, the formation of PAR at Hsp70 loci after HS results in formation of a localized compartment that aids in the local retention of transcription factors, including Pol II, to sustain continued transcription activation of Hsp70 (Zobeck et al., 2010). It is yet to be determined if PAR also aids in the local retention of histones that were previously measured to be lost from Hsp70 after HS (Petesch and Lis, 2008).
The activation of PARP through the acetylation of H2AK5 also ascribes a unique function to dTip60. Like PARP, Tip60 has been studied for both its roles in DNA repair and also transcriptional activation (Sapountzi et al., 2006). In Drosophila, dTip60 is part of a complex containing Domino, an ATPase homologous to the mammalian p400 and SRCAP proteins, which, like Swr1p in S. cerevisiae, catalyzes the exchange of histone variant H2A.Z into H2A-containing nucleosomes (Kusch et al., 2004; Mizuguchi et al., 2004; Talbert and Henikoff, 2010). Drosophila contains only one H2A variant, which has properties of both H2A.Z and the C-terminal extension of H2A.X, and, when phosphorylated, marks sites of DNA damage (Madigan et al., 2002). Before HS, it is known that Hsp70 contains nucleosomes harboring H2Av near the 5′ end of the gene that is lost upon HS (Leach et al., 2000). Recently, the phosphorylation of H2AvS137 was shown to globally regulate PARP activation and is necessary for full transcriptional activation of Hsp70 (Kotova et al., 2011). dTip60 acetylates K5 on H2Av that is already phosphorylated on its C-terminal domain at S137 (Kusch et al., 2004). This acetylation stimulates the dTip60 complex to exchange out the modified H2Av. Additionally, in vitro studies show that the ability of H4 to activate PARP is squelched in the context of a nucleosome due to H2A (Pinnola et al., 2007). Collectively, these studies suggest a model in which the phosphorylation of H2AvS137 stimulates dTip60 to acetylate H2AvK5 following its recruitment upon HS (Figure S7E). These modifications are sufficient to stimulate the dTip60 complex to remove the modified H2Av and expose PARP to H4 and activate its enzymatic activity. The importance of H2A variant exchange has also been documented in Arabidopsis, where the Swr1 complex is also necessary for changes in chromatin structure at HS genes following HS (Kumar and Wigge, 2010).
This proposed model for the order of events that lead to the activation of PARP upon HS raises many questions for future exploration. First, is the H2Av that is present before HS already phosphorylated, and what is the kinase responsible for phosphorylation? Second, is phosphorylation of H2Av necessary for dTip60 acetylation of H2AvK5 upon HS? Third, is H2AvK5Ac by itself or in combination with S137 phosphorylation sufficient for PARP activation in vitro? Fourth, is the ATPase activity of the dTip60 complex to exchange H2Av following HS necessary or sufficient for PARP activation? Finally, is the activity of PARP regulated on a genomic scale at sites with H2Av nucleosomes that are both acetylated at K5 and phosphorylated at S137?
The fact that transcription-independent nucleosome loss following HS at Hsp70 is reliant on factors that respond to DNA damage provokes the question if changes in chromatin at Hsp70 are the result of a response to DNA repair. Indeed, transcriptional activation can occur in response to PARP activation from a topoisomerase II break in DNA (Ju et al., 2006). However, in contrast to this study, we find that PARP is already present at Hsp70 before HS and is not recruited upon HS. Although topoisomerase II mediated breaks have been mapped to sites near the TSS of Hsp70 before HS (Udvardy and Schedl, 1991), these breaks are not sufficient to detect active PARP at Hsp70 before HS and might be more important for the initial deposition of PARP before HS. We propose an alternative mechanism for PARP activation whereby a transcriptional activator hijacks DNA repair proteins to aid transcriptional activation. The fact that PARP is bound near the majority of human TSSs containing Pol II (Krishnakumar et al., 2008), as at Drosophila Hsp70, also hints at the generality for a mechanism whereby activation of prebound PARP leads to changes in chromatin structure and ultimately contributes to gene expression.
EXPERIMENTAL PROCEDURES
All primer sets used are listed in Table S1. The experimental procedures follow those used in (Petesch and Lis, 2008) and are present in the Supplemental Information, with the following exceptions.
ChIP
Heat shocks and ChIP were performed as in (Petesch and Lis, 2008) with the following exceptions. Following resuspension of the crosslinked pellet in sonication buffer, samples to be immunoprecipitated for PAR were TCA precipitated. 100% (w/v) TCA was added to a final concentration of 20% and incubated at 4°C for 10 min. The sample was centrifuged at 4°C for 5 min at 20,000 g and the resulting pellet was washed twice with 250 μl of acetone with the same centrifuging conditions. The final pellet was dried and resuspended in ChIP sonication buffer and the typical ChIP protocol was resumed from the point of sonication.
For ChIP with PARG-treated extracts, chromatin from crosslinked cells was isolated as in the high-resolution MNase assay and resuspended in the sonication buffer not containing SDS. Samples were split and either treated with final concentrations of 0 or 1200 nM PARG for 30 min at 37°C. The reaction was stopped by addition of SDS to a final concentration of 0.5% and the ChIP protocol continued at the step of sonication. The amount of antibody per IP used was: 8 μl rabbit anti-PARP raised to the N terminus (Kim et al., 2004), a gift of W. Lee Kraus; 4 μl mouse anti-PAR (Trevigen 4335); 2 μl rabbit anti-Histone H2A acetyl K5 (Abcam ab45152); and 4 μl rabbit anti-Histone H2A-ChIP grade (Abcam ab13923).
Supplementary Material
Acknowledgments
We would like to thank the members of the Lis lab, Alexei Tulin, Thomas Kusch, and Lee Kraus for comments on the manuscript and Thomas Kusch for personal communication of unpublished results. This work was supported by an NIH grant, GM25232, to J.T.L. and a predoctoral training grant, T32-GM70723, to S. J. P.
Footnotes
Supplemental Information includes seven figures, Supplemental Experimental Procedures, Supplemental References, and one Supplemental Table and can be found with this article online at doi:10.1016/j.molcel.2011.11.015.
References
- Althaus FR, Höfferer L, Kleczkowska HE, Malanga M, Naegeli H, Panzeter PL, Realini CA. Histone shuttling by poly ADP-ribosylation. Mol Cell Biochem. 1994;138:53–59. doi: 10.1007/BF00928443. [DOI] [PubMed] [Google Scholar]
- Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- Das C, Tyler JK, Churchill ME. The histone shuffle: histone chaperones in an energetic dance. Trends Biochem Sci. 2010;35:476–489. doi: 10.1016/j.tibs.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Murcia G, Jongstra-Bilen J, Ittel ME, Mandel P, Delain E. Poly(ADP-ribose) polymerase auto-modification and interaction with DNA: electron microscopic visualization. EMBO J. 1983;2:543–548. doi: 10.1002/j.1460-2075.1983.tb01460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhofer TA, Minoda A, Klugman S, Lee K, Kolasinska-Zwierz P, Alekseyenko AA, Cheung MS, Day DS, Gadel S, Gorchakov AA, et al. An assessment of histone-modification antibody quality. Nat Struct Mol Biol. 2011;18:91–93. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa PO, Covic M, Hasan S, Imhof R, Hottiger MO. The enzymatic and DNA binding activity of PARP-1 are not required for NF-kappa B coactivator function. J Biol Chem. 2001;276:45588–45597. doi: 10.1074/jbc.M106528200. [DOI] [PubMed] [Google Scholar]
- Johnson CA, White DA, Lavender JS, O’Neill LP, Turner BM. Human class I histone deacetylase complexes show enhanced catalytic activity in the presence of ATP and co-immunoprecipitate with the ATP-dependent chaperone protein Hsp70. J Biol Chem. 2002;277:9590–9597. doi: 10.1074/jbc.M107942200. [DOI] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- Karagianni P, Wong J. HDAC3: taking the SMRT-N-CoRrect road to repression. Oncogene. 2007;26:5439–5449. doi: 10.1038/sj.onc.1210612. [DOI] [PubMed] [Google Scholar]
- Kim MY, Mauro S, Gévry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Kimura A, Horikoshi M. Tip60 acetylates six lysines of a specific class in core histones in vitro. Genes Cells. 1998;3:789–800. doi: 10.1046/j.1365-2443.1998.00229.x. [DOI] [PubMed] [Google Scholar]
- Knezetic JA, Luse DS. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1986;45:95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- Kotova E, Lodhi N, Jarnik M, Pinnola AD, Ji Y, Tulin AV. Drosophila histone H2A variant (H2Av) controls poly(ADP-ribose) polymerase 1 (PARP1) activation in chromatin. Proc Natl Acad Sci USA. 2011;108:6205–6210. doi: 10.1073/pnas.1019644108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010a;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell. 2010b;39:736–749. doi: 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- Leach TJ, Mazzeo M, Chotkowski HL, Madigan JP, Wotring MG, Glaser RL. Histone H2A.Z is widely but nonrandomly distributed in chromosomes of Drosophila melanogaster. J Biol Chem. 2000;275:23267–23272. doi: 10.1074/jbc.M910206199. [DOI] [PubMed] [Google Scholar]
- Lebedeva LA, Nabirochkina EN, Kurshakova MM, Robert F, Krasnov AN, Evgen’ev MB, Kadonaga JT, Georgieva SG, Tora L. Occupancy of the Drosophila hsp70 promoter by a subset of basal transcription factors diminishes upon transcriptional activation. Proc Natl Acad Sci USA. 2005;102:18087–18092. doi: 10.1073/pnas.0509063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc Y, de Murcia G, Lamarre D, Poirier GG. Visualization of poly(ADP-ribose) synthetase associated with polynucleosomes by immunoelectron microscopy. Biochim Biophys Acta. 1986;885:248–255. doi: 10.1016/0167-4889(86)90239-9. [DOI] [PubMed] [Google Scholar]
- Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, LaPointe JW, Kornberg RD. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49:203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- Madigan JP, Chotkowski HL, Glaser RL. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res. 2002;30:3698–3705. doi: 10.1093/nar/gkf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Park JM, Werner J, Kim JM, Lis JT, Kim YJ. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol Cell. 2001;8:9–19. doi: 10.1016/s1097-2765(01)00296-9. [DOI] [PubMed] [Google Scholar]
- Pavri R, Lewis B, Kim TK, Dilworth FJ, Erdjument-Bromage H, Tempst P, de Murcia G, Evans R, Chambon P, Reinberg D. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol Cell. 2005;18:83–96. doi: 10.1016/j.molcel.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinnola A, Naumova N, Shah M, Tulin AV. Nucleosomal core histones mediate dynamic regulation of poly (ADP-Ribose) polymerase 1 protein binding to chromatin and induction of its enzymatic activity. J Biol Chem. 2007;282:32511–32519. doi: 10.1074/jbc.M705989200. [DOI] [PubMed] [Google Scholar]
- Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc Natl Acad Sci USA. 1982;79:3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- Sapountzi V, Logan IR, Robson CN. Cellular functions of TIP60. Int J Biochem Cell Biol. 2006;38:1496–1509. doi: 10.1016/j.biocel.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Smith ST, Petruk S, Sedkov Y, Cho E, Tillib S, Canaani E, Mazo A. Modulation of heat shock gene expression by the TAC1 chromatin-modifying complex. Nat Cell Biol. 2004;6:162–167. doi: 10.1038/ncb1088. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. Histone variants–ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- Udvardy A, Schedl P. Chromatin structure, not DNA sequence specificity, is the primary determinant of topoisomerase II sites of action in vivo. Mol Cell Biol. 1991;11:4973–4984. doi: 10.1128/mcb.11.10.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker DA, Ruhl DD, Balagamwala EH, Hope KM, Zhang T, Kraus WL. The DNA binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Mol Cell Biol. 2007;27:7475–7485. doi: 10.1128/MCB.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- Winegarden NA, Wong KS, Sopta M, Westwood JT. Sodium salicylate decreases intracellular ATP, induces both heat shock factor binding and chromosomal puffing, but does not induce hsp 70 gene transcription in Drosophila. J Biol Chem. 1996;271:26971–26980. doi: 10.1074/jbc.271.43.26971. [DOI] [PubMed] [Google Scholar]
- Zobeck KL, Buckley MS, Zipfel WR, Lis JT. Recruitment timing and dynamics of transcription factors at the Hsp70 loci in living cells. Mol Cell. 2010;40:965–975. doi: 10.1016/j.molcel.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.