Abstract
Most malaria drug development focuses on parasite stages detected in red-blood cells even though to achieve eradication next-generation drugs active against both erythrocytic and exo-erythrocytic forms would be preferable. We applied a multifactorial approach to a set of >4,000 commercially available compounds with previously demonstrated blood stage activity (IC50 < 1 μM), and identified chemical scaffolds with potent activity against both forms. From this screen, we identified an imidazolopiperazine scaffold series that was highly enriched among compounds active against Plasmodium liver stages. Our orally bioavailable lead imidazolopiperazine confers complete causal prophylactic protection (15 mg/kg) in rodent models of malaria and shows potent in vivo blood-stage therapeutic activity. The open source chemical tools resulting from our effort provide starting points for future drug discovery programs, as well as opportunities for researchers to investigate the biology of exo-erythrocytic forms.
Malaria continues to present a major health challenge in many of the poorest countries in the world with 225 million cases leading to an estimated 781,000 deaths in 2009 (1). In humans, malaria is caused by Plasmodium falciparum, Plasmodium malariae, Plasmodium ovale, Plasmodium vivax and the simian parasite Plasmodium knowlesi (2). Plasmodium is naturally transmitted by the bite of an infected female Anopheles mosquito. During the bite, the sporozoites are injected with the mosquito’s saliva and find their way to the host liver. There the parasites multiply asexually as exo-erythrocytic forms (EEFs) during an asymptomatic incubation period of ~1 week prior to emerging into the blood stream. This initiates the asexual erythrocytic cycle that is responsible for disease manifestations. While the EEFs of some Plasmodium species have a limited life span, in the case of P. ovale and P. vivax the parasite can persist within the liver as dormant hypnozoite for several months to years (3). Upon hypnozoite reactivation via an unknown mechanism, parasites can re-populate the blood with rapidly multiplying parasites that can cause pathology. As a consequence, the persistence of hypnozoites represents a formidable barrier to the eradication of malaria.
The only drugs with significant activity against proliferating EEFs and hypnozoites are the 8-aminoquinolines, including primaquine, pamaquine and tafenoquine (4, 5). Primaquine is the only treatment recommended by the World Health Organization to eliminate liver stages (4). However, 8-aminoquinolines can all cause dangerous levels of methemoglobinemia as a side effect in patients with glucose-6-phosphate dehydrogenase deficiency (6, 7), a common adaptive genetic condition in malaria-endemic regions. Resistance to this chemical class has also been reported, further increasing the need to find alternative drugs (8).
To identify leads with liver stage activity, we refined an in vitro assay with Plasmodium yoelii sporozoites and HepG2-A16-CD81EGFP cells (9) to screen a library of compounds known to be active against P. falciparum blood stages. The data revealed substantial differences in activity between blood and liver stages of the parasite and identified a chemically diverse group of molecules active against both.
Measuring sporozoite infection ratios and schizont size using high-content imaging
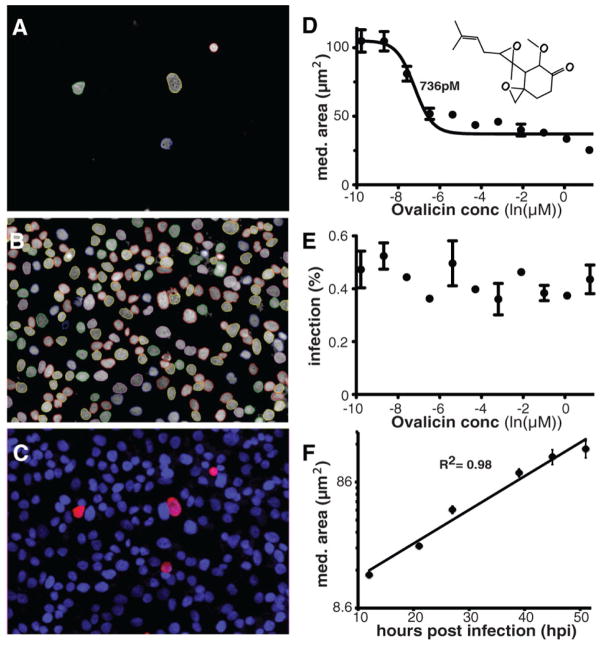
Only 1% of liver cells typically become infected by sporozoites in vitro, and because we needed to screen many thousands of compounds, we created a robust, reproducible assay and image analysis to automate data collection. Eight thousand sporozoites freshly dissected from infected Anopheles stephensi mosquitoes were added to each well of a 384-well plate, containing ~15,000 hepatoma cells per well. After a two-day incubation in the presence of compound the host cell and parasite nuclei were labeled with Hoechst 33342 and the parasites were labeled with αPyHSP70 antibodies. A Perkin Elmer high-content imaging system captured 25 images (Fig. 1) for each well of the 384-well plate. A custom Acapella™ (PerkinElmer) script parameterized for this assay was used to analyze the images on the basis of morphology and fluorescence intensity for each liver cell nucleus and each parasite. The infection ratio was the ratio between parasite number (αPyHSP70 positive) and the number of host nuclei per well.
Fig. 1.
Images of high-content screening of parasite hepatic schizonts and parasite growth dynamics. (A) P. yoelii sporozoites were stained 48 hours post infection (hpi) with (αPyHSP70/goat anti-mouse antibodies. The parasites areas were defined by outlines generated by a custom Acapella script (31), indicating separate objects. (B) Hoechst 33342 nuclear staining of HepG2-A16-CD81EGFP cells. Nuclei are delineated using a script as in part A. (C) False color channel merge of A and B. (D and E) Measuring inhibition using parasite area. The IC50 of ovalicin is 736 pM when measured by median parasite area (D), but no drug effect is seen when the overall infection ratio is measured in (E). (F) Correlation of incubation time and schizont size. P. yoelii EEF growth approximates a linear rate with an R2 of 0.98 in the HepG2-A16-CD81EGFP cell line when no test compounds are added. Data are mean +/− SD based on four experimental replicates of approx. 50 infected cells per time point.
To quantify the effects of the test compounds, we determined median parasite size to be preferable to the infection ratio, because infection ratio may be affected by drug toxicity to host cells and host cell division during the incubation (e.g., a 1% infection ratio may drop to 0.3–0.6% after 48 hours exposure to a cell-toxic compound). Measuring parasite size also enabled the identification of compounds that arrest parasite growth without reducing the infection ratio. For example, the compound ovalicin reduced schizont size (Fig. 1D) with an IC50 of 736 pM, but had no measurable effect on the infection ratio (Fig. 1E). Finally, measuring parasite size allowed us to estimate when a particular compound affected parasite development (Fig. 1F).
Performance of control compounds
For evaluation the assay was first conducted on a set of control compounds, which confirmed that compounds known to affect only blood stages were either inactive [artemisinin and the spiroindolone, NITD609 (10)] or had only marginal activity (mefloquine and chloroquine) against EEFs. Other compounds with known activity against EEFs, including ovalicin, berberine, atovaquone, lasalocid, pyrimethamine and primaquine, were confirmed as active in our assay (Table 1). The control compounds were further classified based on their time of action using schizont size, which is proportional to developmental stage and which ranged from 9 μm2 (the smallest size detected using our imaging software and objective) up to 180 μm2 for a fully developed untreated schizont (Fig. 1F). Because of the resolution of the microscope (~0.43 μm2/pixels), we could not distinguish between compounds that prevent sporozoite infection and those that arrest schizont growth at a size less than 9 μm2 (a value too close to the average background staining). To examine this possibility, cultured liver cells were infected and examined using deconvolution microscopy after 48 hours in the presence of drugs (Fig. S1). Our results showed that liver schizonts formed at the expected rate, but were very small compared with untreated or artemisinin-treated parasites.
Table 1.
Blood-stage and liver-stage (EEF) IC50 values (with standard deviations) of select characterized compounds. Blood stage IC50 values were measured against P. falciparum W2 parasites using a SYBR green assay (30). The median EEF P. yoelii parasite area (in μm2±SD) was calculated from two or more independent experiments with ~50 schizonts each. All drugs were dissolved in DMSO. Median parasite size of control P. yoelii DMSO-treated EEFs ranged from 160 to 180 μm2.
| Compound | P. yoelii EEF IC50 (nM) | HepG21 inhibition IC50 (μM) | Median parasite area (μm2)
|
Blood IC50 (nM) | |
|---|---|---|---|---|---|
| 1 μM | 10 μM | ||||
| Cyclosporin | 71±3 | 2.3±0.7 | < 9 | < 9 | 345±116 |
| Lasalocid | 37±20 | 3.9±1.4 | < 9 | < 9 | 50±8 |
| Pyrimethamine | 2.4±1.6 | 6.3±3.3 | 31.1±0.7 | 29.6±2.0 | 15±2 |
| Berberine | 224±152 | 2.6±0.5 | 46.4±7.0 | 22.4±2.3 | 117±9 |
| Atovaquone | 9.4±15 | 9.5±2.5 | 18.2±1.0 | 18.6±1.0 | 2.2±0.4 |
| Primaquine | 1,162±85 | 6.1±1.9 | 56.8±15.6 | < 9 | 317±60 |
| Chloroquine2 | 1,650±132 | 1.6±0.3 | 65.4±9.5 | < 9 | 84±3 |
| Ovalicin | 0.7±0.3 | >10 | 35.1±3.1 | 19.1±0.9 | >10,000 |
| Artemisinin | >10,000 | >10 | na | na | 3.3±0.4 |
| Mefloquine | 3,270±840 | 3.1±0.6 | 65.4±4.7 | < 9 | 9.5±10 |
| Strobilurin3 | 8.8±0.6 | >1 | 22.3±3 | na | 7±0.1 |
| Cycloheximide | 78±28 | 0.22±0.10 | <9 | <9 | 366±69 |
| Decoquinate | 0.16±0.02 | >10 | 17.7±0.3 | 18.8±4.7 | 0.25±0.06 |
HepG2-A16-CD81EGFP inhibition was calculated by determining the dose response for host nuclei number.
Chloroquine was dissolved in water, not DMSO.
Maximum concentration tested was 1μM.
na, not applicable
A majority of compounds active against blood stages is inactive against liver-stages
We screened a set of antimalarials derived from a collection of commercially available compounds that had previously been tested for activity against blood stage P. falciparum parasites (11, 12). Chemical scaffold analysis based on chemical fingerprint similarities (13) revealed that the library of 5697 compounds comprised of 2715 independent chemical scaffold clusters, some of which were unique to this library and some of which were also hits in independent screens (Fig. S2). Of the 5697 compounds, 275 could be confirmed as active against liver stage parasites (fitted IC50 < 10 μM) in two rounds of screening (Table S1). 229 compounds had an IC50 of less than 1 μM and 86 less than 100 nM.
Scaffold clustering
A major challenge associated with translating compounds into drugs is in determining which of the leads are the most promising for further medicinal chemistry development aimed at increasing a compound’s exposure, physicochemical properties, and potency. The dataset of 275 active compounds contains scaffolds that are distinct from known antimalarial pharmacophores (quinolines, quinones, trioxanes) and reactive functional groups (such as Michael acceptors or nitroarenes). Such filtering still gives more leads than can be reasonably investigated, but the advantage of completing such a large screen of putative antimalarials is that it allows for a system-wide analysis of the data by exploring structure-activity relationships (SAR) within the compound library. SARs are more important than absolute potency because they indicate a chemical space that can be optimized to compounds with the desired potency (13).
To identify which of the 2715 scaffold clusters in the compound library would offer the most promising leads for liver-stage drugs, the statistically enriched clusters comprised of the 655 primary liver hits [compounds causing >50% parasite growth inhibition at 10 μM (Table S2)] were identified using an accumulative hypergeometric mean function (HMF). This identified 62 primary clusters using a cutoff of p<0.05 (Table S3) that were over- or under-represented in the liver-stage hits. For example, the quinazolinone scaffold (cluster 2691) has 68 related members within the entire library and only two of the 68 screened appeared as initial hits in the liver-stage screen (HMF p=9.8×10−4), indicating that a compound developed from this series would be unlikely to provide causal prophylactic activity. By contrast, there were ten members of the pyrazolopyrimidine scaffold (cluster 1766, Fig. S3, Fig. S2B, Table S4), nine of which showed initial activity in the liver assay (HMF p=2.8×10−7). Five of the nine compounds in cluster 1766 were reconfirmed to have activity EEFs (HMF p=5.1×10−4), as were seven of the quinazoline scaffold (cluster 2620, HMF p=41.5×10−6). In addition, three of the nine scaffolds containing an atovaquone-like napthoquinone core (cluster 2037, HMF p=3.0×10−2) (Fig. S3) inhibited parasite development at very early stages (12.5 μm2 to 21 μm2). Similarly, early parasite development was inhibited by 14 compounds (cluster 2714, Fig. S3) predicted to inhibit dihydrofolate reductase (DHFR) based on their similarity to known DHFR inhibitors and their >10× lower activity against Dd2, a strain that carries three point mutations in DHFR (14, 15). In this cluster, 13 of the 14 were active in the initial assay (HMF p=2.0×10−7) and all 13 showed dose-response activity, yielding very similarly sized schizonts with a median area of 24±2.8 μm2 (Table S1).
Developing a drug candidate with liver-stage activity
Although smaller compound clusters are less impressive, they nevertheless provide interesting starting points for medicinal chemistry efforts. The imidazolopiperazine (IP) cluster 1035 is a scaffold class of saturated cyclic amines with a predicted high probability of activity against both liver and blood stage parasites (Fig. S3, Table S5). All three members of the original dataset (Pf-5069, Pf-5179 and Pf-5466) share the same IP saturated cyclic amine chemical core and showed activity in the P. yoelii liver assay (HMF p=0.0035). These compounds also showed submicromolar IC50s against P. falciparum blood stage cultures in vitro, including the multidrug-resistant strain W2. The IP scaffold is attractive as a starting point for medicinal chemistry because it is structurally unrelated to known antimalarial scaffolds and is chemically tractable. In addition, it showed no cross-resistance with strains having mutations making them resistant to drugs acting on liver stages, including atovaquone (Table S6) and pyrimethamine.
To test whether the modified IP scaffolds retained activity against liver-stage parasites, several analogs optimized for blood stage potency (16) were examined in our P. yoelii liver stage assay. As expected, all compounds in the series caused the parasites to arrest near the minimal detectable size (< 9 μm2) in the liver assay (Table S5). In addition, members of the series displayed liver-stage activity that correlated well with their activity against P. falciparum blood stages. For example, the IC50s for Pf-5069 were 454 and 334 nM against the blood stages of strains 3D7 and W2 and 221 nM in the P. yoelii assay using HepG2-A16-CD81EGFP cells (17), while the IC50s for GNF180 in the same assays were 37, 39 and 59 nM.
GNF179, a molecule with drug-like properties
By synthesizing over 1,200 analogs and optimizing their blood stage potency (16), derivatives of the IP lead compounds with suitable properties for testing in animal models of malaria were created. This work led to the design of an optimized 8,8-dimethyl IP analog (GNF179; Fig. 2AB; synthesis described in Supplemental Materials and Methods) that exhibited the potency [4.8 nM against the multidrug resistant strain W2 (Table S5)], in vitro metabolic stability and in vivo oral bioavailability (58%) required for an antimalarial drug development candidate (Table S7). GNF179 exhibits a low clearance (CL=22 ml/min/kg, ~25% of hepatic blood flow in mice), a large volume of distribution (steady-state volume of distribution, Vss=11.8 l/kg), a moderate residence time (MRT=9 hours) and suitable terminal half-life (t1/2=8.9 hours). As expected for a compound with potency in the low nanomolar range and good pharmacokinetic properties, GNF179 reduced Plasmodium berghei parasitemia levels by 99.7% with a single 100 mg/kg oral dose, and prolonged mouse survival by an average of 19 days. In contrast, we found that the average survival times for chloroquine and artesunate in mice infected with P. berghei and given a single oral dose at 100 mg/kg, were 12.5 days and 7 days, respectively (Table S8). To demonstrate that the compound was suitable for testing for causal prophylactic activity, we confirmed that the compound was distributed into the liver. On oral dosing we found a maximum concentration (Cmax) within the liver of 55 μM at 15 mg/kg and 193 μM at 100 mg/kg, with a half-life of 7–9 hours, similar to that in plasma (Table S9). The overall oral exposure (Area Under the Curve) liver to plasma ratio of 55 and 25 at the 15 and 100 mg/kg oral administration (p.o.) doses, respectively, further confirms good liver exposure to the compound.
Testing in an animal prophylaxis model of malaria
To confirm in vivo the tissue schizonticidal activity observed in vitro, we tested GNF179 in a rodent malaria model of causal prophylactic treatment (Table 2). Groups of five mice were given a single dose of atovaquone (positive control), GNF179, or the negative control NITD609 (10) and then injected with 105 P. berghei sporozoites. Although NITD609 is slightly more potent against P. falciparum blood stages than GNF179 (~1 nM versus 7 nM), and is fully curative at a 100 mg/kg oral dose in a blood-stage animal model (10), GNF179 was able to protect against an infectious P. berghei sporozoite challenge with a single oral dose at 15 mg/kg (Fig. 2D) while NITD609 was not (Table 2). Taken together, these data show that the in vitro HepG2-A16-CD81EGFP assay correlates well with in vivo efficacy and that compounds identified in this cellular screening approach offer evidence of causal prophylactic activity.
Table 2.
Prophylaxis testing results with P. berghei. The prepatent period was the average of days for all five mice tested per group. In all cases the drug was administered orally 1–3 hours prior to sporozoite injection, except where indicated by asterisk, where drug administration was six hours after infection. All dosing was in a suspension formulation of 0.5% w/v methylcellulose, 1% (v/v) solutol HS15.
| Dose mg/kg | Average Survival (%) | Prepatent Period (days) | |
|---|---|---|---|
| Untreated | - | 0 | 4 |
| Untreated* | - | 0 | 5.3 |
| GNF179 | 5 p.o. | 20 | 4.2 |
| GNF179 | 15 p.o. | 100 | na |
| GNF179* | 15 p.o. | 80 | 6.2 |
| GNF179 | 20 p.o. | 100 | na |
| Atovaquone | 2.5 p.o. | 100 | na |
| Atovaquone* | 2.5 p.o. | 100 | na |
| NITD609 | 30 p.o. | 0 | 4.2 |
| NITD609 | 15 p.o. | 0 | 4.5 |
p.o., oral administration; na, not applicable due to lack of blood stage infection.
Fig. 2.
The effect of GNF179 on the liver stage parasite and a comparison with lasalocid, pyrimethamine and atovaquone. (A) High-resolution deconvolution microscopy of the GNF179-treated liver-stage parasites. Columns show Hoechst 33342 staining in blue, αPyHSP70 staining in red and a merge with the host plasma membrane marker CD81-GFP in green. Cultures were treated with 1 μM GNF179 for 48 hr. (Insets) DMSO-treated control parasite at the same scale and time point. Scale bar indicates 10 μm. (B) Chemical structure of GNF1.79. (C) The targets of GNF179 and lasalocid appear to be required throughout development. Compounds were added at 1 μM final concentration at the start (lightest shaded bars), 15 hpi (medium shaded bars) and 27 hpi (darkest shaded bars); all samples were incubated up to an end-point of 51 hpi. Dashed lines represent the control DMSO-treated growth levels at 15, 27 and 51 hpi. Data are mean +/− SD from 2 experimental replicats of approx. 50 infected cells per time point. Las, lasalocid; Pyr, pyrimethamine; Atov, atovaquone. (D) In vivo bioluminescence imaging of representative mice infected with P. berghei and treated with GNF179 (15 mg/kg) or vehicle (no compound), at 6 hpi. In the control, luminescence showing the developing parasite was detected in the liver area from 24 hpi and the lung and gastrointestinal track region from 72 hpi. No luminescence signal was detected from GNF179 or atovaquone (2.5 mg/kg) -treated mice even at the maximum sensitivity setting and no blood stage parasitemia was detected from later blood smear examinations. The luminescence intensity color-coding does not represent absolute values and is individually adjusted for each recording, which is why you can see background signal around the muzzle in some of the control pictures.
GNF179 works through a distinct mechanism
Examination of infected cells treated with GNF179 by high-resolution microscopy showed that this compound series causes the parasites to arrest at the earliest stages of development (Fig. 2AC), even earlier than atovaquone and the antifolates. Because IP compounds may act only during a short period of early development, the efficacy of compounds given 15 and 27 hours post-invasion was tested. These data showed that when pyrimethamine was added 27 hours post-invasion, the parasites grew to normal size (Fig. 2C), showing that folates are needed only at a specific time in development. In contrast, GNF179, like lasalocid and atovaquone, remained active later in EEF development and the rapidly induced growth arrest could not be reversed after compound removal. GNF179 may act against sporozoites instead of hepatic stages, thus the compound was also administered to mice six hours after infection. Luciferase imaging showed that no blood stage infection developed (Fig. 2D, Table 2) indicating that the target of GNF179 is needed for continuous growth and development of the parasite in the liver.
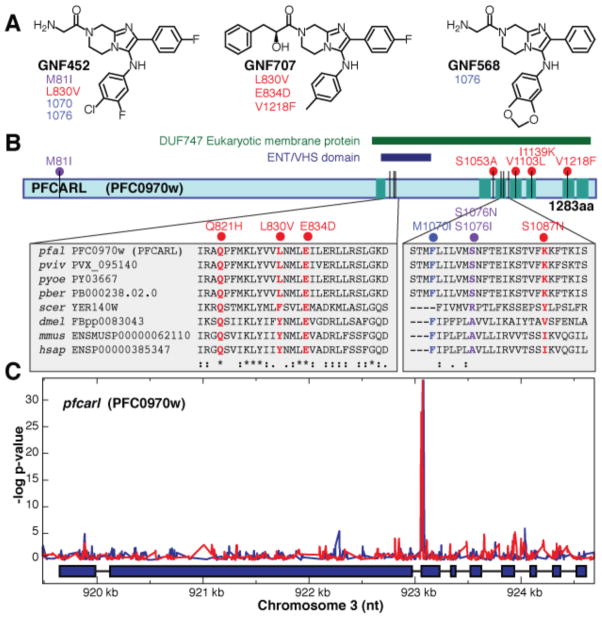
Unlike other compounds that act early in hepatic stage development, such as cycloheximide, GNF179 does not rapidly inhibit parasite protein biosynthesis (Fig. S4); nor is it likely to target parasite cytochrome bc1, which has been validated as a hepatic stage target for atovaquone, because we see no cross-resistance (Table S6). In addition, unlike electron chain transport inhibitors, members of the IP scaffold do not cause a shift in IC50 when tested against transgenic parasites lines expressing S. cerevisiae dihydroorotate dehydrogenase (Table S10) (18, 19). To confirm that the mechanism of action was distinct, P. falciparum Dd2 and 3D7 parasite strains were exposed to increasing concentrations of the IP series (GNF452, GNF707, Pf-5069) until resistance emerged (Fig. 3A). Mutants were cloned and the genomes of the resistant clones were compared with the parental parasites using high-density microarrays or full genome sequencing (Table S11, Fig. 3B). The mutants showed no cross-resistance (i.e., no changes in IC50) for mefloquine (~16 nM), or artemisinin (~6 nM). Microarray analysis revealed only between one and six high probability genetic changes per mutant genome (Fig. 3; Table S11, Table S12). Notably, only a single gene (PFC0970w), which we have named P. falciparum Cyclic Amine Resistance Locus (pfcarl), encoding an uncharacterized protein with seven predicted transmembrane regions, was mutated in all strains. We observed no new genetic changes [either single nucleotide polymorphisms (SNPs) or copy number variations (CNVs)] in pfmdr1, pfcrt, or any other known Plasmodium drug resistance gene. Full-genome sequencing of four of the independently derived drug-resistant clones showed six high quality SNPs that distinguished the drug-pressured parasites from the parental Dd2 clone. Five of those SNPs were in pfcarl. Additional selection of Dd2 parasites with related scaffolds produced three resistant clones (D1, D2, D3), which each harbored a single non-synonymous point mutation in pfcarl (Table S11).
Fig. 3.
SNPs identified in pfcarl by microarray analysis and whole genome sequencing analysis. (A) Stereochemical structures of three compounds (GNF452 (B1, B2, B3), GNF707 (C1, C2, C3), and Pf-5069 (A1, A2) from the IP series (Table S5) used to generate resistant parasite clones. Detected SNP,s indicated below the compound IDs, are detailed in Table S11 (SNPs identified by sequencing analysis are red and SNPs identified by both sequencing and microarray analysis are purple) (B) Schematic of pfcarl, showing the conserved regions, including the transmembrane domains. SNP color-coding is the same as in (A) Two regions of the protein are enlarged to show close SNPs and the amino acid sequence conservation of the region across parasite and other species (pfal, P. falciparum; pviv, P. vivax; pyoe, P. yoelii; pber, P. berghei; scer, S. cerevisiae; dmel, Drosophila melanogaster; mmus, Mus musculus; hsap, Homo sapiens).(C) Microarray SNP analysis across pfcarl from two GNF452-resistant clones (GNF452-1 in blue and GNF452-3 in red). The lines trace the p-values indicating whether mapped probes are different in the resistant clone compared with the Dd2 parental strain.
Homologs of pfcarl are conserved across almost all eukaryotic genera, including humans [OrthoMCL Group Summary (OG4_11754)], and the homolog is essential in C. elegans (20). Analysis of the corresponding yeast deletion strains (21), suggests that it plays a role in protein folding within the endoplasmic reticulum (22), a process that in Plasmodium would presumably be required in both blood and hepatic stages. All mutations observed in our resistant clones affected amino acids that were evolutionarily conserved. Polymorphism data across various different strains in P. falciparum (23) also indicate conservation. Although more work is needed to characterize this gene, our data suggest that the IPs have a mechanism of action distinct from known anti-malarials and that pfcarl could represent a conserved drug target or resistance gene.
Recent efforts to develop novel malaria control measures recognize that ideally antimalarials should be active against liver stages as well as blood stages (24). Compounds with dual activity are rare among the antimalarials that are licensed or in development. Indeed, tests with compounds in clinical development, including OZ277 (25) and OZ439 (26), showed that while most are equally potent against blood stages, only atovaquone and the antifolates showed equivalent potency to GNF179 in the HepG2-A16-CD81EGFP model. It is also worth noting that compounds related to naphthoquinones (cluster 2037) and DHFR inhibitors (cluster 2714) were identified in a previously reported in vivo screening effort aimed at identifying new liver-stage antimalarial drugs using whole animal testing (27). Our dataset reveals compounds that could offer starting points for the development of new dual-stage antimalarial drugs.
One outstanding question is whether these scaffolds can also provide radical cure activity against the dormant liver stages of the P. vivax parasite. To our knowledge, of all chemical classes reported to show causal prophylactic activity in vivo only the 8-aminoquinolines also display radical cure activity (27), suggesting that only a small fraction of the compounds identified in our screen would have similar properties. Nonetheless, it remains to be determined how physiologically distinct a hypnozoite is from an EEF schizont and whether these differences could prevent a drug from acting on both forms. Interestingly, our data show that the known, hypnozoite-targeting compound, primaquine, acts against the earliest EEFs in our P. yoelii cell culture models, suggesting that early acting compounds such as GNF179 may also have similar activity. Nevertheless, as long as these differences are not defined and there are no suitable model systems, the possibility remains that any drug developed with the use of rodent parasite EEF screens may not be efficacious in P. vivax. Nonetheless, a drug that acted on EEFs but not hypnozoites, although potentially unsuitable for development, could become a tool for understanding hypnozoite biology. Although the targets of most of the compounds analyzed here remain uncharacterized, we have developed methods that allow us to associate targets and compounds with liver-stage activity, especially if they are also active against blood stages (10, 28, 29).
Supplementary Material
Acknowledgments
We wish to thank Irwin Sherman for helpful comments on the manuscript, members of the New York University Insectary for providing infected mosquitoes and Christoph Fischli for P. berghei efficacy testing. The whole genome sequencing data has been deposited at the NCBI SRA database (http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?) with the project accession no. SRA045972.1. The microarray data has been deposited at the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/) with the accession no XXX. E.A.W. is supported by the Keck Foundation and by the NIH (grant R01AI090141). S.M. was supported by Deutsche Forschungsgemeinschaft (ME 3528/1-1). Funding for M.C.L. and D.A.F. is provided in part by the Medicines for Malaria Venture and the NIH (grant R01 AI079709). We gratefully acknowledge translational research support (WT078285) from the Wellcome Trust and funding from the Medicines for Malaria Venture to the Genomics Institute of the Novartis Research Foundation, the Swiss Tropical and Public Health Institute and the Novartis Institute for Tropical Diseases.
References
- 1.WHO. World Malaria Report 2010. 2010 http://www.who.int/malaria/world_malaria_report_2010/en/index.html.
- 2.Singh B, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 3.Trampuz A, Jereb M, Muzlovic I, Prabhu RM. Clinical review: Severe malaria. Crit Care. 2003;7:315. doi: 10.1186/cc2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. Guidelines for the Treatment of Malaria. 2011:194. http://www.who.int/malaria/publications/atoz/9789241547925/en/index.html.
- 5.Sweeney AW, Blackburn CRB, Rieckmann KH. Short report: the activity of pamaquine, an 8-aminoquinoline drug, against sporozoite-induced infections of Plasmodium vivax (New Guinea strains) Am J Trop Med Hyg. 2004;71:187. [PubMed] [Google Scholar]
- 6.Cohen RJ, Sachs JR, Wicker DJ, Conrad ME. Methemoglobinemia provoked by malarial chemoprophylaxis in Vietnam. N Engl J Med. 1968;279:1127. doi: 10.1056/NEJM196811212792102. [DOI] [PubMed] [Google Scholar]
- 7.Coleman MD, Coleman NA. Drug-induced methaemoglobinaemia. Treatment issues. Drug Saf. 1996;14:394. doi: 10.2165/00002018-199614060-00005. [DOI] [PubMed] [Google Scholar]
- 8.Talisuna AO, Bloland P, D’Alessandro U. History, dynamics, and public health importance of malaria parasite resistance. Clin Microbiol Rev. 2004;17:235. doi: 10.1128/CMR.17.1.235-254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gego A, et al. New approach for high-throughput screening of drug activity on Plasmodium liver stages. Antimicrobial Agents and Chemotherapy. 2006;50:1586. doi: 10.1128/AAC.50.4.1586-1589.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rottmann M, et al. Spiroindolones, a potent compound class for the treatment of malaria. Science. 2010;329:1175. doi: 10.1126/science.1193225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowther GJ, et al. Identification of inhibitors for putative malaria drug targets among novel antimalarial compounds. Mol Biochem Parasitol. 2011;175:21. doi: 10.1016/j.molbiopara.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gagaring K, et al. Novartis-GNF Malaria Box. ChEMBL-NTD. 2010 http://www.ebi.ac.uk/chemblntd.
- 13.Yan SF, Asatryan H, Li J, Zhou Y. Novel statistical approach for primary high-throughput screening hit selection. Journal of chemical information and modeling. 2005;45:1784. doi: 10.1021/ci0502808. [DOI] [PubMed] [Google Scholar]
- 14.Wang P, Read M, Sims PF, Hyde JE. Sulfadoxine resistance in the human malaria parasite Plasmodium falciparum is determined by mutations in dihydropteroate synthetase and an additional factor associated with folate utilization. Mol Microbiol. 1997;23:979. doi: 10.1046/j.1365-2958.1997.2821646.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang P, et al. Genetic and metabolic analysis of folate salvage in the human malaria parasite Plasmodium falciparum. Mol Biochem Parasitol. 2004;135:77. doi: 10.1016/j.molbiopara.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 16.Wu T, et al. Imidazolopiperazines: Hit to Lead Optimization of New Antimalarial Agents. Journal of Medicinal Chemistry. 2011;54:5116. doi: 10.1021/jm2003359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yalaoui S, et al. Hepatocyte permissiveness to Plasmodium infection is conveyed by a short and structurally conserved region of the CD81 large extracellular domain. PLoS Pathog. 2008;4:e1000010. doi: 10.1371/journal.ppat.1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Painter HJ, Morrisey JM, Mather MW, Vaidya AB. Specific role of mitochondrial electron transport in blood-stage Plasmodium falciparum. Nature. 2007;446:88. doi: 10.1038/nature05572. [DOI] [PubMed] [Google Scholar]
- 19.Nam T-G, et al. A chemical genomic analysis of decoquinate, a Plasmodium falciparum Cytochrome b inhibitor. ACS Chem Biol. 2011 doi: 10.1021/cb200105d. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol. 2001;11:171. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- 21.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 22.Jonikas MC, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.PlasmoDB: The Plasmodium Genomics Resource. http://plasmodb.org/plasmo/
- 24.Mazier D, Rénia L, Snounou G. A pre-emptive strike against malaria’s stealthy hepatic forms. Nature reviews Drug discovery. 2009;8:854. doi: 10.1038/nrd2960. [DOI] [PubMed] [Google Scholar]
- 25.Dong Y, et al. The structure-activity relationship of the antimalarial ozonide arterolane (OZ277) J Med Chem. 2010;53:481. doi: 10.1021/jm901473s. [DOI] [PubMed] [Google Scholar]
- 26.Charman SA, et al. Synthetic ozonide drug candidate OZ439 offers new hope for a single-dose cure of uncomplicated malaria. Proc Natl Acad Sci USA. 2011;108:4400. doi: 10.1073/pnas.1015762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson DE, Jr, et al. New tissue schizontocidal antimalarial drugs. Bulletin of the World Health Organization. 1981;59:463. [PMC free article] [PubMed] [Google Scholar]
- 28.Istvan ES, et al. Validation of isoleucine utilization targets in Plasmodium falciparum. Proc Natl Acad Sci USA. 2011;108:1627. doi: 10.1073/pnas.1011560108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dharia NV, et al. Use of high-density tiling microarrays to identify mutations globally and elucidate mechanisms of drug resistance in Plasmodium falciparum. Genome Biol. 2009;10:R21. doi: 10.1186/gb-2009-10-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plouffe DM, et al. In silico activity profiling reveals the mechanism of action of antimalarials discovered in a high-throughput screen. Proc Natl Acad Sci USA. 2008;105:9059. doi: 10.1073/pnas.0802982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silvie O, et al. Hepatocyte CD81 is required for Plasmodium falciparum and Plasmodium yoelii sporozoite infectivity. Nat Med. 2003;9:93. doi: 10.1038/nm808. [DOI] [PubMed] [Google Scholar]
- 32.Tsuji M, et al. Demonstration of heat-shock protein 70 in the sporozoite stage of malaria parasites. Parasitol Res. 1994;80:16. doi: 10.1007/BF00932618. [DOI] [PubMed] [Google Scholar]
- 33.Cohen SB, Gaskins C, Nasoff MS. Generation of a monoclonal antibody agonist to toll-like receptor 4. Hybridoma (Larchmt) 2005;24:27. doi: 10.1089/hyb.2005.24.27. [DOI] [PubMed] [Google Scholar]
- 34.Kilpatrick KE, et al. Rapid development of affinity matured monoclonal antibodies using RIMMS. Hybridoma. 1997;16:381. doi: 10.1089/hyb.1997.16.381. [DOI] [PubMed] [Google Scholar]
- 35.Rénia L, et al. A malaria heat-shock-like determinant expressed on the infected hepatocyte surface is the target of antibody-dependent cell-mediated cytotoxic mechanisms by nonparenchymal liver cells. Eur J Immunol. 1990;20:1445. doi: 10.1002/eji.1830200706. [DOI] [PubMed] [Google Scholar]
- 36.Galitzer SJ, Oehme FW, Bartley EE, Dayton AD. Lasalocid toxicity in cattle: acute clinicopathological changes. J Anim Sci. 1986;62:1308. doi: 10.2527/jas1986.6251308x. [DOI] [PubMed] [Google Scholar]
- 37.Novilla MN. The veterinary importance of the toxic syndrome induced by ionophores. Vet Hum Toxicol. 1992;34:66. [PubMed] [Google Scholar]
- 38.Pereira GC, et al. Mitochondrially targeted effects of berberine [Natural Yellow 18, 5,6-dihydro-9,10-dimethoxybenzo(g)-1,3-benzodioxolo(5,6-a) quinolizinium] on K1735-M2 mouse melanoma cells: comparison with direct effects on isolated mitochondrial fractions. J Pharmacol Exp Ther. 2007;323:636. doi: 10.1124/jpet.107.128017. [DOI] [PubMed] [Google Scholar]
- 39.Kalla G, Singhi M. Cutaneous leishmaniasis in Jodhpur district. Indian J Dermatol Venereol Leprol. 1996;63:149. [PubMed] [Google Scholar]
- 40.Sriwilaijareon N, et al. Stage specificity of Plasmodium falciparum telomerase and its inhibition by berberine. Parasitol Int. 2002;51:99. doi: 10.1016/s1383-5769(01)00092-7. [DOI] [PubMed] [Google Scholar]
- 41.Sheng WD, Jiddawi MS, Hong XQ, Abdulla SM. Treatment of chloroquine-resistant malaria using pyrimethamine in combination with berberine, tetracycline or cotrimoxazole. East Afr Med J. 1997;74:283. [PubMed] [Google Scholar]
- 42.Bell A, Roberts HC, Chappell LH. The antiparasite effects of cyclosporin A: possible drug targets and clinical applications. Gen Pharmacol. 1996;27:963. doi: 10.1016/0306-3623(95)02148-5. [DOI] [PubMed] [Google Scholar]
- 43.Mahmoudi N, et al. New active drugs against liver stages of Plasmodium predicted by molecular topology. Antimicrobial Agents and Chemotherapy. 2008;52:1215. doi: 10.1128/AAC.01043-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pradines B, et al. In vitro activity of tafenoquine against the asexual blood stages of Plasmodium falciparum isolates from Gabon, Senegal, and Djibouti. Antimicrob Agents Chemother. 2006;50:3225. doi: 10.1128/AAC.00777-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bray PG, et al. Primaquine synergises the activity of chloroquine against chloroquine-resistant P. falciparum. Biochem Pharmacol. 2005;70:1158. doi: 10.1016/j.bcp.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 46.Bates MD, et al. In vitro effects of primaquine and primaquine metabolites on exoerythrocytic stages of Plasmodium berghei. Am J Trop Med Hyg. 1990;42:532. doi: 10.4269/ajtmh.1990.42.532. [DOI] [PubMed] [Google Scholar]
- 47.Hill DR, et al. Primaquine: report from CDC expert meeting on malaria chemoprophylaxis I. Am J Trop Med Hyg. 2006;75:402. [PubMed] [Google Scholar]
- 48.Schlesinger PH, Krogstad DJ, Herwaldt BL. Antimalarial agents: mechanisms of action. Antimicrob Agents Chemother. 1988;32:793. doi: 10.1128/aac.32.6.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suswam E, Kyle D, Lang-Unnasch N. Plasmodium falciparum: the effects of atovaquone resistance on respiration. Exp Parasitol. 2001;98:180. doi: 10.1006/expr.2001.4639. [DOI] [PubMed] [Google Scholar]
- 50.Srivastava IK, et al. Resistance mutations reveal the atovaquone-binding domain of cytochrome b in malaria parasites. Mol Microbiol. 1999;33:704. doi: 10.1046/j.1365-2958.1999.01515.x. [DOI] [PubMed] [Google Scholar]
- 51.Zhou Y, et al. Large-scale annotation of small-molecule libraries using public databases. Journal of chemical information and modeling. 2007;47:1386. doi: 10.1021/ci700092v. [DOI] [PubMed] [Google Scholar]
- 52.Yan SF, et al. Learning from the data: mining of large high-throughput screening databases. J Chem Inf Model. 2006;46:2381. doi: 10.1021/ci060102u. [DOI] [PubMed] [Google Scholar]
- 53.Schuffenhauer A, et al. The scaffold tree--visualization of the scaffold universe by hierarchical scaffold classification. J Chem Inf Model. 2007;47:47. doi: 10.1021/ci600338x. [DOI] [PubMed] [Google Scholar]
- 54.Franke-Fayard B, et al. A Plasmodium berghei reference line that constitutively expresses GFP at a high level throughout the complete life cycle. Mol Biochem Parasitol. 2004;137:23. doi: 10.1016/j.molbiopara.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 55.Ploemen IHJ, et al. Visualisation and quantitative analysis of the rodent malaria liver stage by real time imaging. PLoS ONE. 2009;4:e7881. doi: 10.1371/journal.pone.0007881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 57.Ganesan SM, et al. Yeast dihydroorotate dehydrogenase as a new selectable marker for Plasmodium falciparum transfection. Molecular and biochemical parasitology. 2011;177:29. doi: 10.1016/j.molbiopara.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guiguemde WA, et al. Chemical genetics of Plasmodium falciparum. Nature. 2010;465:311. doi: 10.1038/nature09099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gamo FJ, et al. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465:305. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.