Abstract
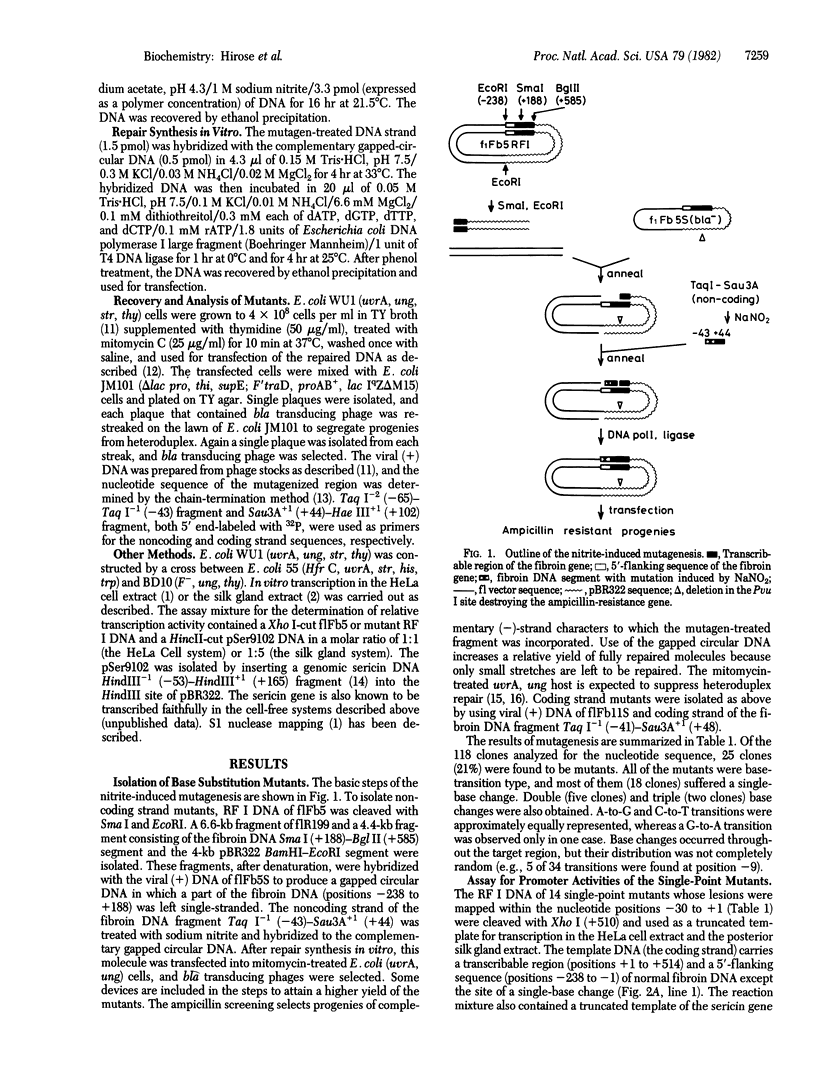
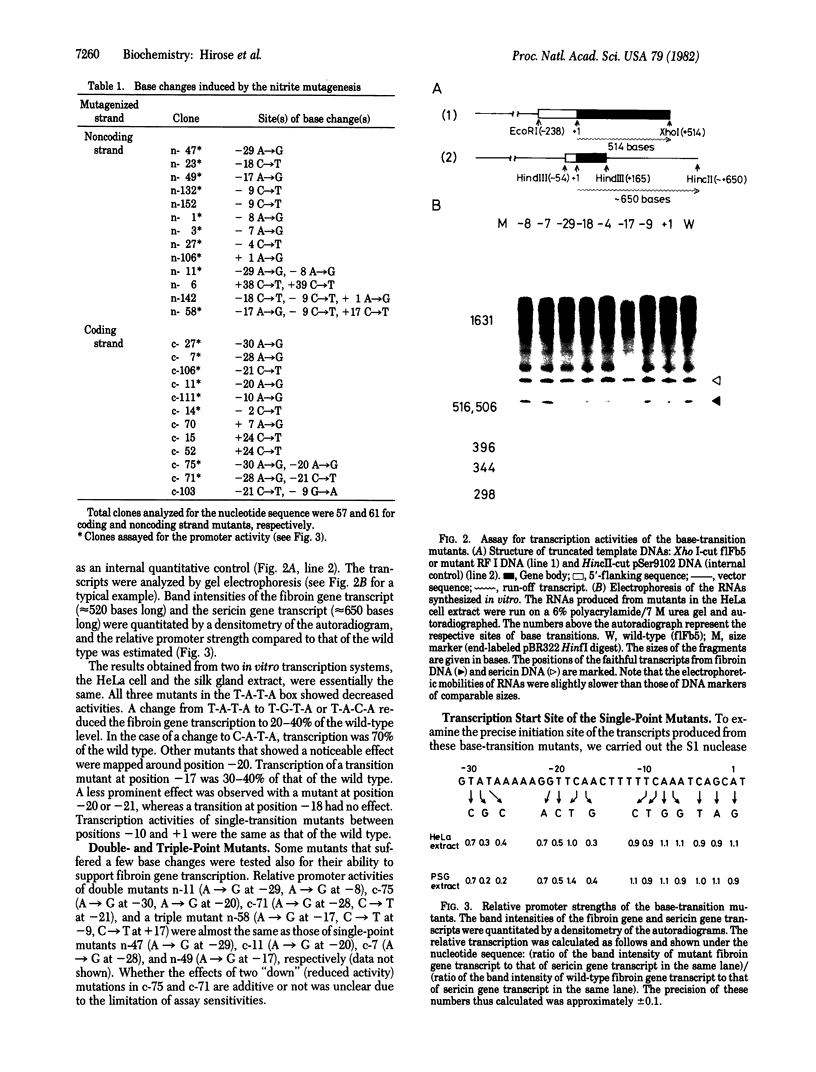
A highly efficient method for segment-directed mutagenesis has been developed. The method relies on the deamination by sodium nitrite of the bases in the separated strands of a small DNA restriction fragment. The mutagen-treated strands produce transition mutations by the following sequence: (i) hybridization with the complementary strand of the wild-type DNA that had been cloned into a phage fl vector, (ii) repair synthesis in vitro, and (iii) transfection of Escherichia coli. Using this method, we have isolated 14 single-point mutants within a 31-base-pair stretch of the fibroin gene (from the T-A-T-A box at the nucleotide position -30 to the cap site at +1). In vitro transcription experiments with the HeLa cell or the silk gland cell extract show that single-base transitions at the T-A-T-A box (T to C at -30, A to G at -29, and T to C at -28) and at the -20 region (G to A at -21, T to C at -20, and A to G at -17) result in decreased promoter activities, whereas those at the cap site and the -10 regions have no effect. The initiation site of transcription is the same for five "down" (reduced activity) mutants (T to C at -30, T to C at -28, G to A at -21, T to C at -20, and A to G at -17), the cap site mutant (A to G at +1), and the wild-type genes--position +1. However, the A-to-G transition at -29 (the second base of the T-A-T-A box) induces an additional transcription start from position +4. Functions of the T-A-T-A box and the -20 regions are discussed.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baas P. D., Jansz H. S. Asymmetric information transfer during phi X174 DNA replication. J Mol Biol. 1972 Feb 14;63(3):557–568. doi: 10.1016/0022-2836(72)90447-0. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Vovis G. F., Zinder N. D. Insertion mutant of bacteriophage f1 sensitive to EcoRI. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2699–2702. doi: 10.1073/pnas.76.6.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Corden J., Wasylyk B., Buchwalder A., Sassone-Corsi P., Kedinger C., Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980 Sep 19;209(4463):1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- Duncan B. K., Rockstroh P. A., Warner H. R. Escherichia coli K-12 mutants deficient in uracil-DNA glycosylase. J Bacteriol. 1978 Jun;134(3):1039–1045. doi: 10.1128/jb.134.3.1039-1045.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Wasylyk B., Chambon P., Birnstiel M. L. Point mutation in the TATA box curtails expression of sea urchin H2A histone gene in vivo. Nature. 1981 Nov 12;294(5837):178–180. doi: 10.1038/294178a0. [DOI] [PubMed] [Google Scholar]
- Grosveld G. C., Shewmaker C. K., Jat P., Flavell R. A. Localization of DNA sequences necessary for transcription of the rabbit beta-globin gene in vitro. Cell. 1981 Jul;25(1):215–226. doi: 10.1016/0092-8674(81)90246-4. [DOI] [PubMed] [Google Scholar]
- Hu S. L., Manley J. L. DNA sequence required for initiation of transcription in vitro from the major late promoter of adenovirus 2. Proc Natl Acad Sci U S A. 1981 Feb;78(2):820–824. doi: 10.1073/pnas.78.2.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison C. A., 3rd, Phillips S., Edgell M. H., Gillam S., Jahnke P., Smith M. Mutagenesis at a specific position in a DNA sequence. J Biol Chem. 1978 Sep 25;253(18):6551–6560. [PubMed] [Google Scholar]
- Kabsch W., Sander C., Trifonov E. N. The ten helical twist angles of B-DNA. Nucleic Acids Res. 1982 Feb 11;10(3):1097–1104. doi: 10.1093/nar/10.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Model P., Zinder N. D. In vitro synthesis of bacteriophage f1 proteins. J Mol Biol. 1974 Feb 25;83(2):231–251. doi: 10.1016/0022-2836(74)90389-1. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Sequence dependence of the helical repeat of DNA in solution. Nature. 1981 Jul 23;292(5821):375–378. doi: 10.1038/292375a0. [DOI] [PubMed] [Google Scholar]
- Razin A., Hirose T., Itakura K., Riggs A. D. Efficient correction of a mutation by use of chemically synthesized DNA. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4268–4270. doi: 10.1073/pnas.75.9.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D., Nathans D. Local mutagenesis: a method for generating viral mutants with base substitutions in preselected regions of the viral genome. Proc Natl Acad Sci U S A. 1978 May;75(5):2170–2174. doi: 10.1073/pnas.75.5.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Talkington C. A., Leder P. Rescuing the in vitro function of a globin pseudogene promoter. Nature. 1982 Jul 8;298(5870):192–195. doi: 10.1038/298192a0. [DOI] [PubMed] [Google Scholar]
- Tsai S. Y., Tsai M. J., O'Malley B. W. Specific 5' flanking sequences are required for faithful initiation of in vitro transcription of the ovalbumin gene. Proc Natl Acad Sci U S A. 1981 Feb;78(2):879–883. doi: 10.1073/pnas.78.2.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M., Ohshima Y., Suzuki Y. Assumed initiation site of fibroin gene transcription. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4872–4876. doi: 10.1073/pnas.76.10.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M., Suzuki Y. Faithful transcription initiation of fibroin gene in a homologous cell-free system reveals an enhancing effect of 5' flanking sequence far upstream. Cell. 1981 Nov;27(1 Pt 2):175–182. doi: 10.1016/0092-8674(81)90371-8. [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y., Hirose S., Tsuda M., Suzuki Y. Promoter sequence of fibroin gene assigned by in vitro transcription system. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4838–4842. doi: 10.1073/pnas.78.8.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto Y., Suzuki Y. Structural analysis of the fibroin gene at the 5' end and its surrounding regions. Cell. 1979 Feb;16(2):425–436. doi: 10.1016/0092-8674(79)90018-7. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Derbyshire R., Guy A., Molko D., Roget A., Téoule R., Chambon P. Specific in vitro transcription of conalbumin gene is drastically decreased by single-point mutation in T-A-T-A box homology sequence. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7024–7028. doi: 10.1073/pnas.77.12.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]