Abstract
Both high- and low-molecular-weight glutenin subunits (LMW-GS) play the major role in determining the viscoelastic properties of wheat (Triticum aestivum L.) flour. To date there has been no clear correspondence between the amino acid sequences of LMW-GS derived from DNA sequencing and those of actual LMW-GS present in the endosperm. We have characterized a particular LMW-GS from hexaploid bread wheat, a major component of the glutenin polymer, which we call the 42K LMW-GS, and have isolated and sequenced the putative corresponding gene. Extensive amino acid sequences obtained directly for this 42K LMW-GS indicate correspondence between this protein and the putative corresponding gene. This subunit did not show a cysteine (Cys) at position 5, in contrast to what has frequently been reported for nucleotide-based sequences of LMW-GS. This Cys has been replaced by one occurring in the repeated-sequence domain, leaving the total number of Cys residues in the molecule the same as in various other LMW-GS. On the basis of the deduced amino acid sequence and literature-based assignment of disulfide linkages, a computer-generated molecular model of the 42K subunit was constructed.
The glutenin fraction of the gluten proteins is primarily responsible for the viscoelastic properties of wheat (Triticum aestivum L.) flour doughs. It consists of various types of protein subunits that are linked together by intermolecular disulfide bonds. These form a polymeric mixture that has a broad molecular-weight distribution, with component polymers ranging from the dimeric forms with molecular weights as low as 60,000, to polymers containing many subunits with molecular weights in the millions (for review, see Kasarda, 1989; Wrigley, 1996). Variations in the types and amounts of subunits correlate with quality variations among wheat cultivars, probably by affecting the molecular-weight distribution of the glutenin polymers (Gupta et al., 1993, 1995). There are two main types of subunits, the HMW-GS and the LMW-GS, with the former having been much more extensively characterized than the latter.
Difficulties in characterization of LMW-GS arose because they derive from many more genes than HMW-GS and because the subunits are somewhat insoluble after reduction of the intermolecular disulfide bonds (which is necessary for their purification, but which also breaks down intramolecular disulfide bonds to expose buried hydrophobic regions). Until recently, almost all attempts at cloning lmw-gs genes led to DNA sequences corresponding to similar protein products that are not representative of the major LMW-GS types; almost all had the apparent N-terminal sequence METSCIPGL-, relatively low molecular weights of about 35,000 or less, and a total of eight Cys residues, including the Cys at position 5 (for review, see Shewry and Tatham, 1997; Cassidy et al., 1998).
In contrast to the apparently single type (with very minor variations) of the LMW-GS indicated by the DNA sequencing, two main types of LMW-GS have been defined on the basis of N-terminal amino acid sequences: the LMW-s and LMW-m types, with the former starting with the sequence SHIPGL-, and the latter represented by the METSHIPGL-, METSRIPGL-, or METSCIPGL- N-terminal sequences (Kasarda et al., 1988; Tao and Kasarda, 1989; Lew et al., 1992). The LMW-s types are predominant. They also tend to have higher molecular weights, in the approximate range of 35,000 to 45,000 relative to the LMW-m types, which seem to fall into the wider molecular-weight range of about 30,000 to 45,000 (Lew et al., 1992). In bread wheat cultivars, the LMW-m type with the METSHIPGL- sequence was the next most abundant type of LMW-GS, followed by the METSRIPGL- type, whereas the METSCIPGL- N-terminal sequence, typical of the cloned sequences, appeared to be somewhat rare among the types defined by direct protein sequencing (Lew et al., 1992).
Both LMW-s and LMW-m types are coded by genes present at the complex Glu-3 loci (Glu-A3, Glu-B3, and Glu-D3 in hexaploid wheat). Only partial sequence information has been available for the LMW-s types because they seemed almost impossible to clone. Recently, a partial DNA sequence that did not show the Cys residue at position 5 was published (accession no. X84960). Soon after, the complete DNA sequence of a lmw-gs gene from durum wheat was achieved that might correspond to either the LMW-s type or the LMW-m type without the Cys at position 5 (D'Ovidio et al., 1997). Both of these DNA-based sequences showed a Cys codon in the repeated-domain region. An apparently homologous Cys was found in glutenin by Köhler et al. (1993) and Keck et al. (1995), although because it was defined by proteolytic digestion of a residue glutenin preparation, peptide purification, and sequencing, the exact position in any defined LMW-GS was not available.
For some years a misunderstanding about the nature of LMW-GS has prevailed as a consequence of many investigators assuming that the cloned LMW-m type with Cys at position 5 was typical of LMW-GS, even if protein studies indicated otherwise. Consequently, we deemed it important to sequence a major LMW-s-type gene and provide extensive direct amino acid sequences for the corresponding protein to avoid similar pitfalls. In particular, we considered it of great importance to define all Cys residues in the primary structure of the subunit so that those likely to form intermolecular disulfide bonds could be determined (by comparison with disulfide-linked peptides described in the literature). This would enable the subunit to be classified as a potential chain extender (having two or more Cys residues that form intermolecular disulfide bonds) or as a chain terminator (having only one Cys available for intermolecular disulfide bond formation) during the not-yet-understood oxidative polymerization process that gives rise to the glutenin polymers in developing endosperm (Kasarda, 1989). A predominance of the chain-extender types in glutenin should lead to strong gluten with good viscoelastic properties, whereas too much of the chain-terminator types would have the opposite effect.
In this paper we report the isolation and characterization of a lmw-gs gene coding for a 42K LMW-s-type protein in the bread wheat cv Yecora Rojo, and show, for the first time to our knowledge, correspondence between a LMW-GS and its encoding gene through a comparison with extensive amino acid sequences of the purified polypeptide. We also present evidence for this particular subunit being very closely related to the 42K subunit found in durum wheat, which plays a major role in determining quality, and discuss the structural organization that might be responsible for this characteristic.
MATERIALS AND METHODS
The bread wheat (Triticum aestivum L. cv Yecora Rojo) used in these studies was obtained from the California Wheat Commission (Woodland). Flour was milled from the wheat with a Quadrumat Senior mill (C.W. Brabender Instruments, Inc., South Hackensack, NJ). The lot was designated CWC-141 and the quality characteristics of CWC-141 flour have been reported previously by MacRitchie et al. (1991).
DNA Extraction
Genomic DNA was isolated from 5 g of leaves from single plants, as reported previously (D'Ovidio et al., 1992).
PCR Analysis
Amplifications of the gene encoding the 42K LMW-GS were performed using primers and conditions reported by D'Ovidio (1993). Aliquots (10 μL) of the amplification products were fractionated on 1.5% agarose gel in 1× Tris-borate-EDTA buffer following standard procedures (Sambrook et al., 1989).
Cloning, Nucleotide Sequencing, and Computer Analysis
The amplification product of about 1.15 kb was purified from an agarose gel using the Gene Clean Kit (Bio101, La Jolla, CA) and ligated into the EcoRV dephosphorylated site of the pGEM-T plasmid vector (Promega) using standard procedures (Sambrook et al., 1989). After transformation into the Escherichia coli strain NM522, the recombinant colonies were analyzed to verify the presence of the 1.15-kb fragment. Several recombinant clones contained an insert but none of them was of the expected size. The insert size in the different clones was about 50 to 200 bp shorter than the expected size. Similar results have also been obtained using different E. coli strains, and it was demonstrated that single deletions of 50 to 200 bp had occurred within the repetitive domain of the different recombinant clones (R. D'Ovidio, unpublished data). To overcome this limitation, the nucleotide sequencing was carried out directly on the 1.15-kb PCR product using the Thermo Sequenase radiolabeled terminator cycle sequencing kit (Amersham). The PC/GENE computer program (Intelligenetics, Mountain View, CA) was used to analyze the sequence data.
Purification of LMW-GS
A fraction enriched in LMW-GS was obtained using a combination of published procedures. HMW-GS and LMW-GS were obtained according to the procedure of Singh et al. (1991) with the exceptions that extraction of glutenin from the residue was performed at room temperature with 50% (v/v) 1-propanol containing 50 mm Tris-HCl, pH 8.0, 1% (w/v) DTT, and 4 m urea, and alkylation was omitted. HMW-GS were precipitated according to the procedure of Melas et al. (1994) by adding acetone up to 40% (v/v). After 10 min at room temperature and 5 min of centrifugation at 40,000g (20°C), LMW-GS were selectively precipitated by bringing the acetone concentration up to 80% (v/v). After 10 min, centrifugation was again carried out as described above, and LMW-GS present in the pellet were resuspended in 25% (v/v) ACN containing 0.05% (v/v) TFA and 4 m urea.
RP-HPLC was carried out with the System Gold apparatus, version 3.11, composed of the solvent delivery module 126 and UV-detector module 166 (Beckman). About 1 mg of the protein preparation was filtered through a 0.45-μm membrane and fractionated onto a semipreparative C8 column (10 mm × 25 cm; Vydac, Hesperia, CA) equipped with an Aquapore guard column (4.6 mm × 3 cm; Applied Biosystems). A linear gradient of 35% to 49% aqueous ACN over 25 min at a flow rate of 1.5 mL/min was used. Solvent A was water plus 0.07% TFA, and solvent B was ACN plus 0.05% TFA. The columns were equilibrated at 50°C and proteins were detected by UV absorbance at 210 nm.
Single peaks were collected and analyzed on a mini SDS-PAGE apparatus (Bio-Rad) according to the instruction manual. The 42K LMW-GS we have analyzed in this paper corresponds to peak 10 described by Lew et al. (1992).
Determination of the Number of Cys Residues in the 42K LMW-GS
The number of Cys residues was determined by comparing the molecular weight of the alkylated 42K LMW-GS with that of the unalkylated polypeptide by MALDI-MS. These analyses were performed by an external service (Charles Evans and Associates, Redwood City, CA) according to procedures described by Wu et al. (1995). Alkylation was performed with 4-VP, as described by Lew et al. (1992). In some cases, 6 m guanidinium hydrochloride was used as an alternative to 4 m urea.
To reduce the experimental error attributable to the MALDI-MS technique by decreasing the mass of the peptide analyzed, we used the same procedure to analyze the peptides obtained from the 42K LMW-GS after digestion with the proteolytic enzyme endoprotease Lys-C (Boehringer Mannheim). Lys-C hydrolyzes specifically peptide bonds at the carboxylic side of Lys residues. The proteolytic digestion was performed according to the manufacturer's instructions.
Peptides obtained after Lys-C digestion were purified by RP-HPLC using the chromatographic system described above and a water-ACN gradient (both solvents A and B contained TFA) ranging from 32% to 46% in 35 min with a flow rate of 1.5 mL/min. The peaks collected were analyzed by mini SDS-PAGE and N-terminal amino acid sequencing. Samples of the resulting peptides were alkylated with 4-VP and submitted to MALDI-MS analysis.
Detection of Cys-Containing Peptides in the 42K LMW-GS
To identify the positions of all of the Cys residues present in the 42K LMW-GS, we initially followed the procedure reported by Egorov (1997). About 10 nmol of the 42K LMW-GS, purified by RP-HPLC and resuspended in a buffer containing 50 mm Tris-HCl, pH 7.6, 5 mm EDTA, and 6 m guanidinium hydrochloride, was attached to the support thiopropyl Sepharose 6B (Pharmacia), which specifically binds peptides with free sulfydryl groups. The support was previously hydrated with the protein resuspension buffer. After attachment of the protein, the support was equilibrated with 50 mm Tris-HCl, pH 8.0, and the protein was digested in situ with the proteolytic enzyme chymotrypsin (sequencing grade, Boehringer Mannheim), according to the manufacturer's instructions. Peptides that did not contain Cys residues were removed from the resin by a washing step, whereas Cys-containing peptides that remained selectively attached to the support were subsequently detached by adding 0.5% DTT, and collected. These latter peptides were fractionated by RP-HPLC with a 5% to 35% ACN gradient (with TFA) for 120 min at a flow rate of 1.5 mL/min. Peaks were collected, alkylated with 4-VP, and repurified by RP-HPLC, and the collected peaks were dried in a Speed-Vac apparatus (Savant, Farmingdale, NY) for further characterization.
The same procedure was applied to the N-terminal portion of the 42K LMW-GS obtained after Lys-C digestion, and this fragment was also subjected to direct chymotrypsin digestion (without attachment to the thiopropyl Sepharose column), under the same conditions as described above, after alkylation with 4-VP. The chymotryptic fragments of the N-terminal peptide were purified by RP-HPLC under the same conditions.
Amino Acid Sequencing and Composition Analysis
Protein/peptide sequencing was performed as described by Lew et al. (1992). The 42K LMW-GS amino acid composition was compared with that of a typical LMW-m-type subunit that had a Cys at position 5, also purified from cv Yecora Rojo (corresponding to peak 14 of Lew et al. [1992]) by a procedure similar to that reported above. Amino acid analyses were performed by the Protein Structure Laboratory (University of California, Davis) using an analyzer (model 6300, Beckman) with methods as described by Ozols (1990).
Computer Molecular Modeling: Structure and Flexibility
A computer molecular model was based on the gene-derived amino acid sequence, the secondary structure prediction, and the putative intramolecular and intermolecular disulfide linkages, as defined by Köhler et al. (1993), Keck et al. (1995), and Müller and Wieser (1997) for homologous LMW-GS and γ-gliadins. The model was constructed largely by methods described previously (Kasarda et al., 1994; D'Ovidio et al., 1995; Köhler et al., 1997). Secondary structure prediction was carried out with the PHD program of Rost and Sander (1993, 1994), version 5.94_317. A Silicon Graphics (Mountain View, CA) Personal Iris workstation running Quanta 4.0 and CHARMm (version 23.1) software (Molecular Simulations, San Diego, CA) was used for the molecular modeling.
To avoid distortion of assigned secondary structure when disulfide connections were patched in, the torsional angles of arbitrarily selected amino acids for which no secondary structure assignment was indicated by the PHD analysis were adjusted in a serial process that included frequent energy minimizations, to bring the Cys residues assigned to a disulfide pair moderately close together (within about 1 nm) before turning on disulfide patching. Because of the large size of the protein and the limitations of the semiempirical force fields in dealing with solvent water, we did not attempt to study the effects of hydration on our structures. Final equilibration was for 30 to 300 ps (various models), followed by energy minimization.
Polypeptide chain flexibility for the 42K LMW-GS was predicted by the method of Karplus and Schulz (1985), in which B-values (individual atomic temperature factors) for the Cα atoms of selected proteins of known three-dimensional structure from the protein data bank were correlated with chain flexibility. Nearest-neighbor effects were included in the predictive method. The software used was part of the MacVector package (Oxford Molecular Group, Campbell, CA).
RESULTS
PCR Amplification and Nucleotide Sequence of the Gene Encoding the 42K LMW-GS
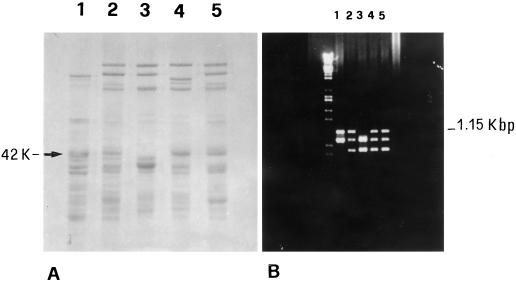
SDS-PAGE patterns of the glutenin subunits found in the bread wheat cv Yecora Rojo showed the presence of a 42K LMW-GS (Fig. 1A, lane 4), the mobility of which corresponds to the main LMW-GS present in the LMW-2 type of durum wheat (Fig. 1A, lane 1), which exhibit good quality relative to the LMW-1 types. This particular component is also present in some other good-quality bread wheats (Fig. 1A, lanes 2 and 5) (Masci et al., 1998). PCR analysis using primers specific for a gene encoding one of the LMW-2 components (D'Ovidio et al., 1996) confirmed this correspondence. The 1.15-kb fragment that has been shown to encode a LMW-GS belonging to the LMW-2 type in durum wheat (D'Ovidio et al., 1996) is also present in those bread wheat cultivars that show the 42K LMW-GS, including Yecora Rojo (Fig. 1B) (Masci et al., 1998). On the basis of these results, the 1.15-kb PCR product from the bread wheat cv Yecora Rojo was sequenced. The nucleotide sequence revealed that the PCR product is 1144 bp long and corresponds to a lmw-gs gene, the complete sequence of which is reported (accession no. Y17845). The deduced amino acid sequence comprises 381 amino acids and includes the complete coding region and part of the signal peptide. The part of the signal peptide included is similar to that reported for other LMW-GS (Fig. 2).
Figure 1.
Comparison between SDS-PAGE (A) and the PCR-amplification pattern (B) of durum wheat cv Lira biotype 45 (lane 1) and bread wheat cvs Red River (lane 2), Cheyenne (lane 3), Yecora Rojo (lane 4), and Solar (lane 5). Both the 42K LMW-GS protein band and the 1.15-kb PCR product (both indicated) were present in all bread wheat cultivars except cv Cheyenne.
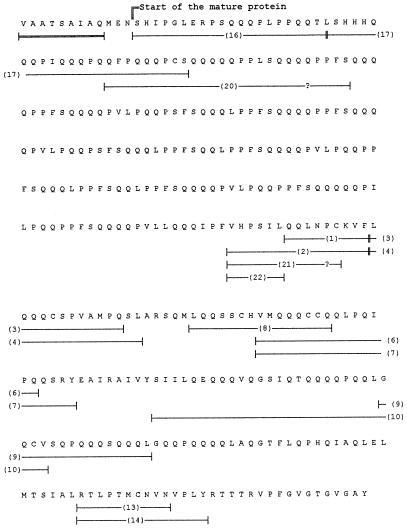
Figure 2.
Deduced amino acid sequence of the lmw-gs gene corresponding to the 1.15-kb PCR product. Underlines show those peptides that confirmed correspondence with the 42K LMW-GS. The peptide numbers are keyed to Figure 5. The double-underlined sequence represents the final part of the signal peptide typical of LMW-GS. Question marks (?) indicate unidentified amino acids. The complete sequence is reported under accession no. Y17845.
The deduced protein sequence was compared with the amino acid sequence obtained by direct sequencing of the 42K LMW-GS from the bread wheat cv Yecora Rojo (see below). The direct sequence covered nearly 50% of the total sequence. On the basis of this comparison, it was possible to determine that the Ser at position 13 is the first amino acid of the mature protein. However, it is noteworthy that the signal peptide includes the MEN sequence (Fig. 2). The presence of an Asn residue instead of a Thr residue, as in the MET sequence, the latter being typical of the LMW-m type, might be the cause of the different signal cleavages between the LMW-s and LMW-m types. Based on the identification of the first amino acid of the mature protein, it was possible to calculate that the 1.15-kb PCR product codes for a mature LMW-GS, with 369 amino acid residues having a molecular weight of 42,111 and a pI of 8.31. The hydropathy profile (data not shown) revealed the hydrophilic character of the repetitive domain and a more hydrophobic character of the C-terminal domain and the short N-terminal region (about 10 amino acids). The repetitive domain is composed of repeats having the consensus sequence PPFSQQQQ. There are 25 repetitions of the consensus sequence or its variants. Although the octapeptide repeat was the most common, hexapeptide, heptapeptide, and nonapeptide repeats were also present, with variations in the number of Gln residues mostly responsible for the differences (Fig. 3).
Figure 3.
Alignment of the amino acid repeats found in the repetitive domain of the 42K LMW-GS encoded by the 1.15-kb PCR product. The figure shows the regular distribution of the repeats from amino acids 13 to 163 of the deduced mature protein. Cons, Consensus sequence.
To date, published nucleotide and amino acid sequence comparisons between the 1.15-kb PCR product and the lmw-gs genes showed a high degree of homology (65%–85%) along the entire sequence, with the main differences being located within the repetitive domain. Similar to the other lmw-gs genes encoded at the Glu-B3 locus (accession no. X84960; D'Ovidio et al., 1996, 1997), the 1.15-kb PCR product showed the presence of a Cys codon in the repetitive domain and seven additional Cys residues in the C-terminal domain.
Characterization of the 42K Protein and Amino Acid Sequencing of the Cys-Containing Chymotryptic Peptides
Direct N-terminal sequencing of the 42K LMW-GS up to about 30 amino acids has been reported (Lew et al., 1992). To analyze in more detail specific portions of the molecule and to verify the possible correspondence with the 42K lmw-gs gene, we characterized primary structural regions of the protein that included the Cys residues.
To compare the purified 42K LMW-GS with a typical LMW-m type having Cys at position 5 and a molecular weight of about 33,000 (peak 14 of Lew et al., 1992), we submitted both to amino acid-compositional analysis. The results are reported in Table I. Because it is difficult to determine the number of Cys residues in a protein with high accuracy by compositional analysis, this approach was not used; however, the compositional data provided useful information about the possible structure of the two proteins. It is evident that the major difference found between the two types of subunits was mainly in the content of Glu/Gln and Pro, amino acids that are especially common to the repeated sequence domain. Differences in molecular weight among LMW-GS are likely to result from variations in the number of repeated units in the repeated sequence domain, as also seems to be true for HMW-GS (D'Ovidio et al., 1994).
Table I.
Amino acid composition of the 42K LMW-GS and of a LMW-m type with the Cys residue at position 5
| Amino Acid | 42K LMW-GS | LMW-m |
|---|---|---|
| mol % | ||
| Ala | 2.6 | 3.8 |
| Arg | 2.2 | 2.0 |
| Asn + Asp | 1.6 | 2.3 |
| Cys | 1.9 | 2.6 |
| Glu + Gln | 37.2 | 34.1 |
| Gly | 3.5 | 4.5 |
| His | 1.8 | 1.4 |
| Ile | 3.7 | 4.5 |
| Leu | 8.6 | 8.6 |
| Lys | 0.5 | 0.8 |
| Met | 1.4 | 1.7 |
| Phe | 4.7 | 4.2 |
| Pro | 15.7 | 13.2 |
| Ser | 7.2 | 6.4 |
| Thr | 2.7 | 3.2 |
| Tyr | 1.5 | 1.8 |
| Val | 4.9 | 5.7 |
We attempted to determine the exact number of Cys residues in the intact 42K subunit by MALDI-MS. Because the molecular weight of the alkylating group was 105, the results shown in Table II indicate only seven Cys residues in the 42K LMW-GS instead of the expected eight. However, the error reported for MALDI-MS is 0.1%. Thus, for a protein with a molecular weight of 42,000, there is the possibility of over- or underestimating the number of Cys residues by about one residue. Alternatively, the absence of one Cys residue after MALDI-MS analysis on the intact protein might be caused by partial alkylation of the protein in the repeated region. The molecular weight of the 42K LMW-GS, as deduced by MALDI-MS, was 42,123, which is practically identical to the value of 42,111 as determined from the deduced amino acid sequence.
Table II.
Comparison of the molecular weights of the 4-VPalkylated 42K LMW-GS versus the unalkylated form, as determined by MALDI-MS
| Subunit | Alkylated Protein | Unalkylated Protein | Difference | No. of Cys Residues |
|---|---|---|---|---|
| 42K LMW-GS | 42,846 | 42,123 | 723 | 7 |
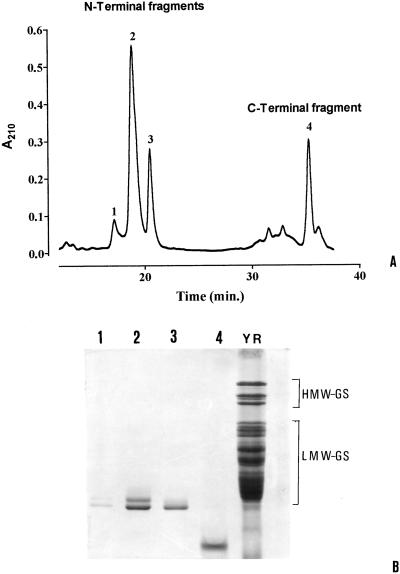
To determine with greater certainty the exact number of Cys residues, we analyzed peptides obtained from the 42K LMW-GS after Lys-C digestion. The decision to use this proteolytic enzyme came from the observation that the 42K LMW-GS has only one Lys residue immediately following the first Cys residue present in the C-terminal domain (Fig. 2), which is characteristic of all Glu-3-coded subunits, based on published sequences. Consequently, Lys-C digestion should result in only two peptides, which, after fractionation, could then be analyzed independently by MALDI-MS with greater accuracy because of the smaller molecular weights. Purification of the resulting Lys-C fragments by RP-HPLC (Fig. 4A), SDS-PAGE (Fig. 4B), and N-terminal amino acid sequencing indicated that the first three peaks shown in Figure 4A all exhibited the N-terminal sequence of the molecule, whereas peak 4 exhibited a sequence corresponding to the amino acids following the single Lys residue (data not shown). Peak 4, then, corresponds approximately to the C-terminal domain of the protein. The presence of three N-terminal fragments after Lys-C digestion might be attributable to minor heterogeneities of the 42K LMW-GS that were not resolved during analyses of the intact molecule. Lew et al. (1992) reported heterogeneity at three positions in the first 30 N-terminal amino acids of peak 10, corresponding to the 42K LMW-GS. In particular, they reported the presence of a Lys residue at position 8, instead of an Arg residue. The SDS-PAGE analysis of the protein corresponding to peak 2 indicated two bands (Fig. 4B, lane 2). However, neither the MALDI-MS analyses of the fragments nor the N-terminal amino acid sequences revealed the presence of a Lys-C fragment lacking the first eight amino acids. Perhaps the two bands in the SDS-PAGE pattern correspond to different conformational forms that are not normalized in SDS solution.
Figure 4.
RP-HPLC separation of the Lys-C digests of the 42K LMW-GS (A) and the SDS-PAGE pattern of the peaks collected (B).
The major peaks 2, 3, and 4 were submitted to MALDI-MS analyses in both the unalkylated and alkylated forms (Table III). Peaks 2 and 3, which corresponded in sequence to the N-terminal part of the molecule and extended slightly into the unique sequence region to include the first Cys of the C-terminal domain (Fig. 2), each showed mass differences for the alkylated and unalkylated forms, corresponding to two pyridylethylated Cys residues. Peak 4, which corresponded (according to its N-terminal sequence) to most of the C-terminal domain, had a mass difference that indicated six Cys residues. Accordingly, the MALDI-MS results clearly show that eight Cys residues were present in the 42K LMW-GS.
Table III.
Comparison of the molecular weights of the 4-VPalkylated Lys-C fragments versus the unalkylated peptides, as determined by MALDI-MS
| Peak | Alkylated Fragment | Unalkylated Fragment | Difference | No. of Cys Residues |
|---|---|---|---|---|
| 2 | 24,822 | 24,612 | 210 | 2 |
| 3 | 24,896 | 24,684 | 212 | 2 |
| 4 | 17,949 | 17,356 | 593 | 6 |
To gain more information about the internal amino acid sequence of the 42K LMW-GS, this protein was analyzed by direct N-terminal amino acid sequencing of the Cys-containing peptides obtained by covalent attachment of the protein to the sulfydryl matrix, followed by in situ chymotryptic digestion and RP-HPLC fractionation of the resulting peptides. The results obtained (Fig. 5A) defined sequences that included all of the seven Cys residues present in the C-terminal domain of the molecule, but failed to define the expected Cys in the repeated sequence domain. Although we could offer some speculation, we are uncertain why we were unable to recover this particular peptide. A few positions in the sequences were found to be heterogeneous in that two amino acids were identified during sequencing. This is in agreement with the earlier work of Lew et al. (1992) and Vensel et al. (1995).
Figure 5.
Amino acid sequences of Cys-containing chymotryptic peptides obtained after covalent chromatography (A) and peptides obtained after chymotryptic digestion of the Lys-C N-terminal fragment of the 42K LMW-GS (B) (peak 2 of Fig. 4A). The alternative amino acids at positions where heterogeneity is present are reported.
To find the missing Cys residue, we carried out chymotryptic digestion of peak 2 (pyridylethylated) from the Lys-C digestion (Fig. 4A), which includes the complete repeated sequence region, and sequenced the resulting peptides. The expected Cys residue was found among the sequences obtained (Fig. 5B, peptides 17–20). Again, some minor heterogeneities were found at a few positions.
All of the peptides sequenced were aligned with the amino acid sequence deduced from the gene sequence (Fig. 2). The directly obtained peptide sequences corresponded to about 50% of the gene-based sequence and showed perfect agreement with it. Even where amino acid heterogeneities were present in a peptide sequence at an occasional cycle (Fig. 5), at least one of the amino acids found at that cycle corresponded to the amino acid defined by the gene sequence.
Computer Molecular Modeling: Structure and Flexibility
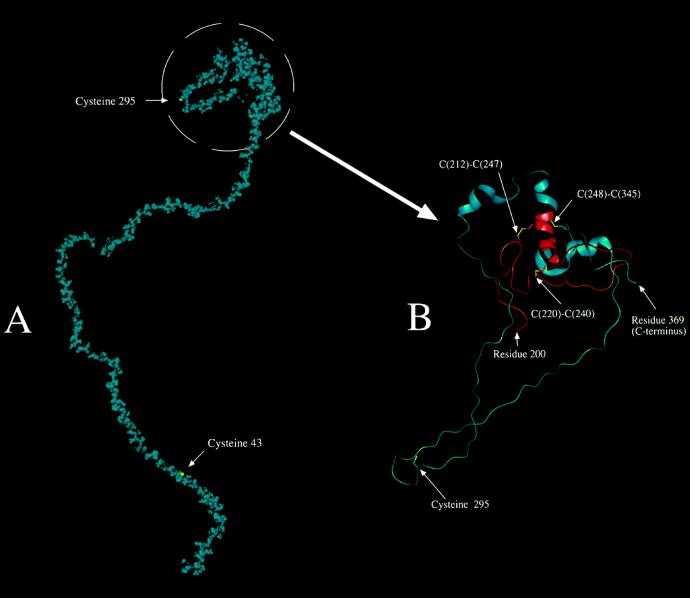
A molecular model was constructed from the primary structure of the 42K LMW-GS, as defined by the gene sequence. The secondary structure prediction indicated 6% or fewer of the residues in the α-helical conformation, a smaller amount of extended/strand structure, and the remainder almost entirely undefined (loop). Consequently, we applied only the predicted α-helical and extended structure to the molecule. The structure was energy minimized, subject to simulated heating to 300 K, and then equilibrated for 30 ps. By adjusting the model (before heating and equilibration) as described in “Materials and Methods,” a final minimized, equilibrated structure with suitable negative energy and low root-mean-square force could be achieved that still retained, for example, nearly all of the assigned α-helical structure. Failure to carry out prior conformational adjustments, as described, resulted in the complete loss of predicted and assigned α-helical structure surrounding the Cys residues during disulfide patching followed by energy minimization. The resulting speculative model is shown in a space-filling format (van der Waals's radii) in Figure 6A for a 30-ps equilibration. The C-terminal domain is expanded and presented in protein cartoon format in Figure 6B to provide a better illustration of the intramolecular disulfide bond arrangements. Although there were small differences for the model in which equilibration had been carried out for 300 ps, important conclusions based on the 30-ps model were not affected.
Figure 6.
Computer-generated molecular model of the 42K subunit. A, Entire 42K LMW-GS shown in space-filling form (van der Waals's radii for atoms) with all atoms shown in blue, except the sulfur atoms of Cys or cystine side chains, which are shown in yellow. B, Simplified model of the region containing the intramolecular disulfide linkages and Cys-295, which presumably forms one of two intermolecular disulfide cross-linkages. Residues 200 to 369 of the 42K subunit are shown in protein cartoon format, which displays the α-helical structure as a helical ribbon. There is also a specific display of selected side chains in a licorice (stick) bond format. The sulfur atoms of the Cys and cystine residues are shown in yellow. The main polypeptide chain is shown in red for residues 200 to 248 and in blue from there on to the C-terminal end at residue 369. The numbers of connected (intramolecular) Cys residues are shown.
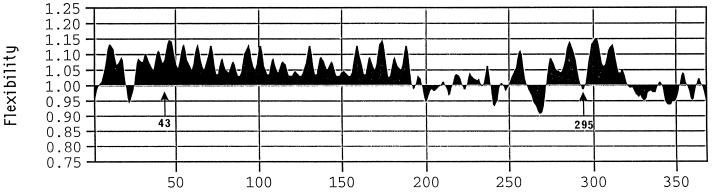
Flexibility modeling (Karplus and Schulz, 1985) indicated a high degree of flexibility for the entire repeating-sequence domain of the protein, particularly for the stretches of Gln residues included in the repeats (Fig. 7). This high degree of flexibility also applied to the repeating sequences surrounding the Cys residue at position 43 of the mature protein sequence (beginning with the SHIP- sequence). In addition, although at Cys-295 of the mature protein sequence the polypeptide chain was predicted to have slightly lower-than-average chain flexibility, this short region consisting of just a few residues was flanked by extensive regions predicted to have considerable flexibility, largely because of a concentration of Gln residues in these flanking regions. Gln residues in various known structures had temperature factors that indicated a high degree of flexibility for this residue, particularly when there was more than one Gln in a series (Karplus and Schulz, 1985).
Figure 7.
Flexibility of the polypeptide chain of the 42K LMW-GS, as predicted by the method of Karplus and Schulz (1985). A coefficient of 1.0 represents an average flexibility, and lower values indicate less-than-average flexibility. Positive values of 1.15 seen for some regions of the 42K LMW-GS are equivalent to highly flexible regions of the polypeptide chain, based on the model proteins used in the development of the predictive method. Arrows indicate the positions of the two Cys residues in the 42K subunit that are supposed to form intermolecular disulfide bonds.
DISCUSSION
LMW-GS are in large excess over HMW-GS in doughs, i.e. glutenin polymers are composed mainly of LMW-GS. Because the LMW-GS are more numerous and more difficult to purify, and because their structural definition through nucleotide sequencing has been more problematical than for HMW-GS, they have been poorly characterized so far. As a consequence, no correspondence between a gene coding for a LMW-GS and a recognized protein has been defined. Here we report the complete nucleotide sequence of a lmw-gs gene from the bread wheat cv Yecora Rojo. We show that peptide sequences corresponding to almost 50% of the total number (369 in the mature protein) of residues in this particular LMW-GS were identical to the equivalent regions of the deduced amino acid sequence from nucleotide sequencing. We also show that the molecular weight obtained by MALDI-MS was virtually identical to that calculated from the corresponding gene product. Furthermore, the protein we characterized from the bread wheat cv Yecora Rojo is strongly homologous to a protein that is clearly correlated with good quality in LMW-2-type durum wheat cultivars.
We have defined all eight Cys residues of our 42K LMW-GS in the context of their adjacent sequences. On the basis of homologies with Cys residues that have been shown to form either intermolecular or intramolecular disulfide bonds (Köhler et al., 1993; Keck et al., 1995), we suggest that the first and seventh Cys residues in the sequence are likely to participate in intermolecular disulfide bond formation, whereas the remaining Cys residues are likely to form intramolecular disulfide bonds (Fig. 6). The sequences that participate in intramolecular disulfide linkages appear to be conserved (Shewry and Tatham, 1997) and are readily recognized compared with the sequences surrounding the disulfide bonds in other glutenin subunits and in gliadins, apparently because stronger evolutionary constraints are required for the formation of intramolecular bonds (Keck et al., 1995). In contrast, the positions and surrounding sequences of the first Cys, located in the N-terminal part of the molecule, and the seventh Cys, located in the C-terminal part of the molecule, are more variable (D'Ovidio et al., 1997; Shewry and Tatham, 1997).
It does not seem likely that conformational structure plays an important role in the formation of intermolecular disulfide cross-linkages. There are bound to be some steric effects on sulfydryl accessibility and reactivity, but our model (see below) suggests that the two Cys residues available for intermolecular disulfide-bond formation are in the flexible regions of the polypeptide chain, making them about equally accessible. These latter two Cys residues would define the 42K subunit as a linear chain extender (Lew et al., 1992) capable of enhancing chain length during glutenin polymer formation. If the 42K subunit is indeed a linear chain extender, this would be in accordance with the correlation of this type of subunit with good quality, which has been well established, at least for the equivalent protein found in durum wheats (Carrillo et al., 1990; Masci et al., 1995).
On the basis of these results and of those in the literature, it seems plausible that both LMW-m and LMW-s types have two Cys residues available for the formation of intermolecular disulfide bonds. Although no work has been carried out on LMW-m types that do not have a Cys residue at position 5 (e.g. those having the N-terminal sequence METSHIPGL-), it seems likely that this type possesses a Cys residue in the repeating sequence domain (the Cb* Cys described by Köhler et al. [1993]) in place of the Cys at position 5. If both main types of LMW-GS act as chain extenders, with just two Cys residues available for intermolecular disulfide-bond formation, the positive correlation between the relative abundance of the LMW-s types and the good end-use quality of flours has to be attributed to their being present in greater amounts in good-quality flours rather than to intrinsic structural characteristics. This correlation between larger amounts of LMW-s-type proteins and good quality is in agreement with the difference in quality found between two biotypes of the Italian durum wheat cv Lira. These biotypes differ at the complex locus Gli-B1/Glu-B3 in having the poor-quality allelic variant of LMW-GS (LMW-1) in one biotype but having the good-quality allelic form LMW-2 in the other. Masci et al. (1995) found that these two biotypes differ mainly in the abundance of a LMW-GS that is highly homologous to the 42K LMW-GS described here. The hypothesis that the amount of LMW-s types is positively correlated with good quality is also supported by the characterization of two allelic lmw-gs genes (D'Ovidio et al., 1996), the products of which have different effects on quality. The deduced protein products from the durum lines show a difference of only 15 amino acids within the repetitive domain, which is insufficient to explain the different effects on quality, whereas these products differ in quantity, with the protein product corresponding to the “best” allele being present in a significantly greater amount.
The 42K LMW-GS has a higher molecular weight than the other LMW-glutenin subunits, presumably because of the presence of a larger number of repeated units. The repeating sequence in this domain consists mainly of the core sequence PPFSQQ (D'Ovidio et al., 1997), which appears about 25 times. This relatively consistent part of the repeat is interrupted by a series of Gln residues (varying from zero to three residues; see Fig. 3). These repeats are fairly regular in character, more so than in other LMW-GS, and this characteristic might also exert a positive influence on gluten quality, as measured by dough strength and elasticity. In this regard, it is notable that a correlation between the length of the repeated-sequence domain and gluten-quality characteristics has already been reported for the HMW-GS (Anderson et al., 1996). We suggest that these longer repeated regions in LMW-GS enhance the contributions to dough strength and elasticity relative to subunits with smaller-sized repeated-sequence domains.
The molecular model illustrated in Figure 6A did not lead to any indication of regular structure for the repeated-sequence region. Secondary-structure prediction left this region in the default (unpredicted) category, although we should point out that the PHD program does not attempt to define turns that are probably dependent on tertiary interactions for stability to a considerable extent (Yang et al., 1996). All secondary structure prediction is somewhat uncertain in the absence of exact three-dimensional structural information from physical methods such as x-ray diffraction or NMR analysis, both of which seem largely inapplicable to gluten proteins, but it seems safe to say that, in general, α-helix prediction has a higher probability of accuracy than β-strand prediction and turn prediction. The situation is complicated further by the absence of more than weak similarities between gluten protein primary structures and proteins of known three-dimensional structures in the protein data bank.
We suggest that the stretches of Gln residues in the repeats will likely be largely unordered and flexible in solution, as was found by Altschuler et al. (1997) for peptides containing strings of Gln residues. The conclusion that the Gln residues in the repeating sequences will be flexible and unordered was also supported by the chain-flexibility prediction (Karplus and Schulz, 1985). The finding that a 10-residue Gln insert into a model protein was unstructured and highly dynamic (Ladurner and Fersht, 1997) is also in accord with our suggestion. The potential for the non-Gln part of the sequence to form turn structures is unlikely to be of any great importance, because tight turns are not highly stable in the absence of significant hairpin character in the adjacent strands or in the absence of favorable tertiary structure (Yang et al., 1996). Also, the dihedral angle limitation for Pro combined with the Pro-Pro motif of the repeats does not favor tight turns, but, rather, a shallow turn angle corresponding to a somewhat extended conformational structure, analogous to the Pro residues in a left-handed polyproline II-type helix (Toumadje and Johnson, 1995).
Our molecular model showed fairly frequent inverse γ-turns that had formed spontaneously during energy minimization and equilibration, but these shallow turns did not promote regular structure in the repeated region. The stability of these three-residue turns in a hydrated structure is unknown.
The irregular amounts of Gln in the repeats, ranging from two to five residues, may be functional by way of a tendency to prevent the polypeptide chain from forming any highly regular intra- and intermolecular interactions. If all repeats had exactly the same number of Gln residues, there would be some possibility of the formation of a large-diameter spiral, but the varying lengths of Gln residues that include both odd and even numbers of residues will tend to diminish the likelihood that such a regular structure might form. For the purposes of storage-protein deposition in the endosperm cells and, ultimately, enzymatic degradation of the storage proteins upon germination of the seed, highly regular repeats involving Gln (with its strong tendency to act as both hydrogen bond donor and acceptor) might lead to strongly interacting aggregates having excessive insolubility and enzyme inaccessibility. There is some indication that highly regular, identical repeats that include a series of adjacent Gln residues tend to be poorly insoluble in aqueous solutions (D.D. Kasarda, unpublished results). These variations in the number of Gln residues per repeat might simply reflect tolerated drift in the DNA-replication process, but there is also the possibility that irregular repeats serve to stabilize the total number of repeats in the domain insofar as exact repeats might be more prone to expansion and contraction during DNA replication.
When high concentrations of proteins are present, as in water-flour doughs, the Gln stretches in the somewhat extended repeated-sequence domain are likely to interact intermolecularly with their counterparts in other protein molecules through side-chain- and main-chain-amide hydrogen bonding. This type of interaction, modulated by interaction with water molecules as the plasticizer and with the monomeric gliadin proteins, would very likely contribute positively to the viscoelasticity of the system (Belton, 1994).
In contrast to the large repeated-sequence domain, the intramolecularly disulfide-bonded C-terminal domain occupies a relatively small, compact part of the overall protein molecule in our model (Fig. 6B). Folding that gives rise to the intramolecular disulfide bonds may be relatively straightforward. Examination of the model suggests the following scenario: folding of the shortest loop (C[220]–C[240]) gives rise to the first disulfide bond, which places C(212) in a position to interact with C(247), leaving three Cys residues of the C-terminal domain unreacted. C(295), which apparently forms an intermolecular disulfide bond, is located in a somewhat flexible loop, perhaps stiffened by some extended structure. Flexibility in the region of C(295) was supported by the flexibility prediction. In any case, we postulate that there is sufficient structure to this loop to make it impossible for C(295) to access conformational space in the vicinity of either C(248) or C(345). Accordingly, these latter two Cys residues, which are homologous to certain Cys residues of related subunits and gliadins, which are known to oxidize to form an intramolecular bond, eventually find one another in conformationally allowed space and form the third intramolecular disulfide bond. To what extent disulfide-bond formation is catalyzed by protein disulfide isomerase, chaperones, or other types of proteins is not well defined. All of the predicted α-helix in the molecule seems to be located in the vicinity of the intramolecular disulfide bonds. There is at least the possibility that helix-helix interactions are involved in guiding the formation of the intramolecular disulfide bonds, although such interactions were not evident in our model.
Finally, it is noteworthy that the nucleotide sequence of the lmw-gs reported here also includes the codon corresponding to an Asn residue instead of the codon for a Thr residue. This gives rise to an MEN sequence in our subunit, which is homologous to the MET sequence typical of the N terminus of the LMW-m-type subunits. So far, no LMW-m type having the MEN sequence instead of the MET sequence has been reported (for review, see Shewry and Tatham, 1997; Cassidy et al., 1998). Because, in the subunit we describe, the MEN sequence is part of the signal peptide rather than corresponding to the N-terminal sequence, as in LMW-m types, but is close to the signal cleavage site, it may be speculated that differential processing occurs between the LMW-m and LMW-s types because of the presence of the Asn in the MEN sequence. This differential processing might give rise to the slightly different N-terminal sequences characteristic of LMW-m versus LMW-s types.
Nevertheless, the classification of LMW-GS as the LMW-m and LMW-s types probably has no essential importance in itself with regard to dough quality. The main difference among the various LMW-GS resides in the presence of the first Cys residue either in the short, nonrepetitive N-terminal region or farther along in the repetitive domain, but all seem likely to have two Cys residues available for intermolecular disulfide-bond formation, are chain extenders, and form linear, as opposed to branched, polymers. The likely similarity of both types in having two Cys residues available for intermolecular disulfide-bond formation, one located near the N terminus and the other located near the C terminus, probably predominates in determining how these similar types of subunits affect properties significant to good quality.
ACKNOWLEDGMENTS
We thank Donald D. Kuzmicky (Western Regional Research Center [WRRC]) for carrying out the amino acid-sequencing analyses, Sam Huang (California Wheat Commission) for supplying and milling the wheat sample, Marco Spigaglia (DABAC, Viterbo, Italy) for technical assistance, Dave Rockhold (WRRC) for carrying out the MacVector program analysis, and Olin D. Anderson and Susan B. Altenbach (WRRC) for helpful discussion.
Abbreviations:
- ACN
acetonitrile
- HMW-GS
high-molecular-weight glutenin subunit(s)
- LMW-GS
low-molecular-weight glutenin subunit(s)
- MALDI-MS
matrix-assisted laser-desorption ionization-MS
- RP-HPLC
reversed-phase HPLC
- TFA
trifluoroacetic acid
- 4-VP
4-vinylpyridine
Footnotes
This research was supported in part by the Italian Ministero delle Risorse Agricole, Alimentarie Forestali, National Research Project “Plant Biotechnology,” the Italian Ministero per l'Universita' e la Ricerca Scientifica e Tecnologica, and the National Research Project “Studio delle proteine dei cereali e loro relazioni con aspetti tecnologici e nutrizionali.”
LITERATURE CITED
- Altschuler EL, Hud NV, Mazrimas JA, Rupp B. Random coil conformation for extended polyglutamine stretches in aqueous soluble peptides. J Peptide Res. 1997;50:73–75. doi: 10.1111/j.1399-3011.1997.tb00622.x. [DOI] [PubMed] [Google Scholar]
- Anderson OD, Békès F, Kuhl J, Tam A (1996) Use of bacterial expression system to study wheat high-molecular-weight (HMW) glutenins and the construction of synthetic HMW-glutenin genes. In CW Wrigley, ed, Proceedings of the 6th International Gluten Workshop. Cereal Chemistry Division, Melbourne, Australia, pp 195–198
- Belton PS (1994) A hypothesis concerning the elasticity of high molecular weight subunits. In Wheat Kernel Proteins: Molecular and Functional Aspects. Università della Tuscia, Viterbo, Italy, pp 159–165
- Carrillo JM, Vazquez JF, Orellana J. Relationship between gluten strength and glutenin proteins in durum wheat cultivars. Plant Breeding. 1990;104:325–333. [Google Scholar]
- Cassidy BG, Dvorak J, Anderson OD. The wheat low-molecular-weight glutenin genes: characterization of six new genes and progress in understanding gene family structure. Theor Appl Genet. 1998;96:743–750. [Google Scholar]
- D'Ovidio R. Single-seed PCR of LMW glutenin genes to distinguish between durum wheat cultivars with good and poor technological properties. Plant Mol Biol. 1993;22:1173–1176. doi: 10.1007/BF00028988. [DOI] [PubMed] [Google Scholar]
- D'Ovidio R, Porceddu E, Lafiandra D. PCR analysis of genes encoding allelic variants of high-molecular-weight glutenin subunits at the Glu-D1 locus. Theor Appl Genet. 1994;88:175–180. doi: 10.1007/BF00225894. [DOI] [PubMed] [Google Scholar]
- D'Ovidio R, Simeone M, Marchitelli C, Masci S, Porceddu E (1996) Isolation and characterisation of members of the LMW glutenin gene family in durum wheat. In CW Wrigley, ed, Proceedings of the 6th International Gluten Workshop. Cereal Chemistry Division, Melbourne, Australia, pp 81–84
- D'Ovidio R, Simeone M, Masci S, Porceddu E. Molecular characterization of a LMW-GS gene located on chromosome 1B and the development of primers specific for the Glu-B3 complex locus in durum wheat. Theor Appl Genet. 1997;95:1119–1126. [Google Scholar]
- D'Ovidio R, Simeone M, Masci S, Porceddu E, Kasarda DD. Nucleotide sequence of a γ-gliadin type gene from a durum wheat: correlation with a γ-type glutenin subunit from the same biotype. Cereal Chem. 1995;72:443–449. [Google Scholar]
- D'Ovidio R, Tanzarella OA, Porceddu E. Isolation of an alpha-type gliadin gene from Triticum durum Desf and genetic polymorphism at the Gli-2 loci. J Genet Breed. 1992;46:41–48. [Google Scholar]
- Egorov TA. Identification of cysteine residues and disulfide bonds in proteins. In: Kamp RM, Choli-Papadopolu T, Wittman-Liebold B, editors. Protein Structure Analysis. Heidelberg, Germany: Springer-Verlag; 1997. pp. 259–268. [Google Scholar]
- Gupta RB, Khan K, MacRitchie F. Biochemical basis of flour properties in bread wheats. I. Effects of variation in the quantity and size distribution of polymeric protein. J Cereal Sci. 1993;18:23–41. [Google Scholar]
- Gupta RB, Popineau Y, Lefebvre J, Cornect M, Lawrence GJ, MacRitchie F. Biochemical basis of flour properties in bread wheats. II. Changes in polymeric protein formation and dough/gluten properties associated with the loss of low Mr or high Mr glutenin subunits. J Cereal Sci. 1995;21:103–116. [Google Scholar]
- Karplus PA, Schulz GE. Prediction of chain flexibility in proteins. Naturwissenchaften. 1985;72:212–213. [Google Scholar]
- Kasarda DD (1989) Glutenin structure in relation to wheat quality. In Wheat Is Unique. American Association of Cereal Chemistry, St. Paul, MN, pp 277–302
- Kasarda DD, King G, Kumosinski TF. Comparison of spiral structure in wheat high molecular weight glutenin subunits and elastin by molecular modeling. In: Kumosinski TF, Liebman MN, editors. Molecular Modeling: From Virtual Tools to Real Problems. Washington, DC: American Chemical Society; 1994. pp. 209–220. [Google Scholar]
- Kasarda DD, Tao HP, Evans PK, Adalsteins AE, Yuen SW. Sequencing of protein from a single step of a 2-D gel pattern: N-terminal sequence of a major wheat LMW-glutenin subunit. J Exp Bot. 1988;39:899–906. [Google Scholar]
- Keck B, Köhler P, Wieser H. Disulfide bonds in wheat gluten: cystine peptides derived from gluten proteins following peptic and thermolytic digestion. Z Lebensm Unters Forsch. 1995;200:432–439. doi: 10.1007/BF01193253. [DOI] [PubMed] [Google Scholar]
- Köhler P, Belitz H-D, Wieser H. Disulphide bonds in wheat gluten: further cystine peptides from high molecular weight (HMW) and low molecular weight (LMW) subunits of glutenin and from γ-gliadins. Z Lebensm Unters Forsch. 1993;196:239–247. doi: 10.1007/BF01202740. [DOI] [PubMed] [Google Scholar]
- Köhler P, Keck-Gassenmeier B, Wieser H, Kasarda DD. Molecular modeling of the N-terminal regions of high molecular weight glutenin subunits 7 and 5 in relation to intramolecular disulfide bond formation. Cereal Chem. 1997;74:154–158. [Google Scholar]
- Ladurner AG, Fersht AR. Glutamine, alanine, or glycine repeats inserted into the loop of a protein have minimal effects on stability and folding rates. J Mol Biol. 1997;273:330–357. doi: 10.1006/jmbi.1997.1304. [DOI] [PubMed] [Google Scholar]
- Lew EJ-L, Kuzmicky DD, Kasarda DD. Characterization of low molecular weight glutenin subunits by reversed-phase high-performance liquid chromatography, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and N-terminal amino acid sequencing. Cereal Chem. 1992;69:508–515. [Google Scholar]
- MacRitchie F, Kasarda DD, Kuzmicky DD. Characterization of wheat protein fractions differing in contributions to breadmaking quality. Cereal Chem. 1991;68:122–130. [Google Scholar]
- Masci S, D'Ovidio R, Spigaglia M, Lafiandra D, Kasarda DD (1998) A 1B-coded low-molecular-weight glutenin subunit associated with pasta-making quality is present also in bread wheat. In Proceedings of the 9th International Wheat Genetics Symposium. University Extension Press, Saskatoon, Saskatchewan, Canada, pp 198–200
- Masci S, Lew EJL, Lafiandra D, Porceddu E, Kasarda DD. Characterization of low-molecular-weight glutenins type 1 and type 2 by RP-HPLC and N-terminal sequencing. Cereal Chem. 1995;72:100–104. [Google Scholar]
- Melas V, Morel M-H, Autran C, Feillet P. Simple and rapid method for purifying low molecular weight subunits of glutenin from wheat. Cereal Chem. 1994;71:234–237. [Google Scholar]
- Müller S, Wieser H. The location of disulphide bonds in monomeric γ-type gliadins. J Cereal Sci. 1997;26:169–176. [Google Scholar]
- Ozols J. Amino acid analysis. Methods Enzymol. 1990;182:587–601. doi: 10.1016/0076-6879(90)82046-5. [DOI] [PubMed] [Google Scholar]
- Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- Rost B, Sander C. Combining evolutionary information and neural networks to predict secondary structure. Comput Appl Biosci. 1994;10:53–60. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shewry PR, Tatham AS. Disulphide bonds in wheat gluten proteins. J Cereal Sci. 1997;25:207–227. [Google Scholar]
- Singh NK, Shepherd KW, Cornish GB. A simplified SDS-PAGE procedure for separating LMW subunits of glutenin. J Cereal Sci. 1991;14:203–208. [Google Scholar]
- Tao HP, Kasarda DD. Two-dimensional gel mapping and N-terminal sequencing of LMW-glutenin subunits. J Exp Bot. 1989;40:1015–1020. [Google Scholar]
- Toumadje A, Johnson WC., Jr Systemin has the characteristics of a poly(L-proline) II type helix. J Am Chem Soc. 1995;117:7023–7024. [Google Scholar]
- Vensel WH, Tarr GE, Kasarda DD. C-terminal and internal sequence of a low molecular weight (LMW-s) type of glutenin subunit. Cereal Chem. 1995;72:356–359. [Google Scholar]
- Wrigley CW. Giant proteins with flour power. Nature. 1996;381:738–739. doi: 10.1038/381738a0. [DOI] [PubMed] [Google Scholar]
- Wu KJ, Marsh EP, Odom RW (1995) MALDI electrostatic analyzer TOF mass spectrometry studies of biopolymers. In Proceedings of the 43rd Conference on Mass Spectrometry and Allied Topics, Atlanta, Georgia, May 21–26. American Society of Mass Spectrometry, Santa Fe, NM, p 998
- Yang A-S, Hitz B, Honig B. Free energy determinants of secondary structure formation. III. β-turns and their role in protein folding. J Mol Biol. 1996;259:873–882. doi: 10.1006/jmbi.1996.0364. [DOI] [PubMed] [Google Scholar]